Abstract
The application of small molecules has played a crucial role in identifying novel components involved in plant signalling. Compared to classic genetic approaches, small molecule screens offer notable advantages in dissecting plant biological processes, such as technical simplicity, low start-up costs, and most importantly, bypassing the problems of lethality and redundancy. To identify small molecules that target a biological process or protein of interest, robust and well-reasoned high-throughput screening approaches are essential. In this review, we present a series of principles and valuable approaches in small molecule screening in the plant model system Arabidopsis thaliana. We also provide an overview of small molecules that led to breakthroughs in uncovering phytohormone signalling pathways, endomembrane signalling cascades, novel growth regulators, and plant defence mechanisms. Meanwhile, the strategies to deciphering the mechanisms of these small molecules on Arabidopsis are highlighted. Moreover, the opportunities and challenges of small molecule applications in translational biology are discussed.
Keywords: Chemical biology, Plant growth and development, Small molecules, Screening, Translational biology
Introduction
There is a long tradition of small molecule screenings to generate starting points (hit compounds) for drug discovery in animal and microbial systems. This requires a screening collection with a large number of compounds that can be analysed for the desired effect. Pharmaceutical companies have access to collections that often amount to a total of several millions of compounds. In addition, the agro-industry has used similar approaches to identify useful agrochemicals. In recent years, diverse compound collections have become available to academic researchers through commercial suppliers. The availability of these commercial chemical libraries allows exploration of their effect in specific pathways and cellular processes in an academic setting [20].
The effect of these compounds can be tested via two types of screening approaches. In pharmaceutical companies, drug discovery often utilizes a target-based approach, by looking at a protein that plays a role in a specific disease process and subsequently identifying compounds that interfere with the function of that protein [34]. But in addition to this, drug discovery can also be approached in a phenotypic way to identify compounds that produce a certain phenotype-of-interest, either in a model organism or in a cell-based system. For this purpose, highly advanced and innovative ways for screening and evaluating compounds have been developed. One such tool that has been extensively used in phenotypic screening is high content imaging. By utilising automated microscopy, scientists can design in-depth qualitative and quantitative paradigms into specific cellular and subcellular processes to discover how these processes respond to certain chemical stimuli [41]. This type of screening has led to significant breakthroughs in the field of neurobiology, for example by the discovery of compound FK506 and its respective immunophilin receptors [27]. Other areas of research that have been significantly advanced include oncology, toxicology, cell cycle research, and protein ligand and receptor identification [1, 5, 8, 24].
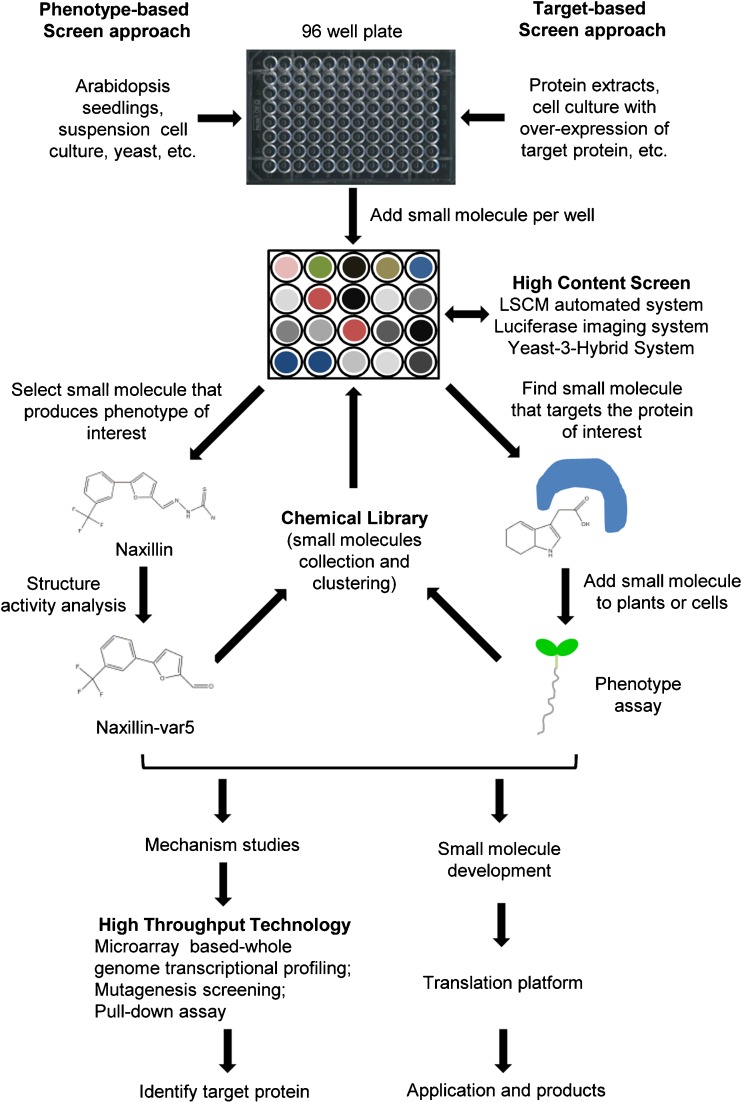
In agricultural research, synthetic molecules have a longstanding tradition to be applied as fungicides, insecticides, and herbicides. Many of them are structural analogues of plant hormones, which are very pleiotropic in nature, and as such are more difficult to apply as research tools to study specific developmental processes. Only recently, the application of chemicals to study biological processes (‘chemical biology’ or ‘chemical genetics’) has found its way into the field of plant sciences (Fig. 1). Many of the general methods and principles of chemical biology can also be utilised in the plant research field. Here, we will review the screening approaches that were used to identify novel chemical research tools and the strategies to identify their specific mode-of-action.
