Abstract
Brassinosteroid (BR) and gibberellin (GA) promote many similar developmental responses in plants; but their relationship remains unclear. Here we show that BR and GA act interdependently through a direct interaction between the BR-activated BZR1 and GA-inactivated DELLA transcription regulators. GA promotion of cell elongation required BR signaling, whereas BR or active BZR1 can suppresssed the GA-deficient dwarf phenotype. DELLAs directly interacted with BZR1 and inhibited BZR1-DNA binding both in vitro and in vivo. Genome-wide analysis defined a BZR1-dependent GA-regulated transcriptome, which is enriched with light-regulated genes and genes involved in cell wall synthesis and photosynthesis/chloroplast. GA promotion of hypocotyl elongation requires both BZR1 and the phytochrome interacting factors (PIFs), as well as their common downstream targets PREs. The results demonstrate that GA releases DELLA-mediated inhibition of BZR1, and that the DELLA-BZR1-PIF4 interaction defines a core transcription module that mediates coordinated growth regulation by GA, BR and light signals.
The remarkable plasticity of plant development is believed to rely on networks of interconnected signal transduction pathways that integrate multiple hormonal and environmental signals coordinately regulating common cellular activities and developmental processes1. However, direct crosstalk between hormone pathways has rarely been observed in plants, although many signalling pathways have been characterized in detail2. BR and GA are two major growth-promoting hormones that have similar effects on a wide range of developmental processes1. Mutants deficient in either BR or GA show various degrees of similar phenotypes, including dwarfism, reduced seed germination, de-etiolation in the dark, and delayed flowering2-6. Despite the physiological evidence for overlapping actions on development7, the relationship between the BR and GA signalling pathways has remained unclear.
Both BR and GA signalling pathways have been studied extensively8, 9. BR is perceived by the receptor kinase Brassinosteroid-Insenstivie 1 (BRI1), and downstream signal transduction leads to activation of the Brassinozale-Resistant 1 (BZR1) family transcription factors, which control BR-responsive gene expression8. When BR levels are low, BZR1 is phosphorylated by the GSK3-like kinase Brassinosteroid-Insensitive 2 (BIN2), and as a result, loses its DNA-bind activity and becomes retained in the cytoplasm8. When BR levels are high, activation of BRI1 leads to sequential phosphorylation and activation of the BR-signalling kinases (BSKs and CDG1) and members of the BRI1 Suppressor 1 (BSU1) family phosphatases8, 10, which inactivates the GSK3s through tyrosine dephosphorylation11, allowing BZR1 activation by PP2A-mediated dephosphorylation12. Dephosphorylated BZR1 translocates into the nucleus to regulate over a thousand target genes13.
GA promotes plant growth by removing the DELLA proteins. Binding of GA to its receptor GA INSENSITIVE DWARF 1 (GID1) induces GID1-DELLA interaction and association with the E3 ubiquitin ligase SCFSLY1/GID2, leading to polyubiquitination and degradation of DELLAs9, 14. When GA levels are low, DELLAs accumulate and directly inactivate a number of transcription factors9, 15-21, including the bHLH factor phytochrome interacting factor4 (PIF4), which promotes cell elongation when plants are in the dark, shade, or high temperature22. Despite their overlapping physiological functions and the extensive knowledge about each signalling pathway, little is known about how BR and GA interact at the molecular level. Previous genome expression analysis of BR and GA responsive genes identified largely nonoverlapping transcriptional responses23, however, such meta-analysis might have biased the results1. Therefore, it is unclear whether and how the actions of BR and GA are coordinated in regulating common developmental processes1, 2. Here we demonstrate an interdependent relationship between BR and GA, mediated by direct interaction between DELLAs and BZR1, and identify a BZR1-dependent GA-regulated transcriptome controlling cell elongation and photosynthesis. These results provide evidence for a central growth-regulating transcription module that integrates BR, GA, and environmental signals for regulating cell elongation and seedling etiolation.
RESULTS
BR or active BZR1/BZR2 is required for GA promotion of cell elongation
To understand the relationship between BR and GA, we examined how defects in one hormone pathway influence the sensitivity to the other in hypocotyl elongation. We found that GA increased hypocotyl length in wild type plants but not in the BR-deficient mutant det2-1 or BR-insensitive mutant bri1-5 and bri1-119 (Fig. 1a, b). BR restored GA response to det2-1 (Fig. 1a, b), and increased the GA sensitivity in a dose-dependent manner (Supplementary Fig. 1a). However, BR cannot restore the GA responsiveness of the BR-insensitive mutants bri1-5 and bri1-119 (Fig. 1a, b), indicating that BR signalling is required for GA-induced hypocotyl elongation. The GA-insensitive phenotypes of bri1-119 and bri1-116 were suppressed by the dominant gain-of-function mutant bzr1-1D (Fig. 1b and Supplementary Fig. 1b, c), in which active BZR1 accumulates as a result of increased interaction with PP2A phosphatase12. The bzr1-1D mutation also partially suppressed the short-hypocotyl phenotype of the GA-deficient ga1-3 mutant and wild type plants treated with a GA biosynthesis inhibitor paclobutrazol (PAC) (Fig. 1c and Supplementary Fig. 1d-f). BZR2/BES1 is BZR1 homolog and have 88% protein identity; its gain-of-function mutant bes1-D showed resistant to PAC and partially suppressed the GA insensitivity of BR-deficient plants (Supplementary Fig. 1g-i). These results suggest that BR or BR-activated BZR1/BZR2 is required for GA promotion of hypocotyl elongation.
