Abstract
Merkel cell carcinoma (MCC) is an aggressive skin cancer with a high mortality rate. The majority of MCC (70–80%) harbor clonally integrated Merkel cell polyomavirus (MCV) in the tumor genome and express viral T antigen oncoproteins. The characterization of an early passage MCV-positive MCC cell line MS-1 is described, and its cellular, immunohistochemical, and virological features to MCV-negative (UISO, MCC13, and MCC26) and MCV-positive cell lines (MKL-1 and MKL-2) were compared. The MS-1 cellular genome harbors integrated MCV, which preserves an identical viral sequence from its parental tumor. Neither VP2 gene transcripts nor VP1 protein are detectable in MS-1 or other MCV-positive MCC cell lines tested. Mapping of viral and cellular integration sites in MS-1 and MCC tumor samples demonstrates no consistent viral or cellular gene integration locus. All MCV-positive cell lines show cytokeratin 20 positivity and grow in suspension. When injected subcutaneously into NOD scid gamma (NSG) mice, MS-1 forms a discrete macroscopic tumor. Immunophenotypic analysis of the MS-1 cell line and xenografts in mice show identical profiles to the parental tumor biopsy. Hence, MS-1 is an early passage cell line that provides a useful in vitro model to characterize MCV-positive MCC.
Keywords: Merkel cell carcinoma (MCC), Merkel cell polyomavirus (MCV), MS-1, Cell line, Xenograft model, T antigen
1. Introduction
Merkel cell carcinoma (MCC) is an aggressive cutaneous neoplasm with a rising incidence rate of ~1500 cases per year in the US (Lemos and Nghiem, 2007; Heath et al., 2008). MCC is more common in elderly, Caucasian populations, and risk factors for MCC include UV exposure as well as immunosuppression (Engels et al., 2002; Popp et al., 2002; Agelli and Clegg, 2003). MCC arises from mechanoreceptor Merkel cells of neuroendocrine origin, which are distributed sparsely in the basal layer of the epidermis (Halata et al., 2003; Van Keymeulen et al., 2009), and are responsible for touch and pressure sensation (Maricich et al., 2009). MCC resembles small cell lung carcinoma (SCLC) morphologically and by some immunophenotypical markers, given that these are both small round cell tumors that display neuroendocrine features (Leonard et al., 1995; Koljonen, 2006). Critical diagnostic markers used to distinguish MCC from SCLC and other neuroendocrine tumors include cytokeratin 20 (CK20) positivity as well as thyroid transcription factor (TTF-1) negativity (Koljonen, 2006). The current recommended treatment includes wide local resection followed by radiation therapy to the site of excision and local lymph nodes (Allen et al., 2005; Veness, 2005). There is a high recurrence rate of 30–40% and primary tumors frequently metastasize to distant body sites (Mercer et al., 1990).
The recently discovered Merkel cell polyomavirus (MCV) is causally associated with 70–80% of MCC as demonstrated in studies world-wide (Feng et al., 2008; Foulongne et al., 2008; Kassem et al., 2008; Becker et al., 2009; Busam et al., 2009; Paolini et al., 2011). MCV-induced MCC development is associated with viral integration into the tumor genome allowing constitutive expression of the viral oncoproteins, large tumor (LT) and small tumor (sT) antigens. T antigen proteins cannot be detected in other tumors or in healthy adjacent skin tissue in MCC patients (Shuda et al., 2009; Loyo et al., 2010; Schrama et al., 2011). In addition to LT and sT, the T antigen locus encodes a 57 kT splice isoform of unknown function with similar splicing pattern to the 17 kT isoform of simian vacuolating virus 40 (SV40) (Shuda et al., 2008). Tumor-derived LT proteins are truncated compared to wild-type LT (Shuda et al., 2008), and are defective in initiating viral origin replication due to loss of the C-terminal ATPase/helicase domain (Shuda et al., 2008; Kwun et al., 2009). MCV-positive MCC cell lines require T antigen expression for survival as assessed by T antigen knockdown studies (Houben et al., 2010) in several MCV-positive cell lines including MS-1, supporting an oncogenic role of MCV T antigens in Merkel cell carcinogenesis. MCV sT transforms rodent cells in vitro as assessed by focus formation and soft agar assays (Shuda et al., 2011). The transforming activity of sT in these assays is dependent on sT targeting of the 4E-BP1 translation regulator (Shuda et al., 2011).
Due to the limited availability of patient samples and the lack of an animal model, MCC cell lines are critical in investigating MCV-dependent Merkel cell carcinogenesis in vitro (Van Gele et al., 2004; Shuda et al., 2008; Fischer et al., 2010; Houben et al., 2010). Historically, MCC cell lines have been divided into classic and variant subtypes based on the expression of immunohistochemical markers including neuron specific enolase (NSE) and chromogranin A, as well as their neurosecretory granule status (Carney et al., 1985; Gazdar et al., 1985). Further division of MCC into subtypes I–IV has been based on SCLC morphology, aggregation, and colony shape in culture (Leonard et al., 1995). Subtypes I and II grow in dense, floating, spherical aggregates, with type I showing central necrosis (Leonard et al., 1995). Type III cells are aggregated loosely with a 2-dimensional appearance, and type IV cells grow in an adherent monolayer (Carney et al., 1985; Van Gele et al., 2004). Most MCV-positive cell lines studied to date are of the classic phenotype and have class III growth morphology in culture, while MCV-negative cell lines mostly fall into the variant class IV phenotype (Van Gele et al., 2004). However, some MCC cell lines diverge from this grouping, and the discovery of MCV in 2008 has provided a more meaningful classification of MCC cell lines to approach the molecular study of MCC.
Although a number of MCC cell lines have been established from MCC tumors (Leonard et al., 1995; Krasagakis et al., 2001; Fischer et al., 2010), detailed characterization of features associated with virus positivity, e.g. integration sites, viral sequence and truncation patterns, as well as viral protein expression in MCV-positive cell lines, has not been performed. In addition, many cell lines derived previously are adapted to cell culture conditions as a result of extended passages, which could lead to the accumulation of spontaneous mutations in the viral or cellular genomes. Such culture-derived changes can only be detected if the sequence of the parental tumor biopsy is known. The establishment of a new cell line allows the comparison of viral sequences as well as protein expression data from the MS-1 cell line to the patient tumor tissue from which it was derived. This aids in determining whether or not this cell line is an appropriate surrogate for its in vivo tumor. Comparison of MS-1 phenotypic and genotypic features to previously derived MCV-positive as well as MCV-negative MCC cell lines in this study supports that MCV-positive MCC cell lines are distinct from MCV-negative cell lines in their immunohistochemical profile as well as cell culture morphology. Additionally, MS-1 is tumorigenic in immunocompromised mice, forming xenograft tumors that retain T antigen expression as well as phenotypic markers of the cell line in vivo.
In summary, the MS-1 cell line is a well-characterized early passage MCC cell line, which manifests characteristics of the parental MCC tumor tissue, providing a valuable tool for studying MCV-induced MCC carcinogenesis in vitro. The MCV genome copy number, viral and host cell integration sites are described, as well as the LT antigen truncation pattern and viral protein expression in this cell line. The establishment of a MS-1 xenograft model that recapitulates features of the original tumor biopsy provides a novel tool for in vivo studies of MCV-positive MCC.