Fig. 1.
Small molecule screen approaches. The phenotype-based approach (left) is analogous to forward genetics and comprises three different steps. The first step is the assembly of a set of mutation equivalents, i.e. a chemical library with 10,000 or more compounds capable of altering protein function. Subsequently, a high-throughput screen is performed to identify compounds that affect a biological process of interest. A high-content screen can be processed by using advanced technologies. Target-based chemical genetics (right) is comparable to reverse genetics and entails overexpressing a protein of interest, screening for compounds that interact with the protein, and finally using this compound to determine the phenotypic consequences of altering the function of this protein in a cellular context. As a final step, the protein targets of these compounds or the potential mechanism are identified. Furthermore, bioactive small molecules would be modified and applied into a translation platform. Naxillin was used as example for the structure-activity analysis
Why do we need to screen in plants?
The significance of using synthetic molecules to disrupt highly specific biological processes in plants is evident when looking at the advantages of this technique compared to classical genetics. In plant and animal systems, the highly conserved nature of, for instance, protein kinases or phosphatases, which constitute large families of signal transduction enzymes, presents a challenging task for the development of chemical inhibitors that target only a subset of these enzymes. RNA interference against non-conserved sequences can be used as a genetic approach to analyse a subset of a large gene family during plant growth and development. However, this approach can become a significant problem when these genes play an essential role in development at the embryonic stage. Mutations in essential genes often lead to embryonic lethality, and thus, prevent the discovery of other roles for that gene later in development. For example, AURORA (AUR) kinases and PROTEIN PHOSPHATASE 2A (PP2A) are comprised of multiple classes or subunits, respectively. Single aur kinase mutants show no obvious macroscopic phenotype, whereas double mutants with strong alleles lead to gametophytic lethality and no plants can be recovered [43]. This makes it difficult to determine the potential roles of these proteins at later stages of development. Unlike genetic approaches, in which mutations at the DNA level perturb gene function, synthetic molecules exert their effect directly at the protein level in a manner which is tunable, reversible, and conditional. Therefore, embryonic lethality can be circumvented, and the effect of the molecules can be assessed in later developmental stages under variable conditions.
Although inhibitors against animal PP2As, such as cantharidin and okadaic acid, and AUR kinases, such as aurora inhibitor II, are available for the research community, they are ineffective in plants because they abolish overall activity, are not very specific, and/or result in pleiotropic effects [4, 6, 14, 31]. For instance, the AUR family consists of two classes [13] of which only α AUR kinases (AUR1/2) are involved in formative division plane orientation [43]. Therefore, general inhibitors affecting the activity of all three Arabidopsis AUR kinases would not be useful when examining the specific process of cell division plane orientation and cell division. Thus, to modulate the activity of individual proteins within a biological process, novel, (plant-)specific molecules are required.
Small molecules are also very useful as they can address the issue of genetic redundancy, a problem often associated with reverse genetic approaches in plants. If interfering with multiple pathways simultaneously is required to influence plant growth and development, multiple molecules can be added, which is analogous to multiple gene modifications. Alternatively, synthetic molecules can target several members of the same protein family (i.e. by interacting at conserved sites) and can consequently overcome genetic redundancy. Additionally, due to the highly conserved nature of major plant protein families, such as receptor-like kinases (RLKs), chemical genetics in model systems (like Arabidopsis thaliana) allows for techniques to be transferred from one species to another, greatly enhancing the significance of a single chemical screen.
Screening procedures
A prerequisite to find new chemicals that interfere with a certain phenotypic response or biological pathway is the availability of a ‘compound screening toolbox’. First, a large collection of compounds needs to be available that, as a whole, is capable of altering the function of a broad range of proteins, including those involved in the biological process of interest. The screening collection can consist of synthetic molecules, natural products, or small signalling peptides (collectively referred to as compounds) [19]. There are several compound collections commercially available that can be used for small molecule screening in Arabidopsis [37]. For example, the ChemBridge DIVERSet library contains in total about 100,000 drug-like low molecular mass molecules designed to maximize structural diversity (http://www.chembridge.com/screening_libraries/). Subsets of this collection have been used previously in Arabidopsis screenings and have yielded interesting hits and tool compounds [12, 15, 23]. Similar diverse collections are also available from other suppliers such as Life Chemicals (http://www.lifechemicals.com/), Asinex (http://www.asinex.com/Libraries.html), and TimTec (http://www.timtec.net/Screening-Compound-Libraries.html). During the assembly process of these collections, compounds are selected via in silico filtering algorithms based upon physico-chemical properties to enhance bio-availability. In addition, substructure analyses are applied to remove unstable and/or toxic compounds [44]. Diversity of the compound collection is essential if no prior knowledge of the protein target is known and the screening aims for the identification of compounds that interfere with a phenotypic response rather than a specific protein. On the other hand, if structural information is known about the protein site(s) to target, a more focused library can be designed in which screening compounds are assembled or synthesized based upon one or several structural scaffolds. Most suppliers allow cherry picking from their collection to assemble custom and/or focused libraries. In some cases, commercial focused libraries are already available such as collections of kinase inhibitors and ion channel inhibitors.
