Abstract
Rare mutations in PARK loci genes cause Parkinson’s disease (PD) in some families and isolated populations. We investigated the association of common variants in PARK loci and related genes with PD susceptibility and age at onset in an outbred population. 1,103 PD cases from the upper Midwest, USA were individually matched to unaffected siblings (n = 654) or unrelated controls (n = 449) from the same region. Using a sequencing approach in 25 cases and 25 controls, single nucleotide polymorphisms (SNPs) in species-conserved regions of PARK loci and related genes were detected. We selected additional tag SNPs from the HapMap. We genotyped a total of 235 SNPs and two variable number tandem repeats (VNTRs) in the ATP13A2, DJ1, LRRK1, LRRK2, MAPT, Omi/HtrA2, PARK2, PINK1, SNCA, SNCB, SNCG, SPR, and UCHL1 genes in all 2,206 subjects. Case-control analyses were performed to study association with PD susceptibility, while cases-only analyses were used to study association with age at onset. Only MAPT SNP rs2435200 was associated with PD susceptibility after correction for multiple testing (OR = 0.74, 95% CI = 0.64 – 0.86, uncorrected P < 0.0001, log additive model); however, 16 additional MAPT variants, seven SNCA variants, and one LRRK2, PARK2, and UCHL1 variants each had significant uncorrected P-values. There were no significant associations for age at onset after correction for multiple testing. Our results confirm the association of MAPT and SNCA genes with PD susceptibility, but show limited association of other PARK loci and related genes with PD.
INTRODUCTION
Parkinson’s disease (PD) affects 2% of all persons during their life.1 In 1997, the first genetic cause of PD was discovered, a mutation in the α-synuclein gene (SNCA)2. The α-synuclein protein was subsequently found to be a major component of Lewy bodies, the pathological hallmark of PD.3 Additional mutations in SNCA and in the other genes have been reported to cause PD. The regions of the genome to which these genes map are known as PARK loci, and they have been designated as: PARK 1 (SNCA), PARK 2 (PARK2),4 PARK 5 (UCHL1),5. PARK 6 (PINK1),6 PARK 7 (DJ-1),7. PARK 8 (LRRK2),8, 9 PARK 9 (ATP13A2),10 PARK 11 (GIGYF2),11 and PARK 13 (Omi/HtrA2).12 While PD cases that cluster in families or in isolated populations are sometimes explained by these mutations, the majority of PD cases that occur in outbred populations have unknown causes.
Previous studies suggested that common variants in some PARK loci genes, e.g., UCHL1 or SNCA, are associated with PD susceptibility worldwide.13, 14 We expanded the scope of those studies to include multiple variants in several PARK loci and related genes, and analyses for age at onset as well as susceptibility, in an outbred US sample.
SUBJECTS AND METHODS
Study Subjects
We conducted a case-control study (PD susceptibility) and a cases-only study (age at onset). All subjects were recruited as part of an ongoing study of the molecular epidemiology of PD. The subjects included in this study are partially overlapping with those included in our previously published studies; the current study included a larger sample than in our previously published studies (due to ongoing study recruitment).15–17 PD cases were referred sequentially to the Department of Neurology of the Mayo Clinic in Rochester, MN, from June 1, 1996 through June 30, 2007. They resided in Minnesota or in a surrounding state (Wisconsin, Iowa, North Dakota, and South Dakota). Controls consisted primarily of unaffected siblings18 of PD cases who screened negative for PD or parkinsonism via telephone interview,19 or siblings who screened positive but were free of parkinsonism at clinical examination. Cases were matched to a single participating sibling first by sex (when possible) and then by closest age. Cases without an available sibling were matched to unrelated controls of same sex, age (year of birth ± 2 years), and residential region (Minnesota, Wisconsin, Iowa, or North and South Dakota pooled together). Controls of age 65 or older were randomly selected from the Centers for Medicare and Medicaid Services (CMS) lists. Controls younger than 65 years were selected using random digit dialing, according to standard techniques.20, 21 All unrelated controls screened negative for PD or parkinsonism via telephone interview; unrelated controls screening positive could not be examined and were excluded from the study. All examinations (cases and siblings screening positive) were performed in a standardized fashion by neurologists specialized in Movement Disorders, and employing a detailed protocol for clinical assessment.
Genotyping
For all subjects, genomic DNA was collected, extracted, and stored as previously described.17, 22 The Institutional Review Board of the Mayo Clinic approved the study, and all subjects provided written informed consent.
Via sequencing in 25 cases and 25 controls, single-nucleotide polymorphisms (SNPs) were detected in species-conserved regions of PARK loci genes (SNCA, PARK2, SPR, UCHL1, PINK1, DJ1, LRRK2, ATP13A2, and Omi/HtrA2) and in related genes (SNCB, SNCG, LRRK1, and MAPT). The species-conserved regions in these genes were defined using the UCSC Genome browser (http://genome.ucsc.edu/) and dbSNP Build 126 (http://www.ncbi.nlm.nih.gov/SNP). The chromosomal positions were identified using gene names and the search was extended to include 10 kb regions upstream of the 5’ end and downstream of the 3’ end of each gene. Evolutionary conservation in 17 vertebrates was measured, using the Vertebrate Multiz Alignment and conservation track of the genome browser with full filters. The PhastCons program computed a conservation score and SNPs with a conservation score of lod > 200 were genotyped in the entire sample.
Additional SNPs were selected for these genes using the HapMap unrelated CEU samples.23 The LDSelect program was used to identify tag SNPs using a linkage disequilibrium (LD) r2 threshold of 0.8 and with minor allele frequencies ≥ 0.05. Two tag SNPs were selected for each LD bin when the number of SNPs in the bin was 10 or more. SNPs with an Illumina platform design scores < 0.4 were excluded, as well as those within 60 bp of another SNP that had already been chosen. The PARK11 locus gene GIGYF2 was not included in this study, because it was mapped after our genotyping was completed. Recent studies have suggested that the mutations originally reported in GIGYF2 are not causal.24, 25
We genotyped 234 SNPs using a bead array platform (Illumina GoldenGate), one SNP using a pyrosequencing platform (Biotage), and two variable number tandem repeats (VNTRs) (SNCA REP1, MAPT H1/H2 haplotype) using a sequencing platform (Applied Biosystems). Table 1 summarizes the number of variants genotyped in each gene. Seven SNPs failed genotyping, while 230 variants were successfully genotyped. Of these 230 variants, nine SNPs were monomorphic and three additional SNPs had minor allele frequencies < 0.01 and were excluded from analyses. The remaining 218 informative variants were included in our analyses.
TABLE 1.
