SUMMARY
To maintain lifelong production of blood cells, hematopoietic stem cells (HSC) are tightly regulated by inherent programs and extrinsic regulatory signals received from their microenvironmental niche. Long-term repopulating HSC (LT-HSC) reside in several, perhaps overlapping, niches that produce regulatory molecules/signals necessary for homeostasis and increased output following stress/injury 1–5. Despite significant advances in specific cellular or molecular mechanisms governing HSC/niche interactions, little is understood about regulatory function within the intact mammalian hematopoietic niche. Recently, we and others described a positive regulatory role for Prostaglandin E2 (PGE2) on HSC function ex vivo 6,7. While exploring the role of endogenous PGE2 we unexpectedly observed hematopoietic egress after nonsteroidal anti-inflammatory drug (NSAID) treatment. Surprisingly, this was independent of the SDF-1/CXCR4 axis. Stem and progenitor cells were found to have differing mechanisms of egress, with HSC transit to the periphery dependent on niche attenuation and reduction in the retentive molecule osteopontin (OPN). Hematopoietic grafts mobilized with NSAIDs had superior repopulating ability and long-term engraftment. Treatment of non-human primates and healthy human volunteers confirmed NSAID-mediated egress in higher species. PGE2 receptor knockout mice demonstrated that progenitor expansion and stem/progenitor egress resulted from reduced EP4 receptor signaling. These results not only uncover unique regulatory roles for EP4 signaling in HSC retention in the niche but also define a rapidly translatable strategy to therapeutically enhance transplantation.
Keywords: NSAID, stem cell, mobilization, niche, osteopontin, prostaglandin, hematopoiesis
Mice were treated with the prototypical NSAID indomethacin (Supplemental Fig. 1a) to reduce endogenous PGE2 production, resulting in a significant increase in hematopoietic progenitor cells (HPC) in the peripheral blood (PB) that was not accompanied by an increase in white blood cell count (Supplemental Fig. 1b,c), likely accounting for the lack of previous detection of this observation despite decades of clinical NSAID use. No increase in HPC egress was seen in mice treated with the lipoxygenase inhibitor baicalein, suggesting a cyclooxygenase (COX) pathway-specific effect. Co-administration of indomethacin with the clinically used mobilizing agent granulocyte-colony stimulating factor (G-CSF), significantly enhanced (~2 fold) HPC mobilization (Supplementary Fig. 1b). NSAIDs with varying COX-1- and COX-2-selectivity demonstrated significant mobilization with indomethacin, aspirin, ibuprofen, and meloxicam (Supplementary Fig. 2). Meloxicam inhibits both COX-1 and COX-2 within the bone marrow microenvironment (Supplementary Fig. 3) and when compared to other dual inhibitors it has a reduced incidence of gastrointestinal discomfort 8 and inhibition of platelet aggregation 9. Therefore, meloxicam was used in the majority of the studies. We did not extensively test the differential roles of COX-1 and COX-2 and, therefore, there may be similar activity of NSAIDs with different COX-1/COX-2 inhibitory profiles when compared to meloxicam.
Meloxicam, similar to indomethacin, increased egress of HPC (Fig. 1a, Supplementary Fig. 4) and the phenotypic HSC-enriched populations Sca-1+ c-kit+ lineage− (SKL) or the highly purified CD150+ CD48− (SLAM) SKL populations (Fig. 1b, Supplementary Fig. 4). Enhanced egress was maintained in 5-ALOX knockout mice (Supplementary Fig. 5), further demonstrating effects are not due to general eicosanoid inhibition. Enhancement in egress was also not specific to G-CSF, as meloxicam enhanced mobilization by the clinically used CXCR4 antagonist AMD3100 (Supplementary Fig. 6).
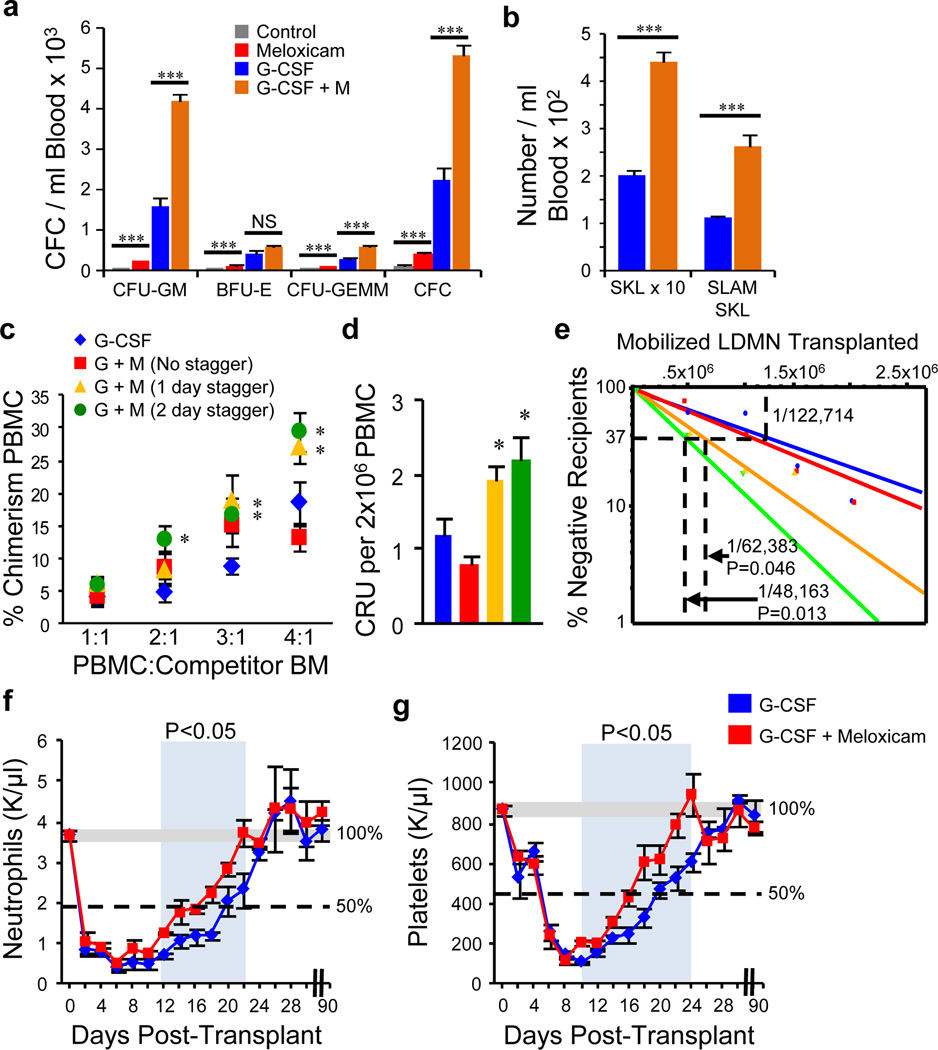
Figure 1. NSAIDs mobilize hematopoietic stem and progenitor cells.