To assess the potential effect of a compound collection on a particular biological process or protein-of-interest, a robust screening assay has to be developed in cell-free systems, cellular systems, or even small model organisms. In the animal field, these are, for example, Danio rerio or Xenopus laevis embryos [22, 39]. In plants, these are mainly A. thaliana seedlings, but also suspension cells [33]. In yeast, Saccharomyces cerevisiae is used as a tool in the yeast-3-hybrid system, allowing for molecule–protein interactions in vivo, which can be used to refute or confirm interactions shown in other model systems [25]. An important aspect during assay development is miniaturization of the assay to 96- or 384-well plates. This significantly reduces reagent costs during screening campaigns and makes the assay compatible with automation and liquid handling systems, which allows the distribution of compounds, reagents, and model systems in a high-throughput fashion. Because in many screening collections compounds are dissolved in DMSO, determining the sensitivity of the model system to DMSO is essential to avoid toxicity due to too high solvent concentrations. In addition, analysis of positive and negative controls during assay development allows to determine the assay window and to calculate a Z’ value, a measure to assess robustness of the screening assay [46]. After assay development and acquisition or synthesis of the screening compounds, the compound collection is applied to the assay system with automated liquid handling platforms, and the assay output is detected by means of automated plate readers or microscopes. Informatics and databases are required to track, analyse, and retrieve screening data. After hit identification, hits are validated with secondary screening assays and chemical characterization including evaluation of chemical structure and initial structure-activity analysis.
Chemical genetics in plant growth
Chemical genetic approaches have been successfully applied to study plant signalling pathways and to modulate plant growth [2, 9, 11, 18, 23, 38, 42]. Initially, chemical screens were mainly applied to gain insight into auxin signal transduction. For example, the small molecule sirtinol was identified because it activated the auxin signal transduction pathway and mimicked auxin-related developmental phenotypes. It led to the identification of SIRTINOL RESISTANT 1 (SIR1), an upstream regulator of auxin signalling pathways [48]. Further studies revealed that the activation of sirtinol required a functional aldehyde oxidase [9]. In addition, inhibitory small molecules of auxin signalling pathways have also been identified by chemical screens [2]. Only recently, phenotype-based small molecule screens in Arabidopsis gave rise to the discovery of various novel signalling pathways in abiotic stress and plant growth development [11, 23, 35, 36]. In this section, well-characterized small molecules which were identified from phenotypic screens will be introduced. Furthermore, the screening methods and the mechanism of these chemicals will be briefly discussed.
Pyrabactin
The identification of the synthetic molecule pyrabactin (4-bromo-N-[pyridin-2-yl methyl]naphthalene-1-sulfonamide) as a selective abscisic acid (ABA) agonist has led to major breakthroughs in understanding ABA perception mechanisms [35]. Although many intermediate signalling components had been described before [16], knowledge at the level of ABA perception was only marginal. This was mainly due to the high genetic redundancy of the ABA receptor gene family. During a screen of a 10,000-membered chemical library, pyrabactin was identified as a synthetic seed germination inhibitor in an A. thaliana seed germination assay [47]. An ABA-hypersensitive Arabidopsis accession was observed to also show hypersensitivity to pyrabactin. Subsequently, pyrabactin-insensitive mutants were identified containing insensitive alleles of PYRABACTIN RESISTANCE1 (PYR1) genes. PYR1 was shown to interact with HYPERSENSITIVE TO ABA1 (HAB1) (homolog of ABI1 and ABI2), a protein phosphatase which is a negative regulator of ABA signalling. Thus, the selectivity of pyrabactin for a subset of ABA receptors allowed to bypass this redundancy, and led to the identification of PYR/REGULATORY COMPONENT OF ABA RECEPTOR (RCAR) proteins as ABA receptors [35]. The PYR/RCAR proteins act together with PP2Cs and SNF1-RELATED PROTEIN KINASE2 (SnRK2s) [17, 45] as negative and positive regulators, respectively, of downstream ABA signalling [28, 35]. This breakthrough, together with further detailed structural and mutational approaches, provided new insights into ABA perception and signalling, and exemplified the need for and use of target-specific agonists in chemical genetics [30, 32].