Fig. 1. BR signalling and BZR1 activity are required for GA promotion of hypocotyl elongation.
(a) Wild type and BR mutants were grown under the constant light for 7 days on medium with or without 1 μM GA3 and 1 μM brassinolide (BL, the most active brassinosteroid). (b) BR-deficient and insensitive mutants show a GA-insensitive phenotype, which is suppressed by bzr1-1D. Wild type (Col) and BR mutant seedlings were grown in the dark for 6 days on medium containing 1 μM PAC and different concentrations of GA3, and with 10 nM BL where indicated (+BL). Error bars, s.e.m. (n=32 plants) (c) bzr1-1D partly suppressed ga1-3 phenotype. (d) BR promotes cell elongation in GA mutants. Wild type (Col), ga1-3, sly1-10 were grown under light for 7 days on medium containing 1 μM PAC or 1 μM BL as indicated. (e) GA enhances the BR sensitivity. Wild type and det2-1 were grown under constant light for 7 days on medium with or without 1 μM GA3 and different concentrations of BL. Error bars, s.e.m. (n=35 plants). (f, g) Removing DELLAs enhances BR sensitivity. Ler, gai-1 and della seedlings were grown for 7 days under constant light on medium containing 2 μM brassinazole (BRZ, an inhibitor of BR synthesis) and 10 nM (+), 1 μM (++) (f), or different concentrations (g) of BL. Error bars, s.e.m. (n=25 plants).
GA-induced DELLA degradation enhances BR response
In contrast to the GA insensitivity of BR mutants, the GA-deficient mutant ga1-3, GA-insensitive mutant sly1-10, and wild type plants treated with PAC were responsive to BR and partly rescued by high concentration of BR (Fig. 1d and Supplementary Fig. 1j). GA and PAC had no effect on hypocotyl elongation of det2-1 and bri1-5 mutants, but enhanced and reduced, respectively, the BR-induced hypocotyl elongation in det2-1 (Fig. 1e and Supplementary Fig. 1k, l), suggesting that GA promotes cell elongation by enhancing the BR-induced response. Such essential role of BR and enhancing role of GA are consistent with the stronger dwarf/de-etiolation phenotypes of BR-deficient than GA-deficient mutants.
GA is known to promote growth by degradation of the growth-repressor proteins DELLAs9. The della pentuple mutant lacking all five members of the DELLA family genes showed dramatically enhanced BR response, whereas the GA-insensitive mutant gai-1, which accumulates high levels of GAI (one of the five DELLA proteins in Arabidopsis)9, 15, showed slightly reduced BR response (Fig. 1f, g). The DELLA protein RGA was degraded normally in response to GA treatment in BR mutants (det2-1 and bri1-116) and BR-treated plants (Supplementary Fig. 2a, b), suggesting that BR is not required for GA induced DELLA degradation and that degradation of DELLA is not sufficient for promoting hypocotyl elongation in the absence of BR. These genetic and physiological results suggest that DELLAs may repress a BR-activated component, likely BZR1 or a downstream component, and GA-induced DELLA degradation releases this repression.
RGA interacts with BZR1 and inhibits BZR1-DNA binding ability
DELLAs are known to inhibit several transcription factors via protein-protein interactions16, 17, 19, 21. The requirement of BR-activated BZR1 for GA/DELLA regulation of hypocotyl elongation raises a possibility that DELLAs may directly repress BZR1. Indeed a yeast two-hybrid screen identified the DELLA protein RGA as a BZR1-interacting protein (Fig. 2a). Additional yeast two-hybrid assays showed that both BZR1 and bzr1-1D interacted with RGA (Fig. 2a). RGA contains the N-terminal DELLA domain, which is required for GA-induced degradation9 and possesses transactivation acitivity24, and the C-terminal GRAS domain, which is important for its repressor function25-27 (Fig. 2b). Deletion of DELLA domain had no effect on BZR1 interaction, indicating that BZR1 interacts with the GRAS domain but not the DELLA domain of RGA (Fig. 2a, c).
Fig. 2. RGA interacts with BZR1 and inhibits BZR1 DNA binding activity in vitro and in vivo.