2. Materials and methods
2.1. Preparation of tumor biopsy and cell culture conditions
The MCC tumor biopsy (R08-05) was minced in RPMI 1640 with (filtered) 10% FBS, 1% Pen-Strep, 1% l-glutamine, and 1% MEM Hepes buffer. Digestion media (0.1% hyaluronidase, 0.02% DNase, 1.0% collagenase in PBS, Sigma–Aldrich, St. Louis, MO) was added, and the specimen was placed at 37 °C for 30–40 min. After incubation, the suspension was passed through a cell strainer to remove undigested tissue. Cells were initially cultured in RPMI medium with 20% human serum, 0.01% Pen Strep, 0.01% fungison, 0.01% insulin–transferrin–selenous-acid, 50 μM bathocuproine disulfate, and 1 M l-cystein. After 5 passages, serum was switched from 20% human to 20% fetal bovine (FBS), and following passage 10, serum concentration was reduced to 10% FBS in RPMI for culture. Experimental analyses in this study were performed using MS-1 cells of passage <40.
2.2. Real time quantitative PCR
Quantitative PCR was performed using primers amplifying the MCV T antigen promoter region (pTAg 98–184 nt; forward primer: 5′-CCC AAG GGC GGG AAA CTG-3′, reverse primer: 5′-GCA GAA GGA GTT TGC AGA AAC AG-3′) and VP2 locus (4563–4472 nt, forward primer: 5′-AGT ACC AGA GGA AGA AGC CAA TC-3′, reverse primer: 5′-GGC CTT TTA TCA GGA GAG GCT ATA TTA ATT-3′) with internal TaqMan probes (pTAg: 5′-CCA CTC CTT AGT GAG GTA GCT CAT TTG C-3′, VP2: 5′-CAG CAG CAA ACT CC-3′) labeled with FAM and BGH quencher for pTAg (Biosearch Technologies, Carlsbad, CA), and MGB quencher for VP2 (Applied Biosystems, Novato, CA). Standard curves for copy number calculations were generated for TAg and VP2 primers separately using values from serial dilutions of known copy numbers of the full-length MCV-HF genomic clone (Feng et al., 2011). Water was used as control and no evidence of PCR template contamination was observed. An RNase P primer probe mixture (Applied Biosystems) was used to determine cell genome copy number. qPCR reaction was performed in triplicates as described previously (Shuda et al., 2009). Results are expressed as numbers of viral copies per cell calculated from Ct values of viral and cellular gene standards (Table 2). To determine T antigen and VP2 mRNA expression levels, the expression of the human RNase P gene was used as a reference, and RNAs from MCV-negative cell lines were used as controls.
Table 2.
MCV genome copy number, and viral RNA expression.
| MCC cell line | MCV genome copies per cell |
Relative MCV RNA expressiona |
|||
|---|---|---|---|---|---|
| TAg | VP2 | TAg | VP2 | ||
| MCV negative | MS-1 | 2.1±0.1b | 3.6±0.9b | 0.069a | NDc |
| MKL-1 | 3.5±0.4b | 8.6±1.2b | 0.073a | NDc | |
| MKL-2 | 1.7±0.5b | 3.0±0.6b | 0.014a | NDc | |
| MCV positive | UISO | NDc | NDc | NDc | NDc |
| MCC13 | NDc | NDc | NDc | NDc | |
| MCC26 | NDc | NDc | NDc | NDc | |
Normalized to RNase P expression.
Standard deviation of three independent experiments.
Not detected.
2.3. MCV genome sequencing
Genomic DNA from MCV-positive MCC cell lines as well as the MS-1 tumor biopsy R08-05 was extracted by the standard phenol chloroform extraction method. 13 PCR primer sets (contig1 to contig13) were used for PCR amplification of the MCV genome as described before (Feng et al., 2008). All PCR reactions were performed with High Fidelity Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA). Standard Sanger sequencing of PCR products was performed using DNA analyzers from Applied Biosystems.
2.4. Phage library screen of MCV integration site
Ten micrograms of genomic DNA extracted from MKL-1 and MS-1 cells were digested with EcoRI (New England Biolabs, Ipswich, MA), and ligated into Lambda Zap II predigested EcoRI/CIAP-treated vector using the Gigapack III Gold Packaging Extract (Stratagene, Santa Clara, CA) to construct the phage library. After titer optimization, 7 μl of a 1:10 dilution of phage library mixed with 600 μl MRF’ cells at an OD600 of 0.500 were plated on NZY agar, and incubated at 37 °C overnight. Two rounds of screens of virus-positive clones were performed with α32P dCTP-labeled MCV DNA probes according to a published protocol (Feng et al., 2008). Positive clones were isolated, and inserts were sequenced.
2.5. RACE analysis
Both 5′-RACE and 3′-RACE were used to identify integration sites in MCC cases with GeneRacer Kit (Invitrogen) according to the manufacturer’s instructions. For MCC347/348 tumor cases, primer sequences were described previously (Feng et al., 2008). For MCC350, 5′-GCT AGA TTT TGC AGA GGT CCT G-3′ and 5′-TTT CCT TGG GAA GAA TAT GGA A-3′ primers were used to identify a viral integration site by 3′-RACE analysis. For MCC345, 5′-AAA CAA CAG AGA AAC TCC TGT TCC-3′ and 5′-TGA TCT TTC TGA TTA TCT TAG CCA TGC TG-3′ primers were used in 3′-RACE analysis. For MCC352, 5′-GGG AGG CTC AGG GGA GGA AA-3′ and 5′-TCC AGA GGA TGA GGT GGG TTC C-3′ primers were used in 5′-RACE analysis. PCR fragments were isolated from 1.2% agarose gel, extracted with QIAEX II Gel Extraction Kit (Qiagen, Valencia, CA), and ligated into pCR2.1 vector (Invitrogen) for DNA sequencing.
2.6. Southern blot
Genomic DNA was extracted by the standard phenol chloroform extraction method. Fifteen micrograms of genomic DNA from cell lines were digested with MCV genome single cut enzymes EcoRI and BamHI separately or together. Digested DNA was separated on 0.7% agarose–TBE gel, followed by denaturation for 45 min (1.5 M NaCl, 0.5 M NaOH), and neutralization for 1 h (3.0 M NaCl, 0.5 M Tris–HCl pH7.4). RNA was extracted using Trizol (Invitrogen). Genomic DNA was transferred onto a Hybond-C nitrocellulose membrane (GE Healthcare) with 10× SSC. To generate α32P dCTP-labeled probes for Southern hybridization, five MCV DNA fragments (contig1, contig3, contig6, contig9 and contig12) described previously (Feng et al., 2008) were used as templates for the Amersham Rediprime II Random Prime Labeling kit (GE Healthcare). Hybridization was performed as described previously (Shuda et al., 2008).