Bikinin
In addition to specific agonists, such as pyrabactin, general antagonists can also be powerful chemical tools. For example, bikinin, (4-[(5-bromo-2-pyridinyl)amino]-4-oxobutanoic acid), was identified as an activator of brassinosteroid (BR) signalling in a screen for small molecules that induce a constitutive BR response [10]. A commercial 10,000-compound library (DIVERSet, ChemBridge Corporation) was used for this screen. The structure-activity analysis identified bikinin as a non-steroidal molecule modulating the BR signalling cascade downstream of the BRASSINOSTEROID-INSENSITIVE1 (BRI1) receptor. A combination of BES1 phosphorylation analysis, kinase assays, surface plasmon resonance binding studies, and microarray analysis showed that bikinin directly targets BRASSINOSTEROID-INSENSITIVE2 (BIN2) protein, which belongs to the group II glycogen synthase kinase 3 family (GSK3s). To assess the binding mode of bikinin, an ATP-competition assay with BIN2 and modelling of the compound into the crystal structure of the human BIN2 homolog, GSK3β, revealed that bikinin acts as an ATP-competitive kinase inhibitor. In A. thaliana, a set of ten GSK3s is present [21]. Interestingly, because bikinin targets several subsets of GSK3s, including a subset of three GSK3s shown to be involved in the negative regulation of BR signalling, the compound could act as a conditional and multiple knockout tool for this subset of GSK3s and therefore induce a BR response [10]. This type of response would never have been observed by single loss-of-function mutants in genes encoding GSK3s or by a selective GSK3 inhibitor. Thus, the specificity of bikinin for a subset of GSK3s offers the opportunity to study other effects of specifically inhibiting GSK3s in A. thaliana.
DFPM
The small molecule [5-(3, 4-dichlorophenyl) furan-2-yl]-piperidine-1-ylmethanethione (DFPM) has been used to determine the coordination and interaction between abiotic stress and plant immunity [23]. DFPM was first selected from a chemical library of ChemBridge’s DIVERSet E library of 9,600 compounds (ChemBridge, San Diego) as a negative regulator of the ABA signalling pathway by using a WT-RAB18 reporter line. Microarray-based whole genome transcriptomic analysis revealed that DFPM downregulated ABA-induced gene expression, but also stimulates the expression of pathogen resistance genes, including PATHOGENESIS-RELATED5 (PRS) and ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1). Interestingly, the inhibitory effects of DFPM on ABA-responsive genes and ABA-induced stomatal closure were impaired in mutants of plant disease resistance pathways, such as eds1, pad4, sgt1b, and rar1, but not in npr1, which is the crucial salicylic acid (SA) response regulator [7]. This indicated that DFPM-dependent ABA signal transduction required early pathogen resistance response regulators rather than SA signalling. Notably, transcriptional activation of defence-related gene expression or Pseudomonas syringae infection can mimic the effect of DFPM on ABA responses, suggesting a negative regulation of ABA signal transduction by activation of plant immunity pathways. Further investigation on the mechanism of DFPM-interfered ABA signal transduction revealed that ABA perception by PYR/RCAR receptors [35] and subsequent activation of the major ABA signalling kinases, SnRK2s, were not affected by DFPM. However, DFPM blocked ABA-induced Ca2+ activated S-type anion channel currents in the wild-type guard cells, but not in pad4-1 background. This indicated a EDS1/PAD4–dependent plant immunity pathway which plays a key role in interrupting early ABA responses by modulation of Ca2+ signalling [23]. Taken together, the synthetic molecule DFPM has provided a comprehensive understanding of cross talk between biotic and ABA signalling networks. DFPM also presents the characteristics of an effective instigator of plant immunity, and could thus be widely applied in abiotic–biotic interaction research.
Naxillin
The non-auxin like probe naxillin was identified as a specific modulator of lateral root development from a marker/phenotype-based small-molecule screen of a commercial 10,000-compound library (DIVERSet, ChemBridge Corporation) in A. thaliana [12]. The plant hormone auxin is known as a regulator of many plant developmental processes, including lateral root development [10]. By contrast, naxillin specifically induces root branching with minimal side effects typical of auxin treatment, such as inhibition of primary root growth. At the transcriptome level, naxillin treatment induced 401 genes, whereas treatment with the synthetic auxin analog naphthalene acetic acid (NAA)-induced 2,581 genes, suggesting a much narrower mechanism of action. As such, naxillin represents a valuable tool to decipher the molecular networks involved in lateral root development. To gain insight into the mode of action of naxillin, an ethyl methane sulfonate-mutagenized population was screened, and a naxillin-resistant mutant allele was selected for further analysis. A positional cloning approach identified a missense mutation in INDOLE-3-BUTYRIC ACID RESPONSE3 (IBR3), which acts on conversion of indole-3-butyric acid (IBA) to indole-3-acetic acid (IAA) [49]. IBA-to-IAA conversion pathway mutants were further checked upon naxillin treatment and demonstrated that naxillin acts at the level of the enoyl-CoA hydratase step of the pathway. Expression pattern analysis of IBA-to-IAA conversion genes INDOLE-3-BUTYRIC ACID RESPONSE1, IRBR1IBR3, IBR10, and ABNORMAL INFLORESCENCE MERISTEM 1 (AIM1) revealed that expression domains of all these genes overlapped in the root tip of the primary root, specifically in root cap cells. This indicated that root cap-specific auxin production might be involved in root branching. The existence of tissue-specific sources of auxin as a mechanism to fine-tune developmental processes, such as root branching, has never been observed by applying auxins or its analogs, which produce the global effects on plant root developmental processes. This breakthrough provides new insights into the function of auxin homeostasis on root development and nicely illustrates how novel chemical tools can be applied to discover biological mechanisms that are involved in specific plant developmental processes.
