Abstract
In this paper we report the molecular profiling, lipidome and proteome, of the plant organelle known as an oil body (OB). The OB is remarkable in that it is able to perform its biological role (storage of triglycerides) whilst resisting the physical stresses caused by changes during desiccation (dehydration) and germination (rehydration). The molecular profile that confers such extraordinary physical stability on OBs was determined using a combination of 31P/1H nuclear magnetic resonance (NMR), high-resolution mass spectrometry and nominal mass-tandem mass spectrometry for the lipidome, and gel-electrophoresis-chromatography-tandem mass spectrometry for the proteome. The integrity of the procedure for isolating OBs was supported by physical evidence from small-angle neutron-scattering experiments. Suppression of lipase activity was crucial in determining the lipidome. There is conclusive evidence that the latter is dominated by phosphatidylcholine (∼60 %) and phosphatidylinositol (∼20 %), with a variety of other head groups (∼20 %). The fatty acid profile of the surface monolayer comprised palmitic, linoleic and oleic acids (2:1:0.25, 1H NMR) with only traces of other fatty acids (C24:0, C22:0, C18:0, C18:3, C16:2; by MS). The proteome is rich in oleosins (78 %) with the remainder being made up of caleosins and steroleosins. These data are sufficiently detailed to inform an update of the understood model of this organelle and can be used to inform the use of such components in a range of molecular biological, biotechnological and food industry applications. The techniques used in this study for profiling the lipidome throw a new light on the lipid profile of plant cellular compartments.
Electronic supplementary material
The online version of this article (doi:10.1007/s12154-012-0090-1) contains supplementary material, which is available to authorized users.
Keywords: Lipidome, Proteome, Lipase inhibition, NMR, MS, SANS
Introduction
Molecular profiling (‘omics’) is beginning to reveal the variety of molecular species in biological systems. A relatively recent innovation is profiling of the lipid fraction, referred to as lipidomics [16, 17, 22, 54]. The applications of this chemical approach are increasing rapidly as a direct result of the powerful insights it provides into determining molecular composition. Lipidomics is currently being used not only to probe the lipid composition of biological systems but also some of the molecular changes during metabolic processes, and disease [2, 46, 62]. Proteomic studies are also used for tracking disease [19, 27], with some work on proteinaceous systems with a mechanical role [10]. Furthermore, as the physical properties and behaviour of the lipids and proteins in biological systems start to become apparent at a molecular level [1, 11, 26, 40, 59, 63, 72], the applicability of molecular profiling data in understanding the molecular construction of organelles increases [18, 73]. However, in order to produce an accurate molecular profile of an organelle, there is a clear need for the use of diagnostic orthogonal chemical techniques.
One cellular compartment found in plants, about which there is some understanding of the biological role but relatively little about the structure or physical properties, is the seed-specific oil body (OB). OBs have evolved the function of a biological repository for triglycerides and typically represent about 40 % of the mass of the cotyledon in (desiccating) oleaginous seeds [43]. They are 0.5–2.0 μm in diameter [25, 29] with a postulated structure of a monolayer of amphiphilic species covering a mass of triglycerides [29, 34]. Evidence to date suggests that the monolayer that forms the surface of the OB comprises phospholipids and proteins from a relatively small group of protein families [28–31, 66]. However, it has not yet been possible to be precise about the molecular composition or the assembly it forms, as no comprehensive studies of molecular composition have been published, nor have there been any physical studies of purified isolated OBs.
The structure of these organelles remains stable and controlled throughout the dehydration associated with desiccation and the rehydration associated with germination [43], suggesting that they are able to resist the well-documented changes that occur in model lipid [37, 38, 50] and protein [20, 58, 71] systems on drastic changes in hydration. This remarkable stability makes OBs a desirable research interest, not only in terms of understanding the molecular biology of processes like germination [43] but also in biotechnological engineering, such as generating recombinant therapeutics [3, 7, 56] and as an adjuvant for vaccines [15]. It has also been possible to use OBs as a means for recovering and recycling immobilised enzymes [9, 35, 36]. Recent research in food science and technology suggests that OBs have potential as an ingredient in industrial production of edible emulsions [23, 24, 39, 47]. This range of interests highlights the importance of understanding the physical behaviour and properties of OBs and their components.
We chose to analyse OBs from Helianthus annuus (common sunflower) in order to provide an insight into this representative plant system. This species is germane to industrial-scale food industry applications as it is a viable crop for industrial applications. Also, the seed material presents a low allergen risk. Importantly, the OBs from this species are not well characterised to date. We used small-angle neutron-scattering (SANS) to assess the purity of the OBs isolated and to inform the details of improved protocols for isolating OBs from raw seeds. Molecular profiling of the organelle comprised lipidomic and proteomic analyses. The lipidome was profiled using 31P nuclear magnetic resonance (NMR) using an 800 MHz magnet, HRMS, and MSn MS/MS. We have also developed a novel method of analysis using NMR that can identify a lipid biomarker even when it is a minor component in a complicated mixture of lipids. We used GeLC-MS/MS to identify components of the protein fraction and densitometry for identifying their relative proportions.
Experimental section
All solvents used were HPLC-grade and were bought from Sigma Aldrich Ltd (Gillingham, Dorset, UK), as were all fine chemicals and deuteriated solvents, unless otherwise stated. Lipase inhibitors, except 2-butoxyphenylboronic acid (BPBA) and n-butanol, were bought from Tocris Biosciences Ltd (Bristol, Somerset, UK), and lipid standards were bought from Avanti Polar lipids (Alabaster, Alabama, USA). All chemicals were used as supplied. Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out using equipment and consumables from Bio-Rad Laboratories Ltd. (Hemel Hempstead, Hertfordshire, UK). Protease inhibitors ‘cOmplete’, in tablet form were purchased from Roche Diagnostics Ltd (Burgess Hill, Sussex, UK). De-hulled H. annuus seeds (2011 season) were bought from the Goldene Mühle GmbH (Garrel, Lower Saxony, Germany). Seed material was stored at ∼10 °C in an airtight container and used within a month of purchase.
Oil body isolation
De-hulled H. annuus seeds (100.0 g) were mixed with a solution of lipase inhibitors (suspension in n-butanol [5 mL] of FIPI, 5 mg; D609, 10 mg; U73122, 10 mg; BPBA, 1 g) and Tris-base (0.1 M, pH 7.5, 200 mL, cOmplete ULTRA protease inhibitor tablets [used as directed]) and ground using a Silverson high-shear mixer L 5 M (Silverson Ltd, Chesham, Buckinghamshire, UK), operated at <2 krpm) with the dilution of the mixture (addition of 200 mL Tris-base 0 · 1 M, pH 7.5) with successively narrowing mesh sizes of the homogeniser work head (5 mm mesh, then 2.5 mm, then 1.5 mm). The resulting suspension was made up to a total volume of 1.4 L (Tris-base 0 · 1 M, pH 7. 5, total volume 1 L, made in advance and chilled to <10 °C) and the mixture fractionated by centrifugation (9,950×g, 20 min, 4 °C). The uppermost layer was removed, resuspended in Tris-base (0.01 M, pH 7.5, ∼4:1 v/v) and fractionated by centrifugation (2,800×g, 20 min, 4 °C). The uppermost layer was removed, resuspended in a series of aqueous solutions (∼4:1 v/v) before centrifugation at each step (2,800×g, 20 min, 4 °C). The aqueous suspensions used were: Tween 20 (1 % v/v, with Tris-base 0.01 M, pH 7.5, ddH2O), NaCl (2 M, with Tris-base 0.01 M, pH 7.5, ddH2O), urea (8 M, with Tris-base 0.01 M, pH 7.5, ddH2O), and finally Tris-base (0.01 M, pH 7.5). The suspensions in aqueous urea were agitated at room temperature for 30 min, before centrifugation.
