Abstract
Hippocampal volumetry derived from structural MRI is increasingly used to delineate regions of interest for functional measurements, assess efficacy in therapeutic trials of Alzheimer’s disease (AD) and has been endorsed by the new AD diagnostic guidelines as a radiological marker of disease progression. Unfortunately, morphological heterogeneity in AD can prevent accurate demarcation of the hippocampus. Recent developments in automated volumetry commonly use multitemplate fusion driven by expert manual labels, enabling highly accurate and reproducible segmentation in disease and healthy subjects. However, there are several protocols to define the hippocampus anatomically in vivo, and the method used to generate atlases may impact automatic accuracy and sensitivity – particularly in pathologically heterogeneous samples. Here we report a fully automated segmentation technique that provides a robust platform to directly evaluate both technical and biomarker performance in AD among anatomically unique labeling protocols. For the first time we test head-to-head the performance of five common hippocampal labeling protocols for multi-atlas based segmentation, using both the Sunnybrook Longitudinal Dementia Study and the entire Alzheimer’s Disease Neuroimaging Initiative 1 (ADNI-1) baseline and 24-month dataset. We based these atlas libraries on the protocols of (Haller et al., 1997; Killiany et al., 1993; Malykhin et al., 2007; Pantel et al., 2000; Pruessner et al., 2000), and a single operator performed all manual tracings to generate de facto “ground truth” labels. All methods distinguished between normal elders, mild cognitive impairment (MCI), and AD in the expected directions, and showed comparable correlations with measures of episodic memory performance. Only more inclusive protocols distinguished between stable MCI and MCI-to-AD converters, and had slightly better associations with episodic memory. Moreover, we demonstrate that protocols including more posterior anatomy and dorsal white matter compartments furnish the best voxel-overlap accuracies (Dice Similarity Coefficient = 0.87–0.89), compared to expert manual tracings, and achieve the smallest sample sizes required to power clinical trials in MCI and AD. The greatest distribution of errors was localized to the caudal hippocampus and alveus-fimbria compartment when these regions were excluded. The definition of the medial body did not significantly alter accuracy among more comprehensive protocols. Voxel-overlap accuracies between automatic and manual labels were lower for the more pathologically heterogeneous Sunnybrook study in comparison to the ADNI-1 sample. Finally, accuracy among protocols appears to significantly differ the most in AD subjects compared to MCI and normal elders. Together, these results suggest that selection of a candidate protocol for fully automatic multi-template based segmentation in AD can influence both segmentation accuracy when compared to expert manual labels and performance as a biomarker in MCI and AD.
Keywords: Automatic Hippocampal Segmentation, Multi-atlas, Alzheimer’s disease, Hippocampal Tracing Protocol
Introduction
The hippocampus is one of the most extensively studied medial temporal lobe (MTL) structures in Alzheimer’s disease (AD), demonstrating early pathological atrophy (den Heijer et al., 2010; Jack et al., 2009) and association with episodic memory decline (Leung et al., 2010; Schuff et al., 2009). Importantly, hippocampal volumetry offers an attractive marker to quantify pathoanatomical changes and delineate functional changes across the continuum of AD progression (Dubois et al., 2007). It has been proposed for use in putative AD-modifying therapeutic trials, as both an in vivo marker of disease progression and to select candidate patients for study enrichment (Hampel et al., 2010). Moreover, radiological assessment of the hippocampus was recently endorsed by the new mild cognitive impairment (MCI) and AD diagnostic guidelines (McKhann et al., 2011).
Recent automated hipppocampal segmentation techniques are commonly based on a priori anatomical characteristics and encompass several strategies, which furnish volumetric and/or surface based information (Collins and Pruessner 2010; Hu et al., 2011; Leung et al., 2010; Lotjonen et al., 2011; Patenaude et al., 2011; Shen et al., 2012; Wang et al., 2011b). In particular, techniques using multi-atlas registration and fusion strategies generate some of the best accuracies among automated methods to-date (Aljabar et al., 2009; Collins and Pruessner 2010; Leung et al., 2010; Lotjonen et al., 2010; Wang et al., 2011b; Wolz et al., 2010a) Multi-atlas techniques use a series of structural MRIs that have been labeled by an expert operator (atlas library), which are then selectively registered to an unseen or query subject’s MRI. The technique is based on three principal steps that include (1) atlas-to-target (query) MRI similarity matching, (2) image registration with binary label mapping to the target MRI and (3) label fusion. This framework requires only a single 3-dimensional high-resolution T1-weighted MRI acquisition, and is suitable for segmenting the hippocampus from large prospective studies and legacy MRI datasets. Template libraries can be customized for pathological studies in epilepsy (Hammers et al., 2007) and AD (Leung et al., 2010), effectively capturing the significant morphological variation that occurs in both disease processes. Moreover, advances in diffeomorphic registration algorithms (Avants et al., 2008; Klein et al., 2009) may provide improved label mapping to target images compared to other registration techniques. Although multiatlas fusion methods are computationally more expensive than some automated techniques, improvements in server memory and parallel processing can significantly expedite segmentation.
There are several degrees of freedom within a multi-template based approach, which may impact accuracy including atlas assembly, template-to-target matching scheme, registration parameters (affine + nonlinear), label hybridization and false positive minimization. A number of recent methods have focused on improving atlas selection, label fusion strategy and post-fusion modifications, showing equivocal outcomes. For example, (Leung et al., 2010) compared accuracy across label combination methods for the Multi-Atlas Propagation Segmentation (MAPS) tool including shape-based average (SBA) (Rohlfing and Maurer 2007), voxel-wise voting and Simultaneous Truths and Probability Label Estimation (STAPLE) (Warfield, Zou, Wells 2004), with STAPLE achieving the best performance, although (Robitaille and Duchesne 2012) reported that SBA frequently outperformed both STAPLE and vote method. Techniques also use graphcuts and morphological operations to improve label mapping (van der Lijn et al., 2008). Other work has compared registration methods for subcortical segmentation, demonstrating that non-linear label propagation methods furnish greater accuracy than rigid and affine normalization (Barnes et al., 2008a; Leung et al., 2010).
