Abstract
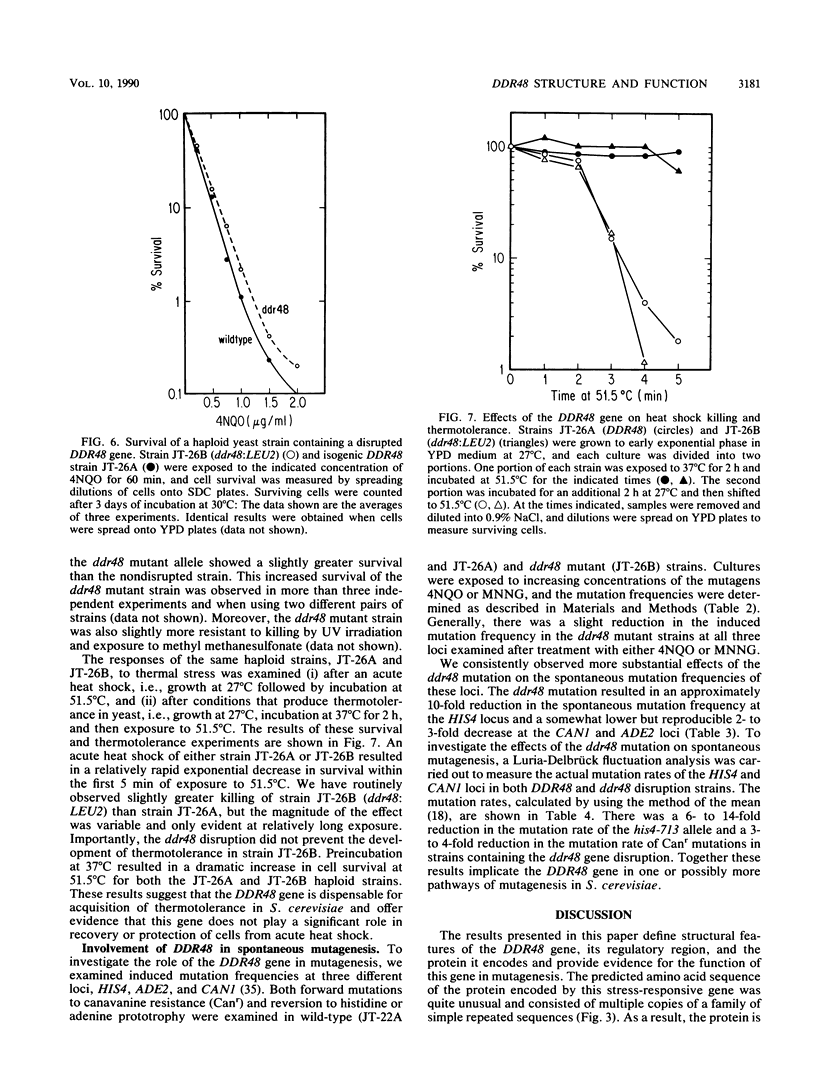
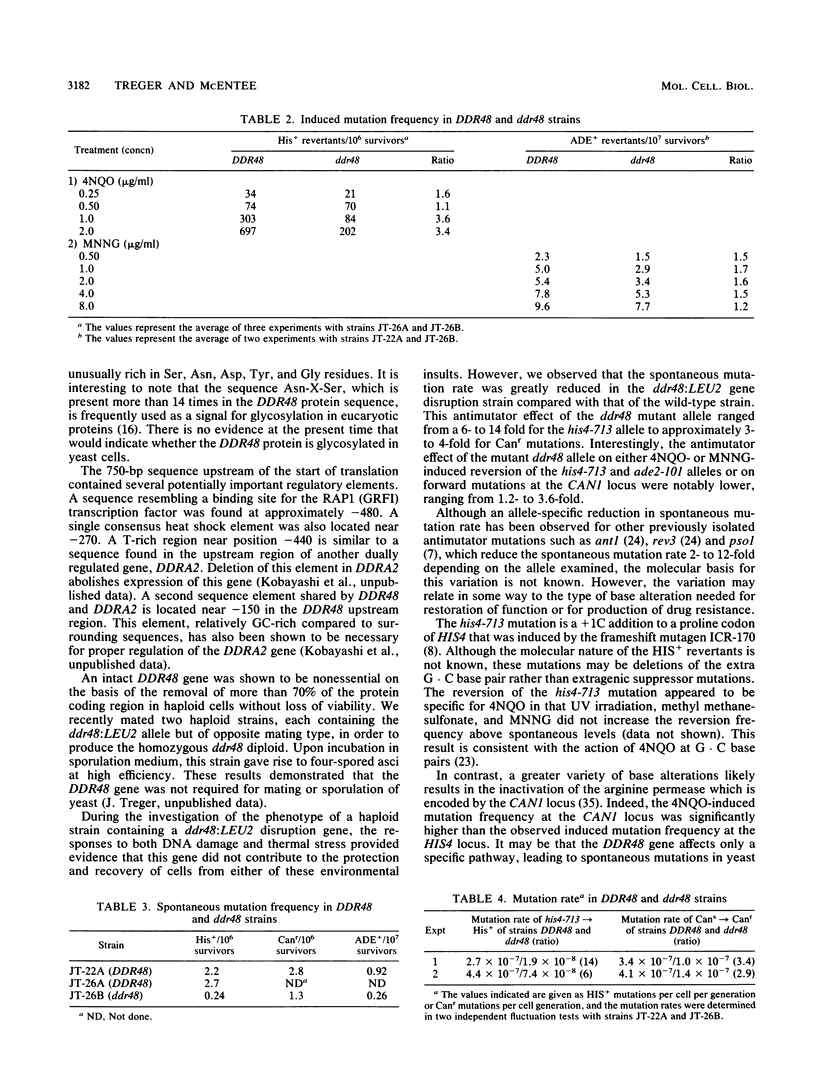
The DDR48 gene of Saccharomyces cerevisiae is a member of a set of genes that displays increased transcription in response to treatments that produce DNA lesions or to heat-shock stress. Other members of this group include the DDRA2 and UBI4 genes. DNA sequence analysis of the DDR48 gene demonstrates the presence of two overlapping open reading frames, each of which has the capacity to encode a protein with a molecular mass of approximately 45 kilodaltons. Fusions of the DDR48 coding sequences to lacZ demonstrates that only one of these frames is expressed in yeast cells. The protein predicted from this sequence is extremely hydrophilic and contains multiple repeats of the peptide sequence Ser-Asn-Asn-X-Asp-Ser-Tyr-Gly where X is either Asn or Asp. Additionally, closely related sequences are found throughout the primary sequence. Primer extension data indicate that, after 4-nitroquinoline-1-oxide and heat-shock treatments, there are three major and two minor transcriptional start sites which are utilized. The function of the DDR48 gene was investigated by disrupting this gene in diploid cells. Viable haploid cells containing the DDR48 gene disruption were isolated after tetrad analysis. Although the ddr48 mutant showed a slightly altered sensitivity to killing by 4-nitroquinoline-1-oxide and to heat shock compared with the DDR48 haploid, the spontaneous mutation rate of reversion of a his4 mutation was reduced 6- to 14-fold in the ddr48 strain. These results implicate the DDR48 gene in the production or recovery of mutations in S. cerevisiae.