Oil body fractionation
Immediately after isolation, the OBs were suspended in ethanol (8:1 v/v) and the internal phosphorus standard added (lyso-phosphatidylglycerol, known mass of 2.5–4.5 mg). The solvents were then removed under reduced pressure (2 × 200 mL ethanol) and the remaining material washed with petroleum spirit (3 × 300 mL, 50 mmHg, glass sinter). The combined solutions were concentrated in vacuo to give the triglyceride (oil) fraction. The retentate was suspended (chloroform/ethanol/triethylamine, 3:1:1 [CET], 300 mL), sonicated (Ultrasonicator USC1700D, VWR (Lutterworth, Leicestershire, UK), power 9, 25 °C, 15 min), and washed (CET, 2 × 100 mL, 50 mmHg, glass sinter). The retentate was washed lightly with chloroform. The combined CET solutions and chloroform wash were concentrated in vacuo to give the lipid fraction. The remaining solid was dried in vacuo to give what is referred to as the protein fraction. The mass of protein was measured using the Bradford Assay. Details of spectral techniques and protein analysis can be found in the Electronic Supplementary Material.
Results
OB isolation and SANS
Traditionally, OBs have been isolated by gentle mechanical grinding of mature seeds in an aqueous medium, followed by centrifugation [66, 70]. A similar procedure was used here, with a number of important adaptations. Several steps are required to remove extraneous protein (particularly 11S storage [helianthinin] and 2S ribosomal proteins) and extraneous lipophilic material satisfactorily. We therefore used a modified version of the previously reported methods. In the preparation used in this study, the OBs are centrifuged immediately after grinding and were then resuspended in a series of aqueous media before centrifugation.
In order to verify the efficacy of these preparation and purification steps for isolating OBs, and to provide an independent check of the proteins observed in GeLC-MS/MS analysis of the OB protein mixtures that are postulated to be extraneous (see below), we elected to use contrast-variation SANS to determine the compartmentalisation and spatial arrangement of system components on the micron scale.
Both the crude and purified OB samples (see Supplementary Information) were prepared in 10 % D2O (to match the SLD of lipid component and highlight any protein signal) 40 % D2O (to match the protein and highlight lipid) and 90 % D2O (in which both species would be visible). The scattering patterns obtained are broadly compatible with a model of large poly-disperse spheres with sizes on the micron range of poly-dispersity index of around 60–70 %.
In the pattern from the crude preparation in 90 % D2O, an additional feature was present, not seen in the 40 % pattern, suggesting a significant contribution from protein (Fig. 1a), that was absent in preparations designed to remove extraneous material (Fig. 1b). The data recorded at 10 % D2O (lipid matched) for the crude preparation suggest a mono-dispersed but small protein species that could be modelled as an isolated ellipse (Fig. 1c). The fits used in Fig. 1 were consistent with a Gaussian polydisperse sphere model with scaled OB volume fractions. These scattering patterns are therefore reliable indicators of the system’s spatial arrangement, even at higher polydispersities. This suggested that either the crude preparation had a contaminant that was a free protein, or that a significant quantity of protein is present in the OBs as isolated free-floating species. The modelled shape is consistent with the structural data for 11S globulin (helianthinin) from formative studies [52, 55].
Fig. 1.
SANS scattering of OB systems containing free protein (a) and not (b). c SANS of OB samples with a lipid-matching concentration of exchangeable deuterium (10 %). The crude preparation (blue) shows a pattern consistent with an elliptical shape approximately two orders of magnitude smaller than the compartmentalised proteins in the purified OB preparation (red)
To determine the source of the signal a sample of more thoroughly purified OBs was analysed at high concentrations with the same three contrasts. In the 90 % D2O sample where both protein and lipid should be visible, the (additional) feature visible in data from the crude preparation was no longer present (Fig. 1b). The 10 % D2O sample suggests that the relative concentration of free protein has been reduced significantly with respect to the crude preparation, as there is no longer evidence for an elliptical protein after purification of the OBs (Fig. 1c). This was in agreement with the reduction in the amount of helianthinin and ribosomal proteins observed (GeLC-MS/MS, see below). This suggested that the oil body preparation protocol was competent to isolate oil bodies with impurities below the level of detection. This was verified using GeLC-MS/MS.
OB fractionation
Only OBs isolated according to the procedure informed by SANS and SDS-PAGE followed by GeLC-MS/MS were used for the profiling data presented in this work. The OBs were isolated and then fractionated into the triglyceride (oil), lipid, and protein fractions for characterisation. The OBs were doped before drying with a known mass of lyso-PG as a quantitative lipid standard before fractionation and dried under reduced pressure. Triglycerides were extracted using petroleum spirit and the lipid fraction using a mixture of chloroform, ethanol and triethylamine (3:1:1, ‘CET’) developed for this application. Triethylamine was used in order to produce triethylammonium cations and thus facilitate dissolving of the most acidic lipid species in an organic solvent system. The remaining matter after these extractions contained the proteome. OB fractionation revealed that they comprised the following by mass: triglycerides (98.63 % ± 0.01 %), lipids (0.31 % ± 0.039 %) and protein (1.06 % ± 0.001 %).
Proteome: peptide separation, tandem MS and peptide identification
Proteins were separated by SDS-PAGE (Supplementary Figure 1) and enzymatically digested with trypsin to peptides before LC-MS/MS and analysis. GeLC-MS/MS revealed a proteome comprising three protein families, namely: oleosins, caleosins and steroleosins [28, 29, 33, 34, 45, 66, 68]. The relative quantities of proteins observed were measured using densitometry of colloidal Coomassie-stained SDS-PAGE gels (n = 3) (Table 1), indicating that oleosins (bands K-M, types A–E) account for around 78 % of the proteome by mass, whereas steroleosins account for around 13 % and caleosins for around 9 %. When present, traces of extraneous proteins were observed as a discrete band with a low staining intensity (helianthinin isoforms, band H, Supplementary Figure 1).
Table 1.