Despite these advances, there remains significant variation in atlas construction among multi-template driven techniques, which are commonly developed and validated using in-house manual tracing datasets. The hippocampus has been historically defined using various cerebrospinal fluid (CSF), white matter (WM), grey matter (GM) and landmark-based boundaries, and can be labeled in various stereotactic spaces (e.g. normalization to brain templates and reorientation along either the anterior commissure – posterior commissure (AC-PC) line or the long hippocampal axis) (Boccardi et al., 2011). In fact a recent literature review by Konrad and colleagues identified 71 hippocampal tracing methods. Indeed, the absolute volume differences between certain protocols may vary by >30 percent (Konrad et al., 2009). Additionally, hippocampal atlas libraries use varying template numbers, combine tracings by multiple operators, include/exclude certain pathologies – all of which prevent direct performance assessments among protocols. In dementia the relative positions of anatomical landmarks can change in the atrophic sub-cortex, which may confound landmark driven delineation. When taken together, these issues complicate direct comparisons among techniques to determine an optimal definition for atlas-driven segmentation.
Accordingly, to directly measure the performance of different anatomical definitions for atlas-based segmentation, a study design should satisfy certain minimum requirements. First, a common pipeline should be used for label generation. Second, hippocampal labels must be derived from a common dataset (library); third, a single expert operator should label all template MRIs for consistency, and finally, these atlases should be validated against common datasets. Only a few studies in AD have compared different automated hippocampal segmentation methods using a common dataset (Holland et al., 2011; Leung et al., 2010; Mouiha, Duchesne, Alzheimer's Disease Neuroimaging Initiative 2011; Wolz et al., 2010b), although these studies were usually based on numeric summary data, unique algorithms and various structural priors. In addition, previous direct volumetric-based comparisons of the hippocampus have been limited by sample size and/or survey only a few methods (Carmichael et al., 2005).
To the authors’ knowledge, there have been no head-to-head morphometric comparisons of template protocols for multi-template hippocampal segmentation techniques that satisfy all of the abovementioned criteria and therefore performance across template protocols remains unclear. Thus, the primary goal of the current study is to directly evaluate whether morphological variation among 5 structurally unique and commonly deployed hippocampal labeling protocols modulates the accuracy and sensitivity of automated multi-atlas segmentation in AD, using the entire baseline and 24-month Alzheimer’s Disease Neuroimaging Initiative-1 (ADNI-1) MRI database (Weiner et al., 2010).
To investigate these relationships, we developed a fully automated multi-atlas segmentation technique that provides a robust platform to evaluate performance among anatomically unique labeling protocols. We refer to this method as the SunnyBrook Hippocampal Volumetry (SBHV) Tool. A single expert operator created 5 template libraries that were selected from 12 protocols investigated by the hippocampal harmonization initiative (Frisoni and Jack 2011): Protocol 1 (P1) (Haller et al., 1997); Protocol 2 (P2) (Killiany et al., 1993); Protocol 3 (P3) (Malykhin et al., 2007); Protocol 4 (P4) (Pruessner et al., 2000), and Protocol 5 (P5) (Pantel et al., 2000). This is a large multi-national project sponsored by the ADNI, the European Alzheimer Disease Consortium (EADC), non-profit organizations and industry partners that are working towards a consensus definition to manually label the hippocampus in-vivo (Frisoni and Jack 2011). A detailed description of the project methodology and results is available from www.hippocampal-protocol.net/SOPs. In the present study, all five template libraries were manually segmented from a set of 50 in-house acquired high-resolution T1-weighted MRI scans that included normal elderly controls (NC), AD, AD with small vessel disease (AD+SVD), vascular dementia (VaD) and mixed dementia (VaD+AD). All previously reported template libraries in AD are based on ADNI-1 data or other pure AD samples (Barnes et al., 2008b; Leung et al., 2010; Wang et al., 2011b; Wolz et al., 2010b). The rationale for including several diagnostic groups was to include more representative morphological variation and improve generalizability to a tertiary memory clinic cohort. Indeed, population-based studies suggest that AD and cerebrovascular disease (CVD) together account for 80 percent of dementia cases, with mixed AD plus superimposed CVD accounting for 38 percent in a community autoposy study (Schneider et al., 2007). Moreover, an ADNI-1 study by Carmichael and colleagues reported compelling evidence that white matter disease predicts 1-year cognitive decline in MCI and AD (Carmichael et al., 2010).
The specific objectives were to directly compare 5 commonly used hippocampal segmentation protocols within an automated multi-template fusion framework by (i) assessment of voxel-wise similarity to ground truth manual labels within and across diagnostic groups, (ii) compare differences in baseline volumes and baseline normalized atrophy rates, (iii) compare neurocognitive-anatomical correlations and (iv) compare differences among samples sizes required to detect a 25% reduction in the rate of MCI- and AD-type hippocampal atrophy in a hypothetical disease-modifying therapeutic trial. Voxel-wise accuracy was evaluated for each automated protocol using a leave-one-out cross-validation (LOOCV) analysis on a large in-house dataset and further cross-validated using a random selection of 30 manual tracings derived from NC, patients with MCI and patients with AD that participated in the ADNI-1 study. Finally, all available baseline and 24-month ADNI-1 datasets were segmented using the 5 automated methods. We chose 24-months as our longitudinal interval, as this has been previously recommend for measuring disease progression in AD (Jack et al., 2011).
Materials and Methods
1.0 Subjects
1.1 Sunnybrook
A total of 50 subjects’ 1.5 Tesla 3D high-resolution MRI scans were used to generate a library of MRI templates and were selected from over 1000 subjects participating in the longitudinal Sunnybrook Dementia Study (Pettersen et al., 2008), here on referred to as the Sunnybrook dataset. All subjects were recruited from the LC Campbell Cognitive Neurology Research Unit, Sunnybrook Health Sciences Centre at the University of Toronto. Patients underwent standardized clinical dementia assessments, including medical history and examination, blood tests, single-photon emission computed tomography, MRI, and neuropsychological testing. Alzheimer’s disease patients were diagnosed according to National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association criteria (McKhann et al., 1984) or for VaD according to the National institute of Neurological Disorders and Stroke-Canadian Stroke Network Vascular Cognitive Impairment Harmonization Standards (Hachinski et al., 2006). Small vessel disease was identified on MRI as silent lacunar infarcts (small hypointense (CSF isointense) regions on T1-weighted MRI), or as white matter hyperintensities that appear as punctate or diffuse regions of hyperintense signal on T2/PD and FLAIR MRI (Ramirez et al., 2011), or microbleeds on gradient echo (T2*) MRI. Normal controls were community-dwelling, healthy elderly volunteers with normal baseline neurocognitive test results. Demographic and co-morbid disease data were acquired on patients, including age, sex, years of education and vascular risk factors. The Sunnybrook Health Sciences Centre research ethics board approved the project and all participants or substitute decision maker provided informed consent.