Meloxicam enhances mobilization of HPC, a, and HSC b, into blood (n=4–5 mice/group/experiment; 3 experiments). c, Chimerism; d, competitive repopulating units (CRU); and e, LT-HSC frequency (Poisson distribution) 36 weeks after limiting dilution competitive transplants of peripheral blood mononuclear cells (PBMC) from mice treated with G-CSF and combination regimens (n=8 mice/group, assayed individually). Mice were treated with G-CSF or a staggered regimen of G-CSF + Meloxicam and PBMC transplanted into lethally irradiated mice. f, Neutrophil and g, platelet recovery were monitored for 90 days. *P<0.05, ** P<0.01, ***P<0.001; unpaired two-tailed t-test. All error bars represent mean ± s.e.m.
Despite significant increases in phenotypic HSC and functional HPC in the PB, two early transplant attempts did not show enhanced HSC engraftment (Supplementary Figs 7a,b). Since we previously showed that PGE2 signaling was a positive regulator of HSC CXCR4 expression and homing to the niche 6, we hypothesized that while HSC/HPC yield was increased in NSAID grafts, CXCR4 expression might be reduced, accounting for apparent lack of enhanced engraftment. To test this hypothesis we staggered the administration of NSAID and G-CSF to allow for hematopoietic mobilization and restoration of normal endogenous PGE2 signaling before transplant (Supplementary Fig. 7c). CXCR4 levels were significantly lower after NSAID treatment and staggered administration allowed for restored receptor levels, while maintaining enhanced HSC egress (Supplementary Fig. 7d,e). We competitively transplanted mobilized grafts from G-CSF, or non-staggered and staggered G-CSF + meloxicam treated mice. Staggered administration resulted in significant enhancement of LT-HSC engraftment, with a 48 hour stagger resulting in a 2.6 fold LT-HSC increase (Figs. 1c,d,e and Supplementary Fig. 8). When grafts were transplanted non-competitively, staggered co-administration of meloxicam resulted in 4-day faster recovery of neutrophils (Fig. 1f) and platelets (Fig. 1g) compared to G-CSF alone. Secondary transplantation confirmed sustained LT-HSC activity with multi-lineage reconstitution 36 weeks post-transplant (Supplementary Fig. 9).
To confirm NSAID-mediated hematopoietic egress in higher species, 4 baboons were treated with a standard regimen of G-CSF, or the combination of G-CSF + meloxicam in a crossover design (Fig. 2a). While individual baboon responses to G-CSF varied, in all cases meloxicam treatment increased CD34+ cells (Fig. 2b) and CFU-GM (Fig. 2c) in PB. Meloxicam treatment on its own also resulted in significant HSC/HPC egress (Figs. 2 d,e). In healthy human volunteers, meloxicam treatment resulted in significant increases in CD34+ cells (Fig. 2f), and functionally defined HPC (Figs. 2 g,h,i), matching hematopoietic egress seen with meloxicam treatment in baboons and mice. Thus, short-term endogenous PGE2 inhibition, closely resembling current clinical NSAID treatment, results in a previously unappreciated increase in HSC and HPC mobilization.
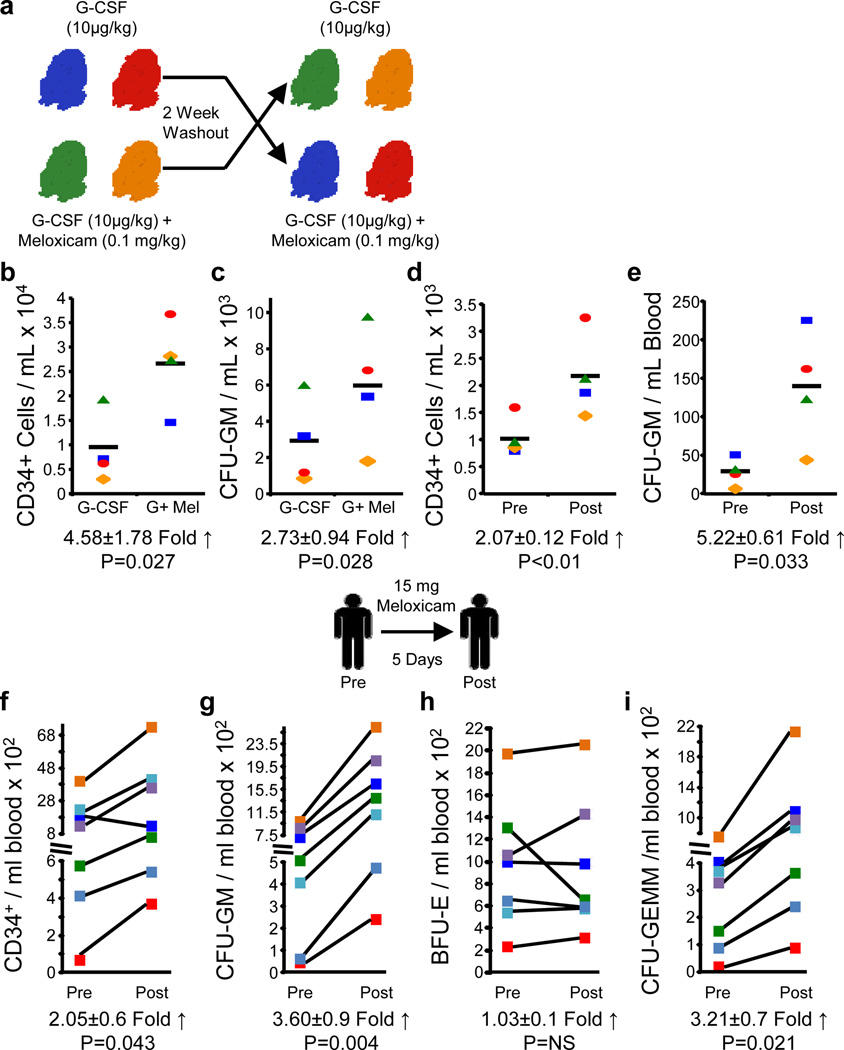
Figure 2. Non-human primates and healthy human volunteers mobilize HSC/HPC in response to NSAID treatment.
a, Four baboons were treated with G-CSF +/- Meloxicam in a cross-over design and b, CD34+ cells and c, CFU-GM in peripheral blood (PB) determined. d, CD34+ cells and e, CFU-GM in PB determined pre- and post-5 days of meloxicam alone treatment. Seven healthy human volunteers were treated with 15 mg/day p.o. for 5 days, and were assessed for f, CD34+ cells; g, CFU-GM; h, BFU-E, and i, CFU-GEMM pre- and post-treatment. Statistics represent paired, two-tailed t-test.