2.7. Immunoblotting
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer containing 50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% Na deoxycholate, 1% NP40, and protease inhibitors (Complete cocktail, Roche, Indianapolis, IN). Lysates were kept on ice for 15 min, and then centrifuged 20,000 × g for 10 min at 4 °C. Supernatants were mixed with 5× sodium dodecyl sulfate (SDS) loading dye, and denatured at 100 °C for 5 min. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), and transferred onto a Hybond-C nitrocellulose membrane (GE Healthcare). Proteins were detected using CM2B4, CM8E6, CM5E1 and 9B2 antibodies as described before (Shuda et al., 2009; Tolstov et al., 2009; Feng et al., 2011; Shuda et al., 2011). Anti alpha tubulin antibody (Sigma) was used to detect Tubulin as a loading control.
2.8. Xenograft model
Triple immunodeficient NOD scid gamma (NSG) mice (Jackson laboratories, Bar Harbor, MN) were housed in a specific pathogen-free area at the Division of Laboratory Animal Research (DLAR) facility at the Hillman Cancer Center, University of Pittsburgh according to University of Pittsburgh Institutional Animal Care and Use Committee (IACUC) regulations (IACUC protocol #1102226). At 6 weeks of age, mice were injected in the flank with 2 × 107 cells in 100 μl PBS subcutaneously. For MS-1 and MKL-1 cell lines, 5 mice were injected each. Mice were euthanized when tumors reached a diameter of 2 cm. Tumor volumes were calculated according to the following formula: (width)2 × (length/2).
2.9. Immunohistochemistry
For immunohistochemical staining of paraffin embedded xenograft tumor tissues, epitope retrieval was performed using 1 mM EDTA buffer pH 8.0 at 125 °C for 3:15 min after deparaffinization and hydrogen peroxide treatment. After blocking with Protein Block (Dako, Carpinteria, CA), primary antibody was applied for 30 min at room temperature with dilutions described below. After washing, samples were incubated with Mouse Envision Polymer (Dako) for 30 min at room temperature for subsequent deaminobenzidine (DAB) reaction. Antibodies and dilutions used for immunohistochemistry were: CM2B4 (1:1000; Santa Cruz, Santa Cruz, CA), CK20 (1:50; Dako), CK7 (1:50; Dako), chromogranin A (1:600; Dako), TTF-1 (1:50; Dako), AE1/3 (1:100; Dako), CAM5.2 (1:10; Becton Dickinson, Franklin Lakes, NJ), synaptophysin (1:100; Biogenex, Fremont, CA), NSE (pre-dilute; Ventana, Tucson, AZ), and LCA (pre-dilute; Ventana). Each cell line was stained once for every marker, and negative human control tissue was stained with the same antibody dilutions for each marker to ensure minimal background reactivity.
3. Results
3.1. MS-1 cell culture morphology and immunohistochemical analysis
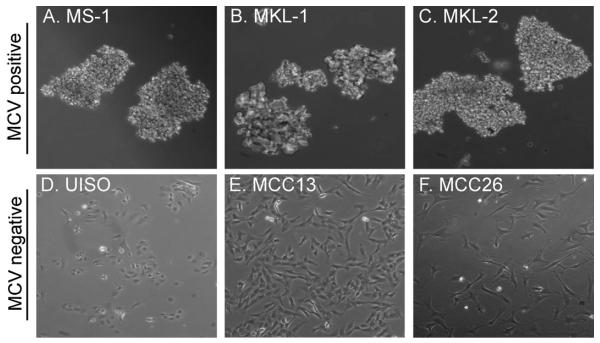
The MS-1 tumor biopsy was obtained from a MCC metastasis to the right adrenal gland of a 59-year-old woman. In culture, the cells form floating aggregates of 3-dimensional cell clusters similar to other MCV-positive MCC cell lines including MKL-1, and MKL-2 (Fig. 1). In contrast, the MCV-negative MCC cell lines UISO, MCC13, and MCC26 grow in an adherent monolayer (Fig. 1). As described before (Houben et al., 2010), immunohistochemical analyses of MCC cell lines reveal differences in various markers diagnostic for MCC between MCV-positive and -negative cell lines (Table 1). In comparison to data published previously (Houben et al., 2010), this immunohistochemical analysis of MCC cell lines is expanded to include MCC26, as well as the negative markers CK-7, TTF-1, and LCA. Cytokeratin 20 (CK20) is a low molecular weight cytokeratin with perinuclear distribution in MCC, which is used to distinguish MCC from other small cell carcinomas (Chan et al., 1997). MCV-positive cell lines MS-1, MKL-1 and MKL-2 are positive for CK20 by immunohistochemistry, whereas the MCV-negative cell lines UISO, MCC13 and MCC26 are not (Table 1). MS-1, MKL-1, MKL-2 and MCC13 are positive for the pan-keratin markers, AE1/3 (anion exchanger 1/3), and CAM5.2 (cytokeratin 8 and 18). Cytokeratin 7 (CK7), which has been reported to stain negative in MCC (Goessling et al., 2002), is positive only in MCC 13 tumor cells. All MCV-positive and –negative cell lines except MCC26 express neuron-specific enolase (NSE), and only MKL-2 expresses chromogranin A (Table 1). All cell lines are negative for leukocyte common antigen (LCA) and thyroid transcription factor 1 (TTF-1) (Table 1), markers that have been described as negative in MCC biopsies (Hanly et al., 2000; Goessling et al., 2002).
Fig. 1. Morphological features of MCC cell lines.
MCV-positive cell lines MS-1 (A), MKL-1 (B), and MKL-2 (C) grow in floating aggregates in cell culture, whereas MCV-negative cell lines UISO (D), MCC13 (E), and MCC26 (F) are adherent cells.
Table 1.
Immunohistochemical features of MCC cell lines.
| Markers | MCC diagnostic profilea,b | MCV positive |
MCV negative |
||||
|---|---|---|---|---|---|---|---|
| MS-1 | MKL-1 | MKL-2 | UISO | MCC13 | MCC26 | ||
| CK20 | + | + | + | + | − | − | − |
| AE1/AE3 | + | + | + | + | − | + | − |
| CAM5.2 | + | + | + | + | − | + | − |
| CK7 | − | − | − | − | − | + | − |
| Chromogranin A | ± c | − | − | + | − | − | − |
| Synaptophysin | ± c | + | − | + | − | − | − |
| NSE | + | + | + | + | + | + | − |
| LCA | − | − | − | − | − | − | − |
| TTF-1 | − | − | − | − | − | − | − |
Abbreviations: CK20, cytokeratin 20; AE1/AE3, pan-cytokeratin; CAM5.2, pan-cytokeratin; CK7, cytokeratin 7; NSE, neuron-specific enolase; LCA, leukocyte common antigen; TTF-1, thyroid transcription factor 1.
±, mostly positive.