1.2 ADNI-1
Certain clinical, demographic and T1-weighted MRI used in the preparation of this article were downloaded by the authors from the ADNI-1 database (adni.loni.ucla.edu) between September and November 2011. The ADNI-1 was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public- private partnership. The primary goal of ADNI-1 has been to test whether serial MRI, positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
2.0 MRI
2.1 Sunnybrook
Sunnybrook participant MRI scans were performed on a 1.5-Tesla Signa system (GE Healthcare, Chalfont St. Giles, England). The acquisition parameters for the T1-weighted 3-dimensional volumetric spoiled gradient echo sequence were 124 slices; matrix, 256 × 192; 22 × 16.5 cm FOV; number of excitations, 1; echo time/repetition time, 35 ms/5 ms; flip angle, 35°, and an in plane resolution of 0.859 × 0.859 mm slice thickness, 1.2–1.4 mm depending on head size.
2.2 ADNI-1 acquisitions
All ADNI-1 participants had high-resolution structural brain MRI scans acquired using a protocol developed for the study by Jack and colleagues (Jack et al., 2008b). Scans were acquired from 59 ADNI sites on 1.5 Tesla GE Health Care, Philips Medical Systems and Siemens MRI scanners. A 3D MP-RAGE scanning protocol was used that captured images in the sagittal plane with the following parameters, repetition time (TR) of 2400 ms, minimum full TE, inversion time (TI) of 1000 ms, flip angle of 8°, 24 cm field of view, 192 × 192 × 166 acquisition matrix (x, y and z dimensions), and a voxel size of 1.25 × 1.25 × 1.2 mm3. All MRI scans were evaluated for quality control.
3.0 MRI pre-processing
3.1 Sunnybrook
A rotation matrix was generated in ANALYZE software (Biomedical Imaging Resource, Mayo foundation, Rochester, MN, USA) by manual placement of landmarks, which were used to align the MRIs along the plane that intersected the anterior and posterior commissures (i.e. AC-PC line) (Ramirez et al., 2011). All raw T1-weighted template images were then reoriented using trilinear interpolation into AC-PC alignment by applying the manually generated rotation matrices and were additionally re-sliced into isotropic 0.86 mm3 voxels. (Ramirez et al., 2011). The open source FSL 4.1 distribution Brain Extraction Tool (BET) (Smith 2002) was used to extract the intra-cranial volume (ICV) for each subject by removal of the skull and infratentorial structures. In addition the –S and -B option were used to improve removal of the eye, optic nerves and to apply a bias field correction. The pre-processed skull stripped MRIs were then used for all further processing steps.
3.2 ADNI-1
All available post-acquisition corrected baseline (screening) and 24-month 1.5 Tesla T1-weighted ADNI-1 MRI data was downloaded from http://www.loni.ucla.edu/ADNI/Data/. Raw images were adjusted using a scheme that performed grad-warp correction of geometric distortion from gradient non-linearity (Jovicich et al., 2006), B1-correction, adjusting for inhomogeneity from B1 field non-uniformity (Jack et al., 2008b), N3 bias field correction (Sled, Zijdenbos, Evans 1998), and geometric scaling to remove scanner calibration errors using a phantom scan acquired for each participant.
4.0 Library creation
4.1 Template library subject selection
To generate a template library, fifty subjects’ T1-weighted MRI scans were selected a priori from the larger Sunnybrook dataset using a combination of each subject’s global Mini Mental State Exam (MMSE) score (Folstein, Folstein, McHugh 1975) and qualitative anatomical evaluation in three orthogonal planes using ANALYZE Software 10.0, Mayo Clinic, Rochester. The goal of the selection process was to generate a library that would deliver sufficient morphological variability within the MTL whilst matching subjects for age, education and gender (Table 1). We included NC, AD, VaD (including a few with hippocampal infarcts), AD + SVD and mixed dementia.
Table 1.
Subject demographics for the Sunnybrook Atlas Library and the total subject pool for the LOOCV (n=35) + LOO (n=15) optimization dataset. MMSE=Mini Mental State Exam, NC = Normal Control, AD = Alzheimer’s Disease, AD+SVD = Alzheimer’s disease and Small Vessel Disease, VaD = Vascular Dementia, Mixed AD = AD + VaD.
| NC | AD | AD+SVD | Mixed AD | VaD | Total | |
|---|---|---|---|---|---|---|
| N | 12 | 21 | 9 | 6 | 2 | 50 |
| Gender (M) | 6 | 12 | 4 | 5 | 1 | 28 |
| Average Age (SD) (years) | 67.6 (7.6) | 68.9 (10.4) | 75.4 (7.6) | 78.3 (7.8) | 79.5 (0.7) | 71.3 (9.5) |
| Average Education (SD) (years) | 17.5 (2.0) | 13.9 (3.7) | 11.2 (3.3) | 14.2 (2.5) | 14 | 14.2 (3.6) |
| Average MMSE Score (SD) (/30) | 29.4 (0.7) | 20.9 (5.8) | 20.9 (5.6) | 21.7 (4.2) | 22 (5.7) | 23.1 (5.9) |
4.2 Template tracing
We selected candidate hippocampal tracing methods from 12 commonly used methods that were previously vetted by the ADNI-EADC hippocampal harmonization project (Frisoni and Jack 2011). First, a detailed set of author endorsed standard operating procedures (SOPs) were downloaded from the harmonization project website: www.hippocampal-protocol.net. These procedures provided a detailed description and slice-by-slice delineation of the anatomical boundaries on a T1-weighted high resolution MRI of a normal elder and person with AD. All protocols in AC-PC space were selected to (1) control for potential confounds between different stereotactic orientations, (2) capture sufficient morphological variation between label methods and (3) reduce the number of manual tracings performed by a single operator (S.N.). For a detailed description of the five hippocampal SOPs refer to www.hippocampal-protocol.net/SOPs. The basic differences between protocols are summarized in Table 1. To improve label accuracy, the (Duvernoy 1998) hippocampal atlas was additionally used for neuroanatomical reference.