Genes and Number of Variants Genotyped
| Genes | Number of Variants Genotyped (n = 237) |
|---|---|
| PARK loci genes | |
| SNCA | 23 |
| PARK2 | 36 |
| SPR | 5 |
| UCHL1 | 11 |
| PINK1 | 15 |
| DJ1 | 5 |
| LRRK2 | 66 |
| ATP13A2 | 10 |
| Omi/HtrA2 | 6 |
| PARK-related genes | |
| SNCB | 10 |
| SNCG | 10 |
| LRRK1 | 3 |
| MAPT | 37 |
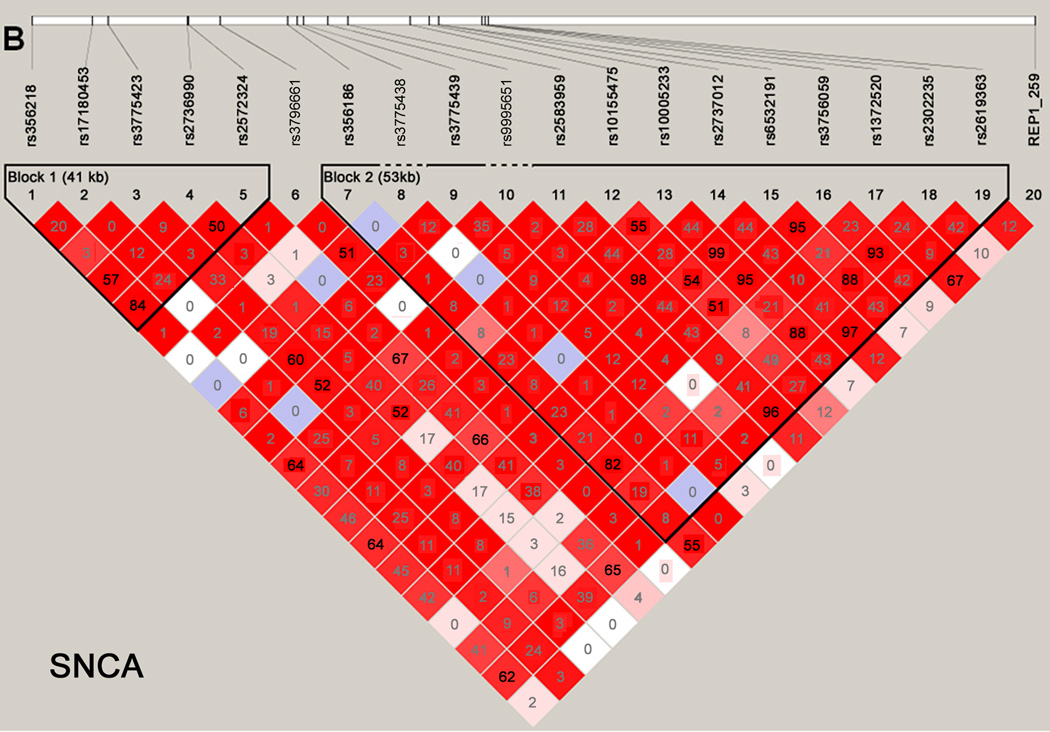
Haploview was used to generate LD maps for each gene using data for the controls.26 Only variants with minor allele frequencies > 0.01 and in Hardy-Weinberg equilibrium (P > 0.001) were included. Figure 1 shows the LD maps for the MAPT (panel A) and SNCA (panel B) genes. Supplementary Figure 1 provides the LD maps for all 13 genes.
Figure 1. Haploview maps.
Linkage disequilibrium (LD) blocks of the MAPT (panel A) and SNCA (panel B) genes. For the SNCA gene, the multiallelic VNTR REP1 was coded as 259 bp vs. others. The LD values as measured using r2 are given by numbers and the LD values as measured by D’ are given by color intensity (red squares indicate strong LD, pink squares indicate intermediate LD, and white squares indicate low LD, with evidence for ancestral recombination; blue indicates limited data).
Statistical Analyses
First, we studied the association of each genetic variant with PD susceptibility in the sample overall, using conditional logistic regression analyses and a log additive coding scheme.27 We also performed analyses using dominant or recessive coding schemes. For analyses of the SNCA REP1 alleles, genotypes were converted to scores ranging from 0 to 4 using a previously described method.17, 22 Each copy of a 259 bp allele was assigned 0 point, each copy of a 261 bp allele was assigned 1 point, and each copy of a 263 bp allele was assigned 2 points; the score for each genotype was then calculated as the sum of the two allele scores. All analyses were adjusted for age at study and sex. For each genetic variant we calculated an odds ratio (OR), a 95% confidence interval (CI), and a two-tailed P-value. We performed similar analyses for each genetic variant in strata of case-unaffected sibling pairs or case-unrelated control pairs separately. We considered strata by sex of the matched pair and by age of the case. For nominally significant variants, we compared log ORs obtained from case-unaffected sibling pairs with log ORs obtained from case-unrelated control pairs.
Second, the association of each genetic variant with age at onset of PD was assessed using Cox proportional hazard models and the same coding schemes.28 All analyses were adjusted for sex. For each genetic variant we calculated a hazard ratio (HR), a 95% CI, and a two-tailed P-value. We performed similar analyses of age at onset of PD in men and women separately.
We estimated our statistical power to detect the association of common variants in PARK locus and related genes with PD susceptibility and age at onset for the available sample (1,103 PD cases, 1,103 controls) using Quanto version 1.2. We assumed a population prevalence of 0.02 and log additive allele effects, and applied a multiple testing correction for 200 independent tests (α = 0.00025). For minor allele frequencies of 0.4 – 0.5, our study had 80% power to detect ORs as small as 1.48 (assuming all case-unaffected sibling pairs) and 1.32 (assuming all case-unrelated control pairs). For minor allele frequencies of 0.05, our study had 80% power to detect ORs as small as 2.14 (assuming all case-unaffected sibling pairs) and 1.75 (assuming all case-unrelated control pairs). We had similar power to detect small HRs for survival free of PD (data not shown).
The statistical packages SAS (version 9.1; SAS Institute Inc., Cary, NC) and S-Plus (version 8.0.1; MathSoft, Seattle, WA) were otherwise used for all analyses. In addition to the uncorrected P-values, a Bonferroni correction and a permutation approach were applied to correct P-values for the number of tests performed.
RESULTS
Sample
There were 1,103 cases and 1,103 controls included in the study (654 case-unaffected sibling pairs and 449 case-unrelated control pairs). The demographic characteristics of the sample are summarized in Table 2.
TABLE 2.
