Abstract
Here, we show that overexpression of fer tyrosine kinase (FER), a non-receptor tyrosine kinase, predicts poor postoperative outcome and might be involved in cancer-cell survival in non-small cell lung cancer (NSCLC). Systematic screening using in silico analyses and quantitative RT-PCR revealed that FER was overexpressed in about 10% of NSCLC patients. Evaluation of FER expression using immunohistochemistry (IHC) on tissue microarrays was consistent with the mRNA level detected using quantitative RT-PCR. In analyses of 135 NSCLC patients who had undergone potential curative resection, we found that FER overexpression detected using IHC had no association with clinicopathological features such as age, sex, smoking history, histological type, disease stage, T factor, N factor, adjuvant chemotherapy history, or EGFR mutation, but was correlated with poor postoperative survival periods. A multivariate Cox regression analysis showed that this prognostic impact was independent of other clinicopathological features. In functional analyses of FER in vitro, FER exhibited a transforming activity, suggesting that it possesses oncogenic functions. We also found that human lung cancer NCI-H661 cells, which exhibited FER-outlier expression, were led to apoptosis by the knockdown of FER using RNA interference. FER overexpression might serve as a prognostic biomarker and be involved in cancer-cell survival in NSCLC.
Keywords: Non-small cell lung cancer, FER overexpression, prognostic factor
Introduction
Lung cancer is the most commonly diagnosed cancer as well as the leading cause of cancer-related mortality worldwide, with over 1 million deaths each year [1], and although significant advances have been made with therapies, the 5-year survival rate of lung cancer is below 20% [2]. The best chance of achieving long-term survival is in complete surgical resection; however, even in resected stage IA patients, 30% succumb from their disease within 5 years [3]. Previous studies have reported several factors associated with poor prognosis in non-small cell lung cancer (NSCLC) patients after surgical resection, such as tumor size, preoperative serum CEA level, visceral pleural invasion, vascular vessel invasion, and histological grade [4-8]. In addition, recently, advances in molecular biology have provided important insights into molecular prognostic biomarkers. The identification of new molecular prognostic parameters will be helpful for planning the treatment of lung cancer patients after surgical resection.
In the present study, we found that fer tyrosine kinase (FER), a non-receptor tyrosine kinase, is overexpressed in a subset of NSCLC patients and is correlated with poor postoperative survival periods. We also showed that FER might play a crucial role in FER-overexpressed lung cancer in vitro. FER is an intracellular tyrosine kinase that has been found to reside in both the cytoplasm and nucleus of mammalian cells [9-11]. It has been shown to interact with proteins such as cortactin and to reorganize the actin cytoskeleton [12,13], or to be linked to the modulation of cell-cell and cell-substrate interactions by associating with adhesion molecules [13-19]. There are also reports that suggest a supportive role of FER in the proliferation and growth of malignant cells [20,21]. Actually, several reports have suggested its roles in cancers [22], including prostate cancer [20,23], colon cancer [24], breast cancer [21], hepatocellular carcinoma [25], malignant mesothelioma [26], or gastric cancer [27]; however, to the best of our knowledge, the present study is the first report to correlate FER with lung cancer.
Materials and methods
Lung cancer patients and samples
Clinical lung cancer samples were collected from patients who had undergone surgical resection at the University of Tokyo Hospital between June 2005 and September 2007. Informed consent was obtained from all the patients, and the study was approved by the Institutional Ethics Review Committee. The diagnoses were based on pathological evidence and were classified according to the TNM classification criteria [28]. To circumvent statistical disruption arising from the heterogeneity of significantly advanced diseases, patients whose tumors were pathologically confirmed as T3 or T4 were excluded.
For all patients, medical records were reviewed to extract data on clinicopathological characteristics. The follow-up data after surgical resection were obtained at our outpatient department. Progression free survival was measured from the date of surgery until disease recurrence or metastasis, and overall survival was measured from the date of surgery until the date of death. Patients without a known date of death were censored at the time of last follow-up.
Cell lines
EBC-1, LK-2, and LUDLU-1 cells were obtained from the Health Science Research Resources Bank (Osaka, Japan). All the other cell lines were obtained from the American Type Culture Collection (Rockville, MD). Cells were cultured according to each manufacturer’s protocol.
Quantitative RT-PCR analysis of mRNA