Full text
PDF


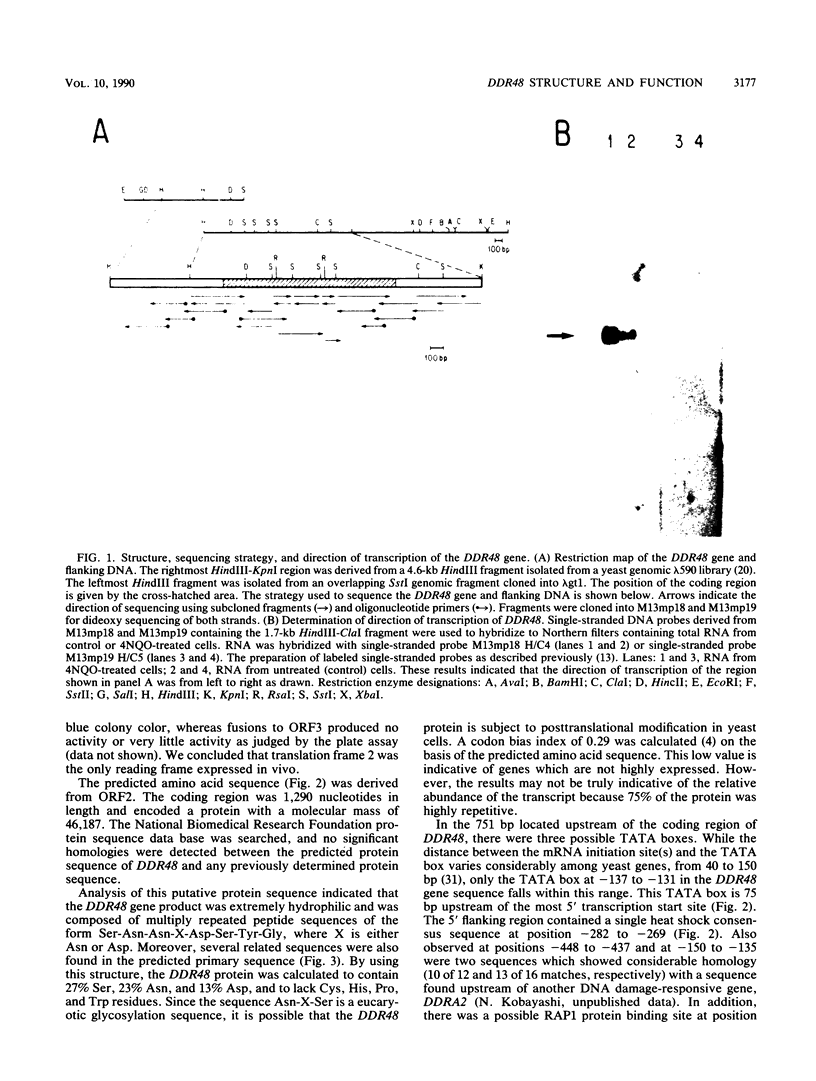

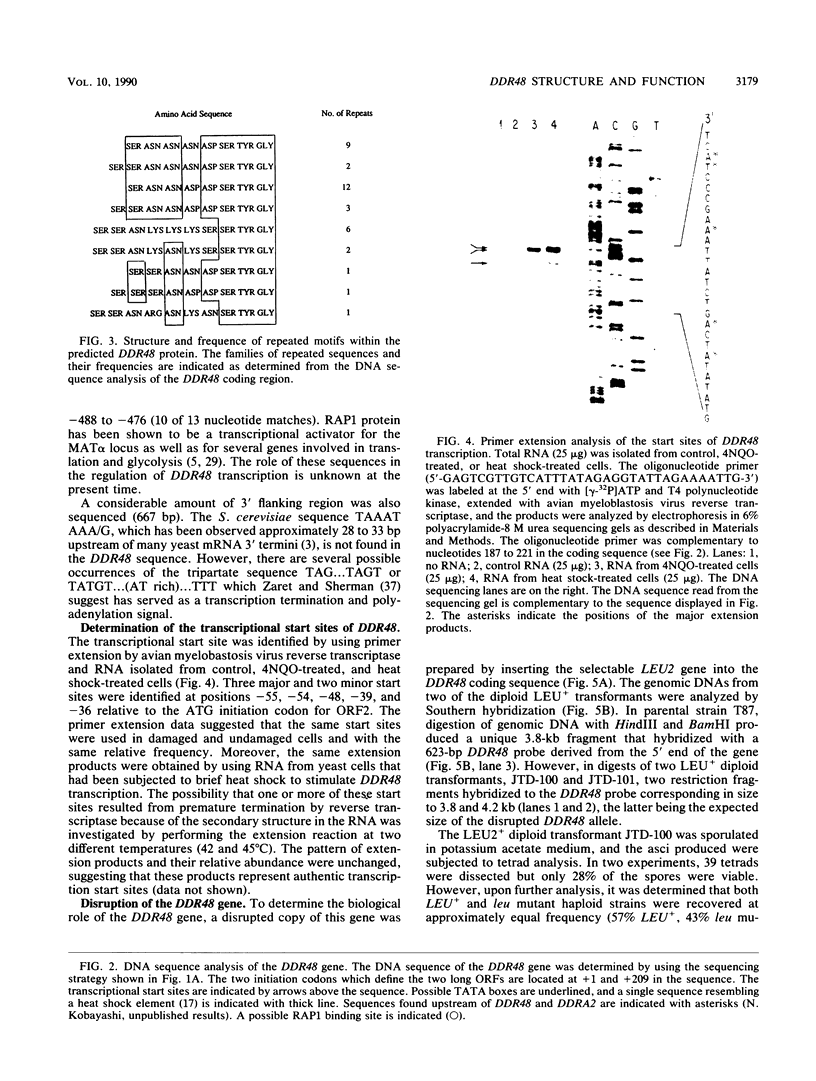



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angulo J. F., Schwencke J., Moreau P. L., Moustacchi E., Devoret R. A yeast protein analogous to Escherichia coli RecA protein whose cellular level is enhanced after UV irradiation. Mol Gen Genet. 1985;201(1):20–24. doi: 10.1007/BF00397980. [DOI] [PubMed] [Google Scholar]
- Bardwell J. C., Craig E. A. Major heat shock gene of Drosophila and the Escherichia coli heat-inducible dnaK gene are homologous. Proc Natl Acad Sci U S A. 1984 Feb;81(3):848–852. doi: 10.1073/pnas.81.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Buchman A. R., Lue N. F., Kornberg R. D. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988 Dec;8(12):5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Cassier C., Chanet R., Henriques J. A., Moustacchi E. The effects of three PSO genes on induced mutagenesis : a novel class of mutationally defective yeast. Genetics. 1980 Dec;96(4):841–857. doi: 10.1093/genetics/96.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. Suppressible four-base glycine and proline codons in yeast. Science. 1981 Apr 24;212(4493):455–457. doi: 10.1126/science.7010605. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. Identification and isolation of the gene encoding the small subunit of ribonucleotide reductase from Saccharomyces cerevisiae: DNA damage-inducible gene required for mitotic viability. Mol Cell Biol. 1987 Aug;7(8):2783–2793. doi: 10.1128/mcb.7.8.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C., Meeks-Wagner D. W., Fangman W. L., Botstein D. A rapid, efficient method for isolating DNA from yeast. Gene. 1986;42(2):169–173. doi: 10.1016/0378-1119(86)90293-3. [DOI] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Hurd H. K., Roberts C. W., Roberts J. W. Identification of the gene for the yeast ribonucleotide reductase small subunit and its inducibility by methyl methanesulfonate. Mol Cell Biol. 1987 Oct;7(10):3673–3677. doi: 10.1128/mcb.7.10.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R., Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClanahan T., McEntee K. DNA damage and heat shock dually regulate genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jan;6(1):90–96. doi: 10.1128/mcb.6.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClanahan T., McEntee K. Specific transcripts are elevated in Saccharomyces cerevisiae in response to DNA damage. Mol Cell Biol. 1984 Nov;4(11):2356–2363. doi: 10.1128/mcb.4.11.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash L., Stewart J. W., Sherman F. Specific induction of transitions and transversions of G-C base pairs by 4-nitroquinoline-1-oxide in iso-1-cytochrome c mutants of yeast. J Mol Biol. 1974 May 5;85(1):51–65. doi: 10.1016/0022-2836(74)90128-4. [DOI] [PubMed] [Google Scholar]