Demographic Characteristics of Parkinson’s Disease (PD) Cases, Siblings, and Unrelated Controls
| PD Case – Sibling Pairs | PD Case-Unrelated Control Pairs | All PD Case- Control Pairs | ||||
|---|---|---|---|---|---|---|
| General characteristics | PD Cases | Sibling Controls | PD Cases | Unrelated Controls | PD Cases | All Controls |
| Total sample, n | 654 | 654 | 449 | 449 | 1,103 | 1,103 |
| Men, n (%) | 417 (63.8) | 329 (50.3) | 290 (64.6) | 290 (64.6) | 707 (64.1) | 619 (56.1) |
| Women, n (%) | 237 (36.2) | 325 (49.7) | 159 (35.4) | 159 (35.4) | 396 (35.9) | 484 (43.9) |
| Age at onset of PD, median year (range) | 60.4 (28.2–86.9) | -- | 64.7 (23.3–88.0) | -- | 62.2 (23.3–88.0) | -- |
| Age at study, median year (range)a | 66.3 (30.8–91.4) | 65.1 (32.0–90.4) | 70.3 (44.5–90.4) | 71.8 (44.9–92.8) | 68.0 (30.8–91.4) | 67.6 (32.0–92.8) |
| Region of origin of parentsb | ||||||
| Both parents of European origin, n (%) | 570 (87.2) | 557 (85.2) | 361 (80.4) | 391 (87.1) | 931 (84.4) | 948 (85.9) |
| Both parents Northern European, n (%) | 154 (27.0) | 148 (26.6) | 116 (32.1) | 126 (32.2) | 270 (29.0) | 274 (28.9) |
| Both parents Central European, n (%) | 233 (40.9) | 219 (39.3) | 119 (33.0) | 124 (31.7) | 352 (37.8) | 343 (36.2) |
| Both parents Southern European, n (%) | 3 (0.5) | 3 (0.5) | 3 (0.8) | 4 (1.0) | 6 (0.6) | 7 (0.7) |
| Both parents European, mixed region, n (%) | 180 (31.6) | 187 (33.6) | 123 (34.1) | 137 (35.0) | 303 (32.5) | 324 (34.2) |
| Only one parent of European origin, n (%)c | 53 (8.1) | 61 (9.3) | 60 (13.4) | 41 (9.1) | 113 (10.2) | 102 (9.2) |
| One parent declared “American”, n (%)d | 2 (0.3) | 1 (0.2) | 1 (0.2) | 4 (0.9) | 3 (0.3) | 5 (0.5) |
| Both parents declared “American”, n (%)d | 19 (2.9) | 20 (3.1) | 13 (2.9) | 7 (1.6) | 32 (2.9) | 27 (2.4) |
| Both parents Asian, n (%) | 3 (0.5) | 3 (0.5) | 5 (1.1) | 0 (0.0) | 8 (0.7) | 3 (0.3) |
| Both parents Mexican, n (%) | 1 (0.2) | 1 (0.2) | 1 (0.2) | 1 (0.2) | 2 (0.2) | 2 (0.2) |
| Unknown, n (%) | 6 (0.9) | 11 (1.7) | 8 (1.8) | 5 (1.1) | 14 (1.3) | 16 (1.5) |
Age at blood draw.
Self-reported by subjects. “Northern European” includes Scandinavian, Swedish, Norwegian, Finnish, Danish, Irish, or British origins. “Central European” includes French, Belgian, Dutch, Swiss, Luxemburgian, German, Austrian, Hungarian, Polish, Czechoslovakian, or Russian origins. “Southern European” includes Italian, Spanish, Portuguese, Greek, or Yugoslavian origins.
Includes subjects for whom origin of one parent is unknown.
These subjects were all Caucasians and not Native Americans
Main Effects of Genetic Variants on PD Susceptibility
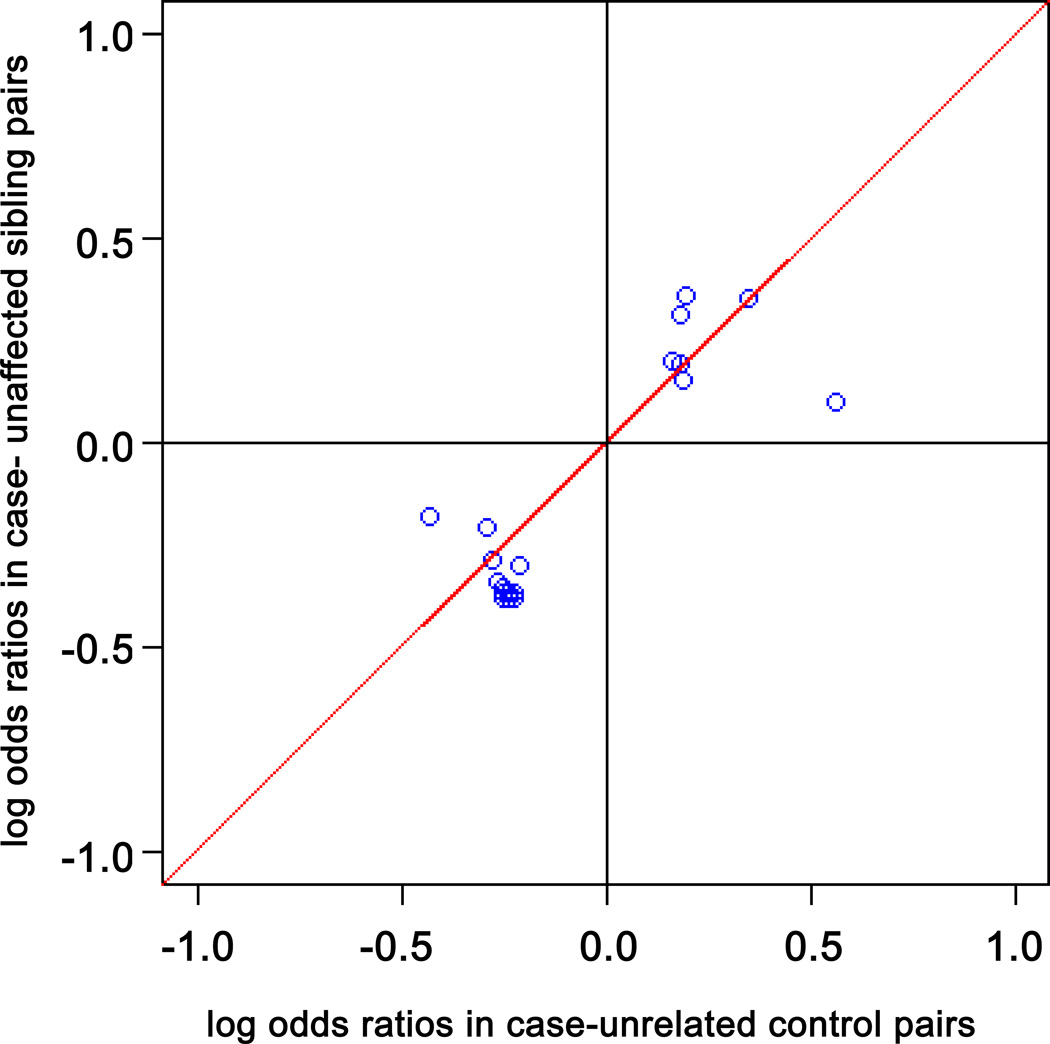
For the sample overall, 27 of the 218 genetic variants were associated with PD susceptibility (uncorrected P < 0.05, log additive model; Table 3). These included 16 MAPT SNPs and the H1/H2 haplotype, six SNCA SNPs and REP1, one LRRK2 SNP, one UCHL1 SNP, and one PARK2 SNP. The main effects of these variants were small, with ORs no less than 0.73 or no greater than 1.43. Only the MAPT SNP rs2435200 was significantly associated with PD susceptibility after Bonferroni or permutation correction for multiple comparisons (OR = 0.74, 95% CI = 0.64 – 0.86, uncorrected P < 0.0001, permutation-corrected P = 0.015). Figure 2 demonstrates that for the 27 genetic variants significantly associated with PD, the log ORs between case-unaffected sibling and case-unrelated control pairs were significantly correlated (r = 0.87; 95% CI = 0.73 – 0.94; P value = 4.75 × 10−9). Supplementary Table 1 provides detailed results for the association with PD susceptibility of all genotyped variants in the 13 PARK loci and related genes, including results for multiple coding schemes in the sample overall, and in subgroups defined by type of control, sex, and age.