Endosidins
The synthetic molecule endosidin1 (ES1) was elected from an automated image-based screen from a chemical library (Microsource Spectrum) containing 2,016 chemicals with known biological activity for inhibitors of pollen germination or effectors of polar growth, and the screen was conducted by using GFP-RIP1, a maker line of apical plasma membrane in Arabidopsis and tobacco pollen tubes [36]. The application of ES1 selectively disrupted the trafficking of PIN-FORMED (PIN) auxin efflux carrier PIN2, AUXIN INSENSITIVE1 (AUX1), and BRI1, and formed intracellular agglomerations termed “endosidin bodies”. Endosidin bodies were further defined as trans-golgi network/endosomal proteins SYP61 and the V-ATPase subunit VHA-a1. This suggested that SYP61 and VHA-a1 act as components of an early endosome compartment in PIN2 and AUX1 mediated endomembrane trafficking processes [36]. To explore more components involved in this pathway, a modified laser scanning confocal microscopy-based high-content intracellular screen was established, which allowed the identification of small molecules that phenocopy ES1 treatment [15]. Meanwhile, more chemical libraries, including Chembridge Diverset library, Chembridge, Novacore library, and Sigma TimTec Myria library, containing 46,418 compounds in total were screened. After two rounds of screening, 123 small molecules were selected as both inhibitors of pollen germination and effectors of plasma membrane markers. The image database was then transformed by a flexible algorithm into a marker-by-phenotype-by-treatment time matrix, and molecules were clustered into groups of endosidins (ESs) depending on the specific profiles of subcellular phenotypes. Although these molecules may induce a similar endomembrane trafficking phenotype, detailed analysis of different plasma membrane makers revealed diverse modes of action of these ESs on early events of endosome trafficking. For example, endosidin3 was found to target Rho GTPases trafficking and exhibited cell polarity defection, whereas endosidin5 was linked to PIN cycling and gravitropism. Thus, the direct discovery of endomembrane-defective phenotypes could then easily be linked to developmental phenotypes, which still poses a challenge for exclusively forward genetic screens. This breakthrough is the first time that an automated microscopy-driven phenotypic molecule screen has been used in plants, suggesting that a high-content small molecule screen could serve as an effective tool to illustrate intracellular signalling pathways in vivo, and also help to set up a comprehensive systems biology view.
Small molecules in translational plant sciences
The above examples illustrate the power of chemical genetics to identify chemical ‘probes’ that can be applied to study biology. From a translational point of view, small molecules could be of great value by forming the starting point in the discovery of new agrochemicals. Evidently, this requires that the compound’s target protein(s) and/or the mechanism of action be conserved between the species in which the activity of the compound was observed (e.g. A. thaliana) and the target crop species.
Based upon analysis of currently available pesticides and herbicides, agrochemicals obey certain structural and physico-chemical rules [40]. This is similar to drug-like properties as illustrated by Lipinski’s Rule-of-Five, which states that poor bioavailability (poor absorption and permeability) is more likely when more than 5 H-bond donors are present, more than 10 H-bond acceptors are present, the molecular weight is greater than 500 Da, and the calculated octanol/water coefficient is greater than 5 [26]. The ranges of these parameters for agrochemicals are similar, except for the lower acceptable number of H-bond donors. However, some important differences exist between agrochemicals and pharmaceuticals regarding the types of functional groups [40]. For example, to be able to protect a crop, a chemical must persist in the field for several weeks to be of practical value. Therefore, alcohols and amines are much less common in agrochemicals than in pharmaceuticals as these groups are less stable in field environments (due to ease of oxidation). Aromatic rings are also more prevalent among agrochemicals because aromatic rings are more likely to be stable in the environment than alicyclic rings. Finally, acidic groups such as carboxylic acids and acylsulfonamides are prevalent among post-emergence agrochemicals. This is because weakly acidic groups promote phloem mobility, which is required to transport the chemical to the growing points of the plant. These structural, functional, and physico-chemical constraints should be considered during the assembly of a compound screening collection with the aim to identify new types of agrochemicals. In view of non-GMO applications, synthetic molecules are required that specifically mimic, disturb and/or enhance protein activities, and that can easily and cost-effectively (potentially as a modified variant) be applied to crops (for instance through addition to fertiliser or water). This will generate tools (synthetic molecules) that can be widely applied to non-related species, without requiring genetic modifications. This translational approach is relevant considering the fact that several key signalling pathways are conserved between species.