The hippocampal standardization project has harmonized semantic differences across methods and distilled the hippocampus into sub-compartments (Boccardi et al., 2011). The current study does not attempt to parcellate the hippocampus. In contrast, we adhered to the author endorsed SOPs to provide accurate delineation and allow comparison to previous studies. However, there were a number of author-supported modifications to the original manual methods that were annotated in the harmonization project SOPs; these modifications were applied to assemble the 5 atlas libraries. Most notably the volumes based on the criteria (i.e. P4) (Pruessner et al., 2000) were not pre-normalized to Talairach space, enabling direct comparison between protocols. Further, the label sets based on (Pantel et al., 2000) (P5) excluded the alveus and fimbria as per the author endorsed SOP.
The 50 T1-weighted MRI atlases were constructed using ANALYZE software. The same window-leveling procedure was used to ensure consistent contrast for labeling. Specifically, for each image the window-level was set until the choroid-plexus within the lateral ventricles was just visible in coronal section. The tracings were viewed in three orthogonal viewports (sagittal, coronal, and axial) to improve segmentation accuracy, prevent partial volume effects and insure inter-slice consistency. However, each template was traced principally in the coronal orientation. The axial plane was also used to assess the amygdalar-hippocampal boundary, and the sagittal plane was also used to detect boundaries where appropriate.
Each template MRI and associated binary hippocampal tracing was flipped along the x-coordinate so that a set of mirror image templates was produced according to previously published methods (Collins and Pruessner 2010). After the first set of tracings, labels were reviewed for quality control by an experienced neuroradiologist (FG).
5.0 Manual labeling inter/intra rater reproducibility
A single expert labeler (SN) produced all 5 100-atlas libraries. To ensure intra-rater label reproducibility, the same author traced the hippocampus bilaterally for each protocol on a random selection of 5 template MRIs. To test inter-rater tracing reproducibility, a second rater (FG) performed bilateral tracing on the same 5 Sunnybrook template MRIs for each protocol for a total of 50 labeled volumes. The second labeler is an experienced neuroradiologist with over 20-years of experience with manual tracing procedures. All intra- and inter-rater reliability coefficients (ICCs) were computed using a random two-way mixed effects design (Fleiss J.L. 1986) in SPSS 12.0 software, SPSS Incorporated. Voxel similarity between label sets was quantified using the Dice Similarity Coefficient (DSC). DSC = 2*((M ⋂ A)/(M + A)). Where M is the manually traced label set, A is the SBHV automatically derived label set and ⋂ is the intersection operator. The DSC provides a measure of voxel correspondence between two label sets and is commonly used to evaluate the accuracy of segmentation techniques. Associated interquartile ranges (IQR) were computed by the Matlab bootci function using 100,000 iterations with replacement, Matlab 14.0, MathWorks Incorporated.
6.0 Automated segmentation method
The SBHV segmentation scheme involved three principal steps including (1) template matching and selection, (2) atlas-to-target image registration with label mapping and (3) generating a consensus label set with intensity thresholding.
6.1 Template Matching and selection
To ensure that all templates comprising the library were aligned to a common space, the 100 BET skull-stripped template MRIs were affinely registered to the freely downloadable MNI 152 template with 1 mm3 isotropic resolution (Fonov et al., 2009; Fonov et al., 2011). Similarly, each query subject’s T1-weighted MRI underwent an affine transformation and was interpolated into MNI 152 template space. A local template matching strategy was applied for assessing similarity between each template and the query image over a predefined right and left volume of interest (VOI). The VOIs encompassed the entire hippocampal formation and adjacent MTL anatomy. Cross-correlation was used locally within the MTL VOIs to compare the voxel intensities of each template to the query image for similarity and ranking, which has been previously demonstrated as an appropriate similarity measure for the hippocampus (Aljabar et al., 2009) and has been applied to ADNI-1 data (Leung et al., 2010). The MNI-152 template VOIs were only used for template-query image similarity assessment and ranking. The highest ordered MNI normalized templates were indexed and the corresponding AC-PC T1-weighted MRIs and binary labels were nonlinearly propagated to the query image.
6.2 Atlas registration and label mapping
The ANTs registration toolkit was used for template-to-target registration and label mapping (Avants et al., 2008). First, an affine registration was applied to move the highest ranked AC-PC templates into the target (query) image space. Next, the ANTs Symmetric Normalization (SyN) algorithm, a deformable (nonlinear) algorithm was initialized on the affine transformed images and used to further register the templates to target space. The affine transform and nonlinear warp files were recovered and used to propagate the atlas hippocampal labels to the skull stripped target image using Nearest Neighbour interpolation. The ANTs software was downloaded from (http://www.picsl.upenn.edu/ANTS/).
The ANTs SyN parameters were optimized for intensity normalization using a multi-step hierarchical-resolution scheme with (60×100×5) number of iterations at each resolution level using histogram matching and cross correlation with a window radius of 2 and Gaussian regularization with sigma of 3 (Avants et al., 2008).
6.3 Label fusion and thresholding
A non-weighted vote-rule was implemented in Matlab 14.0, MathWorks Incorporated, to combine the best 15 intensity normalized and resliced binary templates into target image space. Only odd numbers of templates were selected for registration to target space to exclude potential ties. This method of label fusion has previously demonstrated high accuracy when compared to manual labels (Collins and Pruessner 2010).
A threshold window of 75–115% mean BET derived ICV intensity was used to exclude potential CSF and WM false positive labels based on the skull stripped T1-weighted MRI, and is similar to the method of (Barnes et al., 2008a; Leung et al., 2010). The selected upper mean intensity threshold did not exclude portions of the alveus/fimbria and occasionally excluded hyperintense voxels associated with WM of the parahippocampal gyrus. In a subset of subjects portions of the fornix were excluded. To ensure consistency across labeling methodologies, the same threshold was used for all templates. Finally, volumes for each protocol were computed for both left and right hippocampal volumes by multiplying voxel size by binary label count.