Meloxicam treatment increased functionally defined myeloid progenitors and phenotypically defined granulocyte-macrophage progenitors in the bone marrow, but no differences in phenotypically or functionally defined HSC were observed (Supplementary Fig. 10). Since PGE2 signals through four receptors (EP1-4), each with unique signaling pathways 10, we hypothesized that the myeloid expansion and egress was due to lack of signaling via one or more EP receptors. Only agonists capable of activating the EP4 receptor inhibited myeloid HPC (Supplementary Fig. 11a). To further confirm the specific role of the EP4 receptor, similar assays were performed using knockout mice for each of the EP receptors. Comparison of all knockout strains showed that only HPC from conditional EP4−/− mice had reduced response to inhibition by PGE2 (Supplementary Fig. 11b) and a 2.3 fold increase in marrow CFU-M compared to wild-type (Supplementary Fig. 11c). Co-administration of EP4 antagonists with G-CSF significantly enhanced mobilization, similar to meloxicam, while EP1, 2 and 3 antagonists failed to increase mobilization (Fig. 3a). Furthermore, when a selective EP4 agonist was co-administered with G-CSF + meloxicam, the meloxicam enhancement of mobilization was abrogated, and to the same degree as dmPGE2 co-administration (a long-acting PGE2 analog). Agonists that did not target the EP4 receptor failed to alter meloxicam enhancement. EP4 antagonism with G-CSF enhanced mobilization of LT-HSCs (Figs. 3b,c,d), indicating that the NSAID-mediated effects in hematopoietic egress are due to reduced EP4 receptor signaling. Consistent with pharmacologic data, conditional EP4 deletion increased HPC/HSC egress (Supplementary Fig. 11d,e,f), and enhanced mobilization by meloxicam was abrogated (Figs. 3e,f). These data implicate PGE2/EP4 receptor signaling in mediating the egress effects of NSAIDs, however we did not conduct a comprehensive lipidomic profile and therefore cannot exclude contributions of other eicosanoids.
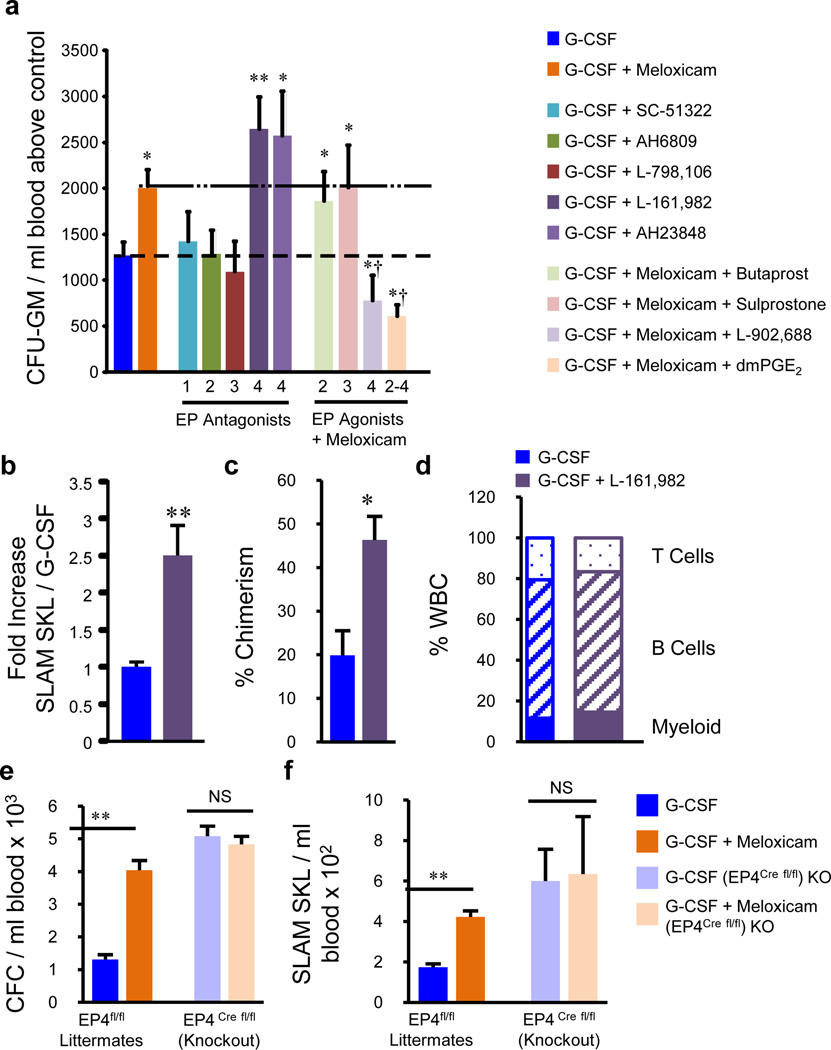
Figure 3. Prostaglandin E2 EP4 receptor antagonism/knockout expands bone marrow HPC and enhances mobilization.
a, HPC mobilization with G-CSF, G-CSF + meloxicam, G-CSF + EP receptor antagonists, or G-CSF + meloxicam + EP receptor agonists (n=5 mice/group, assayed individually). b, The EP4 antagonist L-161,982 enhanced HSC mobilization (n=4 mice/group, assayed individually), and c, long-term reconstitution 16 weeks post-transplant with d, multi-lineage reconstitution (n=5 mice/group, assayed individually). e, Meloxicam enhances mobilization of HPC, and f, SLAM SKL cells in WT littermates, but not in EP4 conditional knockouts (n=3,4 mice/group, assayed individually). *P<0.05, ** P<0.01, ***P<0.001; unpaired two-tailed t-test. †P<0.05 compared to G-CSF + meloxicam. All error bars represent mean ± s.e.m.
In vitro and in vivo results indicate that lack of EP4 signaling drives HPC expansion, possibly elucidating one mechanism responsible for enhanced HPC egress: more marrow HPC allows more to be mobilized to the periphery. However, no alterations in bone marrow HSC content were observed (Supplementary Fig. 10), suggesting that HSC mobilization results from a different mechanism, perhaps acting on the HSC niche. Gross histological analysis of NSAID treated mice over 0–4 days showed a progressive increase in laminarity of endosteal lining osteolineage cells (Supplementary Fig. 12,13), similar to that seen after G-CSF treatment 11. Comparable results were observed in collagen 2.3-GFP reporter mice, showing marked attenuation of osteolineage cells (Fig. 4 a–d), and in mice after conditional EP4 deletion (Supplementary Fig. 14). Dynamic bone formation assays using staggered double calcein labeling and modified Goldner's trichrome staining support significant attenuation of osteolineage cellular function (Supplementary Fig. 15).