3.2. MCV status
The MCV copy number per cell was determined by qPCR in three separate experiments (Table 2) for the MCV-positive cell lines MS-1, MKL-1, and MKL-2. MS-1 harbors an average of 2.1 copies of T antigen per cell, whereas MKL-1 and MKL-2 harbor 3.5 and 1.7 copies per cell respectively. No positivity was detected in the three MCV-negative MCC cell lines UISO, MCC13, and MCC26 used as controls. VP2 specific primers detect consistently higher genomic copy numbers than T antigen specific primers with MS-1 containing an average of 3.6 VP2 copies per cell. MKL-1 and MKL-2 contain 8.6 and 3.0 copies, respectively (Table 2).
In order to determine whether there is any correlation between viral DNA load and viral mRNA expression in the MCV-positive cell lines, total RNA was extracted from the MCC cell lines and treated with DNaseI to eliminate genomic DNA amplification. Quantitative RT-PCR was performed with the same primers used to determine T antigen and VP2 copy numbers per cell. All three MCV-positive cell lines express T antigen mRNA transcripts at different levels, whereas none express VP2 transcripts (Table 2). MS-1 mRNA levels are 0.069-fold RNaseP expression, whereas MKL-1 and MKL-2 are 0.073-, and 0.014-fold respectively (Table 2). There is no apparent evidence for a linear correlation between DNA copy number and mRNA levels.
Southern blot analysis using MCV probes that cover the entire viral genome (Feng et al., 2008), reveals a ~1.6 kb band in the MCV-positive cell lines MS-1, MKL-1, and MKL-2, when digested with the restriction enzymes BamHI and EcoRI. Both enzymes are single cutters within the MCV genome sequence, and double digestion cleaves a ~1.6 kb internal virus sequence (Fig. 2A). Consistent with qPCR results (Table 2), the intensities of viral internal 1.6 kb bands were similar between MS-1 and MKL-2, whereas MKL-1 shows a 1.6 kb band of higher intensity. Single digestions with either BamHI or EcoRI generate multiple MCV-related bands in MS-1 (BamHI), MKL-1 (EcoRI), and MKL-2 (EcoRI) consistent with random clonal integration in different cell lines, as has been shown in MKL-1 previously (Shuda et al., 2008). Single digestion with either BamHI or EcoRI yields a 5.4 kb band representing the whole MCV genome in MKL-1 and MKL-2, demonstrating virus concatenation in these cell lines.
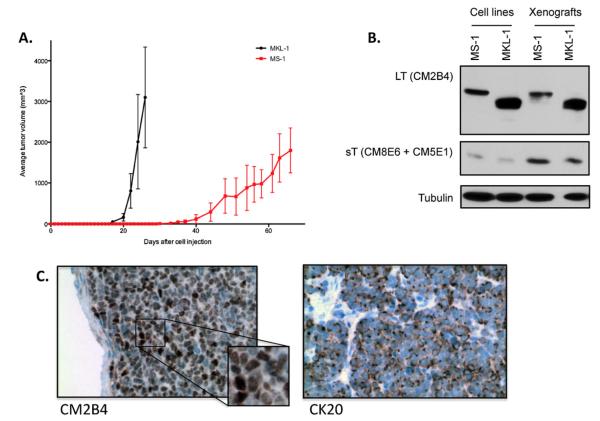
Fig. 2. MS-1 harbors clonally integrated MCV, and expresses viral T antigen proteins.
(A) Southern blot analysis of MCV-positive cell lines shows clonally integrated viral genomes. (B) The MCV-positive MCC cell lines MS-1, MKL-1 and MKL-2 have C-terminal LT truncations as determined by direct sequencing performed on genomic DNA. LT protein sizes correspond to 428 aa, 330 aa, and 275 aa, respectively. (C) Western blotting using antibodies CM2B4 for LT and CM8E6 as well as CM5E1 for sT shows differences in size of LT proteins from different MCV-positive MCC cell lines, whereas sT size is the same. The CM2B4 antibody picks up a faint non-specific band in MCV-negative MCC cell lines (marked *).
3.3. T antigen truncations and corresponding protein sizes
Next, we sought to determine the T antigen truncation pattern in MS-1 as well as MKL-2. The MKL-1 LT truncation (GenBank accession no. FJ173815.1) has been mapped previously (Shuda et al., 2008). Genomic DNA was isolated from MS-1, MKL-1 and MKL-2, and subjected to direct sequencing using MCV contiguous primer pairs spanning the whole MCV genome (Feng et al., 2008). Sequencing of the PCR products revealed premature stop codons in all three cell lines (Fig. 2B). MS-1 LT terminates at nucleotide position 1910, whereas the MKL-1 and MKL-2 LT truncations can be found at positions 1618 and 1495 respectively (Fig. 2B).
The differential LT truncation patterns of MCV-positive cell lines are reflected by varying LT migration on imunobloting (Fig. 2C). MS-1 expresses the largest LT protein among the MCV-positive MCC cell lines in this study, with the truncation occurring after the origin-binding domain (OBD) of LT, yielding a 428 amino acid (aa) long protein. MKL-1 (330 aa) and MKL-2 (275 aa) LT truncations are N-terminal to the MS-1 truncation mutation, while all three truncated LT proteins maintain the LXCXE Rb binding motif (Fig. 2B). This truncation pattern and conservation of the N-terminal motifs is consistent with LT sequences derived from MCC tumor tissues in various studies (Shuda et al., 2008; Sastre-Garau et al., 2009; Nakamura et al., 2010). In agreement with the truncation patterns in Fig. 2B, MS-1 expresses a 60 kDa truncated LT protein as compared to a 105 kDa full length LT expressed exogenously in 293 FT cells (Fig. 2C compare lane 3 to lane 2). MKL-1 and MKL-2 also express shorter forms of LT, which migrate at 50 kDa and 40 kDa, respectively. Even though sequencing results reveal intact splice donor and acceptor sites for the 57 kT isoform, neither MS-1 nor MKL-2 express a 57 kT protein that is detectable by immunoblotting (Fig. 2C). MKL-1 has already been reported to have a deletion in the second splice donor site of 57 kT (Shuda et al., 2008), which explains the absence of this isoform.
All MCV-positive cell lines consistently express an intact 19 kDa sT protein (Fig. 2C). While sT protein size does not differ between cell lines (Fig. 2B and C), sequencing results reveal one single nucleotide polymorphism, which results in a difference in amino acid 20 of sT between cell lines. MS-1 and MKL-1 have an alanine at this position, whereas MKL-2 has a serine.
Viral sequence comparison between the MS-1 cell line and its parental tumor biopsy (R08-05) reveals the same pre-mature stop codon mutation of LT as well as an identical MCV genome sequence with a deletion from nucleotide 1912 to 1951 and a 5 base pair insertion at position 5218 (Fig. S1). As described previously, the MKL-1 viral genome has a deletion at nucleotide position 1612–1657 (FJ173815.1). The MKL-2 genome has two deletions at nucleotide positions 3082–3083 and 4828–5348 (Fig. S1). Complete sequence alignments demonstrating single nucleotide polymorphisms between the different cell lines are included in the supplementary material (Fig. S2). MCV viral genome sequences have been deposited to GenBank with corresponding accession nos. JX045709 (MS-1) and JX045708 (MKL-2).