TABLE 3.
Common Variants in PARK Loci and Related Genes Significantly Associated with PD Susceptibility (n = 27) in Order of Statistical Significance
| Chromosome | SNP | Positiona | Gene | Type of Variantb | Allelec | Minor Allele Frequenciesd (cases/ controls) |
Trend Model OR (95%CI)e |
Trend test p valuef |
|---|---|---|---|---|---|---|---|---|
| 17 | rs2435200 | 41427688 | MAPT | Intronic SNP | A/G | 0.372/0.422 | 0.74 (0.64 – 0.86) | < 0.0001 |
| 4 | rs2736990 | 90897564 | SNCA | Intronic SNP | C/T | 0.490/0.470 | 1.27 (1.09 – 1.47) | 0.0017 |
| 17 | rs17652121 | 41429810 | MAPT | Synonymous | C/T | 0.164/0.196 | 0.76 (0.63 – 0.91) | 0.0035 |
| 17 | rs4792891 | 41329294 | MAPT | 5’ UTR SNP | G/T | 0.284/0.320 | 0.79 (0.68 – 0.93) | 0.0036 |
| 17 | rs17691610 | 41326456 | MAPT | Intronic SNP | G/T | 0.164/0.196 | 0.76 (0.64 – 0.92) | 0.004 |
| 17 | rs1052587 | 41458449 | MAPT | 3’ UTR SNP | C/T | 0.165/0.196 | 0.77 (0.64 – 0.92) | 0.0041 |
| 17 | rs17574361 | 41464049 | MAPT | Conserved | A/G | 0.164/0.197 | 0.77 (0.64 – 0.92) | 0.0041 |
| 17 | rs17651549 | 41417115 | MAPT | Conserved | C/T | 0.163/0.194 | 0.76 (0.63 – 0.92) | 0.0041 |
| 17 | H1/H2 | - | MAPT | Intragenic VNTR | - | - | 0.77 (0.64 – 0.92) | 0.0042 |
| 17 | rs17770343 | 41325948 | MAPT | Intronic SNP | C/T | 0.164/0.196 | 0.77 (0.64 – 0.92) | 0.0046 |
| 17 | rs1052551 | 41424761 | MAPT | Synonymous | A/G | 0.165/0.196 | 0.77 (0.64 – 0.92) | 0.0047 |
| 17 | rs12150242 | 41371645 | MAPT | Intronic SNP | A/G | 0.165/0.197 | 0.77 (0.64 – 0.92) | 0.0048 |
| 17 | rs17574604 | 41467460 | MAPT | Conserved | A/G | 0.164/0.196 | 0.77 (0.64 – 0.92) | 0.0048 |
| 17 | rs17574228 | 41460355 | MAPT | 3’ UTR SNP | C/T | 0.165/0.196 | 0.77 (0.64 – 0.92) | 0.0049 |
| 17 | rs9468 | 41457408 | MAPT | 3’ UTR SNP | C/T | 0.164/0.196 | 0.77 (0.64 – 0.92) | 0.005 |
| 17 | rs17650901 | 41395527 | MAPT | 5’ UTR SNP | A/G | 0.164/0.196 | 0.77 (0.64 – 0.93) | 0.0053 |
| 4 | rs1372520 | 90976528 | SNCA | Intronic SNP | C/T | 0.171/0.198 | 0.77 (0.64 – 0.93) | 0.0056 |
| 17 | rs16940806 | 41459672 | MAPT | 3’ UTR SNP | A/G | 0.165/0.196 | 0.77 (0.64 – 0.93) | 0.0059 |
| 17 | rs1052553 | 41429726 | MAPT | Synonymous | A/G | 0.164/0.195 | 0.78 (0.65 – 0.93) | 0.0072 |
| 4 | rs2572324 | 90897821 | SNCA | Intronic SNP | C/T | 0.338/0.307 | 1.24 (1.05 – 1.45) | 0.009 |
| 4 | rs3775423 | 90876514 | SNCA | Intronic SNP | C/T | 0.099/0.081 | 1.41 (1.09 – 1.82) | 0.009 |
| 4 | REP1 | 91124217 | SNCA | 5’ UTR VNTR | - | - | 1.18 (1.04 – 1.34 | 0.0118 |
| 4 | rs356186 | 90924387 | SNCA | Intronic SNP | A/G | 0.158/0.180 | 0.78 (0.64 – 0.95) | 0.0119 |
| 12 | rs17484286 | 38984953 | LRRK2 | Intronic SNP | A/G | 0.083/0.102 | 0.73 (0.57 – 0.93) | 0.0128 |
| 4 | rs10517002 | 40959306 | UCHL1 | Intronic SNP | A/C | 0.406/0.381 | 1.19 (1.02 – 1.39) | 0.0228 |
| 4 | rs356218 | 90856033 | SNCA | Conserved | A/G | 0.367/0.342 | 1.17 (1.01 – 1.37) | 0.0419 |
| 6 | rs12174410 | 162259158 | PARKIN | Conserved | C/T | 0.052/0.040 | 1.43 (1.01 – 2.04) | 0.0435 |
NCBI build 36 of the human genome.