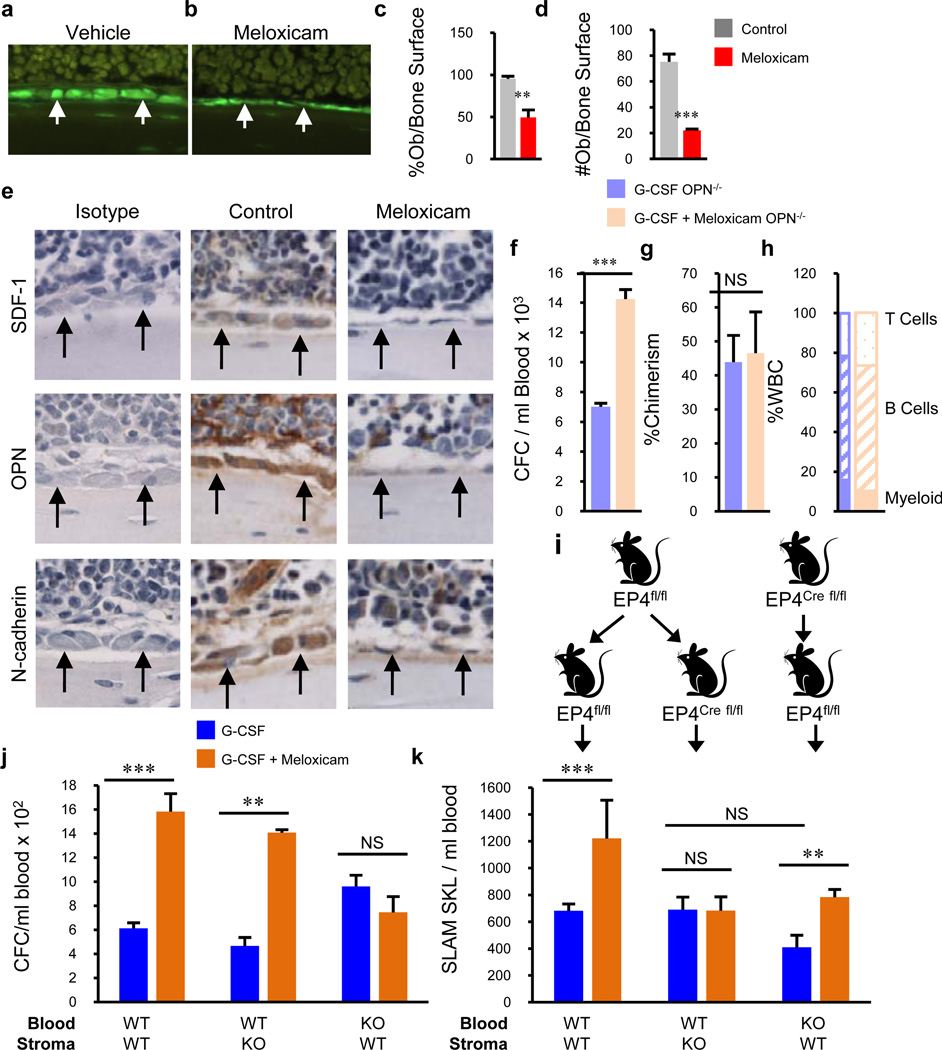
Figure 4. NSAIDs attenuate hematopoietic supportive molecules and differentially mobilize HSC and HPC in OPN knockout and EP4 conditional knockout mice.
a, b, Assessment of Col2.3-GFP cells after vehicle or meloxicam demonstrates reduced c, percentages and d, number of osteolineage cells (n=4 mice/group, assayed individually). e, Immunohistochemical staining of hematopoietic supportive molecules after treatment with meloxicam (400X). f, Meloxicam enhances mobilization of HPC in OPN −/− mice, with g,h, no enhancement in long-term reconstitution 16 weeks post-transplant. i, Representation of chimera generation allowing conditional knockout of donor hematopoietic cells, or recipient stromal cells. EP4 was deleted with tamoxifen 8 weeks post-transplant and mice treated with G-CSF or G-CSF + meloxicam. j, Enhanced mobilization of HPC by meloxicam when EP4 is expressed on hematopoietic cells and k, enhanced mobilization of HSC when EP4 is expressed by stromal cells (n=4 mice/group, assayed individually). *P<0.05, **P<0.01, ***P<0.001; unpaired two-tailed t-test. Error bars represent mean ± s.e.m.
Currently, there is considerable debate regarding direct or indirect roles of osteoclasts (OC) in hematopoietic niche regulation and HSC/HPC retention (reviewed in 12,13). To assess the role of OCs, mice were treated with meloxicam and/or G-CSF with or without zoledronic acid (ZA), a potent inhibitor of OC activity 14. Similar to a recent report 15, ZA resulted in an increase in HSC/HPC mobilization by meloxicam and G-CSF (Supplementary Fig. 16), suggesting that increased OC activity is not a mitigating mechanism for NSAID-mediated hematopoietic egress. Niche attenuation and HSC/HPC mobilization by G-CSF have recently been reported to be mediated by marrow-resident monocyte/macrophage populations 15–17. In contrast to G-CSF 15, immunohistochemical (IHC) analysis demonstrated that meloxicam does not reduce F4/80+ macrophages (Supplementary Fig. 17a), nor is there a reduction in phenotypically defined macrophages assessed by flow cytometry (Supplementary Figs. 17b,c). We observed no changes in sinusoidal endothelial cell number or apoptotic state (Supplementary Fig. 18), nor sinusoid vessels or endothelial cell number by IHC (Supplementary Fig. 19). Similarly, there was no alteration in Nestin+ cell number (Supplementary Fig. 20). No differences in marrow MMP-9 or soluble c-kit, agents reported to regulate HSC motility within the bone marrow niche 18, were observed in NSAID treated mice (data not shown), suggesting other unique HSC retentive molecule(s) are regulated by EP4.
We fractionated osteolineage cells into 3 sub-populations 19,20 (Supplementary Fig. 21a). QRT-PCR analysis revealed that all 3 populations expressed all 4 EP receptors, with EP4 expressed most predominately (Supplementary Fig. 21b). Meloxicam treatment resulted in reductions in mRNA expression of several hematopoietic supportive molecules, including Jagged-1, Runx-2, VCAM-1, SCF, SDF-1, and OPN (Supplementary Fig. 21c). Similarly, IHC staining demonstrated reductions in SDF-1, OPN and N-cadherin expression (Fig. 4e). Analysis in EP4 conditional knockout mice showed a significant reduction in mesenchymal progenitor cells compared to Cre(-) littermates and wild-type controls (Supplementary Fig. 21d), further demonstrating a role for EP4 signaling in hematopoietic niche maintenance.
Since the interaction of SDF-1 with its cognate receptor CXCR4 is a well-known mediator of niche retention we sought to determine whether reduced expression of SDF-1 mediated the hematopoietic egress caused by NSAID treatment. Surprisingly, despite the robust egress of cells in CXCR4 conditional knockout mice, both HPC and HSC trafficking to the periphery were significantly enhanced by meloxicam (Supplementary Fig. 22). Osteopontin has been reported as both a regulator of HSC quiescence 21 and niche retention 22. In contrast to CXCR4, when OPN knockout mice were treated with meloxicam or G-CSF for 6 days, meloxicam enhanced mobilization of HPC (Fig. 4f) but, quite unexpectedly, not HSC (Fig. 5g,h) (additional data in Supplementary Fig. 23), while both HPC and HSC were mobilized by G-CSF in wild-type mice. This surprising result indicates that NSAID-mediated OPN reduction is specifically responsible for the observed HSC niche egress, while increased peripheral HPC results from an independent mechanism(s). To elucidate the differential roles of hematopoietic intrinsic versus stromal niche EP4 signaling in mediating HPC/HSC egress, we created chimeric mice in which we could conditionally delete EP4 from donor hematopoietic cells or recipient stromal cells (Fig. 4i). EP4 expression on hematopoietic cells was required for NSAID-mediated egress of HPC (Fig. 4j), while EP4 on stromal cells was specifically necessary for HSC egress (Fig. 4k). These studies demonstrate that PGE2 signaling differentially regulates HPC and HSC retention in the marrow through both cell intrinsic and extrinsic mechanisms, and future studies should define the relative roles of individual stromal niche cell contributions to EP4-mediated niche retention. To our knowledge, this is the first report of an agent capable of mobilizing both HSC and HPC and doing so through cell stage specific mechanisms.