(a) Wild type and dominant mutant forms of BZR1 and RGA interact in yeast. (b) A diagram of the structure of RGA. (c) The LHR1 domain is necessary for both RGA homodimerization and the interaction with BZR1; the SAW domain is also required for interaction with BZR1. (d, e) MBP and MBP-fusions with BZR1 protein were incubated with GST-RGA bound to glutathione-agarose beads and then eluted and analyzed by anti-MBP immunoblotting. (f, g) BZR1 and RGA interact in plants. (f) The seedling of Col and pBZR1:BZR1-CFP grown in medium containing 100 nM PAC under light for 7 days were treated with 100 nM BL 1hr, and then co-immunoprecipitation was performed using anti-YFP antibody and immunoblotted using anti-RGA and anti-YFP antibodies. (g) Immunoprecipitation was performed using anti-YFP antibody on transgenic Arabidopsis plants expressing 35S::BZR1-Myc only or co-expressing 35S::BZR1-Myc and 35S::RGA-YFP, and immunoblotted using anti-Myc or anti-YFP antibodies. (h) RGA inhibits BZR1 DNA binding in vitro. MBP-BZR1 pre-incubated with MBP or MBP-RGA was incubated with biotinylated DNA fragments from the IAA19 and SAUR-AC1 promoters immobilized on streptavidin beads. The DNA-bound proteins were immunoblotted using anti-MBP antibody. (i) GA increases BZR1-DNA binding in vivo. ChIP was performed using anti-YFP antibodies followed by qPCR analysis. BZR1 binding was calculated as ratio between BZR1-CFP and 35S::YFP control, normalized to that of the control gene CNX5. Error bars mean s.d. of three biological repeats. Significant differences between GA and mock treatment are marked by asterisk (p < 0.01). (j, k) Transient reporter gene assays show RGA inhibition of BZR1 transcription activity. Arabidopsis protoplasts were transformed with the dual luciferase reporter construct containing pPRE::LUC (luciferase) and 35S::REN (renilla luciferase), and constructs overexpressing the indicated effecters. The LUC activity was normalized to REN. Error bars, s.d. of three biological repeats. a: Significant difference compared with BZR1 and BZR1+RGA samples (p < 0.01). b: Significant difference compared with BZR1 and BZR1+GAI samples. (p < 0.05). #: No Significant difference compared with BZR1 and BZR1+RGL2 samples (p > 0.05).
The GRAS domain can be subdivided into five distinct sequence motifs: leucine heptad repeat I (LHRI), the VHIID motif, leucine heptad repeat II (LHRII), the PFYRE motif and the SAW motif25 (Fig. 2b). Deletion of either LHRI or SAW motif abolished the interaction with BZR1 (Fig. 2c). The LHRI domain is required for RGA homodimerization (Fig. 2c), and both LHRI and SAW domains are required for growth-suppression function of DELLAs24, 26, 27. RGA also can heterodimerize with other DELLA proteins (Supplementary Fig. 3a, b). These results thus support a possibility that both dimerization and the SAW domain are required for RGA binding to BZR1 and suppressing plant growth. The GRAS domain is highly conserved in all members of the DELLA family, and both BZR1 and BZR2/BES1 interacted with RGA, GAI, RGL1 and RGL3, but not RGL2, in yeast (Supplementary Fig. 2a).
In vitro pull-down assays showed that GST-RGA interacted strongly with MBP-tagged full-length BZR1 and the N-terminal part of BZR1 (BZR1N), and weakly with the C-terminal part of BZR1 (BZR1C), but not MBP alone (Fig. 2d), suggesting that the N-terminal DNA binding domain of BZR1 has high affinity for RGA. Interestingly, RGA only binds to unphosphorylated BZR1 but not the BIN2-phosphorylated MBP-BZR1 (Fig. 2e). Consistent with the in vitro data, co-immunoprecipitation assays and bimolecular fluorescence complementation (BiFC) assays showed that BZR1 interacts with RGA in vivo and BR-induced BZR1 dephosphorylation increased the interaction (Fig. 2f, g and Supplementary Fig. 3b). These results demonstrate that RGA binds specifically to the BR-activated form of BZR1.