Identities and relative amounts (optical density) of proteins present
| ID | Band | Observed massa (kDa) | Matching peptides found in MS/MS | Database entry number and species | Closest homologue in Viridiplantae | Optical density | ±b |
|---|---|---|---|---|---|---|---|
| Caleosin A | F | 28.1 | VAFDPFGWTAAK | EL461280 | Caleosin; Sesamum indicum, gi|6478218 | 3.1 % | 0.31 % |
| MEWAILYALAK | H. tuberosus | ||||||
| CFDGSLFDYCAK | |||||||
| TDLETHLPKPYLAR | |||||||
| TVPDKLTLGEIWNMTEGNR | |||||||
| Caleosin B | G | 26.3 | MEWAILYALAK | EL460587 | Caleosin; S. indicum, gi|6478218 | 5.7 % | 0.54 % |
| CFDGSLFDYCAK | H. tuberosus | ||||||
| YMPVHFENMFSK | |||||||
| TDLETHLPKPYLAR | |||||||
| GLVAPDTEHPNGTLGHR | |||||||
| TVPDKLTLGEIWNMTEGNR | |||||||
| VMAMADGNALTPEADCQPLTHER | |||||||
| Hydrosteroid 11-β-dehydrogenase A (steroleosin A) | A, B, D, E | 45 (×2), 36.2, 33.4 | AALAQFFETMR | GE513976 | Steroleosin; S. indicum, gi|16033752 | 9.2 % | 1.95 % |
| MVDQTVHHFNR | H. annuus | ||||||
| IIALSSAASWIPLPR | |||||||
| ITLVTPGFIESELTQGK | |||||||
| VVLITGASSGIGEHIAYEYASR | |||||||
| Hydrosteroid 11-β-dehydrogenase B (steroleosin B) | C | 38.2 | AALVNFYETLR | EL457176 | Steroleosin-B; S. indicum, gi|21311775 | 3.5 % | 0.01 % |
| FINETINYFGR | H. tuberosus | ||||||
| LLGAPDVLIMAADVVK | |||||||
| IVVNAAVENWLPLPR | |||||||
| Oleosin A | J | 18.2 | GGHYGTAGVHTGGPGGGVGTYV | CD856507 | Oleosin 1; Prunus dulcis, gi|460803 | 19.1 % | 3.62 % |
| YVTGEQPSGADTSDEASHR | H. annuus | ||||||
| Oleosin B | Mc | 15.1 | YDQQTHSQSTSR | gi|266694 | Oleosin H-isoform; Ficus pumila, (var. Awkeotsang) gi|148283921 | 17.0 %c | 0.28 % |
| QTAGSVPESLDYVK | H. annuus | ||||||
| IQSTAHEMGDQGQVGVHAQVGGGK | |||||||
| LQDLVEYTGQK | |||||||
| RLQDLVEYTGQK | |||||||
| Oleosin C | K | 17.0 | STMSVPDQMDYVK | gi|1345520 | Oleosin 18.2 KDa; Gossypium hirsutum, gi|167363 | 19.8 % | 0.78 % |
| LQDVGEYTGQK | H. annuus | ||||||
| Oleosin D | L | 16.3 | LQDLVEYTGQK | EL512172 H. exilis | Oleosin S1-2; Brassica napus, gi|196122060 | 22.6 % | 5.84 % |
| AHDIGPEGAVHAGSAVGGAK | |||||||
| Oleosin E | Mc | 15.1 | YATGGHPLGSDSLDQAR | EL511252 H. exilis | Oleosin low-molecular-weight isoform; Linum usitatissimum, gi|77737584 | 17.0 %c | 0.28 % |
| Total | 100.0 % |
Gel shown in Supplementary Figure 1
aMeasure estimated from migration on SDS-PAGE
bStandard error
cBand contained peptides for two distinct oleosins and thus the optical density quoted is for the combination of both isoforms
Lipidome: lipase activity
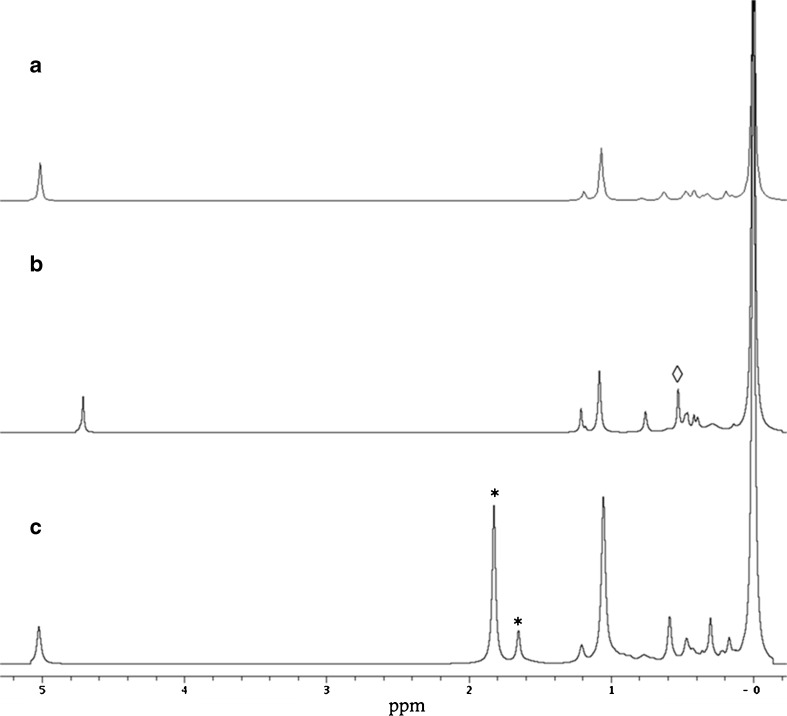
Isolation of the lipid fraction from de-hulled Helianthus seeds in a simple buffered medium showed a surprisingly narrow range of lipid head groups, with only phosphatidylcholine (PC), phosphatidylinositol (PI) and phosphatidic acid (PA) visible (Fig. 2). This is at odds with the compelling evidence that a significantly richer diversity of lipid head groups is present in the few biological systems for which comprehensive lipidomics data currently are available [17, 22, 54]. This led us to hypothesise that lipase activity was affecting the observed lipid profile. The use of the non-selective phospholipase A 1 and 2 (collectively known as PLAxs) inhibitor BPBA succeeded in increasing and making consistent the isolated mass of the lipidome. Although this strongly suggested that PLAxs were indeed present, no further head group types were observed. We therefore examined the possibility that other lipases were active in the system.
Fig. 2.
31P NMR spectra of lipidomes of OBs from helianthus OBs. Trace A is a lipid isolate without added enzyme inhibitors. Trace B is from a lipidome isolation containing BPBA (non-selective PLAx inhibitor) and n-butanol. The signal arising from PB is marked with an open diamond. Trace C is from a lipid fraction isolated with PLAx, PLC and PLD inhibitors, and the quantitative standard (lyso-PG, signals arising from the 1-O and 2-O isomers are marked with an asterisk). There is some variation in the shift of the PA signal due to the formation of triethylammonium adducts with the solvent system and appears to be partly concentration-dependent with respect to the lipid
The presence of PA alerted us to the possible activity of phospholipase D (PLD). PLDs are known to hydrolyse several lipid head groups, invariably producing phosphatidic acid [4, 48, 53]; however, n-butanol has been shown to suppress the formation of PA, with evidence that phosphatidylbutanol (PB, structure shown in Supplementary Figure 20) is produced instead [32, 51]. This has been reported to occur at concentrations of n-butanol as low as 0.01 %, v/v [8, 41, 57]. Thus the presence of PB provides a useful molecular marker for PLD activity. The possible activity of PLDs was therefore tested with a qualitative PLD assay comprising n-butanol.
Lipidome: PLD assay
The change in lipidome according to lipid head group profile in this assay was tracked through the change in lipid profile of ground H. annuus cotyledons suspended in buffered aqueous medium in the presence of n-butanol, agitated at 4 °C over a period of days (Fig. 3). The lipid profiles at given time points (24, 48, 72 h, 9 days) were determined by 31P NMR. The appearance and dominance of a signal at 0.34 ppm (31P NMR) was taken as a strong indication of the presence of a phosphorus-containing lipid that was not found in the lipid composition of commercially available lecithins already reported [6, 13, 14, 42], or in the mixture of lipids from Glycine max used in this project (Supplementary Figure 4), or from lipid species not previously identified in commercially available lipid mixtures (lyso-PS, PI-4-P, lyso-PG; see Supplementary Figures 5–7, respectively, and Table 2 for 31P NMR shifts). The presence of PB was confirmed by both mass spectrometry and 31P NMR from samples of the lipid fraction isolated from OBs subject to 9 days’ agitation, i.e. where the new (PB) resonance dominated most strongly over other lipid species (Fig. 3). The presence of the resonance at 0.34 ppm in isolated lipidomes, attributed to PB, was therefore taken as evidence of PLD activity.
Fig. 3.
31P NMR PLD assay with time points shown. PB remains the dominant species throughout, though the integrations of the signals suggest that the majority of the PB is produced in the first 24 h. The reduction in fraction of all other lipids (especially PA, PC and PI shown here) suggests that PLD acts on a variety of substrates, and/or that other lipases are present
Table 2.