METHODS SUMMARY
C57Bl/6 and OPN−/− mice were purchased from Jackson Laboratories. B6.SJL-PtrcAPep3B/BoyJ mice were bred in-house. CXCR4flox/flox mice were generated as described 23 and were a kind gift from Y. Zou, Columbia University. EP1−/−, EP2−/−, EP3−/−, and EP4flox/flox mice were generated as described 24–26. Conditional mice were bred to Ubc-Cre/ERT2 mice from Jackson. Female olive baboons, Papio anubis, were housed individually in conventional caging of the Biological Resources Laboratory, University of Illinois (UI) at Chicago. Primate research was approved by the UI Animal Care and Use Committee (IACUC). The IACUC of IUSM approved all protocols. The IRB of IUSM approved human subject research and informed consent was acquired from all volunteers.
METHODS
Animals and Subjects
C57Bl/6 (CD45.2) mice were purchased from Jackson Laboratories (Bar Harbor, ME). B6.SJL-PtrcAPep3B/BoyJ (BOYJ) (CD45.1) mice were bred in-house. CXCR4flox/flox mice were generated as described 23 and were a kind gift from Yong-Rui Zou. EP1−/−, EP2−/−, EP3−/−, and EP4flox/flox mice were generated as described 24–26. OPN−/− mice were purchased from Jackson Laboratories. Nestin-GFP 27, Col2.3-GFP 28 and 5-ALOX 29 mice were generated as described. EP4flox/flox mice were bred to Ubc-Cre/ERT2 mice from Jackson to generate conditional EP4 knockout mice. All mice were maintained on a C57Bl/6 background. Female olive baboons, Papio anubis, within the weight range of 16–19 kg, were housed individually in conventional caging and holding rooms of the Biological Resources Laboratory, a centralized animal facility for the University of Illinois at Chicago Medical Center, Chicago, IL. The conducted primate research was approved by the University of Illinois at Chicago Animal Care and Use Committee. The Animal Care and Use Committee of IUSM approved all protocols, and the Institutional Review Board approved human subject research. Informed consent was obtained from all volunteers.
Peripheral blood and bone marrow acquisition and processing
Peripheral blood from mice was obtained by cardiac puncture following CO2 asphyxiation using an ethylenediaminetetraacetic acid (EDTA) rinsed syringe. Blood was transferred to tubes containing EDTA for complete blood cell (CBC) analysis. CBC analysis was performed on a Hemavet 950FS (Drew Scientific, Oxford, CT). Peripheral blood mononuclear cells (PBMC) were prepared by centrifugation over Lympholyte Mammal (Cedarlane Laboratories Ltd, Hunby, Ontario, Canada) at 800g for 30–40 minutes at room temperature, followed by triplicate washes. Bone marrow cells were harvested by flushing femurs with ice-cold PBS and single-cell suspensions prepared by passage through a 26-gauge needle. For baboons, peripheral blood was obtained from the femoral vein of baboons anesthetized with an intramuscular injection of 10 mg/kg ketamine hydrochloride (Bionichepharma, Lakeforest, IL). Blood was collected into 10 ml sterile EDTA vacutainers (Becton, Dickinson and Company, Franklin, NJ) and transported on ice to IUSM for analysis. Complete blood counts with differentials were performed on a Hemavet 950FS. Peripheral blood was then diluted 1:3 with PBS and mononuclear cells were isolated using Ficoll-Paque™ Plus (Amersham Biosciences, Pittsburgh, PA), per manufacturer’s protocol.
Colony assays
Bone marrow cells or PBMC were resuspended in McCoy’s 5A modified media supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin, 0.6 X modified essential medium (MEM) vitamin solution, 1 mM sodium pyruvate, 0.8 X MEM essential amino acids, 0.6 X MEM nonessential amino acids, 0.05% sodium bicarbonate (all from Gibco, Grand Island, NY), serine, asparagine, glutamine mixture and 15% HI-FBS (Hyclone Sterile Systems, Logan, UT) as described 30,31. Cells were mixed with 0.3% agar (Difco Laboratories, Detroit, MI) in McCoy’s 5A medium with 10 ng/ml rhGM-CSF and 50 ng/ml rmSCF (R&D Systems, Minneapolis, MN). PBMC were cultured at 2×105 cells per ml and bone marrow cells at 5×104 cells per ml. All cultures were established in triplicate from individual animals, incubated at 37 °C, 5% CO2, 5% O2 in air for 7 days and colonies quantitated by microscopy. In some experiments, total CFC including CFU-GM, BFU-E and CFU-GEMM were enumerated in 1% methylcellulose/IMDM containing 30% fetal bovine serum, 1 U/ml recombinant human erythropoietin (EPO), 10 ng/ml rhGM-CSF or rmGM-CSF and 50 ng/ml rhSCF or rmSCF as described 32,33. In some experiments, phenotypically defined CMP and GMP were plated at 500 cells per plate and colony growth determined in agar CFC assays with rmGM-CSF + rmSCF or with rmM-CSF. For analysis of CFC in baboons, similar assays were performed using recombinant human growth factors.
Flow cytometry
All antibodies were purchased from BD Biosciences unless otherwise noted. For detection of SKL cells, we used streptavidin conjugated with PE-Cy7 (to stain for biotinylated MACS® lineage antibodies (Miltenyi, Auburn, CA), c-kit-APC, Sca-1-PE or APC-Cy7, CD45.1-PE, CD45.2-FITC. For SLAM SKL, we utilized Sca-1-PE-Cy7, c-kit-FITC, CD150-APC (eBiosciences, San Diego, CA), CD48-biotin (eBiosciences) and streptavidin-PE. CXCR4 expression was analyzed using biotinylated Lineage antibodies, streptavidin-PECy7, c-kit-APC, Sca-1-APC-Cy7, and CXCR4-PE. For baboon CD34 analysis, CD34-PE (Clone 563) was used. For macrophages, antibodies against CD115 (clone AFS98), Gr-1 (clone RB6-8C5), and F4/80 (clone CI:A3-1) were used. Osteolineage populations were identified and sorted as previously described 19. For enumeration of bone marrow endothelial cells, femurs and tibias were crushed in a sterile mortar, and digested in collagenase (0.3%) at 37°C for one hour. Recovered cells were co-stained with fluorochrome-conjugated antibodies to CD45, Ter119, Sca-1, VEGFR3 and CD31 and total number of SECs (CD45−Ter119−Sca-1−VEGFR3+ CD31+) per femur was enumerated by flow-cytometry analysis. To examine endothelial cell apoptosis, gated CD45−Ter119−Sca-1−VEGFR3+ CD31+ cells were stained with Annexin V (BD Biosciences) and LIVE/DEAD staining dye (Invitrogen). For enumeration of myeloid progenitors (CMP, GMP and MEP), femurs and tibias were flushed with 5 ml IMDM containing 2% FBS. Lineage-positive cells were depleted using lineage-cell depletion kit (Miltenyi Biotec) and lineage-negative cells were stained with fluorochrome-conjugated antibodies to Sca-1, c-Kit, IL-7Rα, CD34 and FCRϒll/lll and analyzed by flow cytometry. The Lin− IL-7Rα− Sca-1−c-Kit+ fraction was subdivided into three subpopulation; CMP (FCRϒll/llllowCD34+), MEP (FCRϒll/llllowCD34−), and GMP (FCRϒll/lllhiCD34−) and collected by sorting. All flow cytometry analyses were performed on an LSRII flow cytometer (BD). Cell sorting was performed on a BD Aria or Reflection II or Reflection III sorters.