Total RNA was isolated using RNAiso (TaKaRa, Shiga, Japan) and was reverse transcribed with SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). SYBR green RT-PCR was performed using Thunderbird qPCR Mix (Toyobo, Osaka, Japan) and was analyzed using Applied Biosystems 7500 (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. The PCR conditions and primer sequences are shown in Table 1. Relative expression level was calculated using the ΔΔCt method with the β-actin gene as an internal control. A level more than 16 times higher than the expression level in normal lung tissue was set as the threshold for gene overexpression.
Table 1.
Sequences of primers used for quantitative RT-PCR of candidate genes involved in lung cancer
| Gene Symbol | Primer sequences (top / bottom) | Annealing temperature (°C)1 | Betaine for PCR2 | Length of amplicon (bp) |
|---|---|---|---|---|
| IGF1R | 5’-GCG TGA GAG GAT TGA GTT TC-3’ / 5’-CTC CTT TCC GGT AAT AGT CTG T-3’ | 65 | - | 386 |
| EPHB1 | 5’-TCT GAG TGA GGC GAG CAT-3’ / 5’-GAA GTG AAC TTG CGG TAG G-3’ | 65 | + | 414 |
| EPHA1 | 5’-GGA ACT TCC TTC GAG AGG C-3’ / 5’-GTC CAA CGG ATA GGG ATC TTT-3’ | 65 | - | 376 |
| FGFR3 | 5’-GGT GTC TGA GAT GGA GAT GAT GAA-3’ / 5’-CCG TTG GTC GTC TTC TTG TAG TA-3’ | 65 | + | 396 |
| EPHA2 | 5’-CAG AGA AGC AGC GAG TGG AC-3’ / 5’-GCA GAG GTG AAC TTC CGG TAG-3’ | 70 | - | 433 |
| MET | 5’-TTT CTG ACC GAG GGA ATC ATC-3’ / 5’-CTT TGC ACC TGT TTT GTT GTG TAC-3’ | 65 | - | 363 |
| ITK | 5’-ATA GAG GAG GCT GAA GTA ATG AT-3’ / 5’-GCC TGT GGA ACT GGT GTA C-3’ | 65 3-step | - | 342 |
| EPHA3 | 5’-GAC CCT GAA AGT TGG CTA CAC-3’ / 5’-TCT TCC CTC CTC TTG TTG TAT AAG-3’ | 65 | - | 398 |
| ABL2 | 5’-CTG AAA GAA GCT GCA GTA ATG A-3’ / 5’-TCT CTG GTG CTG TCC ACT TA-3’ | 65 | - | 379 |
| PTK7 | 5’-GGC AGA GAC CCT GGT ACT TG-3’ / 5’-GGC GGA AGT GGT AGT ACT CAC-3’ | 70 | - | 428 |
| AXL | 5’-GAT TTC CTG AGT GAA GCG GTC T-3’ / 5’-TGG CAT CTT GGC GAT ACG TC-3’ | 70 | - | 396 |
| INSR | 5’-GTG AAG ACG GTC AAC GAG TCA-3’ / 5’-CCC CTT TCC GGT AGT AAT CC-3’ | 65 | - | 416 |
| FER | 5’-AAG GGC ACA TTA AAG GAT AAA-3’ / 5’-CCA GAA GAT GAA TAC ACT CCA-3’ | 60 | - | 419 |
| RET | 5’-GAA GAT GCT GAA AGA GAA CGC-3’ / 5’-ATT TAA CTG GAA TCC GAC CCT-3’ | 65 3-step | + | 479 |
| ALK | 5’-GCT CTG AAC AGG ACG AAC TGG-3’ / 5’-GGC ACA GCC TCC CTT TCT ATA GTA-3’ | 65 | - | 401 |
| EPHA5 | 5’-ACG CAG AGA TTT CCT AGG TGA-3’ / 5’-TCC TCC CCT TGT GGT GTA G-3’ | 65 | - | 364 |
| EPHA4 | 5’-CTC TGA AAG CTG GTT ATA CAG AC-3’ / 5’-TGA TGT GAA TTT ACG ATA GGC-3’ | 65 | - | 449 |
| FGFR1 | 5’-GCT GAC TCC AGT GCA TCC AT-3’ / 5’-GCC GTT GGT TGT CTT TTT ATA GTA-3’ | 65 | + | 690 |
| FGFR2 | 5’-AAG ATG TTG AAA GAT GAT GCC ACA-3’ / 5’-CGC CCA TTG GTG GTC TTT T-3’ | 65 | - | 443 |
| FGFR4 | 5’-GTC AAG ATG CTC AAA GAC AAC GC-3’ / 5’-GCC GTT GCT GGT TTT CTT ATA GT-3’ | 65 | + | 444 |
| BTK | 5’-GAA GAA GCC AAA GTC ATG AT-3’ / 5’-GAA ATT TGG AGC CTA CTG AG-3’ | 60 3-step | + | 349 |
| NTRK1 | 5’-GCA GGA CTT CCA GCG TGA G-3’ / 5’-CTC CCA CAC GGT AAT AGT CGG T-3’ | 65 3-step | + | 392 |
| TXK | 5’-AAG AGG CCA AAG TGA TGA T-3’ / 5’-GCT CCA AAA GAA CTG ACA TAC-3’ | 60 | - | 340 |
| ROR1 | 5’-ACA AGA AGC CTC CCT AAT GG-3’ / 5’-GGC AGC AAG GAC TTA CTC TG-3’ | 65 | - | 402 |
| ABL1 | 5’-GAA AGA AGC TGC AGT CAT GAA-3’ / 5’-ATT TCC CAA AGC AAT ACT CCA-3’ | 65 3-step | - | 444 |
| LTK | 5’-TCA GGA TGA GCT GGA TTT CC-3’ / 5’-GGG GCA TCC ACT TGA CTG-3’ | 60 | + | 419 |
| ROS1 | 5’-CAG ACC AGG AGA AGA TTG AAT-3’ / 5’-CCC CTC TCT TTC TAT AGT AAT CAT-3’ | 60 3-step | - | 399 |
| TEC | 5’-TGT GCG AGG AGG ACT TTA TAG A-3’ / 5’-TCT GCC TTC CGT GAA TAC TTC-3’ | 65 3-step | - | 476 |
| NTRK3 | 5’-CCT GGC CGA GTG CTA CAA C-3’ / 5’-CTC CCA CCC TGT AAT AAT CCG-3’ | 65 | - | 482 |
| FES | 5’-CGG TGA AGT CTT GTC GAG AGA-3’ / 5’-GTA GCG GCC GTA GTT AAG GG-3’ | 65 | + | 449 |
| MERTK | 5’-TGA GGC AGC GTG CAT GAA-3’ / 5’-CCA TTT AAC AGG CAT CTT AGC AAT-3’ | 65 | - | 391 |
| MATK | 5’-GGT GGC CGT GAA GAA TAT CA-3’ / 5’-GGT GAA CTT CCC GTG TTT GA-3’ | 65 | + | 433 |
| MUSK | 5’-AGA TAT GCA AGC GGA CTT TC-3’ / 5’-ATA GCG TCG TTT TCA TTA GCT TT-3’ | 65 3-step | + | 441 |
| TEK | 5’-GGA CTT TGC AGG AGA ACT G-3’ / 5’-GTA ACA CAC CAT AGG ACC ATA CAT-3’ | 60 3-step | - | 482 |
PCR was performed basically in a 2-step cycle. For genes with the comment of ‘3-step’, PCR was performed in a 3-step cycle (annealing step was followed by an extension step at 72°C).
Betaine was supplemented at a final concentration of 1M.
Immunohistochemistry of clinical samples
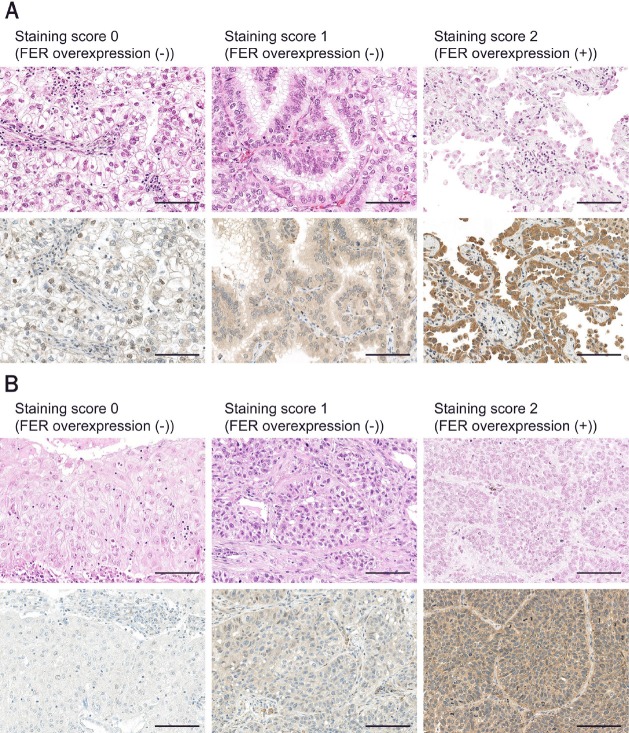
Tissue microarrays were constructed from formalin-fixed, paraffin-embedded lung cancer tissue blocks. Serial tissue sections (4 μm thick) were cut from the tissue microarray, and immunohistochemistry (IHC) was performed with anti-FER monoclonal antibody (diluted 1:1,000, LS-B3467; LS Bio, Seattle, WA) using a Ventana Benchmark XT autostainer (Ventana Medical Systems, Tucson, AZ). Appropriate positive and negative controls were included for each run of immunostaining. All samples were evaluated in a blinded manner by two independent pathologists without knowledge of any other clinicopathological data. The immunostaining score was estimated using a scale of 0 to 2 based on the percentage and intensity of the stained tumor cells as follows: 0, complete absence of staining; 1, low-level staining of the tumor cells or low-to-moderate level staining on less than 50% of tumor cells; 2, moderate-level staining on more than 50% of tumor cells or high-level staining of the tumor cells. Staining with a score of 2 was defined as FER overexpression.