Conclusion
The application of small molecules in plant research has expanded rapidly in the past decade and has made genuine contributions to our comprehensive knowledge of the molecular mechanisms of plant development. However, plant chemical genetics is now at the stage where faster and more efficient ways of screening have to be developed to permit wider accessibility in the plant research field. The establishment of a compound screening platform is of prime importance (Fig. 1) as small molecule use in plant systems has been shown to significantly accelerate and enhance developmental research. This requires development of robust screening assays in plant-based systems and compound collections that are more dedicated for applications in the field of plant sciences. In addition, the application of high-throughput imaging technologies in plant screenings would certainly technically allow us to delve more deeply into complex intracellular networks than previous approaches permitted. In addition, development of small molecules that can modulate protein–protein interactions remains a challenge even in human drug development, and heavily relies on biochemical and biophysical knowledge of the respective target interactions [3]. Such knowledge unfortunately remains scarce in plant biology. Thus, further investigation will not only be emphasized on searching for protein targets but also on the mechanistic level where small molecules act as regulators of, for instance, plant RLK signalling [29]. Importantly, one of the greatest challenges remaining is the generation of useful, applicable small molecules in agricultural production. This requires exploration of small molecules that affect specific protein activities, and that can easily and cost-effectively be applied to crops. This in turn could be a potential solution for the non-GMO, and could ultimately lead to a new green revolution.
Acknowledgments
This work was supported by a Biotechnology and Biological Science Research Council David Phillips Fellowship (BB_BB/H022457/1), a Marie Curie European Reintegration grant (PERG06-GA-2009-256354), the Interuniversity Attraction Poles Programme (IUAP VI/33) initiated by the Belgian State Science Policy Office, VIB, and the Special Research Fund of Ghent University. We thank the School of Biosciences and Malcolm. J. Bennett for studentship funding and acknowledge the University of Nottingham research committee.
Footnotes
Dominique Audenaert and Ive De Smet contributed equally to this study.
Contributor Information
Dominique Audenaert, Phone: +32-93-313840, FAX: +32-93-313809, Email: doaud@psb.vib-ugent.be.
Ive De Smet, Phone: +44-11-59516681, FAX: +44-11-5951629, Email: Ive.De_Smet@nottingham.ac.uk.
References
- 1.Agler M, Prack M, Zhu Y, Kolb J, Nowak K, Ryseck R, Shen D, Cvijic ME, Somerville J, Nadler S, Chen T. A high-content glucocorticoid receptor translocation assay for compound mechanism-of-action evaluation. J Biomol Screen. 2007;12:1029–1041. doi: 10.1177/1087057107309353. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong JI, Yuan S, Dale JM, Tanner VN, Theologis A. Identification of inhibitors of auxin transcriptional activation by means of chemical genetics in Arabidopsis. Proc Natl Acad Sci U S A. 2004;101:14978–14983. doi: 10.1073/pnas.0404312101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arkin MR, Wells JA. Small-molecule inhibitors of protein–protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 4.Bajsa J, Pan Z, Duke SO. Transcriptional responses to cantharidin, a protein phosphatase inhibitor, in Arabidopsis thaliana reveal the involvement of multiple signal transduction pathways. Physiol Plant. 2011;143:188–205. doi: 10.1111/j.1399-3054.2011.01494.x. [DOI] [PubMed] [Google Scholar]
- 5.Barabasz A, Foley B, Otto JC, Scott A, Rice J. The use of high-content screening for the discovery and characterization of compounds that modulate mitotic index and cell cycle progression by differing mechanisms of action. Assay Drug Dev Technol. 2006;4:153–163. doi: 10.1089/adt.2006.4.153. [DOI] [PubMed] [Google Scholar]
- 6.Baskin TI, Wilson JE. Inhibitors of protein kinases and phosphatases alter root morphology and disorganize cortical microtubules. Plant Physiol. 1997;113:493–502. doi: 10.1104/pp.113.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis Npr1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/S0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 8.Chuma M, Sakamoto M, Yasuda J, Fujii G, Nakanishi K, Tsuchiya A, Ohta T, Asaka M, Hirohashi S. Overexpression of cortactin is involved in motility and metastasis of hepatocellular carcinoma. J Hepatol. 2004;41:629–636. doi: 10.1016/j.jhep.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Dai X, Hayashi K, Nozaki H, Cheng Y, Zhao Y. Genetic and chemical analyses of the action mechanisms of sirtinol in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:3129–3134. doi: 10.1073/pnas.0500185102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Rybel B, Audenaert D, Beeckman T, Kepinski S. The past, present, and future of chemical biology in auxin research. ACS Chem Biol. 2009;4:987–998. doi: 10.1021/cb9001624. [DOI] [PubMed] [Google Scholar]