Peripheral blood mobilization
Several different mobilization strategies were employed, with specific details of dosing and schematics of dosing regimens shown on the data figures or included in the figure legends. In general, mice were given subcutaneous treatments of vehicle, NSAID (at varying doses), G-CSF (50µg/kg, twice a day for 4 days), or G-CSF plus NSAID. For studies exploring mobilizing agents other than G-CSF, mice were treated with AMD3100 (5 mg/kg day 5; single injection), and peripheral blood harvested at 1 hour post-AMD3100 treatment. For comparisons of multiple different NSAIDs, all NSAIDs were dosed by oral gavage using an enhanced oral gavage technique 34. Each gavage treatment was given in a 0.2 ml bolus (10 ml/kg) of 0.5% methyl cellulose (Methyl Cellulose M-0512, Sigma- Aldrich, St. Louis, MO) with an NSAID suspended in solution. For EP receptor analysis, mice were mobilized with G-CSF in combination with Meloxicam, AH6809 (EP1-3 antagonist, 10 µg per mouse, ip, 4 days), AH23848 (EP4 antagonist, 10 µg per mouse, ip, 4 days), L-161,982 (EP4 antagonist, 10 µg per mouse, ip, 4 days) or G-CSF plus Meloxicam and an EP2, EP1/3 or EP4 agonist (10 µg per mouse, ip, 4 days) or dmPGE2 (10 µg per mouse, ip, 4 days). For baboon studies, a baseline bleed was performed for CBC, CD34 and CFC analysis. Two days later, 2 baboons were treated with 10µg/kg G-CSF, and 2 baboons were treated subcutaneously with 10µg/kg G-CSF and 0.2 mg/kg Meloxicam on day 1, followed by 0.1 mg/kg Meloxicam subsequent days, for 5 total days. Blood was collected following treatment regimen for CBC, CD34, and CFC analysis. Following a 2 week resting period, the above procedure was repeated, switching treatment groups for individual baboons. Additionally, after another 2 week resting period, blood was collected before and after a 5 day treatment regimen with Meloxicam and CBC, CD34, and CFC were analyzed. For healthy volunteer studies, subjects naive to any medications within 30 days received a baseline bleed, followed by a second bleed after a 5-day regimen of 15 mg of meloxicam per day, orally. CD34 cells were assessed by the ISHAGE procedure 35 performed by the Stem Cell Laboratory of the IUSM Bone Marrow Transplant Program. CFC were assessed as described above.
Limiting dilution competitive transplantation
CD45.1 mice were mobilized with a standard 4 day regimen of G-CSF, or G-CSF plus a 4 day regimen of Meloxicam (6 mg/kg). In some studies designed to evaluate timing and duration of NSAID dosing in combination with G-CSF, initiation of the NSAID regimen preceded G-CSF and was staggered such that NSAID administration ended simultaneous with the G-CSF regimen (no stagger), 1 day prior to G-CSF (1 day stagger) or 2 days prior to G-CSF (2 day stagger) (regimens as depicted in the corresponding data figure). On day 5, PBMC were acquired and transplanted at 1:1, 2:1, 3:1 or 4:1 ratios with 5×105 C57Bl/6J WBM competitors into lethally irradiated C57Bl/6J recipient mice. Peripheral blood chimerism was monitored monthly, and CRU and LT-HSC frequency calculated. Transplants to evaluate LT-HSC mobilized in OPN−/− mice or with EP4 antagonist were performed competitively at a 4:1 ratio; 800,000 PBMC from CD45.2 mice versus 200,000 WBM from CD45.1 mice and peripheral blood chimerism and multilineage reconstitution assessed 16 weeks post-transplant.
Recovery assay
Mice were mobilized with G-CSF or G-CSF plus meloxicam with staggered dosing as described above and 2×106 mobilized PBMC transplanted non-competitively into cohorts of 10 lethally irradiated recipients per group. A cohort of non-irradiated mice was bled on the same schedule as the experimental treated groups of mice. Every other day, 5 mice from each group were bled (~50µl from a tail snip) and neutrophils and platelets in blood enumerated using a Hemavet 950FS. Alternate groups of 5 mice were bled on each successive bleeding time point so that mice were only bled once every 4 days. Recovery of neutrophils and platelets to 50% and 100% were determined by comparison to the average neutrophil and platelet counts in the control group throughout the experimental period. After 90 days, mice were sacrificed, bone marrow harvested, and transplanted at a 2.5:1 ratio with 2×105 congenic competitors into lethally irradiated recipients to determine long-term repopulating ability of the primary mobilized graft.
EP4 Chimera generation and mobilization assay
Chimeras were generated using EP4Cre flox/flox and age and sex matched EP4flox/flox littermate controls. EP4flox/flox mice were lethally irradiated and transplanted with 2 ×106 WBM cells from either EP4flox/flox mice, allowing for generation of a WT:WT chimera, or from EP4Cre flox/flox mice allowing for generation of a KO:WT chimera. Similarly, EP4Cre flox/flox mice were lethally irradiated and transplanted with 2 × 106 WBM cells from EP4flox/flox mice, allowing for generation of a WT:KO chimera. At 8 weeks post-transplant, all mice were treated with 2mg tamoxifen for three consecutive days, rested for 3 days and injected for 3 more days. Mice were then treated with G-CSF or G-CSF + meloxicam starting 10 days after the last treatment, and peripheral blood CFC and SLAM SKL assessed as described. EP4 gene deletion was confirmed by qRT-PCR.
Quantitative RT-PCR
For EP receptor expression on sorted osteolineage cells, total RNA was extracted with Purelink™ RNA micro Kit (Invitrogen, Grand Island, NY). On-column DNase treatment was performed according to the manufacturers' instructions to eliminate contaminating genomic DNA. Conventional reverse transcription was followed with SuperScript™ III First-Strand Synthesis System (Invitrogen). QRT-PCR was performed by using SYBR advantage qPCR Premix kit (Clontech) on MxPro-3000 (Agilent, LaJolla, CA). Primers were synthesized at IDT (Supplementary Table 1). A primer concentration of 250 nM was found to be optimal in all cases. The PCR protocol consisted of one cycle at 95 °C (5min) followed by 45 cycles of 95°C (15s), 55°C (30s) and 72°C (30s). The dissociation curves were determined on each analysis to confirm that only one product was obtained. Expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and hypoxanthine guanine phosphoribosyl transferase (HPRT) were generally used as reference genes. The average threshold cycle number (Ct) for each tested mRNA was used to quantify the relative expression of each gene. For analysis of hematopoietic supportive molecules on sorted osteolineage cells from vehicle treated or NSAID treated mice, quantitative RT-PCR was performed with the TaqMan gene expression assay kit (Life Technologies) (Supplementary Table 2) with cDNA generated from the High Capacity cDNA Reverse Transcription Kit (Life Technologies). Microfluidic quantitative RT-PCR was performed on BioMark Dynamic Arrays according to manufacturer’s instructions (Fluidigm Corporation).