FER expression plasmid vector constructions
To construct the FER expression plasmid vector, full-length human FER was cloned using a previously reported method [29]. Briefly, the 5’ and 3’ sides of FER were first amplified from the 5’ and 3’ RACE products of FER, respectively, using the following primer pairs: 5’-AAA ATG GGG TTT GGG AGT GAC-3’/5’-CCA GAA GAT GAA TAC ACT CCA-3’ for the 5’ side of FER, and 5’-GAC AGC CTG TCT ACA TCA TTA TGG AAC-3’/5’-CGC CAT CCT GTC ACT ATG TGA G-3’ for the 3’ side of FER. The PCR products obtained using these two primer sets had an overlapping region, and the full-length human FER cDNA was constructed by reacting these two products on a Mastercycler (Eppendorf, Hamburg, Germany) as follows: 94°C for 2 min; 30 cycles of 98°C for 10 sec and 68°C for 2.5 min, with holding at 4°C. After ligation into a pGEM-T Easy vector system (Promega, Madison, WI) and sequencing to confirm the absence of unexpected mutations, the resulting product (full-length human FER cDNA) was subcloned into the NotI restriction sites of the mammalian expression vector pcDNA 3.1 Hygro+ (Invitrogen, Carlsbad, CA). E. coli HB101 (TaKaRa, Shiga, Japan) was used as a host cell line to copy the expression plasmid vector.
Focus formation assay
NIH-3T3 cells were transfected by FER expression plasmid vector using HilyMax (Dojindo, Kumamoto, Japan). The cells were cultivated in the presence of hygromycin to select the transfected cells, and whether the surviving cells formed foci after reaching confluence was noted. Pictures were taken after 14 days of culture.
Colony formation assay
A total of 1,000 cells were seeded onto Hydrocell (Cell Seed, Tokyo, Japan), non-adhesive dishes coated with hydrophilic polymers, in a normal medium containing 1.3% methylcel-methylcellulose (Wako, Osaka, Japan). After incubation for 3 days, the formed colonies were quantified using the Cell Counting Kit-8 (CCK-8) assay (Dojindo, Kumamoto, Japan). Ten microliters of CCK-8 reagent was added to each well, and the plate was incubated for 3 hours at 37°C. Then, viable cells forming colonies were evaluated based on absorbance measurements performed at 450 nm using a 1420 Multilabel Counter (Perkin Elmer Life Sciences, Boston, MA).
Western blot analysis
Twenty micrograms of total protein was loaded onto 5%-20% SuperSep Ace (Wako, Osaka, Japan), electrophoresed in Tris-glycine-SDS running buffer, and transferred to Hybond-P (GE Healthcare, Buckinghamshire, UK). The resulting membranes were blocked in 5% skim milk and then incubated in diluted primary antibody. For blotting with the secondary antibody, the SNAP id system (Millipore, Billerica, MA) was used. Antibody against FER was from Cosmo Bio (Tokyo, Japan); Akt, phospho-Akt (Ser473), Erk1/2, phospho-Erk1/2 (Thr202/Tyr204), cortactin, and phospho-cortactin (Tyr421) were from Cell Signaling (Danvers, MA, USA); and β-actin was from Sigma-Aldrich (St. Lois, MO). As for the secondary antibodies, anti-mouse-IgG1-HRP antibody and anti-rabbit-IgG-HRP antibody from Santa Cruz Biotechnology (Santa Cruz, CA) were used. The proteins were detected using ECL plus solution (GE Healthcare, Buckinghamshire, UK).
siRNA and scrambled control vector constructions
siRNA vectors targeting FER and their corresponding scrambled control vectors were constructed as previously described [30]. Briefly, oligo1, which consists of a targeted FER sequence with an AatII recognition site, and oligo2, which consists of a complementary sequence of oligo1 with TTTTT, were ligated sequentially into a TA cloning vector with a U6 promoter. The corresponding scrambled control sequences were designed using the siRNA Wizard web site (http://www.invivogen.com/family.php?ID=236). The sequences of oligo1 and 2 are shown in Table 2.
Table 2.
Oligo1 and oligo2 of siRNA vectors targeting FER and corresponding scrambled control vectors
| Vector | Oligo | Sequence |
|---|---|---|
| siRNA #1 | O1F | 5’-GCA GAA CAG GAC TGG TAC CAT GAC GT-3’ |
| O1R | 5’-CAT GGT ACC AGT CCT GTT CTG C-3’ | |
| O2F | 5’-CAT GGT ACC AGT CCT GTT CTG CCT TTT TGG GCC-3’ | |
| O2R | 5’-CAA AAA GGC AGA ACA GGA CTG GTA CCA TGA CGT-3’ | |
| siRNA #2 | O1F | 5’-GAG ATA CAG TTC AGA GAG TGA GAC GT-3’ |
| O1R | 5’-CTC ACT CTC TGA ACT GTA TCT C-3’ | |
| O2F | 5’-CTC ACT CTC TGA ACT GTA TCT CCT TTT TGG GCC-3’ | |
| O2R | 5’-CAA AAA GGA GAT ACA GTT CAG AGA GTG AGA CGT-3’ | |
| scrambled #1 | O1F | 5’-GCC ATG CGA ACC GAT AGA TGA GAC GT-3’ |
| O1R | 5’-CTC ATC TAT CGG TTC GCA TGG C-3’ | |
| O2F | 5’-CTC ATC TAT CGG TTC GCA TGG CCT TTT TGG GCC-3’ | |
| O2R | 5’-CAA AAA GGC CAT GCG AAC CGA TAG ATG AGA CGT-3’ | |
| scrambled #2 | O1F | 5’-GCG GAT ACG GGA AGA TAT TAA GAC GT-3’ |
| O1R | 5’-CTT AAT ATC TTC CCG TAT CCG C-3’ | |
| O2F | 5’-CTT AAT ATC TTC CCG TAT CCG CCT TTT TGG GCC-3’ | |
| O2R | 5’-CAA AAA GGC GGA TAC GGG AAG ATA TTA AGA CGT-3’ |
Apoptosis assay
The apoptosis levels were assayed by double staining with annexin V-FITC and propidium iodide using the Annexin V-FITC Apoptosis Detection Kit I (BD Pharmingen, San Jose, CA) and were quantified using flow cytometry. The staining procedure was performed according to the manufacturer’s recommendations. Briefly, trypsinized cells were collected and resuspended in 1X annexin V binding buffer at a concentration of 1x106 cells/mL. Next, 5 μL of annexin V-FITC and 2 μL of propidium iodide were added to 100 μL of the solution. After incubation for 15 min at room temperature in the dark, 400 μL of 1X annexin V binding buffer were added to the cell solution, which was then immediately analyzed using flow cytometry. A total of 10,000 events were scored using a FACSCalibur (Becton Dickinson, San Jose, CA) and were analyzed using CellQuest software. Cells exposed to cisplatin at a concentration of 12.5 μg/mL for 24 hours were used as a positive control for apoptosis.
Statistical analysis
A statistical analysis was performed using Dr. SPSS II (SPSS, Chicago, IL). The relation between the immunostaining score for FER and the mRNA level of FER examined using quantitative RT-PCR was analyzed using the Fisher’s exact test. The relation between FER overexpression and the clinicopathological characteristics was analyzed using the chi-square test or the Fisher’s exact test, and a logistic regression analysis was used to analyze independent factors associated with FER overexpression. The progression-free and overall survivals curves were calculated using the Kaplan-Meier method and were compared using a log-rank test. A multivariate Cox regression analysis of factors was performed to identify independent factors related to the prognosis of NSCLC patients. To analyze differences in the colony formation assay, the FER expression levels among lung cancer cell lines, and the percentage of apoptotic cells after FER knockdown in lung cancer cell lines, the Tukey’s honestly significant differences test was used.