DELLAs are known to inhibit the DNA-binding of transcription factors16, 17, 19. We thus tested whether DELLAs block BZR1-DNA binding. MBP-BZR1 can be pulled down by biotinylated DNA fragments of the BZR1 target genes IAA19 or SAUR-AC1 promoter but not by the non-target CNX5 promoter13 (Fig. 2h and Supplementary Fig. 3c), confirming the specific interaction between BZR1 and target promoters. Incubation of MBP-BZR1 with MBP-RGA dramatically reduced BZR1 binding to DNA, whereas incubation with MBP alone had no effect (Fig. 2h, Supplementary Fig. 3c, d), indicating that RGA inhibits BZR1-DNA binding in vitro. To test whether DELLA proteins inhibit BZR1-DNA binding in vivo, we performed chromatin immunoprecipitation followed by quantitative real-time PCR (ChIP-qPCR) assays. The ChIP-qPCR results show that BZR1 binding to the promoters of five BZR1 target genes (PRE1, PRE5, IAA19, SAUR-AC1 and DWF4)13 was enhanced by GA treatment, presumably due to GA-induced degradation of the DELLA proteins (Fig. 2i). GA treatment and GA-signalling mutant (rga-24/gai-t6 and spy-3) did not affect the abundance or phosphorylation status of BZR1 protein (Supplementary Fig. 2c, d), consistent with DELLAs directly blocking DNA binding. In protoplast transient assays, the expression level of luciferase driven by the BR-responsive PRE5 promoter was increased by BZR1 and bzr1-1D, but this increase was abolished by co-expression of RGA, GAI and rga-Δ17, but not by RGL2 and RGAΔSAW (Fig. 2j, k). These results indicate that RGA specifically interacts with BZR1 to inhibit its abilities to bind DNA and regulate transcription.
GA and BR regulate overlapping genomic targets involved in photomorphogenesis and cell elongation
If DELLAs inhibit BZR1 activity in vivo and GA releases DELLA inhibition, GA should affect the expression of BZR1 target genes in similar manners as BR. Indeed the previously identified microarray data sets of genes affected in the BR-insensitive mutant bri1-116 and GA-deficient mutant ga1-3 overlap significantly13, 28 (Fig. 3a). Of the 1194 genes differentially expressed in ga1-3 compared to WT, 419 genes (35%) were also affected in the bri1-116 mutant, of which 296 also affected by light (Fig. 3a and table S1). Among these co-regulated genes, 387 genes (92.3%) were affected in the same way by bri1-116 and ga1-3, with a correlation coefficient R=0.76 (Fig. 3b). The effects of bri1 and ga1 are also similar to that of light on these genes, consistent with BR and GA repressing light responses (Fig. 3b). For 276 (71%) of these genes, the effects of bri1-116 were reversed by the bzr1-1D mutation (compare bzr1-1D/bri1-116 and bri1-116) and the effects of ga1-3 were reversed by loss of DELLA proteins (compare della/ga1-3 and ga1-3) (Fig. 3c). These results show that GA and BR exert similar effects on a large number of common genes through DELLA and BZR1 activities.
Fig. 3. GA and BR co-regulate large number of genes through DELLAs and BZR1.
(a) Venn diagram shows the overlaps between sets of genes differentially expressed in dark-grown bri1-116 versus wild type (bri1-116 D vs WT D), ga1-3 versus wild type (ga1-3 D vs WT D), and light-grown versus dark-grown wild type (WT L vs D). (b) Scatter plot of log2 fold change values for 419 genes differentially expressed of ga1-3 D versus WT D and bri1-116 D versus WT D. Red and blue colors indicate light-activated and light-repressed genes, respectively; black color indicates the genes are not regulated by light. (c) Hierarchical clusters analysis of the genes differentially expressed in bzr1-1D/bri1-116 versus bri1-116 (bzr1-1D/bri1-116), bri1-116 versus WT (bri1-116), ga1-3 versus Ler (ga1-3), and della/ga1-3 versus ga1-3 (della/ga1-3). The gradient bar represents log2 of the ratio. Genes are listed in table S1. (d-h) RNA-Seq analyses of genes affected by GA treatments or by bzr1-1D in BR-deficient plants (grown on 2 μM PPZ medium) (2 μM is right). (d) Venn diagram shows overlaps between sets of genes affected by GA treatment in wild type (Col) plants grown on medium containing PAC (Col GA+/-) or on medium containing PAC and PPZ (Col_PPZ GA+/-), and genes affected by bzr1-1D in the presence of PAC and PPZ (bzr1-1D_PPZ vs Col_PPZ). (e-g) Hierarchical clusters analysis of the expression data of the genes in class A (e), B (f) and C (g) in panel d. (h) Gene Ontology analysis of cellular functions represented by GA up- and down-regulated genes in each gene class (A to D) shown in panel d. All genes detected in RNA-Seq samples were used as control (random).