SNP = single nucleotide polymorphism; UTR = untranslated region; VNTR = variable number tandem repeat.
The REP1 variant is a variable number tandem repeat; common allele lengths are 259 bp, 261 bp, and 263 bp.
Note that REP1 has three common alleles: the frequency of the 259 bp allele was 0.24, of the 261 bp allele was 0.68, and of the 263 bp allele was 0.08. The frequency of the MAPT H1 haplotype was 0.82 and of the H2 haplotype was 0.18.
Log additive model; OR = odds ratio, CI = confidence interval; the OR for REP1 was coded using the score test method.17, 22
Log additive model; only the MAPT gene variant rs2435200 remained significant after Bonferroni or permutation correction for multiple comparisons.
Figure 2. Scatter Plot of Log Odds Ratios.
Log odds ratios (ORs) for the sample stratified as case-unaffected sibling pairs versus case-unrelated control pairs for the 27 genetic variants that were significantly associated with PD susceptibility in the sample overall. There was a significant correlation of log ORs between the two types of controls (r = 0.87; 95% CI = 0.73 – 0.94; P value = 4.75 × 10−9).
Consistent with our previous studies, we observed a significant association of the MAPT H2 haplotype with PD in this expanded sample (OR = 0.77, 95% CI = 0.64 – 0.92, uncorrected P = 0.004).29, 30 The MAPT SNP rs2435200 and the MAPT H2 haplotype were not in strong LD. However, each variant was in LD with several other SNPs that were significantly associated with PD (Table 3, Figure 1).
Although the SNCA SNP rs2736990 was not associated with PD susceptibility in the sample overall after correction for multiple testing, it maps a 3’ block of LD that has been consistently associated with PD in several studies.31–33 SNCA REP1 was also associated with PD susceptibility in the sample overall (uncorrected P = 0.012, OR = 1.18, 95% CI = 1.04 – 1.34, per unit genotype score; OR = 1.94, score 4 vs. score 0). The SNCA SNP rs2736990 and REP1 were not in strong LD (REP1 coded as 261 vs others: D'=0.13, r2=0.01; REP1 coded as 259 vs others: D'=0.09, r2=0; REP1 coded as 263 vs others: D'=0.93, r2=0.07). However, each variant was in LD with several other SNPs that were significantly associated with PD (Table 3, Figure 1).
Main Effects of Genetic Variants on Age at Onset of PD
Eight SNPs (one SNCA SNP: rs1372520; four PINK1 SNPs: rs3738133, rs1043502, rs1043424, and rs2078073; one Omi/HtrA2 SNP: rs17010022; two SNCB SNPs: rs1352303 and rs4868670) were associated with age at onset of PD in the sample overall. None of these associations remained significant after Bonferroni or permutation correction. Supplementary Table 1 provides the details for associations of all genotyped variants in the 13 genes with age at onset of PD, including results for multiple coding schemes in the sample overall, and in subgroups defined by sex.
DISCUSSION
Genetic Interpretation of the Findings
This study confirmed the association of MAPT and SNCA genes with PD susceptibility, but showed limited association of common variations in other PARK loci and related genes with PD in an outbred US population.
Our findings are consistent with the findings from six genome-wide association studies of PD susceptibility.15, 34–38 The study of Maraganore et al. included 775 PD cases and 775 matched controls and genotyped 198,345 informative genomic SNPs, with some suggestive findings for MAPT and SNCA, as well as other PARK loci and related genes. However, none of the findings were significant after correction for multiple testing. The study by Fung and colleagues employed more SNP markers but also failed to observe an association of PARK loci and related genes with PD susceptibility after correction for multiple testing; however, that study included only 276 PD cases and 276 unmatched controls. The study of Pankratz and colleagues employed 857 familial PD cases and 867 controls; and observed suggestive associations for MAPT SNPs (recessive model: OR = 0.56; P = 2.0 × 10−5) and the SNCA SNPs (additive model: OR = 1.35; P = 5.5 × 10−5). Despite enriching their sample for genetic load (familial PD cases), none of the SNPs were significant after correction for multiple testing.
Recently, three larger genome-wide association studies identified that common variants in SNCA and MAPT genes increase PD susceptibility.36–38 The study of Satake and colleagues (2,011 cases and 18,381 controls; Japanese) reported strong associations of SNPs in the SNCA locus (most significant SNP, rs11931074, OR = 1.37, P = 7.35 × 10−17). The study of Simon-Sanchez and colleagues (5,074 cases and 8,551 controls; Caucasian Europeans) observed strong associations of SNPs in SNCA (most significant SNP, rs2736990, OR = 1.23, P = 2.24 × 10−16) and MAPT loci (most significant SNP, rs393152, OR = 0.77, P = 1.95 × 10−16). The study of Edwards and colleagues (1,752 cases and 1,745 controls; Americans) observed strong associations of SNPs in the SNCA (most significant SNP, rs2736990, OR = 1.29, P = 6.7 × 10−8) and MAPT loci (most significant SNP, rs11012, OR = 0.70, P = 5.6 × 10−8). Importantly, the SNCA SNP rs2736990 that was found to be significantly associated with PD in our study is the same SNCA SNP that was found to be associated with PD in the Sanchez-Ramos and Edwards studies. Our study and those two studies included subjects of similar ethnicities (Caucasian European and American). Our study was submitted for publication prior to the publication of those two studies (independent convergence of SNP association findings for our candidate gene study versus their genome-wide association studies).
The six genome-wide association studies of PD employed genotyping platforms that selected SNPs that were haplotype tagging.15, 34–38 We similarly selected SNPs that were haplotype tagging. However, we also sequenced PARK loci and related genes and selected additional SNPs from species-conserved regions. It has been suggested that highly conserved regions of the genome are functionally important.39, 40 Of the conserved-region SNPs included in our study, three MAPT SNPs (rs17574361, rs17651549, rs17574604), one SNCA SNP (rs356218), and one PARK2 SNP (rs12174410) were associated with PD susceptibility in the sample overall. However, none of these SNP associations were significant after correction for multiple testing. The only SNP that was significant in our study after correction for multiple testing (MAPT SNP rs2435200) was not from a species-conserved region.
In addition, we studied the association of variants in PARK loci and related genes with age at onset of PD. We observed suggestive associations for eight SNPs (one SNCA SNP, four PINK1 SNPs, one Omi/HtrA2 SNP, and two SNCB SNPs). We had previously reported an association of the SNCB SNP rs1352303 with delayed age at onset of PD in women.41 In the current study, this SNP was again associated with age at onset of PD in women and also in the expanded sample overall and with the same direction of effect. The subjects for the two studies were partially overlapping.