7.0 Method optimization
The SBHV segmentation pipeline was first trained on a random subset of 15 Sunnybrook subjects with bilateral manual hippocampal labels using a LOOCV design. This training dataset was separate from the larger Sunnybrook dataset used to cross-validate the SBHV method. Template matching, registration and thresholding steps were optimized using the DSC. Supplementary Figure 1 shows the protocol-wise improvement in median DSC value, as the number of best-matching templates fused together increased. Accuracy only incrementally improved after fusing 13–15 templates. Thus, in an effort to optimize processing time, only the highest ranked 15 templates were registered and propagated for all validation studies.
All volumes were processed on a Dell PowerEdge R710 rack-mount server with dual 6-core Intel Xeon X5680 CPUs at 3.33Ghz (12 physical cores – 24 cores with HyperThreading enabled), 16GB (8x2GB) 1333MHz DDR3 RAM, and two 146 GB 15K RPM SAS hard drives. This platform allowed >20 volumes to be computed simultaneously over a period of approximately 7 hours. Processing time was significantly reduced if bilateral target hippocampi generated similar template rankings.
8.0 Method Validation
8.1 Leave-one-out cross-validation dataset
To assess accuracy for each protocol, 35 subjects were selected from the Sunnybrook dataset and the automatically generated labels were compared to expert bilateral manual tracings using a LOOCV. These datasets were independent of the Sunnybrook dataset used to tune the method parameters. Voxel-wise accuracy between manual and automated labels was measured using the DSC. Further, volume-wise agreement between manual and automated volumes was assessed using the Normalized Volume Difference (NVD). NVD= 2*100*abs((MV − AV)/(MV + AV)). Where MV and AV are the manually labeled and SBHV automatically derived volumes respectively. ICCs were also computed to compare measurement agreement between SBHV and manually derived label sets. To ascertain protocol-wise differences between median DSC measurements, a Kruskal-Wallis Signed Rank Test was used in conjunction with post-hoc Mann-Whitney comparisons, Bonferroni corrected for 10 multiple comparisons.
8.2 ADNI-1 cross-validation dataset
To test the reproducibility of the automated protocols on an external dataset, two authors (SN and FG) traced a subset of randomly selected ADNI-1 participants including 10 NEC, 10 MCI and 10 AD. To reduce the number of manual tracings, three of the most morphologically different protocols from the Sunnybrook experiment, were used to cross-validate automated label accuracy including protocols 1, 2 and 4. The right hippocampus was traced for each subject/protocol (90 total manual segmentations). To ensure manual segmentation accuracy, both tracers reviewed all manual labels and corrected manual segmentations where appropriate. To ascertain voxel-wise similarity DSC was measured, and volume-wise similarity was determined using NVD. For both DSC and NVD associated 95% bias accelerated confidence intervals (BACI) were computed by the Matlab bootci function using 100,000 iterations with replacement, Matlab 14.0, MathWorks Incorporated. Kruskal-Wallis Signed Rank Tests were performed to test (1) group-wise DSC differences within each protocol, (2) protocol-wise DSC differences by group (NC, MCI and AD) and (3) differences among protocol-wise DSCs when collapsing across all groups. Exploratory Mann-Whitney post-hoc comparisons were performed when appropriate. In addition, Bland-Altman plots were constructed to test for volume-biases.
8.3 Qualitative analysis of automatic segmentation error maps
To specifically assess the voxel-wise distribution of label errors between protocols for SBHV versus manual labeling, false positive (FP) and false negative (FN) error maps were generated in a standardized template space. Briefly, a standard template was computed using SyN nonlinear registration, ANTs software, from 100 T1 MRIs selected from the Sunnybrook Longitudinal Dementia study (Pettersen et al., 2008). Subjects included both healthy elders and persons with AD. All AC-PC T1 MRIs from the LOOCV and ADNI-1 validation studies were nonlinearly registered to the Sunnybrook average brain atlas. False positive and FN binary images were generated for each subject and resliced into Sunnybrook template space using the nonlinear warp files. For each protocol, FP and FN error maps were generated in template space using a voxel counting method implemented in MatLab (Mathworks). Protocol-wise error maps were generated for the LOOCV dataset and were computed for each diagnostic group in the ADNI-1 validation sample. Only voxels with error counts >1 were visualized. Finally, a single observer (SN) visually assessed the maps.
9.0 Protocol-wise hippocampal biomarker performance applied to the entire ADNI-1 dataset
9.1 ADNI-1 cross-sectional and longitudinal group-wise comparisons
We reported results using absolute hippocampal volumes to allow comparison to previously published data. Hippocampal rates of atrophy were calculated by normalizing to baseline volume and scan interval using the following formula, ΔV=100*(baseline volume − 24 month volume)/(baseline volume)/(month scan interval/24). Results were not annualized, as dividing rates by 12-months did not affect sample sizes.
To test the effect of diagnostic group across all 5 protocols for both baseline volume and 24-month rate of change, a Multivariate Analysis of Covariance (MANCOVA) was calculated in SPSS 12.0 software, SPSS Incorporated. Group was entered as a fixed factor and the baseline volume or baseline-normalized 24-month rate of change for each protocol were used as dependent variables in the model. For group-wise analyses, the MCI group was trichotomized into persons with MCI that remained stable through 24-months (sMCI), persons with MCI that had a clinical conversion to AD through 24- months (cMCI) and persons with MCI that reverted from MCI to NC through 24-months (rMCI). For baseline volumes, ICV, age and gender were entered as nuisance variables, and age and gender were entered to adjust 24-month rate of change comparisons. Post-hoc one-way general linear models were computed to explore group-wise differences for each atlas protocol for baseline volume (corrected for ICV, gender and age) and 24-month percent change from baseline (corrected for gender and age). Post-hoc tests were treated as exploratory and not corrected for multiple comparisons.
9.2 ADNI-1 hippocampal volume and episodic memory associations
Baseline and 24-month longitudinal test scores were downloaded from ADNI-1 for two commonly utilized neuropsychological tests that are putatively associated with hippocampal-mediated episodic memory, including the Auditory Verbal Learning Test (AVLT) (Rey 1964) and the Logical Memory 1 (LM) exam (Wechsler 1981). The AVLT score was computed as the sum of trials 1–4 and the LM immediate recall total score was collected for each participant. Multiple-linear regressions using the enter method were calculated in SPSS 12.0 to compare the relationships between baseline and longitudinal memory measures and total hippocampal volumetry for the five protocols. For baseline regressions, age, gender and ICV were entered as nuisance variables, while age and gender were entered for longitudinal calculations.