Micro-computed tomography µCT
Formalin fixed tibiae and femora were imaged with micro-CT using a microCT-viva 40 (Scanco Medical AG, Bassersdorf, Switzerland) using a voxel size of 10.5 um in all dimensions (N=5). The bones were mounted in a cylindrical specimen holder to be captured in a single scan. Bones were secured in the specimen holder with gauze and were completely submerged in 70% ethanol. The region of interest comprised 100 transverse CT slices. Scans with an isotropic resolution of 10.5 µm were made using a 55-kV peak voltage X-ray beam. Fractional bone volume (BV/TV, Fraction) and architectural properties of trabecular reconstructions, apparent trabecular thickness (Tb.Th.), trabecular number (Tb.N.), trabecular spacing (Tb.Sp.), and connectivity density (Conn.D.) were calculated.
Dynamic and Static bone histomorphometry
Dynamic bone formation assays using staggered double calcein labeling, as we described 36. Bone histomorphometry was performed on 7 µm thick sections of undecalcified femurs embedded in methylmethacrylate using standard procedures. The mineral apposition rate (MAR, mm/day), mineralizing surface (MS/BS) and bone formation rate (BFR/BS, mm3/mm2/day) were measured on femora. Modified Goldner's Trichrome staining procedure was performed on 7 µm thick sections of undecalcified femurs embedded in methylmethacrylate. The osteoid surfaces as well as quiescent surfaces were measured on the tissue sections. Bone marrow sinusoids were visualized with Anti-VEFGFR3 on 3.5 um section. Vessels were identified by the positive staining around the vessel walls and vessel areas were measured using automated measuring system and expressed as a percentage / tissue volume. Vessel surface was traced with the same automated system. Vessel wall that showed an intact epithelial surface was expressed as endothelial surface over total vessel surface. For Col2.3 GFP analysis, 3.5 um thick sections were obtained from treated Col2.3 GFP mice. Sections were visualized under fluorescent microscope (Leica D100) using a FITC filter. Images were captured at 400× magnification at 4 different areas in the mid shaft of the femur. GFP+ osteoblasts were counted on endocortical bone surface and data was expressed at number of osteoblasts/endocortical bone surface. Osteoblast surface defined as endocortical bone surface covered by osteoblasts were measured and expressed and osteoblasts surface over endocortical bone surface. All histomorphometry was done on images captured using a Leica microscope outfitted with Q-imaging camera (W. Nuhsbaum Inc., McHenry, IL) and the histomorphometry was done using Bioquant Osteo software automated measuring system (Bioquant imaging corporation, Nashville, TN). All histomorphometry values were expressed according to the standard nomenclature 37,38.
Immunohistochemistry
Immunohistochemical analysis was performed on decalcified paraffin-embedded tissue sections. Antibodies against N-Cadherin (Abcam Inc., Cambridge, MA) primary: Rabbit polyclonal to N-Cadherin and SDF1 N-terminal respectively, Secondary : anti-Rabbit from Vector Laboratories (Burlingame, CA). Osteopontin (OPN) Ab was purchased from R&D Systems primary: Anti-mouse OPN, Secondary: Biotynylated anti-goat HRP conjugate, HRP-DAB System and DAB Chromogen. Rat IgG2 isotype was used as a primary antibody negative control for SDF-1, OPN and N-Cadherin in the concentration of 1:50. Isotype staining control was performed under the same conditions as the antibody staining.
COX metabolite and activity analysis
Mice were treated with vehicle control or meloxicam s.c. bid. One hour after the last treatment, femurs were pulled and flushed with 1 ml of ice cold PBS, quickly brought to single cell suspension and the flash frozen. COX-1 and COX-2 derived metabolites were assessed by GC/MS as we have previously described 39,40. The second femur was processed in an identical way and COX-1 and COX-2 activity determined using a fluorescent COX activity assay following the manufacturer’s instructions (Cayman Chemicals, kit #700200).
Supplementary Material
ACKNOWLEDGEMENTS
These studies were supported by NIH Grants HL096305 (LMP), CA143057, CA069158 (TG, KSM), HL100402 (DTS) and DK37097 (RMB). JH was supported by NIH training grants DK07519, HL07910 and HL087735. Flow cytometry was performed in the Flow Cytometry Resource Facility of the Indiana University Simon Cancer Center (NCI P30 CA082709). Additional core support was provided by a Center of Excellence in Hematology grant P01 DK090948. The authors would like to thank Hal E. Broxmeyer and Borja Saez for critically reading the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
All authors assisted in writing of the manuscript. J.H. analyzed data, wrote the manuscript, designed all experiments and implemented all experiments with assistance from P.S., A.F.H., B.R.C., J.M.S., P.H., B.A.P., K.N.S., F.F., L.S., F.K.W. K.S.M., M.C. and T.G. performed histologic analyses and assisted with corresponding study designs. G.L.M. and R.M.B. performed eicosanoid analysis and generated EP knockout mice, and C.H.S. assisted with 5-ALOX mice and experiments. D.T.S. and E.F.S. assisted with experimental design and data analyses. L.M.P. designed and performed experiments, analyzed and evaluated all data, and wrote the manuscript.
AUTHOR INFORMATION
Reprints and permissions information is available at www.nature.com/reprints. J.H. and L.M.P. have filed patent applications based on these findings. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
REFERENCES
- 1.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 2.Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raaijmakers MH, Mukherjee S, Guo S, Zhang S, Kobayashi T, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Niu C, Ye L, Huang H, He X, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 6.Hoggatt J, Singh P, Sampath J, Pelus LM. Prostaglandin E2 enhances hematopoietic stem cell homing, survival, and proliferation. Blood. 2009;113:5444. doi: 10.1182/blood-2009-01-201335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed M, Khanna D, Furst DE. Meloxicam in rheumatoid arthritis. Expert. Opin. Drug Metab Toxicol. 2005;1:739. doi: 10.1517/17425255.1.4.739. [DOI] [PubMed] [Google Scholar]
- 9.Rinder HM, Tracey JB, Souhrada M, Wang C, Gagnier RP, et al. Effects of meloxicam on platelet function in healthy adults: a randomized, double-blind, placebo-controlled trial. J. Clin. Pharmacol. 2002;42:881. doi: 10.1177/009127002401102795. [DOI] [PubMed] [Google Scholar]
- 10.Breyer RM, Bagdassarian CK, Myers SA, Breyer MD. Prostanoid receptors: subtypes and signaling. Annu. Rev. Pharmacol. Toxicol. 2001;41:661. doi: 10.1146/annurev.pharmtox.41.1.661. [DOI] [PubMed] [Google Scholar]
- 11.Katayama Y, Battista M, Kao WM, Hidalgo A, Peired AJ, et al. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell. 2006;124:407. doi: 10.1016/j.cell.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 12.Bethel M, Srour EF, Kacena MA. Hematopoietic cell regulation of osteoblast proliferation and differentiation. Curr. Osteoporos. Rep. 2011;9:96. doi: 10.1007/s11914-011-0048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoggatt J, Pelus LM. Many mechanisms mediating mobilization: an alliterative review. Curr. Opin. Hematol. 2011;18:231. doi: 10.1097/MOH.0b013e3283477962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mundy GR, Yoneda T, Hiraga T. Preclinical studies with zoledronic acid and other bisphosphonates: impact on the bone microenvironment. Semin. Oncol. 2001;28:35. doi: 10.1016/s0093-7754(01)90263-5. [DOI] [PubMed] [Google Scholar]