Lipidome of OBs from H. annuus
| Lipid | Fraction (mol., n = 3) | ± | 31P NMR shift (ppm) | Ion mass (es)b |
|---|---|---|---|---|
| PA | 3.00 % | 0.81 % | 4.82–5.10 | 699.5 (C18:2/C18:0, C18:1/C18:1) |
| 671.5 (C16:0/C18:2) | ||||
| 673.5 (C16:0/C18:1) | ||||
| 695.5 (C18:2/C18:2) | ||||
| 697.5 (C18:2/C18:1) | ||||
| lyso-PG | 0.04 % | 0.01 % | 1.72 | 509 (C18:1/0:0) |
| lyso-PI | 0.72 % | 0.65 % | 1.53 | 571.5 (C16:0/0:0) |
| lyso-PE | 0.24 % | 0.13 % | 1.23 | 452 (C16:2/0:0) |
| 476 (C18:2/0:0) | ||||
| PG | 1.81 % | 0.33 % | 1.21 | 745.5 (C18:2/C16:0) |
| 769.5 (C18:2/C18:2) | ||||
| 771.5 (C18:2/C18:1) | ||||
| 773.5 (C18:0/C18:2) | ||||
| PI | 19.97 % | 2.18 % | 1.02 | 833.5 (C18:2/C16:0) |
| 857.5 (C18:2/C18:2) | ||||
| 861.5 (C18:2/C18:0) | ||||
| 917 (C18:2/C22:0) | ||||
| U1 | 0.73 % | 1.09 % | 0.77 | – |
| PE | 4.04 % | 0.42 % | 0.65–0.58 | 714.5 (C18:2/C16:0) |
| 716.5 (C16:0/C18:1) | ||||
| 738.5 (C18:2/C18:2) | ||||
| 742 (C18:3/C18:0) | ||||
| 826.5 (C18:2/C24:0) | ||||
| PS | 0.77 % | 0.96 % | 0.52 | – |
| lyso-PC | 2.38 % | 0.47 % | 0.45–0.49 | 496.5 (C16:0/0:0) |
| 520.5 (C18:2/0:0) | ||||
| U2 | 1.14 % | 0.32 % | 0.42 | – |
| U3 | 0.34 % | 0.60 % | 0.38 | – |
| U4 | 1.14 % | 1.10 % | 0.36 | – |
| U5 | 2.57 % | 1.53 % | 0.30 | – |
| U6a | 0.57 % | 0.31 % | 0.28–0.25 | – |
| U7 | 1.00 % | 0.26 % | 0.24 | – |
| U8a | 1.22 % | 0.81 % | 0.17 | – |
| PC | 58.59 % | 4.08 % | 0.00 | 782.5 (C18:2/C18:2) |
| 784.5 (C18:2/C18:1) | ||||
| 758.5 (C18:1/C16:0) | ||||
| 733.5 (C16:0/C16:0) | ||||
| Total | 100.00 % | |||
Quantitative standard lyso-PG, n = 3. U1–8 represents the resonances of lipids that are at present unknown. There was no evidence for the formation of the biomarker PB in the samples used to compile this data.
Fatty acids: 24:0 lignoceric acid, 22:0 behenic acid, 18:3 α-linolenic acid, 18:2 linoleic acid, 18:1 oleic acid, 18:0 stearic acid, 16:0 palmitic acid.
aResonances for which the shifts are consistent with known lipid species (U6, DMPE; U8, PC plasmalogen) but for which no appropriate ion fragments were observed in MS.
bFatty acid profile consistent with mass given in parentheses, identified by MSn.
Lipase activity in this system cannot be attributed to microbial activity, as no evidence of either spore-forming or Pseudomonas species could be found. We ascribe this apparent sterility to a combination of the bactericidal n-butanol concentration, to the bacteriostatic temperature of the assay (≤4 °C), and to the bacteriostatic PLAx inhibitor.
We noted that the suppression of PLA activity and the redirection of PLD activity to give PB instead of PA (with n-butanol) allowed the activity of phospholipase Cs (PLCs) to be observed qualitatively. A peak in PB concentration after around 48 h suggests that it too is being consumed in this system. Bacterial activity may be ruled out (see above), as is a change in lipid profile by destruction of the organelles; they remain intact as no release of triglycerides was observed. The reduction in total PB cannot be ascribed to PLD activity without the formation of PA, which is not observed. Thus, we suspected that the hydrolysis of PB hinted at the activity of one or more phospholipase C isoforms (PLCs) on this lipid species. PLCs are known to hydrolyse inositides (PI is also hydrolyzed in this system) and with some evidence for hydrolysis of PC [49, 51] (also hydrolyzed in this assay). PLC activity on lipids gives rise to DAGs and releases the (hydrophilic) head group in the aqueous medium, reducing the total amount of phosphorus in the lipid fraction. Thus, in order to determine the lipidome of this organelle, we used comprehensive lipase inhibition designed to suppress PLAx, PLC and PLD activity.
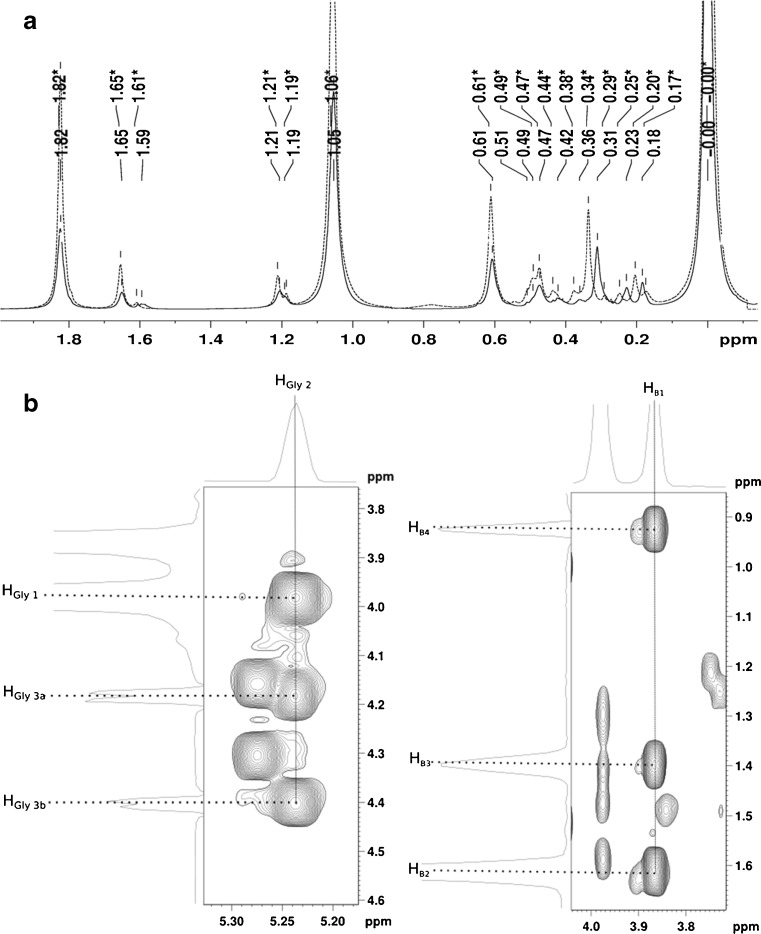
Lipidome isolation The evident necessity of suppressing PLAx, PLC and PLD activities in order to isolate a lipidome with confidence led us to use a cocktail of known, commercially available, lipase inhibitors [5, 21, 57, 64, 65] (see “Experimental section”). No resonance in the NMR spectra or MS ions/fragments associated with PB was observed in lipidomes isolated under conditions including comprehensive lipase inhibition, despite the presence of n-butanol. Signals at 0.36 and 0.31 ppm (Fig. 4a) can be ruled out as indicators of PB, as a combination of 31P HSQC, 13C HSQC, 13C HMBC and TOCSY spectra have allowed us to identify PB resonances unambiguously (Figure 4b, assignment list in Supplementary Information). This has allowed us to demonstrate that the connectivity of the species identified is consistent with PB (Supplementary Figure 20). As this work has identified a number of characteristic resonances in this biomarker, this approach to identifying phosphorus-containing species has the scope for demonstrating the presence of PB in complicated mixtures of phosphorus-containing molecules, even where PB is a minor component.