9.3 ADNI-1 hippocampal derived sample size measures
Power calculations were generated to test the sensitivity of each protocol to measure a hypothetical reduction in the rate of atrophy (disease progression) in MCI and AD studies. Specifically, sample sizes were derived from MCI and AD ADNI-1 data to detect a 25 percent reduction in the 24-month rate of hippocampal atrophy in comparison to a hypothetical placebo group. Sample size = (u + v)2 (2σ2)/(Δμ)2, where u=0.841 (80% power), v=1.96 (5% significance level), Δμ is the change in baseline and scan interval normalized atrophy between groups, and σ is the SD of rates of atrophy in the treatment and placebo groups (Fox et al., 2000). A further calculation accounted for the average rate of hippocampal atrophy in normal aging (adjusted sample sizes) by subtracting the protocol-wise ADNI-1 NC baseline-normalized rate of atrophy from corresponding MCI and AD rates. Holland et al. recently demonstrated NC-adjusted power calculations to be a more valid estimate of sample size estimates for trials assessing amyloid lowering therapies (Holland et al., 2011). We report both non-adjusted and NC-adjusted sample size calculations. For each sample size, 95% BACIs (100,000 iterations with replacement) were computed.
Results
Demographics for the Sunnybrook LOOCV are available in Table 2, and for the ADNI-1 experiment baseline demographic data is reported in Table 3. There was considerable range in the period between baseline/screening and 24-month follow-up, although we normalized all volumetric rates of change for scan interval.
Table 2.
A basic outline of major anatomical hippocampal boundaries for 5/12 protocols surveyed by (Boccardi et al., 2011) and used to generate 5 SBHV multi-atlas libraries. Landmarks definitions were adapted from Standard Operating Procedures (www.hippocampal-protocol.net/SOPs) and the original protocol manuscripts.
| Protocol (P) Code |
Segmentation Method |
Hippocampal Boarder | Excluded Anatomy |
|||||
|---|---|---|---|---|---|---|---|---|
| Superior | Inferior | Medial | Lateral | Posterior | Anterior | |||
| P1 | (Haller et al., 1997) | Fx- Fb- or Av-GM interface, IH, AC | WM of PHG and anteriorly: PHG WM/US* | Tail/Body of the Hc: contour of PHG WM extended by horizontal line towards the AC. Anterior: following incline of PHG WM | Fx, IH, WM of MTL | First slice where Hc appears adjacent to VT | Separation of Hc and Ag (use axial and sagittal views to distinguish) | Fx, Fb, Av, PHG (includes some of the superior-medial PHG) |
| P2 | (Killiany et al.,1993) | Fx- Fb- or Av-GM boundary, IH, AC | WM of PHG and anteriorly: PHG WM/US* | Posterior/body Hc an oblique line along GM-WM of PHG extending to the AC, and at head of HC following incline of PHG WM | Fx, IH, WM of MTL | Longest length of the Crus of the Fx in coronal view | Slice where the Av differentiates Ag from Hc | Fx, Fb, Av, PHG and portions of medial Sb at the level of the body |
| P3 | (Malykhin et al., 2007) | Fx, Th, AC, Av (most anterior slices) | WM of PHG and anteriorly: PHG WM/US* | WM of PHG, and when PHG not a horizontal line used an oblique line along PHG WM extending to the AC | Fx, VT, IH, WM of MTL | First slice where an ovoid mass of Hc GM is inferiomedial to VT | Slice along anterior-posterior axis (sagitally) where PHG WM is first visible | Fx and portions of both the Sb and PHG (when PHG WM not a straight line) |
| P4 | α(Pruessner et al., 2000) | Posterior: Horizontal line that follows the superior PHG WM extending to the AC. Otherwise CSF of QC, IH or Av (most anterior slices) | WM of PHG and anteriorly: PHG WM/US* | Posterior Hc: Vertical line that follows medial edge of VT, otherwise CSF of AC. Body Hc: 45° line from inferior body to the AC. Head Hc: CSF of AC. | Fx, VT, IH, WM of MTL | First ovoid mass of GM inferiomedial to VT | Slice where either the IH, Ag or Av is present (used axial view to help interpret) | Portions of the medial Sb, Medial GM of PHG at level of body, Fx |
| P5 | β(Pantel et al.,2000) | Th, AC, Av (most anterior slices) | WM of PHG and anterio WM/US* | Posterior Hc: QC, Body Hc: Contour of PHG-WM. Anterior Hc: oblique line following PHG WM | Fx, VT, IH, WM of MTL | First ovoid mass of GM inferiomedial to VT | Slice at which head of Hc appears as oval shape below Ag | |
A pre-processing step in the protocol normalizes brains to Talairach space, which was not performed in the current study.
The updated standardized operating procedure for the hippocampal harmonization procedure includes the alveus and fimbria, which are excluded in the original protocol.
The US commonly widens and becomes a visible landmark as a hypointense band on T1-weighted MRI in atrophic brains.
Key: PHG = parahippocampal gyrus, CSF = cerebrospinal fluid, WM = white matter, Av = alveus, AC = Ambient Cistern, EC = Entorhinal Cortex, IH = Inferior horn of lateral ventricle, Ag=Amygdala, Hc = Hippocampus, MTL = Medial Temporal Lobe, VT = Lateral Ventricular Trigone (also includes atrium of lateral ventricle), Uncal Sulcus = US, Vertical Digitation = VD, Fornix = Fx, Fimbria = Fb, Thalamus = Th, QC= Quadrigeminal Cistern
Table 3.