Results
Selection and expression level analyses of candidate genes involved in lung cancer
To identify potential molecular events involved in NSCLC, we first focused on ALK. We hypothesized that genes analogous to ALK might be involved in lung cancer, since ALK has been shown to have very strong transforming activities in many cancers including lung cancer [31-35]. Therefore, we performed an in silico search for genes with a structure analogous to the ALK gene and listed the top 34 gene results.
We performed quantitative RT-PCR and examined the expression levels of these genes using cDNAs from surgically resected lung adenocarcinoma specimens (n = 30). A level more than 16 times higher than the expression level in normal lung tissue was set as the threshold for gene overexpression, and we found that IGF1R, the EPH family, the FGFR family, AXL, FER, RET, and ALK were each overexpressed in some of the NSCLC samples (Figure 1).
Figure 1.
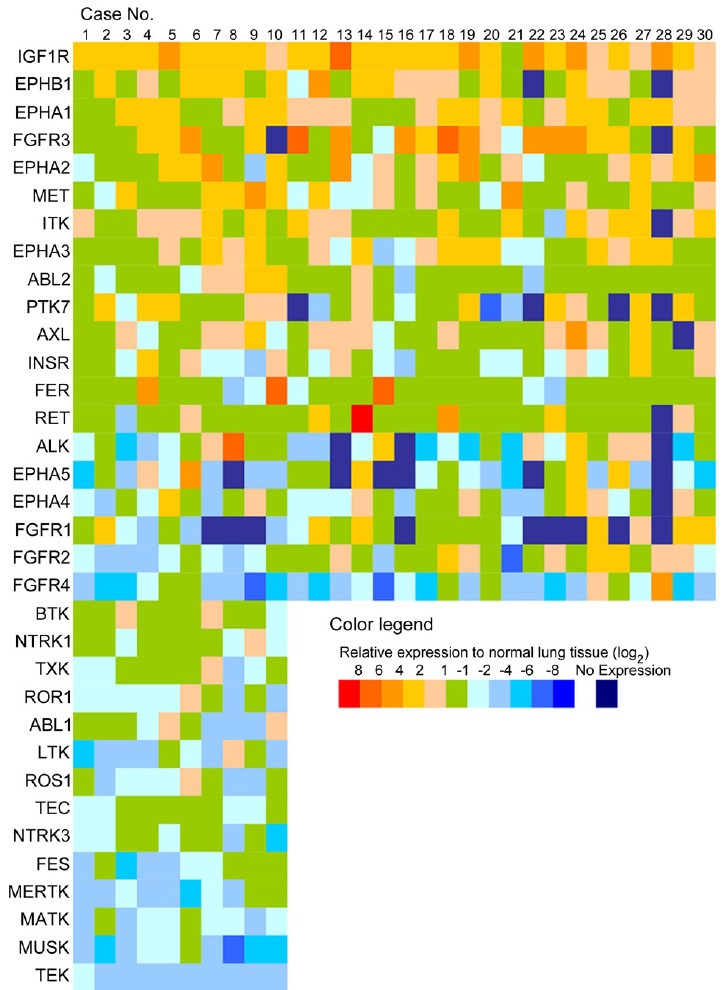
Relative expression level of the candidate genes in NSCLC clinical specimens, compared with normal lung tissue. The relative expression levels of the genes in NSCLC clinical specimens, compared with that in normal lung tissue, are shown according to the color legend.
Among these genes, we selected FER, which was overexpressed in 10% of the cases (3/30 cases), for further analysis because the correlation of FER with NSCLC would be a novel discovery, whereas the involvement of the other overexpressed genes has already been reported in lung cancer [31,35-39].
Immunohistochemical analysis of FER expression in lung cancer
To analyze FER expression in a broader population of NSCLC patients, we performed IHC using tissue microarrays. We scored FER staining based on the percentage and intensity of the stained tumor cells as described in Materials and Methods (Figure 2). These immunostaining scores were consistent with the mRNA level detected using quantitative RT-PCR, and the mRNA level more than 16 times higher than that in normal lung tissue, which was set as the threshold for gene overexpression at the initial screening, corresponded to immunostaining score 2 (Table 3).
Figure 2.
Immunohistochemical analysis of FER on NSCLC tumor microarrays. Representative images of each staining score in adenocarcinoma samples (A) and squamous cell carcinoma samples (B) are shown. The upper panels are hematoxylin and eosin staining, and the lower panels are immunostaining for FER. Scale bar, 100 μm.
Table 3.
Correlation between immunostaining score and expression level analyzed using quantitative RT-PCR for FER
| Relative expression to normal lung tissue analyzed using quantitative RT-PCR (log2) | Total | ||||
|---|---|---|---|---|---|
|
| |||||
| < -4 | ≥ -4, ≤ 0 | > 0, ≤ 4 | > 4 | ||
| Staining score 2, n | 0 | 0 | 1 | 14 | 15 |
| Staining score 1, n | 4 | 62 | 40 | 2 | 108 |
| Staining score 0, n | 12 | 0 | 0 | 0 | 12 |
n, number of cases. Fisher’s exact test P < 0.001.
Then, we counted the cases with immunostaining score 2 as FER-overexpressed cases and found that FER was overexpressed in 10.1% (24/236 cases) of the lung adenocarcinoma cases and 5.5% (7/127 cases) of the squamous cell lung cancer cases (Table 4).
Table 4.
Immunohistochemistry of primary lung cancer staining for FER
| Adenocarcinoma | Squamous cell carcinoma | Small cell lung carcinoma | Large cell neuroendocrine carcinoma | Pleomorphic carcinoma | |
|---|---|---|---|---|---|
| Staining score 2, n (%) | 24 (10.1) | 7 (5.5) | 0 (0) | 1 (16.6) | 0 (0) |
| Staining score 1, n | 194 | 115 | 5 | 5 | 4 |
| Staining score 0, n | 18 | 5 | 1 | 0 | 2 |
| Total, n | 236 | 127 | 6 | 6 | 6 |
n, number of cases.
Correlation between FER overexpression and postoperative survival periods of NSCLC patients
Next, we investigated whether FER overexpression has any association with clinicopathological characteristics of lung cancer. To this end, 135 primary NSCLC patients (108 patients of adenocarcinoma and 27 patients of squamous cell carcinoma), who had undergone potential curative resection and whose follow-up data for their clinical outcome were available, were analyzed. The baseline characteristics of the patient cohort are shown in Table 5. The median follow-up period was 58.5 months.
Table 5.