Fig. 4.
a31P NMR showing the comparison of lipidome samples containing PB (unbroken line, unstarred labels) and not containing it (dashed line, starred labels); b1H, 1H TOCSY showing through bond coupling identifying glyceryl–sn-1-3 (panel, left) and butyl (B1-B4) protons (panel, right). Projections shown opposite the x and y axes indicate protons coupled to 31P and are taken from a slice through a 31P,1H HSQC experiment at the 31P chemical shift of 0 · 34 ppm (characteristic of PB in a 1D 31P experiments, (a)). The subscript notations used above are the same as the notations in Supplementary Figure 20
The inclusion of PLC inhibitors D609 and U73122 in grinding media indicated measurable changes to the PI and PC fractions (viz. traces 2 and 3, Fig. 2), supporting the hypothesis of activity consistent with members of the PLC family.
The lipidome of the OB of mature H. annuus seeds is presented in Table 2. In order to overcome the inherent variation in biological systems, we repeated the isolation (n = 3), using a quantitative phosphorus (lipid) standard. The lipidomes isolated without a quantitative standard show that the presence of lyso-phosphatidylglycerol (lyso-PG) is 0.04 % (±0.01 %), and analysis of this lipid species in vitro shows that the shift of its resonances (1.82 and 1.64 ppm, for the 1-O- and 2-O-fatty acid derivatives, respectively) are conveniently distant from the resonances arising from other species, to make quantification reliable. The head group identification has been made both in NMR spectra and in MS traces; however, 31P NMR suggest the presence of traces of lipids that have not yet been identified (U1–8). Unfortunately, it was not possible to use MS signals from spectra of the lipidomes (Supplementary Figure 9) to inform this further.
Fatty acid fraction
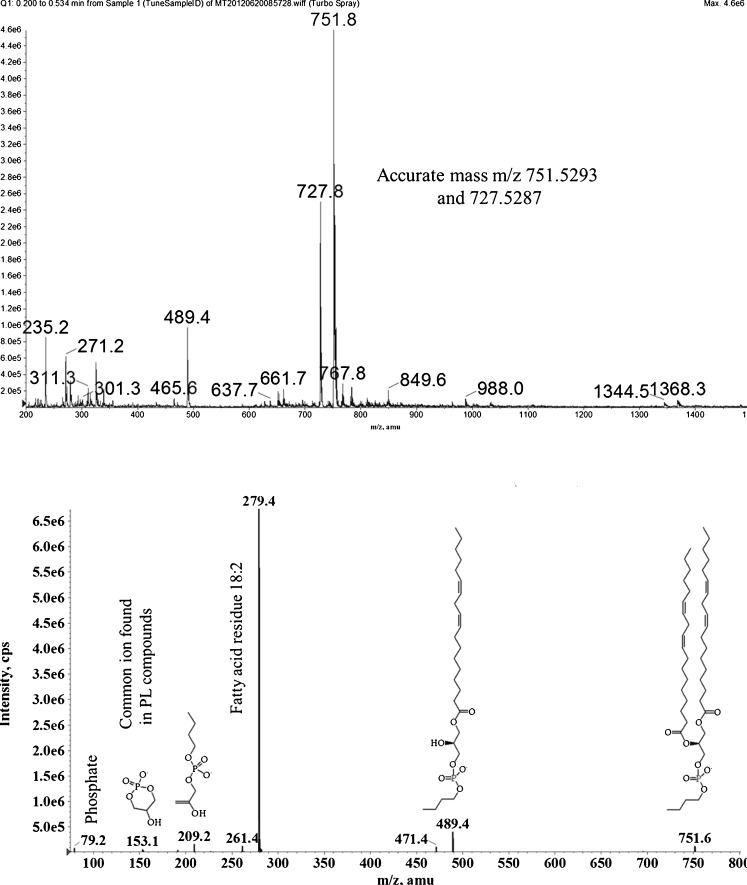
Strikingly, the fatty acid fraction from lipidome samples (1H NMR) was remarkably simple; the mixture it comprises is dominated by palmitic, linoleic and oleic acids (2:1:0.25, respectively, 1H NMR of hydrolytic products, Supplementary Figures 2 and 3). This select group of fatty acids is also observed in the fragmentation of PB under electrospray ionisation (Fig. 5). These data support the hypothesis that the PB is formed not only from the bulk of the lipid present in what appears to be a non-selective manner; it is in agreement with the fatty acid fragmentation of the principle lipid components of the lipidome, PC (Supplementary Figures 10 and 11), PA (Supplementary Figure 12), PG (Supplementary Figure 13), PI (Supplementary Figure 14) and PE (Supplementary Figure 15). This suggests that, although there is some richness in the head group profile of the lipid fraction of the OB, a wide variety of fatty acid residues is not useful to the organism and may even be undesirable.
Fig. 5.
Mass spectrum of PB (from PLD assay). m/z 751–5,293 = dilinoleoylphosphatitdylbutanol, m/z 727–5,287 = palmitoyllinoleoylphosphatidylbutanol
Lipidome: oil The principle component of an OB in terms of mass is undoubtedly its triglyceride fraction [29, 66, 69]. In the OBs examined in the present study, this fraction represented 98.63 % ± 0.01 % of the mass of the organelle (see above). The triglycerides were characterised in this study using NMR (Supplementary Figure 8) and MS (Supplementary Figure 16–19). 1H NMR indicates that there is an average of 1.5 olefin bonds per fatty acid residue (Supplementary Figure 8). MS suggests that, within this average, there is a mixture of triglycerides with 0, 2, 3 and 4 double bonds, with evidence for an abundant triglyceride species containing only linoleoyl residues (two olefin bonds, Supplementary Figure 17). There is evidence for the presence of trace amounts of dihomo-γ-linolenic (20:3) and 7,10,13,16-docosatetraenoic (22:4) acid residues (Supplementary Figure 16).
Discussion
The results of this study indicate that the lipidome of OBs from H. annuus (common sunflower) is much broader than previously expected [66, 67]. Additionally, the use of small-molecule lipase inhibitors is crucial in determining the composition of that lipid fraction accurately. This work also shows that the protein fraction is larger in mass than the lipid fraction by a factor of ∼3.4, suggesting an important structural role for the proteins.
A comparison of the molecular profile of Helianthus OBs with recent physical evidence also suggests an important structural influence of the protein fraction. Recent physical evidence indicates that the lipid fraction in isolation would be likely to exhibit a different surface curvature to that of naturally occurring OBs. Mixtures of PC and PI (4:1) have been shown to produce surfaces that curve towards the cytosol [40] rather than away from it, as in the OB surface. This suggests that another component of the OBs’ surface, not found in the lipid fraction, is able to direct the topology of the surface of this organelle. The evidence from this work shows that a protein fraction containing amphiphilic proteins is a large and integral part of the monolayer that makes forms the surface. Importantly, the proteome of this organelle is dominated by oleosin isoforms (∼78 % by densitometry, Supplementary Figure 1).
The hypothesis that the physical properties of the proteins dominate that of the surface is supported by the observation that OBs stay intact when the relatively rich lipidome found in vivo is converted into a lipid fraction that is dominated by PB (>90 %, 31P NMR). This strongly suggests that the lipid fraction of the surface does not dominate the physical properties of the OB structure under the hydrating conditions used in this study. It is clear from physical studies that a surface composed entirely of any combination of known biological lipids would be unable to sustain the surface topology of OBs through these changes in hydration [40, 60, 61, 63]. The oleosin-rich proteome, whatever the biological function of the individual proteins, therefore seems to be an important player in maintaining the remarkable physical stability of this organelle. Not only are the proteins amphiphilic [44, 69], but this evidence suggests that they also have an implicit thermodynamic role in maintaining the integrity of the organelle, alongside the lipid fraction, as part of its monolayer surface.
We suggest that the evidence presented here supports an updating of the current model for the assembled structure of naturally occurring OBs, a fragment of which is shown schematically in Fig. 6.
Fig. 6.