- 15.Winkler IG, Sims NA, Pettit AR, Barbier V, Nowlan B, et al. Bone marrow macrophages maintain hematopoietic stem cell (HSC) niches and their depletion mobilizes HSCs. Blood. 2010;116:4815. doi: 10.1182/blood-2009-11-253534. [DOI] [PubMed] [Google Scholar]
- 16.Chow A, Lucas D, Hidalgo A, Mendez-Ferrer S, Hashimoto D, et al. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J. Exp Med. 2011;208:261. doi: 10.1084/jem.20101688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christopher MJ, Rao M, Liu F, Woloszynek JR, Link DC. Expression of the G-CSF receptor in monocytic cells is sufficient to mediate hematopoietic progenitor mobilization by G-CSF in mice. J. Exp Med. 2011;208:251. doi: 10.1084/jem.20101700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heissig B, Hattori K, Dias S, Friedrich M, Ferris B, et al. Recruitment of stem and progenitor cells from the bone marrow niche requires MMP-9 mediated release of kit-ligand. Cell. 2002;109:625. doi: 10.1016/s0092-8674(02)00754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chitteti BR, Cheng YH, Poteat B, Rodriguez-Rodriguez S, Goebel WS, et al. Impact of interactions of cellular components of the bone marrow microenvironment on hematopoietic stem and progenitor cell function. Blood. 2010;115:3239. doi: 10.1182/blood-2009-09-246173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura Y, Arai F, Iwasaki H, Hosokawa K, Kobayashi I, et al. Isolation and characterization of endosteal niche cell populations that regulate hematopoietic stem cells. Blood. 2010;116:1422. doi: 10.1182/blood-2009-08-239194. [DOI] [PubMed] [Google Scholar]
- 21.Stier S, Ko Y, Forkert R, Lutz C, Neuhaus T, et al. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J. Exp. Med. 2005;201:1781. doi: 10.1084/jem.20041992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grassinger J, Haylock DN, Storan MJ, Haines GO, Williams B, et al. Thrombin-cleaved osteopontin regulates hemopoietic stem and progenitor cell functions through interactions with alpha9beta1 and alpha4beta1 integrins. Blood. 2009;114:49. doi: 10.1182/blood-2009-01-197988. [DOI] [PubMed] [Google Scholar]
- 23.Nie Y, Waite J, Brewer F, Sunshine MJ, Littman DR, et al. The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J. Exp. Med. 2004;200:1145. doi: 10.1084/jem.20041185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy CR, Zhang Y, Brandon S, Guan Y, Coffee K, et al. Salt-sensitive hypertension and reduced fertility in mice lacking the prostaglandin EP2 receptor. Nat. Med. 1999;5:217. doi: 10.1038/5583. [DOI] [PubMed] [Google Scholar]
- 25.Guan Y, Zhang Y, Wu J, Qi Z, Yang G, et al. Antihypertensive effects of selective prostaglandin E2 receptor subtype 1 targeting. J. Clin. Invest. 2007;117:2496. doi: 10.1172/JCI29838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider A, Guan Y, Zhang Y, Magnuson MA, Pettepher C, et al. Generation of a conditional allele of the mouse prostaglandin EP4 receptor. Genesis. 2004;40:7. doi: 10.1002/gene.20048. [DOI] [PubMed] [Google Scholar]
- 27.Mignone JL, Kukekov V, Chiang AS, Steindler D, Enikolopov G. Neural stem and progenitor cells in nestin-GFP transgenic mice. J. Comp Neurol. 2004;469:311. doi: 10.1002/cne.10964. [DOI] [PubMed] [Google Scholar]
- 28.Kalajzic Z, Liu P, Kalajzic I, Du Z, Braut A, et al. Directing the expression of a green fluorescent protein transgene in differentiated osteoblasts: comparison between rat type I collagen and rat osteocalcin promoters. Bone. 2002;31:654. doi: 10.1016/s8756-3282(02)00912-2. [DOI] [PubMed] [Google Scholar]
- 29.Chen XS, Sheller JR, Johnson EN, Funk CD. Role of leukotrienes revealed by targeted disruption of the 5-lipoxygenase gene. Nature. 1994;372:179. doi: 10.1038/372179a0. [DOI] [PubMed] [Google Scholar]
- 30.King AG, Horowitz D, Dillon SB, Levin R, Farese AM, et al. Rapid mobilization of murine hematopoietic stem cells with enhanced engraftment properties and evaluation of hematopoietic progenitor cell mobilization in rhesus monkeys by a single injection of SB-251353, a specific truncated form of the human CXC chemokine GRObeta. Blood. 2001;97:1534. doi: 10.1182/blood.v97.6.1534. [DOI] [PubMed] [Google Scholar]
- 31.Pelus LM, Broxmeyer HE, Kurland JI, Moore MA. Regulation of macrophage and granulocyte proliferation. Specificities of prostaglandin E and lactoferrin. J. Exp. Med. 1979;150:277. doi: 10.1084/jem.150.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broxmeyer HE, Mejia JA, Hangoc G, Barese C, Dinauer M, et al. SDF-1/CXCL12 enhances in vitro replating capacity of murine and human multipotential and macrophage progenitor cells. Stem Cells Dev. 2007;16:589. doi: 10.1089/scd.2007.0044. [DOI] [PubMed] [Google Scholar]
- 33.Fukuda S, Bian H, King AG, Pelus LM. The chemokine GRObeta mobilizes early hematopoietic stem cells characterized by enhanced homing and engraftment. Blood. 2007;110:860. doi: 10.1182/blood-2006-06-031401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoggatt AF, Hoggatt J, Honerlaw M, Pelus LM. A spoonful of sugar helps the medicine go down: a novel technique to improve oral gavage in mice. J. Am. Assoc. Lab Anim Sci. 2010;49:329. [PMC free article] [PubMed] [Google Scholar]
- 35.Sutherland DR, Anderson L, Keeney M, Nayar R, Chin-Yee I. The ISHAGE guidelines for CD34+ cell determination by flow cytometry. International Society of Hematotherapy and Graft Engineering. J. Hematother. 1996;5:213. doi: 10.1089/scd.1.1996.5.213. [DOI] [PubMed] [Google Scholar]
- 36.Mohammad KS, Chen CG, Balooch G, Stebbins E, McKenna CR, et al. Pharmacologic inhibition of the TGF-beta type I receptor kinase has anabolic and anti-catabolic effects on bone. PLoS. One. 2009;4:e5275. doi: 10.1371/journal.pone.0005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowe PS, Matsumoto N, Jo OD, Shih RN, Oconnor J, et al. Correction of the mineralization defect in hyp mice treated with protease inhibitors CA074 and pepstatin. Bone. 2006;39:773. doi: 10.1016/j.bone.2006.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, et al. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J. Bone Miner. Res. 1987;2:595. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- 39.Murali G, Milne GL, Webb CD, Stewart AB, McMillan RP, et al. Fish oil and indomethacin in combination potently reduce dyslipidemia and hepatic steatosis in LDLR−/−) mice. J. Lipid Res. 2012;53:2186. doi: 10.1194/jlr.M029843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu T, Laidlaw TM, Feng C, Xing W, Shen S, et al. Prostaglandin E2 deficiency uncovers a dominant role for thromboxane A2 in house dust mite-induced allergic pulmonary inflammation. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12692. doi: 10.1073/pnas.1207816109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.