- 11.De Rybel B, Audenaert D, Vert G, Rozhon W, Mayerhofer J, Peelman F, Coutuer S, Denayer T, Jansen L, Nguyen L, Vanhoutte I, Beemster GT, Vleminckx K, Jonak C, Chory J, Inze D, Russinova E, Beeckman T. Chemical inhibition of a subset of Arabidopsis thaliana Gsk3-like kinases activates brassinosteroid signaling. Chem Biol. 2009;16:594–604. doi: 10.1016/j.chembiol.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Rybel B, Audenaert D, Xuan W, Overvoorde P, Strader LC, Kepinski S, Hoye R, Brisbois R, Parizot B, Vanneste S, Liu X, Gilday A, Graham IA, Nguyen L, Jansen L, Njo MF, Inze D, Bartel B, Beeckman T. A role for the root cap in root branching revealed by the non-auxin probe naxillin. Nat Chem Biol. 2012;8:798–805. doi: 10.1038/nchembio.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demidov D, Van Damme D, Geelen D, Blattner FR, Houben A. Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell. 2005;17:836–848. doi: 10.1105/tpc.104.029710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deruere J, Jackson K, Garbers C, Soll D, Delong A. The Rcn1-encoded a subunit of protein phosphatase 2a increases phosphatase activity in vivo. Plant J. 1999;20:389–399. doi: 10.1046/j.1365-313x.1999.00607.x. [DOI] [PubMed] [Google Scholar]
- 15.Drakakaki G, Robert S, Szatmari AM, Brown MQ, Nagawa S, Van Damme D, Leonard M, Yang Z, Girke T, Schmid SL, Russinova E, Friml J, Raikhel NV, Hicks GR. Clusters of bioactive compounds target dynamic endomembrane networks in vivo. Proc Natl Acad Sci U S A. 2011;108:17850–17855. doi: 10.1073/pnas.1108581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14(Suppl):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayashi K, Tan X, Zheng N, Hatate T, Kimura Y, Kepinski S, Nozaki H. Small-molecule agonists and antagonists of F-box protein–substrate interactions in auxin perception and signaling. Proc Natl Acad Sci U S A. 2008;105:5632–5637. doi: 10.1073/pnas.0711146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huggins DJ, Venkitaraman AR, Spring DR. Rational methods for the selection of diverse screening compounds. ACS Chem Biol. 2011;6:208–217. doi: 10.1021/cb100420r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iorio F, Isacchi A, di Bernardo D, Brunetti-Pierri N. Identification of small molecules enhancing autophagic function from drug network analysis. Autophagy. 2010;6:1204–1205. doi: 10.4161/auto.6.8.13551. [DOI] [PubMed] [Google Scholar]
- 21.Jonak C, Hirt H. Glycogen synthase kinase 3/shaggy-like kinases in plants: an emerging family with novel functions. Trends Plant Sci. 2002;7:457–461. doi: 10.1016/S1360-1385(02)02331-2. [DOI] [PubMed] [Google Scholar]
- 22.Kalin RE, Banziger-Tobler NE, Detmar M, Brandli AW. An in vivo chemical library screen in Xenopus tadpoles reveals novel pathways involved in angiogenesis and lymphangiogenesis. Blood. 2009;114:1110–1122. doi: 10.1182/blood-2009-03-211771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim TH, Hauser F, Ha T, Xue S, Bohmer M, Nishimura N, Munemasa S, Hubbard K, Peine N, Lee BH, Lee S, Robert N, Parker JE, Schroeder JI. Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr Biol. 2011;21:990–997. doi: 10.1016/j.cub.2011.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi H, Harada H, Nakamura M, Futamura Y, Ito A, Yoshida M, Iemura S, Shin-Ya K, Doi T, Takahashi T, Natsume T, Imoto M, Sakakibara Y. Comprehensive predictions of target proteins based on protein–chemical interaction using virtual screening and experimental verifications. BMC Chem Biol. 2012;12:2. doi: 10.1186/1472-6769-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Licitra EJ, Liu JO. A three-hybrid system for detecting small ligand–protein receptor interactions. Proc Natl Acad Sci U S A. 1996;93:12817–12821. doi: 10.1073/pnas.93.23.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 27.Liu D, McIlvain HB, Fennell M, Dunlop J, Wood A, Zaleska MM, Graziani EI, Pong K. Screening of immunophilin ligands by quantitative analysis of neurofilament expression and neurite outgrowth in cultured neurons and cells. J Neurosci Methods. 2007;163:310–320. doi: 10.1016/j.jneumeth.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 28.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of Pp2c phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 29.Marshall A, Aalen RB, Audenaert D, Beeckman T, Broadley MR, Butenko MA, Cano-Delgado AI, de Vries S, Dresselhaus T, Felix G, Graham NS, Foulkes J, Granier C, Greb T, Grossniklaus U, Hammond JP, Heidstra R, Hodgman C, Hothorn M, Inze D, Ostergaard L, Russinova E, Simon R, Skirycz A, Stahl Y, Zipfel C, De Smet I. Tackling drought stress: receptor-like kinases present new approaches. Plant Cell. 2012;24:2262. doi: 10.1105/tpc.112.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melcher K, Xu Y, Ng LM, Zhou XE, Soon FF, Chinnusamy V, Suino-Powell KM, Kovach A, Tham FS, Cutler SR, Li J, Yong EL, Zhu JK, Xu HE. Identification and mechanism of ABA receptor antagonism. Nat Struct Mol Biol. 2010;17:1102–1108. doi: 10.1038/nsmb.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortlock AA, Keen NJ, Jung FH, Heron NM, Foote KM, Wilkinson RW, Green S. Progress in the development of selective inhibitors of Aurora kinases. Curr Top Med Chem. 2005;5:807–821. doi: 10.2174/1568026054637719. [DOI] [PubMed] [Google Scholar]