Functional Interpretation of the Findings
While the most significant finding was for the MAPT SNP rs2435200, it remains unknown whether this intronic SNP has a functional effect or whether it is a marker of another functional variant. Although the association of the MAPT H1/H2 haplotype with PD in our study was less significant, this variant has recognized functional effects. Persons who are homozygous for the H1 haplotype express higher levels of the tau protein,42 and tau overexpression may promote fibrillization of the alpha-synuclein protein.43
Although the most significant finding for SNCA (SNP rs2736990) and PD susceptibility lost significance after correction for multiple testing, the SNP is located in an LD block at the 3’ end of the gene that has been shown to regulate expression.32, 44 Similarly, the findings for SNCA REP1 are consistent with functional studies demonstrating that genotypes associated with longer alleles are associated with higher gene expression levels and increased risk for PD.44–48.
The functional UCHL1 S18Y (rs5030732) and PARK2 promoter (rs9347683) variants that we had previously reported to be associated with PD in our smaller sample were no longer associated with PD in this expanded sample.13, 49 Furthermore, additional SNPs that we studied in the UCHL1 and PARK2 loci were not associated with PD. This large candidate gene study and the six published genome-wide association studies lead us to conclude that UCHL1 and PARK2 are not susceptibility genes for PD. 15, 34–38.
Strengths and Limitations
This study has several strengths. First, we had a large sample size (1,103 PD cases and 1,103 controls). This provided good statistical power to detect a range of ORs (and HRs) for a range of minor allele frequencies (log additive model). Second, we studied 13 PARK loci and related genes that have strong plausibility as candidate genes for PD. Third, we studied several variants in each gene, including tag SNPs that we selected from haplotype maps and species-conserved SNPs that we sequenced. Fourth, we observed a strong correlation of log ORs of each variant associated with PD susceptibility between case-unaffected sibling and case-unrelated control pairs (internal replication). Fifth, we studied the association of genetic variants with age at onset of PD as well as with susceptibility.
Our study also has some limitations. First, our sample was not population-based. However, population-based incidence cohorts of PD are often not large enough to detect the small effects of common genetic variants. We tried to limit sampling bias by recruiting cases prospectively from a defined geographic region. We previously showed that for approximately half of our cases (residing within 120 miles of the Mayo Clinic in Rochester, MN), the demographic characteristics are similar to those of an incidence cohort of PD defined for Olmsted County, MN. By contrast, for the other half of our cases (residing within a broader five-state region), the cases were of younger ages at study (possibly increasing the genetic load).50–52 Second, our controls were primarily unaffected siblings because we intended to limit possible population stratification. However, unaffected sibling controls can be overmatched for genetic and environmental factors, leading to false negative findings (reduced statistical power). For this reason, we performed a priori power calculations and determined that we had adequate power to detect a range of small ORs for a range of minor allele frequencies. We also performed sensitivity analyses, which showed that the ORs for the genetic variants were similar when restricting the sample to case-unaffected sibling pairs or to case-unrelated control pairs only. Third, we only sequenced selected regions of PARK loci and related genes in a subset of cases and controls. We did not sequence the entire genes in all subjects to detect rare sequence variants or copy number variants that were associated with PD. Such studies are expected to become feasible in the coming years as the costs come down for next-generation sequencing technologies.53, 54 However, we sequenced species-conserved regions, which is a novel approach for the study of PARK loci and related genes and PD. It was our hypothesis that common variants in species-conserved regions may be more functionally important than common variants in unconserved regions. Our findings did not support this hypothesis. Fourth, we performed multiple statistical tests, increasing the likelihood of chance findings. Therefore, we employed Bonferroni and permutation corrections for our primary analysis to identify genetic associations that exceeded chance expectations. Fifth, we did not study gene-gene or gene-environment interactions (beyond the scope of this study). There is only limited evidence for gene-gene interactions for PARK locus.30, 33, 55 There is only limited evidence for interactions of PARK loci genes and environmental exposures in PD.17, 22 Future studies of PARK loci and related genes and PD should also consider copy number and DNA methylation variations.
Supplementary Material
Supplementary Figure 1. Haploview maps. Haploview maps for the 13 PARK loci and related genes that we studied (ATP13A2, DJ1, LRRK1, LRRK2, MAPT, Omi/HtrA2, PARK2, PINK1, SNCA, SNCB, SNCG, SPR, and UCHL1). The LD values as measured using r2 are given by numbers and the LD values as measured by D’ are given by color intensity (red squares indicate strong LD, pink squares indicate intermediate LD, and white squares indicate low LD, with evidence for ancestral recombination; blue indicated limited data).
Supplementary Table 1. Case-control analyses for susceptibility and cases-only analyses for age at onset for all of the 230 genetic variants in 13 PARK loci and related genes in the sample overall and in strata, and using multiple coding SNPs (log additive, dominant, and recessive).
ACKNOWLEDGEMENTS
The study was funded by the NIH grant 2R01ES10751. We thank the many members of Mayo’s Molecular Epidemiology of Parkinson’s Disease research team for their efforts, and especially our Mayo Clinic patients and their families for their participation.
Footnotes
- Sun Ju Chung, MD, PhD; 1) Research project: C. Execution; 2) Statistical Analysis: C. Review and Critique; 3) Manuscript: A. Writing of the first draft
- Sebastian M. Armasu, MS; 1) Statistical Analysis: A. Design, B. Execution; 2) Manuscript: B. Review and Critique.
- Joanna M. Biernacka, PhD; 1) Statistical Analysis: A. Design, C. Review and Critique; 2) Manuscript: B. Review and Critique.
- Timothy G. Lesnick, MS; 1) Statistical Analysis: A. Design, C. Review and Critique; 2) Manuscript: B. Review and Critique.
- David N. Rider, MSE; 1) Research project: C. Execution; 2) Statistical Analysis: C. Review and Critique; 3) Manuscript: B. Review and Critique
- Sarah J. Lincoln, BS; 1) Research project: C. Execution; 2) Manuscript: B. Review and Critique
- Alexandra I. Ortolaza; 1) Research project: C. Execution; 2) Manuscript: B. Review and Critique
- Matthew J. Farrer, PhD; 1) Research project: A. Conception, B. Organization; 2) Manuscript: B. Review and Critique.
- Julie M. Cunningham, PhD; 1) Research project: B. Organization; 2) Manuscript: B. Review and Critique.
- Walter A. Rocca, MD, MPH; 1) Research project: A. Conception; 2) Statistical Analysis: C. Review and Critique; 3) Manuscript: B. Review and Critique.
- Demetrius M. Maraganore, MD; 1) Research project: A. Conception, B. Organization, C. Execution; 2) Statistical Analysis: A. Design, C. Review and Critique; 3) Manuscript: B. Review and Critique.