Baseline characteristics of 135 analyzed NSCLC patients
| Characteristics | Patient cohort (n=135) |
|---|---|
| Age | |
| Median, years | 66 |
| Interquartile range, years | 34-85 |
| >65 years, n (%) | 77 (57.0) |
| <65 years, n (%) | 58 (43.0) |
| Sex | |
| Male, n (%) | 80 (59.3) |
| Female, n (%) | 55 (40.7) |
| Smoking habit | |
| Never, n (%) | 38 (28.1) |
| Ever, n (%) | 97 (71.9) |
| Histological type | |
| Adenocarcinoma, n (%) | 108 (80) |
| Squamous cell carcinoma, n (%) | 27 (20) |
| Disease stage | |
| IA, n (%) | 61 (45.2) |
| IB, n (%) | 37 (27.4) |
| IIA, n (%) | 4 (3.0) |
| IIB, n (%) | 11 (8.1) |
| IIIA, n (%) | 22 (16.3) |
| T factor | |
| T1, n (%) | 71 (52.6) |
| T2, n (%) | 64 (47.4) |
| N factor | |
| N0, n (%) | 101 (74.8) |
| N1, n (%) | 15 (11.1) |
| N2, n (%) | 19 (14.1) |
| Adjuvant chemotherapy | |
| Yes, n (%) | 26 (19.3) |
| No, n (%) | 109 (80.7) |
| EGFR mutation | |
| Negative, n (%) | 87 (64.4) |
| Positive, n (%) | 41 (30.4) |
| Unknown, n (%) | 7 (5.2) |
n, number of cases.
When we evaluated FER expression in these patients using IHC, FER overexpression was observed in 15 cases out of 135 cases. In a univariate analysis, there was no significant correlation between FER overexpression and various clinicopathological features, such as age, sex, smoking history, histological type, disease stage, T factor, N factor, adjuvant chemotherapy history, or EGFR mutation (Table 6). However, we found that FER overexpression was significantly associated with a poor progression-free and overall survival period after surgical resection in Kaplan-Meier analyses (Figure 3). A multivariate Cox regression analysis further revealed that FER overexpression was an independent factor for both the progression-free and overall survival periods as well as age, disease stage, T factor and N factor (Table 7).
Table 6.
Correlation between FER overexpression and clinicopathological features
| FER overexpression | P | ||
|---|---|---|---|
|
| |||
| (+), n | (-), n | ||
| Overall (n = 135) | 15 | 120 | |
| Age | |||
| >65 years (n = 77) | 8 | 69 | |
| <65 years (n = 58) | 7 | 51 | 0.788 |
| Sex | |||
| Male (n = 80) | 10 | 70 | |
| Female (n = 55) | 5 | 50 | 0.590 |
| Smoking habit | |||
| Never (n = 38) | 4 | 34 | |
| Ever (n = 97) | 11 | 86 | 1.000 |
| Histological type | |||
| Adenocarcinoma (n = 108) | 13 | 96 | |
| Squamous cell carcinoma (n = 27) | 2 | 25 | 0.735 |
| Disease stage | |||
| IA, IB (n = 98) | 12 | 86 | |
| IIA-IIIA (n = 37) | 3 | 34 | 0.759 |
| T factor | |||
| T1 (n = 71) | 7 | 64 | |
| T2 (n = 64) | 8 | 56 | 0.785 |
| N factor | |||
| N0 (n = 101) | 12 | 89 | |
| N1, N2 (n = 34) | 3 | 31 | 0.760 |
| Adjuvant chemotherapy | |||
| Yes (n = 26) | 3 | 23 | |
| No (n = 109) | 12 | 97 | 1.000 |
| EGFR mutation | |||
| Negative (n = 87) | 10 | 77 | |
| Positive (n = 41) | 5 | 36 | 1.000 |
n, number of cases.
Figure 3.
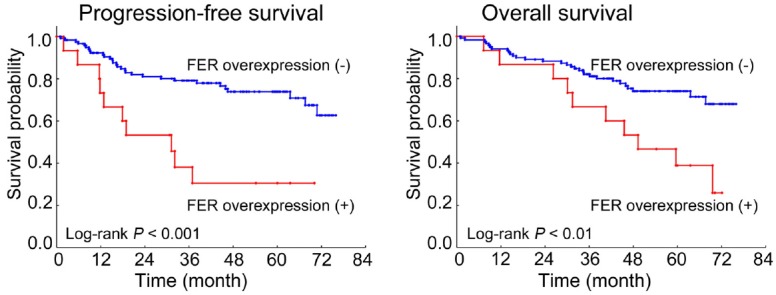
Analysis of FER overexpression and postoperative prognosis of NSCLC patients. Kaplan-Meier analyses of progression-free and overall survival periods among 135 curatively resected NSCLC patients are shown stratified according to FER expression.
Table 7.
Multivariate Cox regression analysis of various factors for prognosis of lung cancer patients
| Progression-free survival | |||
|
| |||
| Variables | Hazard ratio (95% CI) | Unfavorable / Favorable | P |
|
| |||
| FER overexpression | 2.335 (1.043-5.232) | Positive / Negative | 0.033* |
| Age (years) | 2.250 (1.082-4.681) | ≥65 / < 65 | 0.030* |
| Sex | 1.311 (0.840-2.046) | Male / Female | 0.332 |
| Smoking habit | 1.346 (0.464-3.904) | Ever / Never | 0.585 |
| Histological type | 1.142 (0.356-3.664) | Adeno / Squamous | 0.823 |
| Disease stage | 2.570 (0.963-6.859) | IIA-IIIA / IA, IB | 0.059 |
| T factor | 3.258 (1.533-6.922) | T2 / T1 | 0.002* |
| N factor | 1.745 (0.587-5.183) | N1, N2 / N0 | 0.316 |
| Adjuvant chemotherapy | 1.343 (0.757-2.381) | No / Yes | 0.323 |
| EGFR mutation | 1.493 (0.635-3.512) | Negative / Positive | 0.358 |
|
| |||
| Overall survival | |||
|
| |||
| Variables | Hazard ratio (95% CI) | Unfavorable / Favorable | P |
|
| |||
| FER overexpression | 2.220 (1.064-4.631) | Positive / Negative | 0.039* |
| Age (years) | 2.266 (1.104-4.652) | >65 / < 65 | 0.026* |
| Sex | 1.392 (0.699-2.775) | Male / Female | 0.264 |
| Smoking habit | 1.509 (0.531-4.286) | Ever / Never | 0.440 |
| Histological type | 1.350 (0.422-4.316) | Adeno / Squamous | 0.613 |
| Disease stage | 4.267 (1.474-12.353) | IIA-IIIA / IA, IB | 0.007* |
| T factor | 2.618 (1.221-5.615) | T2 / T1 | 0.013* |
| N factor | 3.743 (1.191-11.768) | N1, N2 / N0 | 0.024* |
| Adjuvant chemotherapy | 1.183 (0.445-3.139) | No / Yes | 0.741 |
| EGFR mutation | 1.329 (0.554-3.189) | Negative / Positive | 0.524 |
CI, confidence interval.
P < 0.05.
Transforming activity of FER in vitro
FER has been previously reported to cause transformation in cell cultures [15,40]. To verify the transforming potential of FER, we generated an expression plasmid vector for FER and introduced it into NIH-3T3 cells. The successful introduction of FER was confirmed by a western blot analysis (Figure 4A). We used mutant KRAS as a positive control for a gene harboring transforming activity.
Figure 4.