Demographic details of the complete baseline Alzheimer’s Disease Neuroimaging Initiative-1 dataset used to compute hippocampal volumes. Twenty-four month follow-up data are also reported. MCI = Mild Cognitive Impairment, MCI converters (cMCI) are ADNI1 participants that converted from a diagnosis of MCI to AD through a 24-month study window, while MCI reverters (rMCI) are participants that reverted from a clinical evaluation of MCI to being normal elders. MCI stable or sMCI are subjects that had a diagnosis of MCI that did not change over a 24-month period. MMSE=Mini Mental State Exam, LM1 – Logical Memory Test 1 Immediate Recall, AVLT = Auditory Verbal Learning Test (Sum of trials I–IV), ICV = intracranial volume.
| NC | MCI Total | MCI Converters | MCI Reverters | MCI Stable | AD | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N (baseline) | 227 | 412 | 134 | 13 | 262 | 200 | ||||||
| N (24 month follow-up) | 173 | 254 | 95 | 10 | 148 | 111 | ||||||
| Average Age (SD) (Years) | 76.0 | (5.0) | 74.7 | (7.4) | 74.4 | (7.2) | 73.5 | (9.0) | 75.0 | (7.5) | 75.6 | (7.7) |
| Average Education (SD) (years) | 16.1 | (2.8) | 15.7 | (3.0) | 15.7 | (2.9) | 15.8 | (2.5) | 15.7 | (3.1) | 14.7 | (3.2) |
| Gender (M) | 118 | 262 | 81 | 9 | 172 | 103 | ||||||
| Average MMSE Score (SD) (/30) | 29.12 | (1.00) | 27.07 | (1.89) | 26.65 | (1.73) | 27.54 | (1.39) | 27.17 | (1.81) | 23.28 | (2.04) |
| Average Scan Interval (SD) (months) | 24.7 | (1.3) | 24.6 | (1.1) | 24.6 | (1.1) | 24.4 | (1.8) | 24.6 | (1.0) | 24.7 | (1.6) |
| Average ICV SD) (ml) | 1329.8 | (132.3) | 1346.3 | (137.0) | 1327.2 | (147.0) | 1398.1 | (164.1) | 1353.4 | (129.4) | 1311.9 | (146.6) |
1.0 Manual labeling inter/intra-rater reproducibility
The inter- and intra-rater reproducibility of each hippocampal protocol was excellent and is reported in Table 4. Intra-labeler reproducibility was better than inter-labeler absolute agreement, with lower variation among datasets. This was also reflected by high DSC values across protocols with slightly better intra-labeler DSC values. There were no major differences in reproducibility across protocols with the exception of P2, which slightly underperformed when compared to the other protocols. Although P1 required additional delineation to excise the dorsal WM compartment, it demonstrated comparable results to P3–P5, which included these structures. The reproducibility of the fully automated multi-atlas method for each protocol was unity.
Table 4.
Inter- (n=5) and intra-rater (n=10) Dice Similarity Coefficients (DSC) and interquartile ranges (IQR), intraclass correlation coefficients (ICC) and 95% confidence intervals (CI) for bilateral manual hippocampal tracings on Sunnybrook 1.5T MRI scans. P1= (Haller et al., 1997), P2= (Killiany et al., 1993), P3= (Malykhin et al., 2007), P4= (Pruessner et al., 2000) and P5= (Pantel et al., 2000).
| Atlas Protocol |
Inter-rater | Intra-rater | ||||||
|---|---|---|---|---|---|---|---|---|
| DSC | IQR | ICC | 95% CI | DSC | IQR | ICC | 95% CI | |
| P1 | 0.90 | 0.01 | 0.96 | (0.72, 0.99) | 0.92 | 0.01 | 0.97 | (0.88, 0.99) |
| P2 | 0.91 | 0.01 | 0.92 | (0.51, 0.99) | 0.92 | 0.01 | 0.95 | (0.81, 0.99) |
| P3 | 0.91 | 0.01 | 0.94 | (0.47, 0.99) | 0.92 | 0.01 | 0.97 | (0.90, 0.99) |
| P4 | 0.91 | 0.01 | 0.95 | (0.52, 0.99) | 0.93 | 0.02 | 0.96 | (0.74, 0.99) |
| P5 | 0.91 | <0.01 | 0.95 | (0.70, 0.99) | 0.92 | 0.01 | 0.96 | (0.76, 0.99) |
2.0 Method validation
2.1 DSC manual Vs. automatic segmentation accuracy
2.1.1 Sunnybrook optimization dataset and LOOCV dataset
No manual corrections were performed to the automated segmentations for any of the analyses in the current study. Figure 1 shows 3D renderings of P1–5 manual and corresponding automated hippocampal volumes for a single subject’s right hippocampus, acquired from the Sunnybrook study. Table 5 shows voxel-wise DSC accuracy and ICC results across protocols for the algorithm-training step (N=30 templates) used to optimize all parameters in the automatic pipeline. For the LOOCV experiment (Table 5), the median DSCs (IQR) in order of protocol inclusivity (greatest to least inclusive) were P5= 0.88 (0.02), P3= 0.88 (0.02), P4=0.88 (0.03), P1=0.86 (0.04) and P2=0.85 (0.04). ICCs were high for all protocols for the LOOCV (Table 5). After correction for 10 nonparametric protocol-wise comparisons, P2 and P1 had significantly lower DSCs in comparison to the more inclusive P3–P5 (p<0.05), while there were no significant differences between P1 and P2 DSC measurements. Moreover, P1 and P2 demonstrated higher variation in DSC when compared to the more inclusive atlas methods P3–P5.
Figure 1.
3D rendered right hippocampal volumes for protocols 1–5, of a single Sunnybrook Longitudinal Dementia Study participant with a clinical diagnosis of AD, displaying in dorsomedial orientation with anterior/head of the hippocampus (forward), medial surface (right) and superior surface (top). The top panel shows the manually labeled hippocampus whereas the bottom hippocampus corresponds to the SBHV automatically derived volume using 15 fused templates per protocol. P1=(Haller et al., 1997), P2= (Killiany et al., 1993), P3= (Malykhin et al., 2007), P4= (Pruessner et al., 2000) and P5 = (Pantel et al., 2000). Image rendered in ITK-Snap (Yushkevich et al., 2006).
Table 5.