- Sun Ju Chung, MD, PhD; None
- Sebastian M. Armasu, MS; None
- Joanna M. Biernacka, PhD; None
- Timothy G. Lesnick, MS; None
- David N. Rider, MSE; None
- Sarah J. Lincoln, BS; None
- Alexandra I. Ortolaza; None
- Matthew J. Farrer, PhD; Dr. Farrer reports a US provisional patent application for a device that treats neurodegenerative diseases, which has been licensed to Alnylam Pharmaceuticals Inc. Dr Farrer reports a US patent application for Parkinson disease markers and European provisional patent applications for identification of mutations in PARK8, a locus for familial Parkinson disease and identification of a novel LRRK2 mutation, 6055G>A (G2019S), linked to autosomal dominant parkinsonism in families from several European populations. [(a)International Publication Number WO 2006/045392 A2; (b) International Publication Number WO 2006/068492 A1; (c) US publication Number US-2008-0009454-A1; and (d) Norwegian patent 323175]. In the last 12 months Dr Farrer reports salary and royalty payment from Lundbeck Pharmaceuticals, and an honorarium for a seminar from Genzyme.
- Julie M. Cunningham, PhD; None
- Walter A. Rocca, MD, MPH; Dr. Rocca is funded by NIH grants AR030582, AG006786, and ES010751.
- Demetrius M. Maraganore, MD; NIH grant ES10751. Dr. Maraganore also reports a US provisional patent application for a method to treat Parkinson’s disease, which has been licensed to Alnylam Pharmaceuticals Inc.
REFERENCES
- 1.Elbaz A, Bower JH, Maraganore DM, et al. Risk tables for parkinsonism and Parkinson's disease. J Clin Epidemiol. 2002;55(1):25–31. doi: 10.1016/s0895-4356(01)00425-5. [DOI] [PubMed] [Google Scholar]
- 2.Polymeropoulos MH, Lavedan C, Leroy E, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276(5321):2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 3.Spillantini MG, Schmidt ML, Lee VM, et al. Alpha-synuclein in Lewy bodies. Nature. 1997;388(6645):839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 4.Kitada T, Asakawa S, Hattori N, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 5.Leroy E, Boyer R, Auburger G, et al. The ubiquitin pathway in Parkinson's disease. Nature. 1998;395(6701):451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 6.Valente EM, Abou-Sleiman PM, Caputo V, et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304(5674):1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 7.Bonifati V, Rizzu P, van Baren MJ, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299(5604):256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 8.Paisan-Ruiz C, Jain S, Evans EW, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44(4):595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 9.Zimprich A, Biskup S, Leitner P, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44(4):601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Ramirez A, Heimbach A, Grundemann J, et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet. 2006;38(10):1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 11.Lautier C, Goldwurm S, Durr A, et al. Mutations in the GIGYF2 (TNRC15) gene at the PARK11 locus in familial Parkinson disease. Am J Hum Genet. 2008;82(4):822–833. doi: 10.1016/j.ajhg.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strauss KM, Martins LM, Plun-Favreau H, et al. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson's disease. Hum Mol Genet. 2005;14(15):2099–2111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- 13.Maraganore DM, Lesnick TG, Elbaz A, et al. UCHL1 is a Parkinson's disease susceptibility gene. Ann Neurol. 2004;55(4):512–521. doi: 10.1002/ana.20017. [DOI] [PubMed] [Google Scholar]
- 14.Maraganore DM, de Andrade M, Elbaz A, et al. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. JAMA. 2006;296(6):661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 15.Maraganore DM, de Andrade M, Lesnick TG, et al. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77(5):685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Facheris M, Strain KJ, Lesnick TG, et al. UCHL1 is associated with Parkinson's disease: a case-unaffected sibling and case-unrelated control study. Neurosci Lett. 2005;381(1–2):131–134. doi: 10.1016/j.neulet.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Brighina L, Frigerio R, Schneider NK, et al. Alpha-synuclein, pesticides, and Parkinson disease: a case-control study. Neurology. 2008;70(16 Pt 2):1461–1469. doi: 10.1212/01.wnl.0000304049.31377.f2. [DOI] [PubMed] [Google Scholar]
- 18.Maraganore DM. Blood is thicker than water: the strengths of family-based case-control studies. Neurology. 2005;64(3):408–409. doi: 10.1212/01.WNL.0000152585.76852.9C. [DOI] [PubMed] [Google Scholar]
- 19.Rocca WA, Maraganore DM, McDonnell SK, Schaid DJ. Validation of a telephone questionnaire for Parkinson's disease. J Clin Epidemiol. 1998;51(6):517–523. doi: 10.1016/s0895-4356(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 20.Hartge P, Brinton LA, Rosenthal JF, et al. Random digit dialing in selecting a population-based control group. Am J Epidemiol. 1984;120(6):825–833. doi: 10.1093/oxfordjournals.aje.a113955. [DOI] [PubMed] [Google Scholar]
- 21.Potthoff RF. Telephone sampling in epidemiologic research: to reap the benefits, avoid the pitfalls. Am J Epidemiol. 1994;139(10):967–978. doi: 10.1093/oxfordjournals.aje.a116946. [DOI] [PubMed] [Google Scholar]
- 22.Brighina L, Schneider NK, Lesnick TG, et al. Alpha-synuclein, alcohol use disorders, and Parkinson disease: a case-control study. Parkinsonism Relat Disord. 2009;15(6):430–434. doi: 10.1016/j.parkreldis.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bras J, Simon-Sanchez J, Federoff M, et al. Lack of replication of association between GIGYF2 variants and Parkinson disease. Hum Mol Genet. 2009;18(2):341–346. doi: 10.1093/hmg/ddn340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichols WC, Kissell DK, Pankratz N, et al. Variation in GIGYF2 is not associated with Parkinson disease. Neurology. 2009;72(22):1886–1892. doi: 10.1212/01.wnl.0000346517.98982.1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 27.Breslow NE, Day NE. Statistical methods in cancer research. Volume I - The analysis of case-control studies. IARC Sci Publ. 1980;(32):5–338. [PubMed] [Google Scholar]
- 28.Cox DR. Regression models of life-tables (with discussion) J R Stat Soc [Ser B] 1972;34:187–220. [Google Scholar]
- 29.Maraganore DM, Hernandez DG, Singleton AB, et al. Case-Control study of the extended tau gene haplotype in Parkinson's disease. Ann Neurol. 2001;50(5):658–661. doi: 10.1002/ana.1228. [DOI] [PubMed] [Google Scholar]