To further define the BR/BZR1-dependent genomic targets of GA signalling, we analyzed the effects of BR-deficiency and bzr1-1D mutation on GA-induced gene expression changes using RNA-sequencing (RNA-Seq). RNA-Seq analysis identified 3570 genes affected >1.5-fold by GA treatment in wild type plants grown without propiconazole (PPZ, a specific inhibitor of BR biosynthesis29), 1629 genes affected by GA in wild type plants grown on PPZ medium (BR-deficient plants), and 4306 genes affected in bzr1-1D compared to wild type plants when grown on the PAC+PPZ medium (deficient of both GA and BR) (Fig. 3d). Of the 3570 genes regulated by GA in BR-sufficient plants, only 1187 genes (33%) were affected by GA in the BR-deficient plants, suggesting that GA-responsive expression of most genes requires BR. About half (549 genes) of the BR-independent GA-regulated genes were not affected by bzr1-1D in any significant amount or pattern (Class A, Fig. 3d, e), whereas the other half (638 genes) was affected by bzr1-1D, mostly in similar manners as GA treatment (Class B, Fig 3d, f). It is likely that Class A genes are regulated by other DELLA-interacting transcription regulators, such as JAZ1 and EIN3 (Ref. 18, 19), independent of BR/BZR1, and the Class B genes are regulated through both BR/BZR1-dependent and independent mechanisms. Of the BR-dependent GA-responsive genes, 1027 were affected by bzr1-1D (Class C, Fig. 3d), mostly in similar manners as GA treatment (Fig. 3g), and their GA responses were quantitatively reduced in the BR-deficient plants (Fig. 3g); these genes apparently are regulated by GA through a BR/BZR1-dependent mechanism.
Gene Ontology analyses showed that cell wall-related genes are dramatically enriched in the GA-induced Class B (10%) and Class C genes (10%) compared to the random control (3%) (Fig. 3h), suggesting that GA promotes cell elongation preferentially through the BZR1-dependent mechanism but also through BR-independent mechanisms. In contrast, photosynthesis/chloroplast genes are dramatically enriched in the GA-repressed Class B (44%) and C (62%) but not in Class A or D (13-17%) compared to control (15%) (Fig. 3h), indicating that GA represses photosynthetic genes mainly through a BZR1-dependent mechanism. These genomic data thus provide direct evidence for the important role of BZR1 in GA regulation of genome expression, particularly the transcriptomes that promote cell elongation and repress photosynthetic development.
GA-promotion of hypocotyl elongation requires BZR1, PIF4, and their common downstream targets PREs
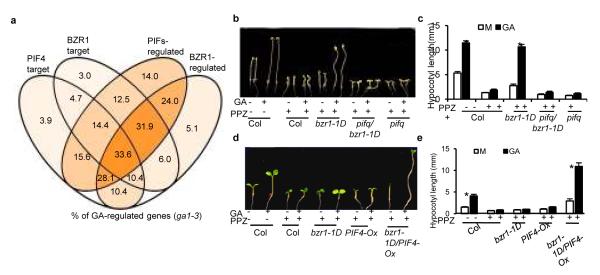
Both DELLAs and BZR1 also interact with PIF4 (Ref. 16, 17, 30), and PIF4 and BZR1 together bind to a large number of common promoters in the genome30. To determine whether the common targets by both BZR1 and PIF4 were preferentially regulated by GA, we grouped genes based on of the chromatin-immunoprecipitation data of BZR1 and PIF4 and the microarray expression profiling data of pifq and bzr1-1D/bri1 mutants13, 30, 31, and we calculated the percentage of GA-regulated genes (based on expression data of the ga1-3 mutant28) of each group. The genes regulated by PIFs and/or BZR1 included higher percentages of GA-regulated genes than genes unregulated by BZR1 and PIFs, and the genes that are common targets co-regulated by BZR1 and PIF4 included the highest percentage of GA-regulated genes (Fig. 4a and Supplementary Fig. 4), indicating that the genome targets shared by BZR1 and PIF4 tend to be regulated by GA.
Fig. 4. BZR1 and PIF4 are required for the GA promotion of hypocotyl elongation.
(a) Venn diagram shows the percentage of GA regulated genes among the gene sets that BZR1 and PIF4 bind to and/or regulate. PIF4 and BZR1 targets were identified by PIF4 ChIP-Seq and BZR1 ChIP-chip, respectively; PIFs- and BZR1-regulated genes were differentially expressed in pifq versus WT and in bzr1-1D/bri1-116 versus bri1-116 grown in the dark. GA-regulated genes were differentially expressed in ga1-3 versus WT in the dark. Numbers show the percentages of each gene set that are GA-regulated genes. (b, c) PIFs are required for BZR1 mediated GA promotion of hypocotyl elongation. Seedlings were grown in the dark for 5 days on medium containing 0.5 μM PAC, 10 μM PPZ with or without 1 μM GA3. Error bars, s.d. (n=10 plants). Asterisks mark significant differences between GA and mock treatments (p < 0.01). (d, e), Both BZR1 and PIFs are required for GA promotion of hypocotyl elongation in light. Seedlings were grown under red light for 5 days on medium containing 0.1 μM PAC, 2 μM PPZ, and 0 (M) or 1 μM GA3 (GA). Error bars, s.d. (n=10 plants). Asterisks mark significant differences between GA and mock treatments (p < 0.01).