Schematic diagram of the OB, given the composition described above, and known thermodynamic behaviour [29, 61] of amphiphilic proteins, lipids and triglycerides. A Neutral surface [61] of the monolayer of the OB; b oleosin; c, triglyceride matrix (one molecule shown for clarity); d, phospholipid. Scale: the lipid molecules (d) are 1.8–2.0 nm long [12], and the oleosin helix that extends into the triglyceride matrix is 12 nm long [29]. NB the length of the lipid molecules represents around 0.1 % of the width of the organelle itself
In summary, the evidence from this study about the molecular profile of OBs from H. annuus shows a monolayer dominated by proteins in terms of mass and perhaps also in terms of its physical properties. The molecular profile generated can be used not only to inform our understanding of the physical properties of this organelle but also its metabolism and general behaviour in vivo. This can also be applied to the preparation of synthetic OBs in applications such as biotechnological engineering (e.g. adjuvants for vaccines and recombinant therapeutics) and in the food industry (e.g. preparing emulsions and controlling flavour). The chemical techniques used have enabled us to identify a range of species in the proteome and lipidome, as well as some that may not have been observed to date.
Electronic supplementary material
(DOCX 879 kb)
Acknowledgements
The authors would like to thank the Department for the Environment, Farming and Rural Affairs (DEFRA) and the LINK scheme for funding (Sustainable Emulsions through Bio-innovation Project) for funding this work. The authors would like to thank Prof. Christine Dodd and Dr. Philip Richards for assistance with microbiological assessment, Dr. Adrienne Davis and Mr. Kevin Butler for assistance with NMR and Dr. Kenneth Davis for assistance with protein densitometry. The authors would like to thank Drs. G. Campbell, C. B. Harvey and N. Wenta for helpful discussions.
Funding sources
The authors would like to thank the Department for the Environment, Farming and Rural Affairs (DEFRA) and the LINK scheme for funding (Sustainable Emulsions through Bio-innovation Project) for funding this work.
Abbreviations
- CET
Mixture of chloroform, ethanol and triethylamine [3:1:1]
- DAG
Diacylglyceride
- HMBC
Hetero-nuclear multiple-bond correlation
- HMQC
Hetero-nuclear single quantum spectroscopy
- HRMS
High-resolution mass spectrometry
- GeLC-MS/MS
Gel-electrophoresis-chromatography-tandem mass spectrometry
- MSn MS/MS
Nominal mass-tandem mass spectrometry
- NMR
Nuclear magnetic resonance
- MS
Mass spectrometry
- PA
Phosphatidic acid
- PB
Phosphatidylbutanol
- PC
Phosphatidylcholine
- PE
Phosphatidylethanolamine
- PG
Phosphatidylglycerol
- PI
Phosphatidylinositol
- PI-4-P
Phosphatidylinositol 4-phosphate
- PS
Phosphatidylserine
- PT
Phosphatidylthreonine
- SANS
Small-angle neutron-scattering
- SLD
Scattering length density
- TOCSY
Total correlation spectroscopy
- TG
Triglyceride
Contributor Information
Samuel Furse, Phone: +44-115-9516147, FAX: +44-115-9516142, Email: Samuel.Furse@nottingham.ac.uk, Email: samuel@samuelfurse.com.
David A. Gray, Phone: +44-115-9516147, FAX: +44-115-9516142, Email: David.Gray@nottingham.ac.uk
References
- 1.Alley SH, Ces O, Templer RH, Barahona M. Biophysical regulation of lipid biosynthesis in the plasma membrane. Biophys J. 2008;94(8):2938–2954. doi: 10.1529/biophysj.107.118380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bain JR, Stevens RD, Wenner BR, Ilkayeva O, Muoio DM, Newgard CB. Metabolomics applied to diabetes research. Diabetes. 2009;58(11):2429–2443. doi: 10.2337/db09-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatla SC, Kaushik V, Yadav MK. Use of oil bodies and oleosins in recombinant protein production and other biotechnological applications. Biotechnol Adv. 2010;28(3):293–300. doi: 10.1016/j.biotechadv.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Billah MM, Pai JK, Mullmann TJ, Egan RW, Siegel MI. Regulation of phospholipase D in HL-60 granulocytes. Activation by phorbol esters, diglyceride, and calcium ionophore via protein kinase-independent mechanisms. J Biol Chem. 1989;264(15):9069–9076. [PubMed] [Google Scholar]
- 5.Bleasdale JE, Thakur NR, Gremban RS, Bundy GL, Fitzpatrick FA, Smith RJ, Bunting S. Selective inhibition of receptor-coupled phospholipase C-dependent processes in human platelets and polymorphonuclear neutrophils. J Pharmacol Exp Ther. 1990;255(2):756–768. [PubMed] [Google Scholar]
- 6.Bosco M, Culeddu N, Toffanin R, Pollesello P. Organic solvent systems for P-31 nuclear magnetic resonance analysis of lecithin phospholipids: applications to two-dimensional gradient-enhanced H-1-detected heteronuclear multiple quantum coherence experiments. Anal Biochem. 1997;245(1):38–47. doi: 10.1006/abio.1996.9907. [DOI] [PubMed] [Google Scholar]
- 7.Capuano F, Bond NJ, Gatto L, Beaudoin F, Napier JA, Benvenuto E, Lilley KS, Baschieri S. LC-MS/MS methods for absolute quantification and identification of proteins associated with chimeric plant oil bodies. Anal Chem. 2011;83:9267–9272. doi: 10.1021/ac201733m. [DOI] [PubMed] [Google Scholar]
- 8.Chen YG, Siddhanta A, Austin CD, Hammond SM, Sung TC, Frohman MA, Morris AJ, Shields D. Phospholipase D stimulates release of nascent secretory vesicles from the trans-Golgi network. J Cell Biol. 1997;138(3):495–504. doi: 10.1083/jcb.138.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang C-J, Chen CY-P, Tzen JTC. Efficient system of artificial oil bodies for functional expression and purification of recombinant nattokinase in Escherichia coli. J Agric Food Chem. 2005;53(12):4799–4804. doi: 10.1021/jf050264a. [DOI] [PubMed] [Google Scholar]
- 10.Clutterbuck AL, Smith JR, Allaway D, Harris P, Liddell S, Mobasheri A. High throughput proteomic analysis of the secretome in an explant model of articular cartilage inflammation. J Proteome. 2011;74(5):704–715. doi: 10.1016/j.jprot.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corsi J, Hawtin RW, Ces O, Attard GS, Khalid S. DNA lipoplexes: formation of the inverse hexagonal phase observed by coarse-grained molecular dynamics simulation. Langmuir. 2010;26(14):12119–12125. doi: 10.1021/la101448m. [DOI] [PubMed] [Google Scholar]
- 12.Costigan SC, Booth PJ, Templer RH. Estimations of lipid bilayer geometry in fluid lamellar phases. Biochim Biophys Acta (BBA)-Biomembranes. 2000;1468(1–2):41–54. doi: 10.1016/S0005-2736(00)00220-0. [DOI] [PubMed] [Google Scholar]
- 13.Cremonini MA, Laghi L, Placucci G. Investigation of commercial lecithin by P-31 NMR in a ternary CUBO solvent. J Sci Food Agric. 2004;84(8):786–790. doi: 10.1002/jsfa.1683. [DOI] [Google Scholar]
- 14.Culeddu N, Bosco M, Toffanin R, Pollesello P. P-31 NMR analysis of phospholipids in crude extracts from different sources: improved efficiency of the solvent system. Magn Reson Chem. 1998;36(12):907–912. doi: 10.1002/(SICI)1097-458X(199812)36:12<907::AID-OMR394>3.0.CO;2-5. [DOI] [Google Scholar]