3.4. Viral genome integration site
To determine the integration site of the MCV genome within the cellular genome of MS-1, a lambda phage library was constructed from MS-1 genomic DNA and used for library screening. The MCV genome integration in MS-1 maps to a non-coding DNA region on human chromosome 5 (5q11.2). In the MCV genome, the integration event occurs after base pair 2843, which maps to the ATPase/helicase domain of LT (Table 3). Four MCC tumor samples (347/348, 345, 350, and 352), identified previously as MCV-positive (Feng et al., 2008), were tested for MCV integration sites by RACE, and no common cellular or viral integration sites were found (Table 3), consistent with previous studies (Sastre-Garau et al., 2009; Laude et al., 2010; Martel-Jantin et al., 2012).
Table 3.
Chromosomal integration sites in MCC.
| MCC | Type | Methods | Chromosome | Locusa | Gene | MCV breakpoint |
|---|---|---|---|---|---|---|
| MS-1 | Cell line | Phage library | 5q11.2 | 2,282,589 | Non-coding region | 2843 |
| 347/348 | Tumor tissue | 3’RACE | 3p14.2 | 61,561,985 | PTPRG (intron1) | 2140 |
| 345 | Tumor tissue | 3’RACE | 6p24.3 | 9,842,760 | MRDS1 (intron6) | 1836 |
| 350 | Tumor tissue | 3’RACE | 3q26.33 | 88,824,524 | Non-coding region | 596 |
| 352 | Tumor tissue | 5’RACE | 10q23.1 | 36,041,150 | Unidentified ORF | 491 |
GRCh37.5.
3.5. Tumorigenicity of MS-1 cells in vivo
The MCV-positive cell line MKL-1 (Rosen et al., 1987) has been shown to induce tumors in immunocompromised mice. The established MS-1 cell line was tested in NSG mice for in vivo growth. After injecting MS-1 cells subcutaneously in the flank, tumor formation was observed at injection sites after 30 days (Fig. 3A). This represents a 14-day delay in tumor onset as compared to MKL-1, and a slower growth behavior subsequently (Fig. 3A). The mice did not develop metastasis during the 80-day observation period, and were killed when the tumor reached a diameter of 2 cm. LT and sT protein sizes were unchanged in vivo as demonstrated by immunoblot (Fig. 3B). Nuclear localization of LT protein expression was maintained, as shown by immunohistochemistry (Fig. 3C), as was perinuclear distribution of CK 20 in the MS-1 xenograft tissue (Fig. 3C).
Fig. 3. MS-1 xenograft model.
Tumor volume graph represents average tumor volumes of 5 animals for each MS-1 and MKL-1 cell lines. (B) Immunoblot analysis of MS-1 and MKL-1 xenograft tumors shows LT as well as sT protein expression in vivo. Three separate xenograft tumor lysates were prepared for each cell line; one representative is shown here. (C) Immunohistochemical analysis of MS-1 xenograft tumors validates LT protein expression and nuclear localization (CM2B4), as well as CK20 positivity in vivo.
4. Discussion
Tumor-derived MCV-positive MCC cell lines represent a valuable tool for in vitro analyses of MCV related MCC carcinogenesis. For example, Houben et al. (2012) demonstrated that T antigen knockdown in MCV-positive MCC cell lines results in growth arrest and cell death underscoring the critical role of viral T antigen expression in MCC tumorigenesis.
The MS-1 cell line is a newly established MCC cell line that has been maintained in culture for few passages (<40 passages). MS-1 is positive for MCV, and displays a tumor specific C-terminal LT truncation pattern. Using RNase P as a cellular reference gene, T antigen copy number in MS-1 was determined by qPCR, to be 2.1 copies per cell, which could be explained by either multiple viral integration events or virus concatenation. At this point however, there is no experimental evidence that would suggest multiple integration events nor virus concatenation in MS-1. MCV-positive MCC cell lines MKL-1 and MKL-2 also have T antigen copy numbers >1 (Table 2), and Southern blot data (Fig. 2A) supports the presence of concatenated viral genomes in these cell lines. In addition, Southern blot analysis confirms higher genome copies in MKL-1 cells than MS-1 and MKL-2 cells, which is consistent with qPCR results. In agreement with our results, Fischer et al. have also described variations in MCV copy number per cell in other MCC cell lines (Fischer et al., 2010). However, the use of different reference genes for qPCR copy number determination has to be taken into account since cancer cells often harbor chromosomal aberrations. In MCC, specific chromosomal aberrations for chromosomes 1, 3–8, 10, 13, and 17 have been reported (Van Gele et al., 2002; Paulson et al., 2009). However, there is no reported data that suggest deletion or amplification of the RNase P reference gene, located on chromosome 14, in MCC.
T antigen copy number per cell corresponds to LT protein expression by immunoblot, which shows a stronger protein band for MKL-1 LT as compared to MS-1, which has a lower copy number of T antigen per cell compared to MKL-1. One explanation for the variations in LT expression levels observed between cell lines may be due to a difference in protein stability caused by different truncations. However, further studies are needed in order to investigate this. Even though VP2 copy numbers in the three MCV-positive MCC cell lines are higher than the T antigen, neither VP1 protein immunoblot (data not shown) nor quantitative RT-PCR on VP2 transcripts (Table 2) detect expression of late gene products. These results do not support one reported observation of viral particle production in MCC by electron microscopy (Wetzels et al., 2009). Instead, the results of this study suggest that late gene expression or viral particle production of MCV is absent in MCC tumors.
LT proteins expressed in the MCV-positive MCC cell lines migrate at higher molecular weights than the predicted sizes from their individual truncations. MS-1 LT is truncated after 428 amino acids and has a predicted molecular weight of 48 kDa, but it migrates at ~60 kDa on SDS-PAGE (Fig. 2B and C). MKL-1 and MKL-2 LT protein have predicted molecular weights of 37 kDa and 31 kDa, respectively, but they migrate at ~50 kDa, and ~40 kDa. Explanations for this difference in migration may include aberrant migration due to charge as well as post-translational modifications of LT such as phosphorylation or acetylation, both of which have been reported previously for T antigens of other polyomaviruses including murine polyomavirus middle T antigen and SV40 LT (Jansen et al., 1999b,c). Phosphorylation of LT in MKL-1 has been confirmed experimentally (data not shown), and results in higher molecular weight of LT on SDS-PAGE. Other modifications are likely, and require further experimental analyses.
The MCV integration site within the cellular genome of MS-1 as well as several other MCC tumor samples was determined. A common integration site could point toward the disruption of a cellular gene as a factor in MCC tumorigenesis. However, the MCC tumor samples including MS-1, did not show any common integration sites nor any interruptions of tumor suppressor genes (Table 3). Therefore, it is less likely that the interruption of a common host cell gene by viral integration contributes to tumorigenesis in most MCV-associated MCC.