We then tested whether GA-induced cell elongation also requires both BZR1 and PIFs. The dominant bzr1-1D mutation rescued GA response in wild type background but not in the pifq background when seedlings were grown on BR biosynthesis inhibitor PPZ in the dark (Fig. 4b, c), indicating that PIFs are required for BZR1-mediated GA response. When grown on medium containing PPZ under light (which causes degradation of PIFs), only bzr1-1D/PIF4-Ox plants showed robust GA response, whereas PIF4-Ox, bzr1-1D, and wild-type plants were all insensitive to GA (Fig. 4d, e). These results indicate that GA promotion of hypocotyl elongation requires both BZR1 and PIF4, and is thus likely mediated by the BZR1-PIF4 heterodimer.
Previous studies have shown that GA, BR and auxin induce the expression of the PRE family helix-loop-helix factors32-34, which promote cell elongation by antagonizing several inhibitory HLH factors33, 35, 36. Several PRE family members, including PRE1, PRE5 and PRE6/KIDARI, are direct targets of both BZR1 and PIF4 (Ref. 30). Consistent with GA acting through the DELLA-BZR1-PIF4 module, the expression of PRE1, PRE2, PRE5 and PRE6 was induced by GA treatment in wild type, but their GA induction was decreased in the bri1-119 mutant and recovered in the bri1-119 bzr1-1D double mutant, indicating that GA induction of these genes requires active BZR1 (Fig. 5a). Similarly, the BR-induction of PRE1, PRE5, and PRE6 was reduced in the dominant gain-of-function gai-1 mutant compared to wild type (Fig. 5b), indicating that BR induction was negated by accumulation of GAI. Two genes, EXP1 and EXP8, encoding cell wall proteins expansins that loosen cell wall37, are affected similarly to PREs by bri1-119 and gai-1 (Fig. 5a, b), suggesting that these expansins might mediate downstream response in cell elongation. The role of PREs in GA response was confirmed using the pre-amiR transgenic line, in which four PRE members are suppressed by artificial microRNA30, and a transgenic line that overexpresses IBH1, the antagonizing partner of PRE1 (Ref. 33). Both pre-amiR and IBH1-overexpression lines had GA-insensitive hypocotyls compared to wild-type (Fig. 5c). Consistent with GA-insensitivity of pre-amiR, PRE1 overexpression reduced plants’ sensitivity to the biosynthesis inhibitors of GA and BR (Fig. 5d, e). ChIP-reChIP with transgenic Arabidopsis expressing both BZR1-myc and PIF4-YFP showed that BZR1 and PIF4 co-occupy in the promoter of PRE1, PRE6, EXP1 and EXP8 (Fig. 5f). These results demonstrate that PREs are essential positive regulators for GA-promoted hypocotyl elongation, acting downstream of the DELLA-BZR1-PIF4 module.
Fig. 5. GA promotion of cell elongation requires the BZR1 and PIF4 targets PREs.
(a, b) Quantitative RT-PCR analyses of gene expression after GA treatment in wild type, bri1-119 and bzr1-1D/bri1-119 (a) and BL treatment in wild type (Col) and gai-1 (b). Total RNAs were extracted from 7-day-old seedlings treated with mock solution, 10 μM GA3, or 100 nM BL for 3 hr. The PP2A gene was analyzed as an internal control. Error bars are s.d. of three biological replicates. (c) Suppressing PREs (pre-amiR) and overexpression of IBH1 (IBH1-Ox) reduce hypocotyl elongation response to GA. Error bars, s.d. (n=25 plants). Asterisks indicate significant difference between GA and mock treatments (p < 0.01). (d, e) The 35S:PRE1-YFP transgenic plants (PRE1-Ox) show reduced sensitivities to the GA biosynthesis inhibitor PAC (d) and BR biosynthesis inhibitor BRZ (e). Seedlings were grown on medium containing different concentrations of PAC (d) or BRZ (e) for 7 days under light. Relative hypocotyl lengths were measured from at least 30 plants and normalized to the untreated plants. Error bars represent s.d. (n=33) (f) ChIP-reChIP analyses show that PRE1, PRE6, EXP1 and EXP8 are the common targets of BZR1 and PIF4. Chromatin from transgenic Arabidopsis expressing both 35S::BZR1-myc and 35S::PIF4-YFP was immunoprecipitated sequentially using anti-myc and anti-YFP antibodies, and then analyzed by qPCR. BZR1 and PIF4 binding was calculated as ratio between BZR1-myc/PIF4-YFP transgenic plants and Col control, normalized to that of the control gene PP2A. Error bars are s.d. of three biological replicates. Asterisks indicate significant difference to PP2A gene (p < 0,01) (g) The model for the signalling network integrating BR, GA and light signals. BZR1 and PIF4 form a functional complex to regulate a large number of genes that contribute to hypocotyl elongation; these include PREs, which in turn inactivates IBH1, leading to cell elongation. DELLAs interact with BZR1 and PIFs to inhibit their DNA binding ability. Signal transduction activated by BR, GA and light/phytochrome modulates the levels of BZR1, DELLAs, and PIFs, respectively, thereby controlling the activity of BZR1-PIF complex and cell elongation.