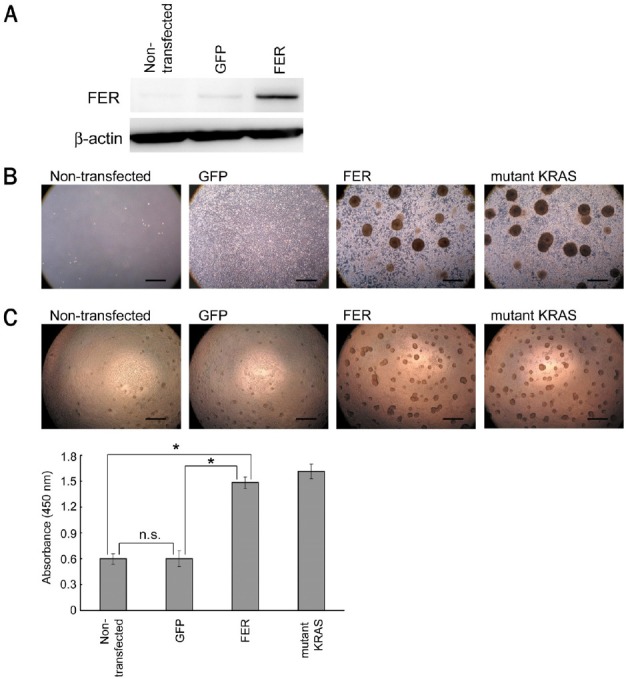
Transforming activity of FER in vitro. A. Western blot analysis after the introduction of FER into NIH-3T3 cells. B. Focus formation assay. Representative images from non-transfected or transfected NIH-3T3 cells after selection using hygromycin are shown. Scale bar, 500 μm. C. Colony formation assay. The formed colonies were quantified according to the absorbance using WST-8 and are shown as a bar graph. The error bars indicate the standard deviations. Scale bar, 500 μm. n. s., not significant. *P < 0.001.
First, to examine whether FER had any potential to lead cells toward a loss of contact inhibition (a hallmark of cell transformation), we performed a focus formation assay and observed the cell growth under the selection of successfully transfected cells with hygromycin. The non-transfected NIH-3T3 cells stopped growing and died after 2 weeks of culture under the selection conditions. The GFP-transfected NIH-3T3 cells grew but stopped proliferation without the generation of any foci when they became confluent. In contrast, the FER-transfected NIH-3T3 cells continued to undergo proliferation even after they reached confluence and formed foci similar to the mutant-KRAS-transfected NIH-3T3 cells after about 2 weeks of culture (Figure 4B).
Next, to evaluate the potential role of FER in anchorage-independent growth, another characteristic of cell transformation, we performed a colony formation assay and quantified the formed colonies using WST-8. The FER-transfected NIH-3T3 cells were revealed to form a significantly larger number of colonies under culture conditions without any anchorage, compared with the GFP-transfected NIH-3T3 cells or the non-transfected NIH-3T3 cells (Figure 4C).
Taken together, these results confirm that FER possesses oncogenic functions in vitro.
Functional analyses of FER in FER-overexpressed NSCLC
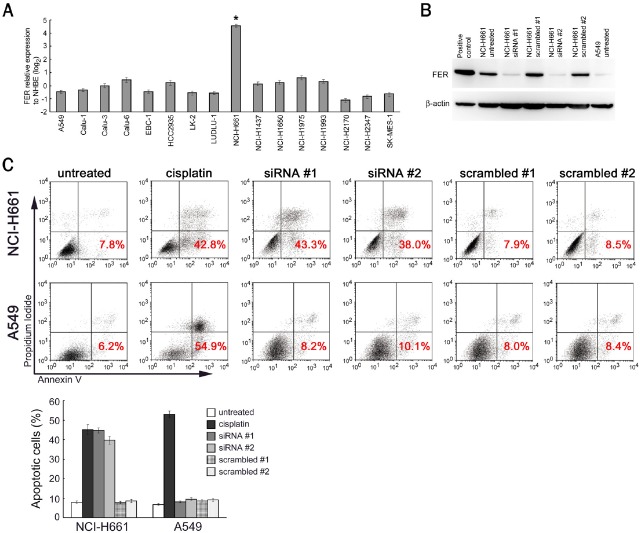
We next analyzed the functional significance of FER overexpression in NSCLC. According to NCBI Gene Expression Omnibus data, human lung cancer NCI-H661 cells exhibit a considerably higher expression level of FER than normal human bronchial epithelial cells (NHBE) or other lung cancer cell lines. Using quantitative RT-PCR, we ascertained that the expression level of FER in NCI-H661 cells was more than 16 times higher than that in NHBE, meeting the criteria for overexpression used in the initial screening for overexpressed genes in the clinical samples, whereas the other examined cancer cell lines showed almost the same level of FER expression as that seen in NHBE (Figure 5A).
Figure 5.
Induction of NCI-H661 apoptosis by FER knockdown. A. Relative expression level of FER in lung cancer cell lines, compared with that in NHBE. The error bars indicate the standard deviations. *P < 0.001. B. Western blot analysis after the knockdown of FER by siRNA vectors. Protein from HEK293T transfected with FER expression vectors was used as a positive control. C. Double staining with annexin V-FITC and propidium iodide, followed by flow cytometry analysis to detect apoptotic cells. The percentages in the dot plot results indicate the percentages of apoptotic cells and are shown as a bar graph after triplicate analyses. Cells treated using cisplatin were used as positive controls for apoptosis. The error bars indicate the standard deviations.
Using NCI-H661 cells as a model, we examined the effect of FER knockdown in FER-overexpressed NSCLC. We constructed two distinct siRNA plasmid vectors directed against FER (siRNA #1, #2) and their corresponding scrambled control vectors (scrambled #1, #2) and introduced these constructed vectors into NCI-H661 cells. We used A549 cells as control cancer cells that did not overexpress FER. Using a western blot analysis, we ascertained that the FER protein level was significantly decreased by the siRNA vectors, but not by the scrambled control vectors (Figure 5B). The FER expression level of untreated A549 cells was as low as that of the siRNA-introduced NCI-H661 cells (Figure 5B).
When we knocked down FER in NCI-H661 cells, we found that most of the cells did not survive. To analyze the cell deaths objectively, we performed an apoptosis assay using double staining with annexin V and propidium iodide. The results showed a significant increase in apoptosis in NCI-H661 cells after FER knockdown, while the scrambled controls did not have any effect (Figure 5C). In contrast, FER knockdown had no impact on A549 cells, which do not have an intrinsically high FER expression level (Figure 5C). We also examined the impact of FER knockdown on other lung cancer cell lines, which are shown as not overexpressing FER in Figure 5A, and found that they were not affected by FER knockdown either (data not shown). These results suggest that FER might be involved in cancer-cell survival specifically in FER-overexpressed NSCLC.
Analysis of downstream of FER signaling pathway
To elucidate the mechanism of how FER is involved in the cell survival of NSCLC, we examined downstream signaling alterations following FER overexpression in NIH-3T3 cells or following FER knockdown in NCI-H661 cells.
We performed western blot analyses and checked the phosphorylation of Akt and MAPK, which play critical roles in many cellular programs, such as cell proliferation and survival. We also checked the phosphorylation of cortactin, which is involved in actin reorganization and is reportedly phosphorylated by FER. However, we did not observe any obvious alterations in these signaling pathways by the overexpression or knockdown of FER (data not shown).
Discussion
FER, a 94-kDa nonreceptor tyrosine kinase, was initially discovered in 1988 during studies on the protooncogene protein Fes/Fps [9,15]. FER and FES/FPS have close structural similarities and compose one subfamily. The exact function of FER is still unknown, but its involvement in integrin/E-cadherin-mediated signaling pathways and a role in regulating crosstalk between cadherin-catenin complexes and focal adhesions have been proposed [13,14,16,17,19,41]. FER is also thought to be involved in growth-promoting pathways and to be activated as the downstream of epidermal growth factor receptor and platelet-derived growth factor receptor in fibroblast cells [13].