Sequence comparison between the MS-1 cell line and its parental tumor biopsy reveals identical viral sequences and LT truncation patterns (Fig. S1). This suggests that additional mutations of the viral genome acquired during cell culture are not necessary to maintain the tumorigenic phenotype of MS-1 neither in culture nor in an in vivo xenograft setting. All three MCV-positive cell lines conserve the LXCXE Rb binding motif in LT as observed in MCC tumor-derived LT (Shuda et al., 2008). Experimentally, the importance of LT binding to Rb has been demonstrated by Houben et al. in vitro and in an in vivo xenograft model using the MCV-positive MCC cell lines WaGa and MKL-1 (Houben et al., 2012). However, another report demonstrates that MCV LT cannot transform rodent NIH 3T3 and Rat-1 cells in contrast to MCV sT, which alone is sufficient to transform rodent cells (Shuda et al., 2011). Therefore, LT’s Rb-targeting function may play an essential role in human cell transformation in concert with sT’s oncogenic function, which is expressed intact in all three MCC cell lines (Fig. 2C).
Morphological comparison between MCV-positive and - negative cell lines demonstrates a significant difference in appearance as has been described before (Houben et al., 2010). Immunohistochemical analysis of the MCV-positive and MCV-negative MCC cell lines shows that only the MCV-positive lines conform to diagnostic criteria that distinguish MCC tumors from other neuroendocrine cancers (Table 1). These results are in concordance with published data (Houben et al., 2010) for CK20, pan-CK as well as NSE markers in the MCC cell lines studied. In contrast, the immunohistochemical data for chromogranin, as well as synaptophysin presented in this study, show differences from published results (Houben et al., 2010) for the cell lines MS-1, MKL-1, and MCC13. These differences may be a result of different experimental techniques used, as Houben et al. determined their results by qPCR instead of immunohistochemistry.
Despite a report by Fischer et al. (2010) of the adherent MCV-positive MCC cell line MCCL12, all MCV-positive MCC cell lines tested in this study demonstrate a suspension growth pattern in culture. The MCV-negative cell lines used in this study are not only immunophenotypically different from MCC they also grow adherently in vitro. In SCLC, adherent cell growth is associated with loss of neuroendocrine marker expression and adherent growth (Gazdar et al., 1985). Despite the data on UISO, MCC13 and MCC26, the MCV-negative cell line MaTi, was found to grow in suspension similar to MCV-positive cell lines. However, MaTi neither expresses CK20 nor other cytokeratin markers similar to the MCV-negative cell lines studied by us. Among the MCV-negative MCC cell lines analyzed, one cell line, MCC26, lacks expression of all neuroendocrine markers examined. Those markers could be attenuated by de-differentiation of cells associated with tumor progression. However, the difference in growth morphology and the lack of CK20 positivity in UISO, MCC13, MCC26, and MaTi raises concern whether they were originally derived from correctly diagnosed MCC tumors. Alternatively, outgrowth of another cell population during the process of culturing is possible, and could explain the difference in morphology as well as phenotypic markers by immunohistochemistry. This is an important factor to take into account when using MCV-negative putative MCC cell lines as controls for MCV-positive MCC cell lines.
In vivo analysis of the MS-1 cell line confirms its tumorigenic growth potential by forming tumors in immunocompromised mice as has been shown in other MCC xenograft studies before (Jansen et al., 1999a; Schlagbauer-Wadl et al., 2000; Houben et al., 2012) MS-1 cells maintain viral T antigen expression and LT nuclear localization, as well as CK20 positivity in vivo, demonstrating the validity of this xenograft model with regard to MCC tumorigenesis. In vivo, MS-1 xenografts are less tumorigenic compared to MKL-1, with a 14-day delayed onset of tumor growth as well as a slower growth rate after tumor onset. In concurrence, xenograft models described in the literature for different cancer cell lines show varying onsets of tumor growth as well as differential levels of metastasis (Gerritsen et al., 1999; Hooijberg et al., 1999). It is possible that higher protein expression of LT in MKL-1 compared to MS-1 (Fig. 2C) is a factor contributing to increased tumorigenicity in vivo. Surprisingly, no metastatic growth was observed in mice injected with MS-1 despite the aggressive nature of MCC in humans, and the fact that MS-1 was derived from a metastasis.
The data presented in this study emphasizes that MS-1 and MCV-positive MCC cell lines in general, represent important tools to understand MCV biology and potential mechanisms of viral tumorigenesis in MCC. MS-1 in particular is a good cell culture model due to its low passage number, and well-characterized viral features. Importantly, MS-1 has identical features to the parental MCC tumor from which it was derived. To conduct translational research focusing on the viral etiology of MCC, having an accurate model system that reproduces the virus-associated features of clinical disease is essential. MS-1 is a cell line that provides these benefits and shows great potential in pre-clinical applications including drug screening of chemotherapeutic agents.
Supplementary Material
Acknowledgements
We thank the UPCI Skin Cancer SPORE for providing us with the MS-1 tumor biopsy, Marie Acquafondata, Susan Sharp and the UPMC Presbyterian Clinical IPEX Laboratory for immunohistochemical staining, Megan Lambert and Katie Leschak for help with animal care, Chrissie Usher for administrative assistance, and Reety Arora, Lindsay Dresang, Frank Jenkins and the Merrill Egorin Writing Group of UPCI for valuable discussion and review. This work was supported by SPORE P50CA121973 from the National Cancer Institute to J.M. Kirkwood, and National Institute of Health grants CA136363 and CA120726 to P.S. Moore and Y. Chang, who are also supported as American Cancer Society Research Professors. This project used the UPCI Animal Facility and was supported in part by award P30CA047904.
Abbreviations
- CK20
cytokeratin 20
- LCA
leucocyte common antigen
- LT
large T antigen
- MCC
Merkel cell carcinoma
- MCV
Merkel cell polyomavirus
- NSE
neuronspecific enolase
- NSG
NOD/Scid gamma
- SCLC
small cell lung carcinoma
- sT
small T antigen
- SV40
Simian vacuolating virus 40
- TAg
T antigen
- TTF-1
thyroid transcription factor
Footnotes
Appendix A. Supplementary data Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jviromet.2012.10.001.