DISCUSSION
How plant growth is controlled by the wide range of environmental signals and endogenous cues is a fundamental question about plant biology. This study demonstrates a major mechanism of crosstalk between two hormonal signals and illustrates a central network that integrates signalling and growth regulation in plants. While previous studies suggested that BR and GA act independently on highly overlapping developmental responses1, this study reveals a much closer relationship between these two hormones. Strong evidence from genetic, biochemical, and genomic analyses support a model that GA and BR crosstalk through direct interaction between the key components of each pathway, DELLAs and BZR1 (Fig. 5g). BR signalling through the BRI1 receptor kinase pathway leads to dephosphorylation and subsequent nuclear accumulation of BZR1 (Ref. 8). However, the activity of BZR1 is attenuated by DELLA proteins when GA levels are low. GA-induced degradation of DELLAs frees BZR1 for DNA binding and transcriptional regulation. BR is required for GA promotion of cell elongation, because when BZR1 is inactivated by phosphorylation and unable to interact with DELLAs in the absence of BR, GA-induced DELLA degradation cannot significantly increase the nuclear BZR1 activity. As such, BR seems to be essential for GA-induced hypocotyl growth, whereas GA quantitatively enhances BR-potentiated growth.
The DELLA-BZR1 interaction is a critical link in the photomorphogenic regulation system. Previous studies have shown similar DELLA interaction with members of the PIF family of phytochrome interacting bHLH factors, and our accompanying study showed that BZR1 also interacts with PIFs to co-regulate large numbers of common target genes, while they also each regulate unique target genes30. Thus DELLAs can potentially inhibit BZR1 and PIF4 individually and modulate their actions of unique targets, and/or inhibit the BZR1-PIF4 heterodimer to modulate their common targets. The higher percentage of GA-responsive genes in the BZR1-PIF4 co-regulated than uniquely regulated genes suggests that DELLAs preferentially targets the BZR1-PIF4 heterodimmer. Interestingly, the downstream genes controlled by the interdependent actions of BR/BZR1, light/PIF4, and GA/DELLAs are enriched with cell wall- and photosynthesis/chloroplast-related genes, which are affected in ways that are consistent with the actions of these pathways on cell elongation and photomorphogenesis. These observations demonstrated that the interdependent interactions among DELLAs, BZR1, and PIFs regulate a core transcription module that mainly controls cell elongation and chloroplast development.
Considering that the levels of PIFs are controlled by light, circadian clock, and temperature22, 38-42, and the level of DELLAs is affected by not only GA but also auxin, abscisic acid, ethylene, jasmonate, and abiotic stresses1, 38, 43-46, the interdependent interactions of BZR1 with PIFs and DELLAs would allow BR to modulate the growth responses to all these other signals, consistent with an ancient and central role of steroid hormones in regulating growth. We propose that DELLAs, BZR1/2, and PIFs form the central command system that controls key growth processes and integrates all major growth-regulating hormonal and environmental signals (Fig. 5g). This command system seems to not only accept numerous inputs but also send out branched outputs, as each component act not only interdependently on shared targets but also independently on unique sets of target genes, possibly through additional interacting partners. Such non-exclusive relationships potentially provide a flexible system that allows both cooperative and independent actions of these signalling pathways on different cellular and developmental processes. These results demonstrate a complex central transcription network that integrates multiple signaling pathways, contains multiple layers of regulators, and controls major plant growth and developmental processes.
Supplementary Material
ACKNOWLEDGEMENT
We thank Dr. Xing Wang Deng for providing seeds of the della pentuple mutant. This study was supported by grants from NIH (R01GM066258 to Z.Y.W.) and from NSF (MCB-0923723 to T.P.S.). J.S. was supported by the China Scholarship Council.
Footnotes
AUTHOR CONTRIBUTIONS
M.B and Z.W together designed the experiments. M.B performed statistical analysis of plant growth, chromatin immunoprecipitation qPCR, DNA binding assay, RNA-Seq, RT-qPCR and together with E.O and Y.B analyzed microarray and RNA-Seq data. J.S performed the yeast two-hybrid, co-immunoprecipitation, protein-protein pull down, transient expression assays and generated bzr1-1D/ga1-3. J.H performed yeast two-hybrid screening and found RGA is a BZR1-interacting protein. E.O analyzed bzr1-1D/PIF4-Ox and bzr1-1D/pifq. M.F analyzed GA-responses of IBH1-Ox. R.Z performed RGA protein degradation studies. T.P.S provided sly1-10 seeds and helped with critical discussion on the work. M.B performed all other experiments. M.B and Z.W wrote the manuscript.
The authors declare no competing financial interests.