Some reports suggest that FER has a regulatory role in the progression and growth of malignant tumors. For example, FER is reportedly highly expressed in numerous malignant cell lines [10,24], and the levels of FER in malignant prostate tumors are significantly higher than those detected in benign prostate tumors [20]. Furthermore, the downregulation of FER impairs the proliferation of prostate carcinoma cells and abolishes their ability to form colonies in soft agar [20]. FER was also found to be involved in the progression of breast cancer and to play a regulatory role during cell cycle progression in breast cancer cells [21]. In hepatocellular carcinoma, tumor tissue cells with distant pulmonary metastasis were shown to have higher FER expression levels than those without distant metastasis [25].
In the present study, we have shown that FER is overexpressed at both transcriptional and translational levels in a subset of NSCLC patients and is positively correlated with poor progression-free and overall survival periods after surgical resection, independent of the other clinicopathological features. Although the data presented here just represents a single patient cohort and it would be important to know whether this could be reproduced in larger independent series, our findings may add some useful information for planning the treatment of lung cancer patients after surgical resection. For example, early-stage patients might warrant an aggressive treatment when FER is overexpressed, or FER overexpression in resected specimens may be an index for the application of adjuvant therapy.
In the functional analyses of FER in vitro, apoptosis was induced after FER knockdown in a FER-overexpressed cancer cell line. While findings in one cell line cannot be generalized, our finding suggests at least the possibility that some FER-overexpressed NSCLC may depend on FER for cancer-cell survival. So far, there are no biological or small molecule inhibitors that selectively inhibit FER, but their development would be useful for confirming this possibility. As for the signal pathway of how FER is involved in cancer-cell survival in FER-overexpressed NSCLC, we did not observe any phosphorylation in the Akt or MAPK pathways, which are the representative pathways that contribute to cancer-cell proliferation or survival. We also checked the phosphorylation of cortactin, which is reportedly phosphorylated by FER, but its obvious phosphorylation was not observed either. Other pathways, such as the ones related to cell-cycle modification, might play important roles. Broader phosphoproteome analyses will be required in the future to investigate the precise mechanism of how FER is involved in cancer-cell survival.
In conclusion, we have shown FER overexpression may have a prognostic impact on the survival of surgically treated NSCLC patients and might be a requisite for cancer-cell survival in vitro. This report is the first to suggest the involvement of FER in lung cancer, and further studies, such as functional analyses performed in vivo or larger-scale statistical analyses of clinical samples, are worth performing.
Acknowledgements
This work was supported by grants from the Mitsui Life Social Foundation, the Takeda Science Foundation, and MEXT/JSPS KAKENHI (24501339, 23390147, 23249045, 2465-9267, 24800019).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Tsuchiya T, Akamine S, Muraoka M, Kamohara R, Tsuji K, Urabe S, Honda S, Yamasaki N. Stage IA non-small cell lung cancer: vessel invasion is a poor prognostic factor and a new target of adjuvant chemotherapy. Lung Cancer. 2007;56:341–348. doi: 10.1016/j.lungcan.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 4.Ichinose Y, Yano T, Asoh H, Yokoyama H, Yoshino I, Katsuda Y. Prognostic factors obtained by a pathologic examination in completely resected non-small-cell lung cancer. An analysis in each pathologic stage. J Thorac Cardiovasc Surg. 1995;110:601–605. doi: 10.1016/S0022-5223(95)70090-0. [DOI] [PubMed] [Google Scholar]
- 5.Okada M, Sakamoto T, Nishio W, Uchino K, Tsubota N. Characteristics and prognosis of patients after resection of nonsmall cell lung carcinoma measuring 2 cm or less in greatest dimension. Cancer. 2003;98:535–541. doi: 10.1002/cncr.11530. [DOI] [PubMed] [Google Scholar]
- 6.Iwasaki A, Shirakusa T, Yoneda S, Makimoto Y, Enatsu S, Hamada T. Results of surgical treatment for non-small cell lung cancer of 20 mm or less in diameter. Thorac Cardiovasc Surg. 2004;52:293–297. doi: 10.1055/s-2004-821155. [DOI] [PubMed] [Google Scholar]
- 7.Shimizu K, Yoshida J, Nagai K, Nishimura M, Yokose T, Ishii G, Nishiwaki Y. Visceral pleural invasion classification in non-small cell lung cancer: a proposal on the basis of outcome assessment. J Thorac Cardiovasc Surg. 2004;127:1574–1578. doi: 10.1016/j.jtcvs.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 8.Sun Z, Aubry MC, Deschamps C, Marks RS, Okuno SH, Williams BA, Sugimura H, Pankratz VS, Yang P. Histologic grade is an independent prognostic factor for survival in non-small cell lung cancer: an analysis of 5018 hospital and 712 population-based cases. J Thorac Cardiovasc Surg. 2006;131:1014–1020. doi: 10.1016/j.jtcvs.2005.12.057. [DOI] [PubMed] [Google Scholar]
- 9.Letwin K, Yee SP, Pawson T. Novel protein-tyrosine kinase cDNAs related to fps/fes and eph cloned using anti-phosphotyrosine antibody. Oncogene. 1988;3:621–627. [PubMed] [Google Scholar]
- 10.Hao QL, Heisterkamp N, Groffen J. Isolation and sequence analysis of a novel human tyrosine kinase gene. Mol Cell Biol. 1989;9:1587–1593. doi: 10.1128/mcb.9.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben-Dor I, Bern O, Tennenbaum T, Nir U. Cell cycle-dependent nuclear accumulation of the p94fer tyrosine kinase is regulated by its NH2 terminus and is affected by kinase domain integrity and ATP binding. Cell Growth Differ. 1999;10:113–129. [PubMed] [Google Scholar]
- 12.Kim L, Wong TW. The cytoplasmic tyrosine kinase FER is associated with the catenin-like substrate pp120 and is activated by growth factors. Mol Cell Biol. 1995;15:4553–4561. doi: 10.1128/mcb.15.8.4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim L, Wong TW. Growth factor-dependent phosphorylation of the actin-binding protein cortactin is mediated by the cytoplasmic tyrosine kinase FER. J Biol Chem. 1998;273:23542–23548. doi: 10.1074/jbc.273.36.23542. [DOI] [PubMed] [Google Scholar]
- 14.Rosato R, Veltmaat JM, Groffen J, Heisterkamp N. Involvement of the tyrosine kinase fer in cell adhesion. Mol Cell Biol. 1998;18:5762–5770. doi: 10.1128/mcb.18.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greer P. Closing in on the biological functions of Fps/Fes and Fer. Nat Rev Mol Cell Biol. 2002;3:278–289. doi: 10.1038/nrm783. [DOI] [PubMed] [Google Scholar]