References
- Agelli M, Clegg LX. Epidemiology of primary Merkel cell carcinoma in the United States. J. Am. Acad. Dermatol. 2003;49:832–841. doi: 10.1016/s0190-9622(03)02108-x. [DOI] [PubMed] [Google Scholar]
- Allen PJ, Bowne WB, Jaques DP, Brennan MF, Busam K, Coit DG. Merkel cell carcinoma: prognosis and treatment of patients from a single institution. J. Clin. Oncol. 2005;23:2300–2309. doi: 10.1200/JCO.2005.02.329. [DOI] [PubMed] [Google Scholar]
- Becker JC, Houben R, Ugurel S, Trefzer U, Pfohler C, Schrama D. MC polyomavirus is frequently present in Merkel cell carcinoma of European patients. J. Invest. Dermatol. 2009;129:248–250. doi: 10.1038/jid.2008.198. [DOI] [PubMed] [Google Scholar]
- Busam KJ, Jungbluth AA, Rekthman N, Coit D, Pulitzer M, Bini J, Arora R, Hanson NC, Tassello JA, Frosina D, Moore P, Chang Y. Merkel cell polyomavirus expression in Merkel cell carcinomas and its absence in combined tumors and pulmonary neuroendocrine carcinomas. Am. J. Surg. Pathol. 2009;33:1378–1385. doi: 10.1097/PAS.0b013e3181aa30a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney DN, Gazdar AF, Bepler G, Guccion JG, Marangos PJ, Moody TW, Zweig MH, Minna JD. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985;45:2913–2923. [PubMed] [Google Scholar]
- Chan JK, Suster S, Wenig BM, Tsang WY, Chan JB, Lau AL. Cytokeratin 20 immunoreactivity distinguishes Merkel cell (primary cutaneous neuroendocrine) carcinomas and salivary gland small cell carcinomas from small cell carcinomas of various sites. Am. J. Surg. Pathol. 1997;21:226–234. doi: 10.1097/00000478-199702000-00014. [DOI] [PubMed] [Google Scholar]
- Engels EA, Frisch M, Goedert JJ, Biggar RJ, Miller RW. Merkel cell carcinoma and HIV infection. Lancet. 2002;359:497–498. doi: 10.1016/S0140-6736(02)07668-7. [DOI] [PubMed] [Google Scholar]
- Feng H, Kwun HJ, Liu X, Gjoerup O, Stolz DB, Chang Y, Moore PS. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS ONE. 2011;6:e22468. doi: 10.1371/journal.pone.0022468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Brandner J, Fuchs F, Moll I, Grundhoff A. Detection of Merkel cell polyomavirus (MCPyV) in Merkel cell carcinoma cell lines: cell morphology and growth phenotype do not reflect presence of the virus. Int. J. Cancer. 2010;126:2133–2142. doi: 10.1002/ijc.24877. [DOI] [PubMed] [Google Scholar]
- Foulongne V, Kluger N, Dereure O, Brieu N, Guillot B, Segondy M. Merkel cell polyomavirus and Merkel cell carcinoma, France. Emerg. Infect. Dis. 2008;14:1491–1493. doi: 10.3201/eid1409.080651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Carney DN, Nau MM, Minna JD. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res. 1985;45:2924–2930. [PubMed] [Google Scholar]
- Gerritsen M, Jansen JA, Lutterman JA. Performance of subcutaneously implanted glucose sensors for continuous monitoring. Neth. J. Med. 1999;54:167–179. doi: 10.1016/s0300-2977(99)00006-6. [DOI] [PubMed] [Google Scholar]
- Goessling W, McKee PH, Mayer RJ. Merkel cell carcinoma. J. Clin. Oncol. 2002;20:588–598. doi: 10.1200/JCO.2002.20.2.588. [DOI] [PubMed] [Google Scholar]
- Halata Z, Grim M, Bauman KI. Friedrich Sigmund Merkel and his “Merkel cell”, morphology, development, and physiology: review and new results. Anat. Rec. A. 2003;271:225–239. doi: 10.1002/ar.a.10029. [DOI] [PubMed] [Google Scholar]
- Hanly AJ, Elgart GW, Jorda M, Smith J, Nadji M. Analysis of thyroid transcription factor-1 and cytokeratin 20 separates Merkel cell carcinoma from small cell carcinoma of lung. J. Cutan. Pathol. 2000;27:118–120. doi: 10.1034/j.1600-0560.2000.027003118.x. [DOI] [PubMed] [Google Scholar]
- Heath M, Jaimes N, Lemos B, Mostaghimi A, Wang LC, Penas PF, Nghiem P. Clinical characteristics of Merkel cell carcinoma at diagnosis in 195 patients: the AEIOU features. J. Am. Acad. Dermatol. 2008;58:375–381. doi: 10.1016/j.jaad.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooijberg JH, Broxterman HJ, Kool M, Assaraf YG, Peters GJ, Noordhuis P, Scheper RJ, Borst P, Pinedo HM, Jansen G. Antifolate resistance mediated by the multidrug resistance proteins MRP1 and MRP2. Cancer Res. 1999;59:2532–2535. [PubMed] [Google Scholar]
- Houben R, Adam C, Baeurle A, Hesbacher S, Grimm J, Angermeyer S, Henzel K, Hauser S, Elling R, Brocker EB, Gaubatz S, Becker JC, Schrama D. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int. J. Cancer. 2012;130:847–856. doi: 10.1002/ijc.26076. [DOI] [PubMed] [Google Scholar]
- Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, Moore PS, Becker JC. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J. Virol. 2010;84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen B, Heere-Ress E, Schlagbauer-Wadl H, Halaschek-Wiener J, Waltering S, Moll I, Pehamberger H, Marciano D, Kloog Y, Wolff K. Farnesylthiosalicylic acid inhibits the growth of human Merkel cell carcinoma in SCID mice. J. Mol. Med. (Berl.) 1999a;77:792–797. doi: 10.1007/s001099900052. [DOI] [PubMed] [Google Scholar]
- Jansen MP, Hopman AH, Bot FJ, Haesevoets A, Stevens-Kroef MJ, Arends JW, Jox A, Wolf J, Ramaekers FC, Schouten HC. Morphologically normal CD30-negative B-lymphocytes with chromosome aberrations in classical Hodgkin’s disease: the progenitor cell of the malignant clone? J. Pathol. 1999b;189:527–532. doi: 10.1002/(SICI)1096-9896(199912)189:4<527::AID-PATH488>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Jansen O, Dorfler A, Forsting M, Hartmann M, von Kummer R, Tronnier V, Sartor K. Endovascular therapy of arteriovenous fistulae with electrolytically detachable coils. Neuroradiology. 1999c;41:951–957. doi: 10.1007/s002340050875. [DOI] [PubMed] [Google Scholar]
- Kassem A, Schopflin A, Diaz C, Weyers W, Stickeler E, Werner M, Zur Hausen A. Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res. 2008;68:5009–5013. doi: 10.1158/0008-5472.CAN-08-0949. [DOI] [PubMed] [Google Scholar]
- Koljonen V. Merkel cell carcinoma. World J. Surg. Oncol. 2006;4:7. doi: 10.1186/1477-7819-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasagakis K, Almond-Roesler B, Geilen C, Fimmel S, Krengel S, Chatzaki E, Gravanis A, Orfanos CE. Growth and characterization of a cell line from a human primary neuroendocrine carcinoma of the skin (Merkel cell carcinoma) in culture and as xenograft. J. Cell. Physiol. 2001;187:386–391. doi: 10.1002/jcp.1086. [DOI] [PubMed] [Google Scholar]