REFERENCES
- 1.Depuydt S, Hardtke CS. Hormone signalling crosstalk in plant growth regulation. Curr Biol. 2011;21:R365–373. doi: 10.1016/j.cub.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Jaillais Y, Chory J. Unraveling the paradoxes of plant hormone signaling integration. Nat Struct Mol Biol. 2010;17:642–645. doi: 10.1038/nsmb0610-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alabadi D, Gil J, Blazquez MA, Garcia-Martinez JL. Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 2004;134:1050–1057. doi: 10.1104/pp.103.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szekeres M, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 6.Steber CM, McCourt P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001;125:763–769. doi: 10.1104/pp.125.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka K, et al. Physiological Roles of Brassinosteroids in Early Growth of Arabidopsis: Brassinosteroids Have a Synergistic Relationship with Gibberellin as well as Auxin in Light-Grown Hypocotyl Elongation. J Plant Growth Regul. 2003;22:259–271. [Google Scholar]
- 8.Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- 9.Sun TP. The Molecular Mechanism and Evolution of the GA-GID1-DELLA Signaling Module in Plants. Curr Biol. 2011;21:R338–345. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 10.Kim T-W, Guan S, Burlingame AL, Wang Z-Y. The CDG1 Kinase Mediates Brassinosteroid Signal Transduction from BRI1 Receptor Kinase to BSU1 Phosphatase and GSK3-like Kinase BIN2. Mol Cell. 2011;43:561–571. doi: 10.1016/j.molcel.2011.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim TW, et al. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang W, et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol. 2011;13:124–131. doi: 10.1038/ncb2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murase K, Hirano Y, Sun TP, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- 15.Harberd NP, Belfield E, Yasumura Y. The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell. 2009;21:1328–1339. doi: 10.1105/tpc.109.066969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 18.Hou X, Lee LY, Xia K, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell. 2010;19:884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 19.An F, et al. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012;22:915–927. doi: 10.1038/cr.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnaud N, et al. Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 2010;24:2127–2132. doi: 10.1101/gad.593410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang ZL, et al. Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:2160–2165. doi: 10.1073/pnas.1012232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 24.Hirano K, et al. The Suppressive Function of the Rice DELLA Protein SLR1 is Dependent on its Transcriptional Activation Activity. Plant J. 2012 doi: 10.1111/j.1365-313X.2012.05000.x. [DOI] [PubMed] [Google Scholar]
- 25.Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- 26.Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell. 2002;14:57–70. doi: 10.1105/tpc.010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, et al. Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell. 2009;21:2378–2390. doi: 10.1105/tpc.108.065433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheminant S, et al. DELLAs Regulate Chlorophyll and Carotenoid Biosynthesis to Prevent Photooxidative Damage during Seedling Deetiolation in Arabidopsis. Plant Cell. 2011 doi: 10.1105/tpc.111.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekimata K, et al. A specific and potent inhibitor of brassinosteroid biosynthesis possessing a dioxolane ring. J Agric Food Chem. 2002;50:3486–3490. doi: 10.1021/jf011716w. [DOI] [PubMed] [Google Scholar]
- 30.Oh E, et al. Interaction between BZR1 and PIF4 integrates brassinosteroid and environment response. Co-submitted. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leivar P, et al. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell. 2009;21:3535–3553. doi: 10.1105/tpc.109.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S, et al. Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006;47:591–600. doi: 10.1093/pcp/pcj026. [DOI] [PubMed] [Google Scholar]
- 33.Zhang LY, et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 2009;21:3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapman EJ, et al. Hypocotyl Transcriptome Reveals Auxin Regulation of Growth-Promoting Genes through GA-Dependent and -Independent Pathways. PLoS One. 2012;7:e36210. doi: 10.1371/journal.pone.0036210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Zhu Y, Fujioka S, Asami T, Li J. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell. 2009;21:3781–3791. doi: 10.1105/tpc.109.072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyun Y, Lee I. KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol Biol. 2006;61:283–296. doi: 10.1007/s11103-006-0010-2. [DOI] [PubMed] [Google Scholar]
- 37.Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- 38.Achard P, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 39.Bauer D, et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 2004;16:1433–1445. doi: 10.1105/tpc.021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koini MA, et al. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 41.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 42.Park E, et al. Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 2004;45:968–975. doi: 10.1093/pcp/pch125. [DOI] [PubMed] [Google Scholar]
- 43.Achard P, Renou JP, Berthome R, Harberd NP, Genschik P. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol. 2008;18:656–660. doi: 10.1016/j.cub.2008.04.034. [DOI] [PubMed] [Google Scholar]
- 44.Achard P, Vriezen WH, Van Der Straeten D, Harberd NP. Ethylene regulates arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell. 2003;15:2816–2825. doi: 10.1105/tpc.015685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu X, Harberd NP. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature. 2003;421:740–743. doi: 10.1038/nature01387. [DOI] [PubMed] [Google Scholar]
- 46.Yang DL, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci U S A. 2012;109:E1192–1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.