- 32.Mosquna A, Peterson FC, Park SY, Lozano-Juste J, Volkman BF, Cutler SR. Potent and selective activation of abscisic acid receptors in vivo by mutational stabilization of their agonist-bound conformation. Proc Natl Acad Sci U S A. 2011;108:20838–20843. doi: 10.1073/pnas.1112838108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noutoshi Y, Okazaki M, Kida T, Nishina Y, Morishita Y, Ogawa T, Suzuki H, Shibata D, Jikumaru Y, Hanada A, Kamiya Y, Shirasu K. Novel plant immune-priming compounds identified via high-throughput chemical screening target salicylic acid glucosyltransferases in Arabidopsis. Plant Cell. 2012;24:3795–3804. doi: 10.1105/tpc.112.098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandey UB, Nichols CD. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol Rev. 2011;63:411–436. doi: 10.1124/pr.110.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robert S, Chary SN, Drakakaki G, Li S, Yang Z, Raikhel NV, Hicks GR. Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor Bri1 and the auxin transporters Pin2 and Aux1. Proc Natl Acad Sci U S A. 2008;105:8464–8469. doi: 10.1073/pnas.0711650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robert S, Raikhel NV, Hicks GR. Powerful partners: Arabidopsis and chemical genomics. Arabidopsis Book. 2009;7:e0109. doi: 10.1199/tab.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Savaldi-Goldstein S, Baiga TJ, Pojer F, Dabi T, Butterfield C, Parry G, Santner A, Dharmasiri N, Tao Y, Estelle M, Noel JP, Chory J. New auxin analogs with growth-promoting effects in intact plants reveal a chemical strategy to improve hormone delivery. Proc Natl Acad Sci U S A. 2008;105:15190–15195. doi: 10.1073/pnas.0806324105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y, Dong Z, Khodabakhsh H, Chatterjee S, Guo S. Zebrafish chemical screening reveals the impairment of dopaminergic neuronal survival by cardiac glycosides. PLoS One. 2012;7:e35645. doi: 10.1371/journal.pone.0035645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tice CM. Selecting the right compounds for screening: does Lipinski’s Rule of 5 for pharmaceuticals apply to agrochemicals? Pest Manag Sci. 2001;57:3–16. doi: 10.1002/1526-4998(200101)57:1<3::AID-PS269>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 41.Trask OJ. Nuclear factor kappa B (Nf-Kappab) translocation assay development and validation for high content screening. In: Sittampalam GS, Gal-Edd N, Arkin M, Auld D, Austin C, Bejcek B, Glicksman M, Inglese J, Lemmon V, Li Z, McGee J, McManus O, Minor L, Napper A, Riss T, Trask OJ, Weidner J, editors. Assay guidance manual. Bethesda, MD: Eli Lilly & Company and the National Center for Advancing Translational Sciences; 2004. [PubMed] [Google Scholar]
- 42.Tsuchiya Y, Vidaurre D, Toh S, Hanada A, Nambara E, Kamiya Y, Yamaguchi S, McCourt P. A small-molecule screen identifies new functions for the plant hormone strigolactone. Nat Chem Biol. 2010;6:741–749. doi: 10.1038/nchembio.435. [DOI] [PubMed] [Google Scholar]
- 43.Van Damme D, De Rybel B, Gudesblat G, Demidov D, Grunewald W, De Smet I, Houben A, Beeckman T, Russinova E. Arabidopsis alpha Aurora kinases function in formative cell division plane orientation. Plant Cell. 2011;23:4013–4024. doi: 10.1105/tpc.111.089565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vert JP, Jacob L. Machine learning for in silico virtual screening and chemical genomics: new strategies. Comb Chem High Throughput Screen. 2008;11:677–685. doi: 10.2174/138620708785739899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K. Aba-activated Snrk2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- 46.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y, Chow TF, Puckrin RS, Alfred SE, Korir AK, Larive CK, Cutler SR. Chemical genetic interrogation of natural variation uncovers a molecule that is glycoactivated. Nat Chem Biol. 2007;3:716–721. doi: 10.1038/nchembio.2007.32. [DOI] [PubMed] [Google Scholar]
- 48.Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J. SIR1, an upstream component in auxin signaling identified by chemical genetics. Science. 2003;301:1107–1110. doi: 10.1126/science.1084161. [DOI] [PubMed] [Google Scholar]
- 49.Zolman BK, Nyberg M, Bartel B. Ibr3, a novel peroxisomal Acyl-Coa dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol Biol. 2007;64:59–72. doi: 10.1007/s11103-007-9134-2. [DOI] [PubMed] [Google Scholar]