- Kwun HJ, Guastafierro A, Shuda M, Meinke G, Bohm A, Moore PS, Chang Y. The minimum replication origin of Merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J. Virol. 2009;83:12118–12128. doi: 10.1128/JVI.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude HC, Jonchere B, Maubec E, Carlotti A, Marinho E, Couturaud B, Peter M, Sastre-Garau X, Avril MF, Dupin N, Rozenberg F. Distinct Merkel cell polyomavirus molecular features in tumour and non tumour specimens from patients with Merkel cell carcinoma. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Nghiem P. Merkel cell carcinoma: more deaths but still no pathway to blame. J. Invest. Dermatol. 2007;127:2100–2103. doi: 10.1038/sj.jid.5700925. [DOI] [PubMed] [Google Scholar]
- Leonard JH, Dash P, Holland P, Kearsley JH, Bell JR. Characterisation of four Merkel cell carcinoma adherent cell lines. Int. J. Cancer. 1995;60:100–107. doi: 10.1002/ijc.2910600115. [DOI] [PubMed] [Google Scholar]
- Loyo M, Guerrero-Preston R, Brait M, Hoque MO, Chuang A, Kim MS, Sharma R, Liegeois NJ, Koch WM, Califano JA, Westra WH, Sidransky D. Quantitative detection of Merkel cell virus in human tissues and possible mode of transmission. Int. J. Cancer. 2010;126:2991–2996. doi: 10.1002/ijc.24737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricich SM, Wellnitz SA, Nelson AM, Lesniak DR, Gerling GJ, Lumpkin EA, Zoghbi HY. Merkel cells are essential for light-touch responses. Science. 2009;324:1580–1582. doi: 10.1126/science.1172890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel-Jantin C, Filippone C, Cassar O, Peter M, Tomasic G, Vielh P, Briere J, Petrella T, Aubriot-Lorton MH, Mortier L, Jouvion G, Sastre-Garau X, Robert C, Gessain A. Genetic variability and integration of Merkel cell polyomavirus in Merkel cell carcinoma. Virology. 2012;426:134–142. doi: 10.1016/j.virol.2012.01.018. [DOI] [PubMed] [Google Scholar]
- Mercer D, Brander P, Liddell K. Merkel cell carcinoma: the clinical course. Ann. Plast. Surg. 1990;25:136–141. doi: 10.1097/00000637-199008000-00012. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Sato Y, Watanabe D, Ito H, Shimonohara N, Tsuji T, Nakajima N, Suzuki Y, Matsuo K, Nakagawa H, Sata T, Katano H. Nuclear localization of Merkel cell polyomavirus large T antigen in Merkel cell carcinoma. Virology. 2010;398:273–279. doi: 10.1016/j.virol.2009.12.024. [DOI] [PubMed] [Google Scholar]
- Paolini F, Donati P, Amantea A, Bucher S, Migliano E, Venuti A. Merkel cell polyomavirus in Merkel cell carcinoma of Italian patients. Virol. J. 2011;8:103. doi: 10.1186/1743-422X-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson KG, Lemos BD, Feng B, Jaimes N, Penas PF, Bi X, Maher E, Cohen L, Leonard JH, Granter SR, Chin L, Nghiem P. Array-CGH reveals recurrent genomic changes in Merkel cell carcinoma including amplification of L-Myc. J. Invest. Dermatol. 2009;129:1547–1555. doi: 10.1038/jid.2008.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp S, Waltering S, Herbst C, Moll I, Boukamp P. UV-B-type mutations and chromosomal imbalances indicate common pathways for the development of Merkel and skin squamous cell carcinomas. Int. J. Cancer. 2002;99:352–360. doi: 10.1002/ijc.10321. [DOI] [PubMed] [Google Scholar]
- Rosen ST, Gould VE, Salwen HR, Herst CV, Le Beau MM, Lee I, Bauer K, Marder RJ, Andersen R, Kies MS, et al. Establishment and characterization of a neuroendocrine skin carcinoma cell line. Lab. Invest. 1987;56:302–312. [PubMed] [Google Scholar]
- Sastre-Garau X, Peter M, Avril MF, Laude H, Couturier J, Rozenberg F, Almeida A, Boitier F, Carlotti A, Couturaud B, Dupin N. Merkel cell carcinoma of the skin: pathological and molecular evidence for a causative role of MCV in oncogenesis. J. Pathol. 2009;218:48–56. doi: 10.1002/path.2532. [DOI] [PubMed] [Google Scholar]
- Schlagbauer-Wadl H, Klosner G, Heere-Ress E, Waltering S, Moll I, Wolff K, Pehamberger H, Jansen B. Bcl-2 antisense oligonucleotides (G3139) inhibit Merkel cell carcinoma growth in SCID mice. J. Invest. Dermatol. 2000;114:725–730. doi: 10.1046/j.1523-1747.2000.00937.x. [DOI] [PubMed] [Google Scholar]
- Schrama D, Peitsch WK, Zapatka M, Kneitz H, Houben R, Eib S, Haferkamp S, Moore PS, Shuda M, Thompson JF, Trefzer U, Pfohler C, Scolyer RA, Becker JC. Merkel cell polyomavirus status is not associated with clinical course of Merkel cell carcinoma. J. Invest. Dermatol. 2011;131:1631–1638. doi: 10.1038/jid.2011.115. [DOI] [PubMed] [Google Scholar]
- Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernández-Figueras M, Tolstov Y, Gjoerup O, Mansukhani MM, Swerdlow SH, Chaudhary PM, Kirkwood JM, Nalesnik MA, Kant JA, Weiss LM, Moore PS, Chang Y. Human Merkel cell polyomavirus infection I MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int. J. Cancer. 2009;125:1243–1249. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J. Clin. Invest. 2011;121:3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, Chang Y, Buck CB, Moore PS. Human Merkel cell polyomavirus infection II MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int. J. Cancer. 2009;125:1250–1256. doi: 10.1002/ijc.24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gele M, Boyle GM, Cook AL, Vandesompele J, Boonefaes T, Rottiers P, Van Roy N, De Paepe A, Parsons PG, Leonard JH, Speleman F. Gene-expression profiling reveals distinct expression patterns for Classic versus Variant Merkel cell phenotypes and new classifier genes to distinguish Merkel cell from small-cell lung carcinoma. Oncogene. 2004;23:2732–2742. doi: 10.1038/sj.onc.1207421. [DOI] [PubMed] [Google Scholar]
- Van Gele M, Leonard JH, Van Roy N, Van Limbergen H, Van Belle S, Cocquyt V, Salwen H, De Paepe A, Speleman F. Combined karyotyping CGH and M-FISH analysis allows detailed characterization of unidentified chromosomal rearrangements in Merkel cell carcinoma. Int. J. Cancer. 2002;101:137–145. doi: 10.1002/ijc.10591. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Mascre G, Youseff KK, Harel I, Michaux C, De Geest N, Szpalski C, Achouri Y, Bloch W, Hassan BA, Blanpain C. Epidermal progenitors give rise to Merkel cells during embryonic development and adult homeostasis. J. Cell Biol. 2009;187:91–100. doi: 10.1083/jcb.200907080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veness MJ. Merkel cell carcinoma: improved outcome with the addition of adjuvant therapy. J. Clin. Oncol. 2005;23:7235–7236. doi: 10.1200/JCO.2005.02.2814. (author reply 7237–7238) [DOI] [PubMed] [Google Scholar]
- Wetzels CT, Hoefnagel JG, Bakkers JM, Dijkman HB, Blokx WA, Melchers WJ. Ultrastructural proof of polyomavirus in Merkel cell carcinoma tumour cells and its absence in small cell carcinoma of the lung. PLoS ONE. 2009;4:e4958. doi: 10.1371/journal.pone.0004958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.