- 16.Craig AW, Greer PA. Fer kinase is required for sustained p38 kinase activation and maximal chemotaxis of activated mast cells. Mol Cell Biol. 2002;22:6363–6374. doi: 10.1128/MCB.22.18.6363-6374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kogata N, Masuda M, Kamioka Y, Yamagishi A, Endo A, Okada M, Mochizuki N. Identification of Fer tyrosine kinase localized on microtubules as a platelet endothelial cell adhesion molecule-1 phosphorylating kinase in vascular endothelial cells. Mol Biol Cell. 2003;14:3553–3564. doi: 10.1091/mbc.E03-02-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miravet S, Piedra J, Castano J, Raurell I, Franci C, Dunach M, Garcia de Herreros A. Tyrosine phosphorylation of plakoglobin causes contrary effects on its association with desmosomes and adherens junction components and modulates beta-catenin-mediated transcription. Mol Cell Biol. 2003;23:7391–7402. doi: 10.1128/MCB.23.20.7391-7402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piedra J, Miravet S, Castano J, Palmer HG, Heisterkamp N, Garcia de Herreros A, Dunach M. p120 Catenin-associated Fer and Fyn tyrosine kinases regulate beta-catenin Tyr-142 phosphorylation and beta-catenin-alpha-catenin Interaction. Mol Cell Biol. 2003;23:2287–2297. doi: 10.1128/MCB.23.7.2287-2297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allard P, Zoubeidi A, Nguyen LT, Tessier S, Tanguay S, Chevrette M, Aprikian A, Chevalier S. Links between Fer tyrosine kinase expression levels and prostate cell proliferation. Mol Cell Endocrinol. 2000;159:63–77. doi: 10.1016/s0303-7207(99)00205-1. [DOI] [PubMed] [Google Scholar]
- 21.Pasder O, Shpungin S, Salem Y, Makovsky A, Vilchick S, Michaeli S, Malovani H, Nir U. Downregulation of Fer induces PP1 activation and cell-cycle arrest in malignant cells. Oncogene. 2006;25:4194–4206. doi: 10.1038/sj.onc.1209695. [DOI] [PubMed] [Google Scholar]
- 22.Condorelli F, Stec-Martyna E, Zaborowska J, Felli L, Gemmi I, Ponassi M, Rosano C. Role of the non-receptor tyrosine kinase fes in cancer. Curr Med Chem. 2011;18:2913–2920. doi: 10.2174/092986711796150522. [DOI] [PubMed] [Google Scholar]
- 23.Zoubeidi A, Rocha J, Zouanat FZ, Hamel L, Scarlata E, Aprikian AG, Chevalier S. The Fer tyrosine kinase cooperates with interleukin-6 to activate signal transducer and activator of transcription 3 and promote human prostate cancer cell growth. Mol Cancer Res. 2009;7:142–155. doi: 10.1158/1541-7786.MCR-08-0117. [DOI] [PubMed] [Google Scholar]
- 24.Orlovsky K, Theodor L, Malovani H, Chowers Y, Nir U. Gamma interferon down-regulates Fer and induces its association with inactive Stat3 in colon carcinoma cells. Oncogene. 2002;21:4997–5001. doi: 10.1038/sj.onc.1205624. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Ren Z, Kang X, Zhang L, Li X, Wang Y, Xue T, Shen Y, Liu Y. Identification of tyrosine-phosphorylated proteins associated with metastasis and functional analysis of FER in human hepatocellular carcinoma cells. BMC Cancer. 2009;9:366. doi: 10.1186/1471-2407-9-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menges CW, Chen Y, Mossman BT, Chernoff J, Yeung AT, Testa JR. A Phosphotyrosine Proteomic Screen Identifies Multiple Tyrosine Kinase Signaling Pathways Aberrantly Activated in Malignant Mesothelioma. Genes Cancer. 2010;1:493–505. doi: 10.1177/1947601910375273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zang ZJ, Ong CK, Cutcutache I, Yu W, Zhang SL, Huang D, Ler LD, Dykema K, Gan A, Tao J, Lim S, Liu Y, Futreal PA, Grabsch H, Furge KA, Goh LK, Rozen S, Teh BT, Tan P. Genetic and structural variation in the gastric cancer kinome revealed through targeted deep sequencing. Cancer Res. 2010;71:29–39. doi: 10.1158/0008-5472.CAN-10-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sobin LH, Wittekind C, editors. TNM Classification of Malignant Tumours (Uicc International Union Against Cancer) 6th edn. New York , NY: Wiley-Liss; 2002. [Google Scholar]
- 29.Sunohara M, Kawakami M, Kage H, Watanabe K, Emoto N, Nagase T, Ohishi N, Takai D. Polymerase reaction without primers throughout for the reconstruction of full-length cDNA from products of rapid amplification of cDNA ends (RACE) Biotechnol Lett. 2011;33:1301–1307. doi: 10.1007/s10529-011-0580-1. [DOI] [PubMed] [Google Scholar]
- 30.Matsukura S, Jones PA, Takai D. Establishment of conditional vectors for hairpin siRNA knockdowns. Nucleic Acids Res. 2003;31:e77. doi: 10.1093/nar/gng077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, Wang L, Soda M, Kikuchi A, Igarashi T, Nakagawara A, Hayashi Y, Mano H, Ogawa S. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 33.Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V, Raynal V, Puisieux A, Schleiermacher G, Pierron G, Valteau-Couanet D, Frebourg T, Michon J, Lyonnet S, Amiel J, Delattre O. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 34.Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P, Laureys G, Speleman F, Kim C, Hou C, Hakonarson H, Torkamani A, Schork NJ, Brodeur GM, Tonini GP, Rappaport E, Devoto M, Maris JM. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, Takada S, Ueno T, Yamashita Y, Satoh Y, Okumura S, Nakagawa K, Ishikawa Y, Mano H. KIF5B-ALK, a novel fusion onco-kinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–3149. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 36.Ju YS, Lee WC, Shin JY, Lee S, Bleazard T, Won JK, Kim YT, Kim JI, Kang JH, Seo JS. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012;22:436–445. doi: 10.1101/gr.133645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, Sakamoto H, Tsuta K, Furuta K, Shimada Y, Iwakawa R, Ogiwara H, Oike T, Enari M, Schetter AJ, Okayama H, Haugen A, Skaug V, Chiku S, Yamanaka I, Arai Y, Watanabe S, Sekine I, Ogawa S, Harris CC, Tsuda H, Yoshida T, Yokota J, Shibata T. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, Curran JA, Balasubramanian S, Bloom T, Brennan KW, Donahue A, Downing SR, Frampton GM, Garcia L, Juhn F, Mitchell KC, White E, White J, Zwirko Z, Peretz T, Nechushtan H, Soussan-Gutman L, Kim J, Sasaki H, Kim HR, Park SI, Ercan D, Sheehan CE, Ross JS, Cronin MT, Janne PA, Stephens PJ. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, Asaka R, Hamanaka W, Ninomiya H, Uehara H, Lim Choi Y, Satoh Y, Okumura S, Nakagawa K, Mano H, Ishikawa Y. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 40.Paulson R, Jackson J, Immergluck K, Bishop JM. The DFer gene of Drosophila melanogaster encodes two membrane-associated proteins that can both transform vertebrate cells. Oncogene. 1997;14:641–652. doi: 10.1038/sj.onc.1200875. [DOI] [PubMed] [Google Scholar]
- 41.Lilien J, Arregui C, Li H, Balsamo J. The juxtamembrane domain of cadherin regulates integrin-mediated adhesion and neurite outgrowth. J Neurosci Res. 1999;58:727–734. doi: 10.1002/(sici)1097-4547(19991215)58:6<727::aid-jnr1>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]