- 15.Deckers H, Van Rooijen G, Boothe J, Goll J, Moloney M, Schryvers A, Alcantara J, Hutchins W (2004) Immunogenic formulations comprising oil bodies. United States Patent
- 16.Dennis EA. Lipidomics joins the omics evolution. Proc Natl Acad Sci. 2009;106(7):2089–2090. doi: 10.1073/pnas.0812636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennis EA, Deems RA, Harkewicz R, Quehenberger O, Brown HA, Milne SB, Myers DS, Glass CK, Hardiman G, Reichart D, Merrill AH, Sullards MC, Wang E, Murphy RC, Raetz CRH, Garrett TA, Guan Z, Ryan AC, Russell DW, McDonald JG, Thompson BM, Shaw WA, Sud M, Zhao Y, Gupta S, Maurya MR, Fahy E, Subramaniam S. A mouse macrophage lipidome. J Biol Chem. 2010;285(51):39976–39985. doi: 10.1074/jbc.M110.182915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443(7112):651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 19.Dubey JP, Liddell S, Mattson D, Speer CA, Howe DK, Jenkins MC. Characterization of the Oregon isolate of Neospora hughesi from a horse. J Parasitol. 2001;87(2):345–353. doi: 10.1645/0022-3395(2001)087[0345:COTOIO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 20.Ebbinghaus S, Kim SJ, Heyden M, Yu X, Gruebele M, Leitner DM, Havenith M. Protein sequence- and pH-dependent hydration probed by terahertz spectroscopy. J Am Chem Soc. 2008;130(8):2374–2375. doi: 10.1021/ja0746520. [DOI] [PubMed] [Google Scholar]
- 21.Ebdrup S, Jacobsen P, Farrington AD, Vedsø P. Structure–activity relationship for aryl and heteroaryl boronic acid inhibitors of hormone-sensitive lipase. Bioorg Med Chem. 2005;13(6):2305–2312. doi: 10.1016/j.bmc.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 22.Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fisk I, Linforth R, Taylor A, Gray D. Aroma encapsulation and aroma delivery by oil body suspensions derived from sunflower seeds (<i > Helianthus annuus</i>) Eur Food Res Technol. 2011;232(5):905–910. doi: 10.1007/s00217-011-1459-z. [DOI] [Google Scholar]
- 24.Fisk ID, White DA, Lad M, Gray DA. Oxidative stability of sunflower oil bodies. Eur J Lipid Sci Technol. 2008;110(10):962–968. doi: 10.1002/ejlt.200800051. [DOI] [Google Scholar]
- 25.Frandsen GI, Mundy J, Tzen JTC. Oil bodies and their associated proteins, oleosin and caleosin. Physiol Plant. 2001;112(3):301–307. doi: 10.1034/j.1399-3054.2001.1120301.x. [DOI] [PubMed] [Google Scholar]
- 26.Furse S, Tate EW, Woscholski R, Templer RH, Gaffney PRJ, Ces O. Lipid membrane curvature induced by distearoyl phosphatidylinositol 4-phosphate. Soft Matter. 2012;8:3090–3093. doi: 10.1039/c2sm07358g. [DOI] [Google Scholar]
- 27.Hill DE, Liddell S, Jenkins MC, Dubey JP. Detection of Neospora caninum oocysts in fecal samples from experimentally-infected dogs using the polymerase chain reaction. J Parasitol. 2001;87(2):395–398. doi: 10.1645/0022-3395(2001)087[0395:SDONCO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 28.Huang AHC. Oil bodies and oleosins in seeds. Annu Rev Plant Phys. 1992;43:177–200. [Google Scholar]
- 29.Huang AHC. Structure of plant seed oil bodies. Curr Opin Struc Biol. 1994;4(4):493–498. doi: 10.1016/S0959-440X(94)90210-0. [DOI] [Google Scholar]
- 30.Jolivet P, Boulard C, Bellamy A, Larré C, Barre M, Rogniaux H, d’Andréa S, Chardot T, Nesi N. Protein composition of oil bodies from mature Brassica napus seeds. Proteomics. 2009;9(12):3268–3284. doi: 10.1002/pmic.200800449. [DOI] [PubMed] [Google Scholar]
- 31.Jolivet P, Roux E, Andrea S, Davanture M, Negroni L, Zivy M, Chardot T. Protein composition of oil bodies in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem. 2004;42(6):501–509. doi: 10.1016/j.plaphy.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Kravets VS, Kolesnikov YS, Kretynin SV, Getman IA, Romanov GA. Rapid activation of specific phospholipase(s) D by cytokinin in Amaranthus assay system. Physiol Plant. 2010;138(3):249–255. doi: 10.1111/j.1399-3054.2009.01324.x. [DOI] [PubMed] [Google Scholar]
- 33.Lin LJ, Tai SSK, Peng CC, Tzen JTC. Steroleosin, a sterol-binding dehydrogenase in seed oil bodies (vol 128, pg 1200, 2002) Plant Physiol. 2002;129(4):1930–1930. doi: 10.1104/pp.010982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin LJ, Tzen JTC. Two distinct steroleosins are present in seed oil bodies. Plant Physiol Biochem. 2004;42(7–8):601–608. doi: 10.1016/j.plaphy.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Liu J-H, Selinger LB, Cheng K-J, Beauchemin KA, Moloney MM. Plant seed oil-bodies as an immobilization matrix for a recombinant xylanase from the rumen fungus Neocallimastix patriciarum. Mol Breed. 1997;3(6):463–470. doi: 10.1023/A:1009604119618. [DOI] [Google Scholar]
- 36.Liu J-R, Duan C-H, Zhao X, Tzen J, Cheng K-J, Pai C-K. Cloning of a rumen fungal xylanase gene and purification of the recombinant enzyme via artificial oil bodies. Appl Microbiol Biotechnol. 2008;79(2):225–233. doi: 10.1007/s00253-008-1418-1. [DOI] [PubMed] [Google Scholar]
- 37.Ludwig R, Appelhagen A. Calculation of Clathrate-like water clusters including Buckminsterfullerene. Angew Chem Int Ed. 2005;44(5):811–815. doi: 10.1002/anie.200460899. [DOI] [PubMed] [Google Scholar]
- 38.Marcelja S, Radic N. Repulsion of interfaces due to boundary water. Chem Phys Lett. 1976;42(1):129–130. doi: 10.1016/0009-2614(76)80567-2. [DOI] [Google Scholar]
- 39.McCaskill DR, Zhang F. Use of rice bran oil in foods. Food Technol. 1999;53(2):50–52. [Google Scholar]
- 40.Mulet X, Templer RH, Woscholski R, Ces O. Evidence that phosphatidylinositol curved membrane interfaces. Langmuir. 2008;24(16):8443–8447. doi: 10.1021/la801114n. [DOI] [PubMed] [Google Scholar]
- 41.Munnik T, Arisz SA, deVrije T, Musgrave A. G protein activation stimulates phospholipase D signaling in plants. Plant Cell. 1995;7(12):2197–2210. doi: 10.1105/tpc.7.12.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murgia S, Mele S, Monduzzi M. Quantitative characterization of phospholipids in milk fat via P-31 NMR using a monophasic solvent mixture. Lipids. 2003;38(5):585–591. doi: 10.1007/s11745-003-1500-3. [DOI] [PubMed] [Google Scholar]
- 43.Murphy DJ. Biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40:325. doi: 10.1016/S0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- 44.Murphy DJ, Keen JN, Osullivan JN, Au DMY, Edwards EW, Jackson PJ, Cummins I, Gibbons T, Shaw CH, Ryan AJ. A class of amphipathic proteins associated with lipid storage bodies in plants—possible similarities with animal serum apolipoproteins. Biochim Biophys Acta. 1991;1088(1):86–94. doi: 10.1016/0167-4781(91)90156-G. [DOI] [PubMed] [Google Scholar]
- 45.Naested H, Frandsen GI, Jauh GY, Hernandez-Pinzon I, Nielsen HB, Murphy DJ, Rogers JC, Mundy J (2000) Caleosins: Ca2 + −binding proteins associated with lipid bodies. Plant Mol Biol 44(4):463–476 [DOI] [PubMed]