- Quah S. K., von Borstel R. C., Hastings P. J. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics. 1980 Dec;96(4):819–839. doi: 10.1093/genetics/96.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Szostak J. W., Murray A. W. Cloning regulated yeast genes from a pool of lacZ fusions. Methods Enzymol. 1983;101:253–269. doi: 10.1016/0076-6879(83)01019-8. [DOI] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Simon J. R., McEntee K. A rapid and efficient procedure for transformation of intact Saccharomyces cerevisiae by electroporation. Biochem Biophys Res Commun. 1989 Nov 15;164(3):1157–1164. doi: 10.1016/0006-291x(89)91790-7. [DOI] [PubMed] [Google Scholar]
- Struhl K. Molecular mechanisms of transcriptional regulation in yeast. Annu Rev Biochem. 1989;58:1051–1077. doi: 10.1146/annurev.bi.58.070189.005155. [DOI] [PubMed] [Google Scholar]
- Treger J. M., Heichman K. A., McEntee K. Expression of the yeast UB14 gene increases in response to DNA-damaging agents and in meiosis. Mol Cell Biol. 1988 Mar;8(3):1132–1136. doi: 10.1128/mcb.8.3.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- Walker G. C., Marsh L., Dodson L. A. Genetic analyses of DNA repair: inference and extrapolation. Annu Rev Genet. 1985;19:103–126. doi: 10.1146/annurev.ge.19.120185.000535. [DOI] [PubMed] [Google Scholar]
- Whelan W. L., Gocke E., Manney T. R. The CAN1 locus of Saccharomyces cerevisiae: fine-structure analysis and forward mutation rates. Genetics. 1979 Jan;91(1):35–51. doi: 10.1093/genetics/91.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]