- 30.Mamah CE, Lesnick TG, Lincoln SJ, et al. Interaction of alpha-synuclein and tau genotypes in Parkinson's disease. Ann Neurol. 2005;57(3):439–443. doi: 10.1002/ana.20387. [DOI] [PubMed] [Google Scholar]
- 31.Mueller JC, Fuchs J, Hofer A, et al. Multiple regions of alpha-synuclein are associated with Parkinson's disease. Ann Neurol. 2005;57(4):535–541. doi: 10.1002/ana.20438. [DOI] [PubMed] [Google Scholar]
- 32.Mizuta I, Satake W, Nakabayashi Y, et al. Multiple candidate gene analysis identifies alpha-synuclein as a susceptibility gene for sporadic Parkinson's disease. Hum Mol Genet. 2006;15(7):1151–1158. doi: 10.1093/hmg/ddl030. [DOI] [PubMed] [Google Scholar]
- 33.Goris A, Williams-Gray CH, Clark GR, et al. Tau and alpha-synuclein in susceptibility to, and dementia in, Parkinson's disease. Ann Neurol. 2007;62(2):145–153. doi: 10.1002/ana.21192. [DOI] [PubMed] [Google Scholar]
- 34.Fung HC, Scholz S, Matarin M, et al. Genome-wide genotyping in Parkinson's disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2006;5(11):911–916. doi: 10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- 35.Pankratz N, Wilk JB, Latourelle JC, et al. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum Genet. 2009;124(6):593–605. doi: 10.1007/s00439-008-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Satake W, Nakabayashi Y, Mizuta I, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat Genet. 2009;41(12):1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 37.Simon-Sanchez J, Schulte C, Bras JM, et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet. 2009;41(12):1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwards TL, Scott WK, Almonte C, et al. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann Hum Genet. 2010;74(2):97–109. doi: 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Brien SJ, Menotti-Raymond M, Murphy WJ, et al. The promise of comparative genomics in mammals. Science. 1999;286(5439):458–462. 479–481. doi: 10.1126/science.286.5439.458. [DOI] [PubMed] [Google Scholar]
- 40.Murphy WJ, Stanyon R, O'Brien SJ. Evolution of mammalian genome organization inferred from comparative gene mapping. Genome Biol. 2001;2(6) doi: 10.1186/gb-2001-2-6-reviews0005. REVIEWS0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brighina L, Okubadejo NU, Schneider NK, et al. Beta-synuclein gene variants and Parkinson's disease: a preliminary case-control study. Neurosci Lett. 2007;420(3):229–234. doi: 10.1016/j.neulet.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwok JB, Teber ET, Loy C, et al. Tau haplotypes regulate transcription and are associated with Parkinson's disease. Ann Neurol. 2004;55(3):329–334. doi: 10.1002/ana.10826. [DOI] [PubMed] [Google Scholar]
- 43.Giasson BI, Forman MS, Higuchi M, et al. Initiation and synergistic fibrillization of tau and alpha-synuclein. Science. 2003;300(5619):636–640. doi: 10.1126/science.1082324. [DOI] [PubMed] [Google Scholar]
- 44.Fuchs J, Tichopad A, Golub Y, et al. Genetic variability in the SNCA gene influences alpha-synuclein levels in the blood and brain. FASEB J. 2008;22(5):1327–1334. doi: 10.1096/fj.07-9348com. [DOI] [PubMed] [Google Scholar]
- 45.Chiba-Falek O, Nussbaum RL. Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum Mol Genet. 2001;10(26):3101–3109. doi: 10.1093/hmg/10.26.3101. [DOI] [PubMed] [Google Scholar]
- 46.Chiba-Falek O, Touchman JW, Nussbaum RL. Functional analysis of intra-allelic variation at NACP-Rep1 in the alpha-synuclein gene. Hum Genet. 2003;113(5):426–431. doi: 10.1007/s00439-003-1002-9. [DOI] [PubMed] [Google Scholar]
- 47.Bonsch D, Lederer T, Reulbach U, et al. Joint analysis of the NACP-REP1 marker within the alpha synuclein gene concludes association with alcohol dependence. Hum Mol Genet. 2005;14(7):967–971. doi: 10.1093/hmg/ddi090. [DOI] [PubMed] [Google Scholar]
- 48.Cronin KD, Ge D, Manninger P, et al. Expansion of the Parkinson disease-associated SNCA-Rep1 allele upregulates human alpha-synuclein in transgenic mouse brain. Hum Mol Genet. 2009;18(17):3274–3285. doi: 10.1093/hmg/ddp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West AB, Maraganore D, Crook J, et al. Functional association of the parkin gene promoter with idiopathic Parkinson's disease. Hum Mol Genet. 2002;11(22):2787–2792. doi: 10.1093/hmg/11.22.2787. [DOI] [PubMed] [Google Scholar]
- 50.Bower JH, Maraganore DM, McDonnell SK, Rocca WA. Incidence and distribution of parkinsonism in Olmsted County, Minnesota, 1976–1990. Neurology. 1999;52(6):1214–1220. doi: 10.1212/wnl.52.6.1214. [DOI] [PubMed] [Google Scholar]
- 51.Rocca WA, Peterson BJ, McDonnell SK, et al. The Mayo Clinic family study of Parkinson's disease: study design, instruments, and sample characteristics. Neuroepidemiology. 2005;24(3):151–167. doi: 10.1159/000083612. [DOI] [PubMed] [Google Scholar]
- 52.Bower JH, Maraganore DM, McDonnell SK, Rocca WA. Influence of strict, intermediate, and broad diagnostic criteria on the age- and sex-specific incidence of Parkinson's disease. Mov Disord. 2000;15(5):819–825. doi: 10.1002/1531-8257(200009)15:5<819::aid-mds1009>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 53.Kahvejian A, Quackenbush J, Thompson JF. What would you do if you could sequence everything? Nat Biotechnol. 2008;26(10):1125–1133. doi: 10.1038/nbt1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24(3):133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 55.Maraganore DM, de Andrade M, Lesnick TG, et al. Complex interactions in Parkinson's disease: a two-phased approach. Mov Disord. 2003;18(6):631–636. doi: 10.1002/mds.10431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Haploview maps. Haploview maps for the 13 PARK loci and related genes that we studied (ATP13A2, DJ1, LRRK1, LRRK2, MAPT, Omi/HtrA2, PARK2, PINK1, SNCA, SNCB, SNCG, SPR, and UCHL1). The LD values as measured using r2 are given by numbers and the LD values as measured by D’ are given by color intensity (red squares indicate strong LD, pink squares indicate intermediate LD, and white squares indicate low LD, with evidence for ancestral recombination; blue indicated limited data).
Supplementary Table 1. Case-control analyses for susceptibility and cases-only analyses for age at onset for all of the 230 genetic variants in 13 PARK loci and related genes in the sample overall and in strata, and using multiple coding SNPs (log additive, dominant, and recessive).