- 46.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy WS, Jr, Eisenson H, Musante G, Surwit RS, Millington DS, Butler MD, Svetkey LP. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nikiforidis CV, Kiosseoglou V. Physicochemical stability of maize germ oil body emulsions as influenced by oil body surface−xanthan gum interactions. J Agric Food Chem. 2009;58(1):527–532. doi: 10.1021/jf902544j. [DOI] [PubMed] [Google Scholar]
- 48.Pai JK, Siegel MI, Egan RW, Billah MM. Phospholipase D catalyzes phospholipid metabolism in chemotactic peptide-stimulated HL-60 granulocytes. J Biol Chem. 1988;263(25):12472–12477. [PubMed] [Google Scholar]
- 49.Paris L, Cecchetti S, Spadaro F, Abalsamo L, Lugini L, Pisanu ME, Iorio E, Natali PG, Ramoni C, Podo F. Inhibition of phosphatidylcholine-specific phospholipase C downregulates HER2 overexpression on plasma membrane of breast cancer cells. Breast Cancer Res. 2010;12(3):R27. doi: 10.1186/bcr2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perera L, Essmann U, Berkowitz ML. Role of water in the hydration force acting between lipid bilayers. Langmuir. 1996;12(11):2625–2629. doi: 10.1021/la9515534. [DOI] [Google Scholar]
- 51.Plevin R, Stewart A, Paul A, Wakelam MJO. Vasopressin-stimulated H-3 inositol phosphate and H-3 phosphatidylbutanol accumulation in A10 vascular smooth muscle cells. Br J Pharmacol. 1992;107(1):109–115. doi: 10.1111/j.1476-5381.1992.tb14471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plietz P, Damaschun G, Müller JJ, Schwenke K-D (1983) The structure of 11-S globulins from sunflower and rape seed. Eur J Biochem 130(2):315–320. doi:10.1111/j.1432-1033.1983.tb07154.x [DOI] [PubMed]
- 53.Potocky M, Elias M, Profotova B, Novotna Z, Valentova O, Zarsky V. Phosphatidic acid produced by phospholipase D is required for tobacco pollen tube growth. Planta. 2003;217(1):122–130. doi: 10.1007/s00425-002-0965-4. [DOI] [PubMed] [Google Scholar]
- 54.Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, Sullards CM, Wang E, Murphy RC, Barkley RM, Leiker TJ, Raetz CRH, Guan Z, Laird GM, Six DA, Russell DW, McDonald JG, Subramaniam S, Fahy E, Dennis EA. Lipidomics reveals a remarkable diversity of lipids in human plasma. JLipid Res. 2010;51(11):3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reichelt R, Schwenke K-D, König T, Pähtz W, Wangermann G. Electron microscopic studies for estimation of the quaternary structure of the 11S globulin (helianthinin) from sunflower seed (Helianthus annuus L.) Biochem Physiol Pflanz. 1980;175(7):653–663. [Google Scholar]
- 56.Rooijen GJH, Motoney MM. Plant seed oil-bodies as carriers for foreign proteins. Nat Biotech. 1995;13(1):72–77. doi: 10.1038/nbt0195-72. [DOI] [PubMed] [Google Scholar]
- 57.Rosivatz E, Woscholski R. Removal or masking of phosphatidylinositol(4,5)bisphosphate from the outer mitochondrial membrane causes mitochondrial fragmentation. Cell Signal. 2011;23(2):478–486. doi: 10.1016/j.cellsig.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rüegg M, Moor U, Blanc B. Hydration and thermal denaturation of β-lactoglobulin. A calorimetric study. Biochim Biophys Acta Protein Struct. 1975;400(2):334–342. doi: 10.1016/0005-2795(75)90188-9. [DOI] [PubMed] [Google Scholar]
- 59.Schmid AC, Wise HM, Mitchell CA, Nussbaum R, Woscholski R. Type II phosphoinositide 5-phosphatases have unique sensitivities towards fatty acid composition and head group phosphorylation. Fed Eur Biochem Soc Lett. 2004;576(1–2):9–13. doi: 10.1016/j.febslet.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 60.Seddon JM. Structure of the inverted hexagonal (Hii) phase, and non-lamellar phase-transitions of lipids. Biochim Biophys Acta. 1990;1031(1):1–69. doi: 10.1016/0304-4157(90)90002-T. [DOI] [PubMed] [Google Scholar]
- 61.Seddon JM, Templer RH (1995) Polymorphism of lipid-water systems. In: Lipowsky R, Sackman E (eds) The handbook of biological physics, vol I. Amsterdam, Elsevier Science
- 62.Shah SH, Bain JR, Muehlbauer MJ, Stevens RD, Crosslin DR, Haynes C, Dungan J, Newby LK, Hauser ER, Ginsburg GS, Newgard CB, Kraus WE. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events/clinical perspective. Circ: Cardiovasc Genet. 2010;3(2):207–214. doi: 10.1161/CIRCGENETICS.109.852814. [DOI] [PubMed] [Google Scholar]
- 63.Shearman GC, Ces O, Templer RH, Seddon JM. Inverse lyotropic phases of lipids and membrane curvature. J Phys Condens Matter. 2006;18(28):S1105–S1124. doi: 10.1088/0953-8984/18/28/S01. [DOI] [PubMed] [Google Scholar]
- 64.Smith RJ, Justen JM, McNab AR, Rosenbloom CL, Steele AN, Detmers PA, Anderson DC, Manning AM. U-73122: a potent inhibitor of human polymorphonuclear neutrophil adhesion on biological surfaces and adhesion-related effector functions. J Pharmacol Exp Ther. 1996;278(1):320–329. [PubMed] [Google Scholar]
- 65.Su W, Yeku O, Olepu S, Genna A, Park J-S, Ren H, Du G, Gelb MH, Morris AJ, Frohman MA. 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol Pharmacol. 2009;75(3):437–446. doi: 10.1124/mol.108.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tzen JTC, Cao YZ, Laurent P, Ratnayake C, Huang AHC. Lipids, proteins, and structure of seed oil bodies from diverse species. Plant Physiol. 1993;101(1):267–276. doi: 10.1104/pp.101.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tzen JTC, Huang AHC. Surface-structure and properties of plant seed oil bodies. J Cell Biol. 1992;117(2):327–335. doi: 10.1083/jcb.117.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tzen JTC, Lai YK, Chan KL, Huang AHC. Oleosin isoforms of high and low-molecular weights are present in the oil bodies of diverse seed species. Plant Physiol. 1990;94(3):1282–1289. doi: 10.1104/pp.94.3.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tzen JTC, Lie GC, Huang AHC. Characterization of the charged components and their topology on the surface of plant seed oil bodies. J Biol Chem. 1992;267(22):15626–15634. [PubMed] [Google Scholar]
- 70.Tzen JTC, Peng C-C, Cheng D-J, Chen ECF, Chiu JMH. A new method for seed oil body purification and examination of oil body integrity following germination. J Biochem. 1997;121(4):762–768. doi: 10.1093/oxfordjournals.jbchem.a021651. [DOI] [PubMed] [Google Scholar]
- 71.Wood K, Frölich A, Paciaroni A, Moulin M, Härtlein M, Zaccai G, Tobias DJ, Weik M. Coincidence of dynamical transitions in a soluble protein and its hydration water: direct measurements by neutron scattering and MD simulations. J Am Chem Soc. 2008;130(14):4586–4587. doi: 10.1021/ja710526r. [DOI] [PubMed] [Google Scholar]
- 72.Zhao Y, Yan A, Feijo J, Furutani M, Takenawa T, Hwang I, Fu Y, Yang Z (2010) Phosphoinositides regulate clathrin-dependent endocytosis at the tip of pollen tubes in Arabidopsis and tobacco. The Plant Cell Online. doi:10.1105/tpc.110.076760 [DOI] [PMC free article] [PubMed]
- 73.Zoncu R, Perera RM, Sebastian R, Nakatsu F, Chen H, Balla T, Ayala G, Toomre D, De Camilli PV. Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proc Natl Acad Sci. 2007;104(10):3793–3798. doi: 10.1073/pnas.0611733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 879 kb)