Voxel similarity measured using the Dice Similarity Coefficient (DSC) and associated interquartile range (IQR) between manual and SBHV automatic bilateral hippocampal labels for the Sunnybrook automated pipeline leave-one-out optimization dataset (template n=30), and Sunnybrook LOOCV dataset (template n=70). Values reported are based on the automated labels that were generated by nonlinear registration to the target space and label fusion of the 15 best matching MRI templates. P1= (Haller et al., 1997), P2=(Killiany et al., 1993), P3=(Malykhin et al., 2007), P4=(Pruessner et al., 2000) and P5=(Pantel et al., 2000).
| Atlas Protocol |
Median DSC | IQR | ICC | 95% CI |
|---|---|---|---|---|
| Sunnybrook Optimization Experiment | ||||
| P1 | 0.87 | 0.05 | 0.93 | (0.56 – 0.98) |
| P2 | 0.85 | 0.04 | 0.86 | (0.42 – 0.96) |
| P3 | 0.89 | 0.03 | 0.94 | (0.79 – 0.98) |
| P4 | 0.88 | 0.03 | 0.94 | (0.60 – 0.98) |
| P5 | 0.89 | 0.03 | 0.93 | (0.76 – 0.98) |
| Sunnybrook LOOCV Experiment | ||||
| P1 | 0.86 | 0.04 | 0.92 | (0.72 – 0.96) |
| P2 | 0.85 | 0.04 | 0.88 | (0.40 – 0.96) |
| P3 | 0.88 | 0.02 | 0.94 | (0.80 – 0.99) |
| P4 | 0.88 | 0.03 | 0.91 | (0.54 – 0.97) |
| P5 | 0.88 | 0.02 | 0.93 | (0.72 – 0.97) |
2.1.2 ADNI-1 cross-validation dataset
Median DSCs were modestly improved by approximately 1–3 percentage points across all protocols for all groups in comparison to the LOOCV (Table 6). Moreover, ICC values were comparable to the LOOCV when volumes were pooled across groups (Table 6), and ICCs were lower in both NC and MCI versus AD. When groups were pooled, the median DSCs for all protocols were significantly different (p<0.001) and post-hoc comparisons ranked accuracy: P4>P1>P2 (P4 vs. P1: p<0.024, P1 vs. P2: p<0.001 and P4 vs. P2: p<0.001). Interestingly, when protocol-wise comparisons were performed within each diagnostic group, only the AD group showed significant differences among all protocols (p<0.001). In NC, post-hoc tests showed only significant median DSC differences between P2 and P4 (p<0.001), while in MCI significant differences were only realized between P2 and P4 (p<0.001)..For each protocol individually, there were no significant differences for DSC measurements between NC, MCI or AD groups, with the exception of P4 (p=0.008). For P4, the AD group had significantly higher DSC measurements than normal elders (p<0.001).
Table 6.
Median voxel similarity measured using the Dice Similarity Coefficient (DSC) and interquartile range (IQR) between manual and optimized automated right hippocampal labels for the ADNI-1 cross-validation experiment (N=30). Manual and automated labels were generated for P1, P2 and P4 from a random sample of ADNI-1 normal controls (NC) (N=10), persons with mild cognitive impairment (MCI) and Alzheimer’s Disease (AD). P1= (Haller et al., 1997) P2= (Killiany et al., 1993) and P4= (Pruessner et al., 2000).
| Dx Group | Atlas Protocol |
DSC (Median) |
IQR | ICC | 95% CI |
|---|---|---|---|---|---|
| NC (n=10) | P1 | 0.88 | 0.02 | 0.88 | (0.32–0.97) |
| P2 | 0.86 | 0.02 | 0.87 | (0..03–0.98) | |
| P4 | 0.89 | 0.02 | 0.86 | (0.24–0.97) | |
| MCI (n=10) | P1 | 0.89 | 0.04 | 0.81 | (0.01–0.96) |
| P2 | 0.88 | 0.02 | 0.72 | (0.06–0.94) | |
| P4 | 0.90 | 0.03 | 0.89 | (0.02–0.98) | |
| AD (n=10) | P1 | 0.90 | 0.02 | 0.97 | (0.66–0.99) |
| P2 | 0.88 | 0.03 | 0.94 | (0.06–0.99) | |
| P4 | 0.91 | 0.01 | 0.95 | (0.50–0.99) | |
| All Groups | P1 | 0.89 | 0.02 | 0.93 | (0.49–0.98) |
| (n=30) | P2 | 0.87 | 0.03 | 0.89 | (0.05–0.97) |
| P4 | 0.90 | 0.02 | 0.93 | (0.43–0.98) | |
2.2 Manual Vs. automatic segmentation volumetric differences
2.2.1 LOOCV dataset
The average SBHV hippocampal volumes for P1–5 are reported in Table 7, and when protocols were ordered from greatest to least, were ranked P5>P3>P4>P1>P2, which was congruous with the anatomical definitions described in Table 1. The Bland-Altman plots in Figure 2 show that there was a small volume-bias in the LOOCV across atlas protocols. Specifically, SBHV modestly underestimated the ground truth volume on average. Moreover, there appeared to be a small bias towards the mean, as the smaller hippocampi were consistently larger than the manual segmentations, with the exception of P4. Table 7 shows that the median NVD was low across all protocols for the LOOCV analysis (range: 5.05–7.96%). There was a trend towards lower NVDs as protocols integrated more hippocampal anatomy. Indeed, for the LOOCV dataset median (IQR) NVDs when ranked from the greatest to the least inclusive protocol were P5 = 5.14 (7.58), P3 = 5.05 (6.96), P4 = 6.43 (9.82), P1 = 6.23 (7.25) and P2 = 7.96 (8.77).
Table 7.
Summery of median absolute total (left + right) SBHV automatic (Auto) and manually (Man) derived volumes with associated interquartile regions (IQR) for the Sunnybrook LOOCV. Volume differences between automated and “ground-truth” manual labels are reported as median Normalized Volume Differences (NVD) with a bootstrap 95% bias accelerated confidence interval (CI) and IQRs. P1=(Haller et al., 1997), P2= (Killiany et al., 1993), P3= (Malykhin et al., 2007), P4=(Pruessner et al., 2000) and P5 = (Pantel et al., 2000).
| Atlas Protocol |
Method | Absolute Volume (mm3) | % Normalized Volume Difference | ||
|---|---|---|---|---|---|
| Median | IQR | Median | IQR | ||
| P1 | Auto | 2045.00 | 627.69 | 6.23 | 7.25 |
| Man | 2175.69 | 709.42 | |||
| P2 | Auto | 1588.97 | 447.11 | 7.96 | 8.77 |
| Man | 1706.00 | 551.53 | |||
| P3 | Auto | 2325.22 | 732.65 | 5.05 | 6.96 |
| Man | 2455.01 | 843.59 | |||
| P4 | Auto | 2263.51 | 841.15 | 6.43 | 9.82 |
| Man | 2428.72 | 847.91 | |||
| P5 | Auto | 2329.20 | 726.07 | 5.14 | 7.58 |
| Man | 2495.74 | 859.42 | |||
Figure 2.
Protocol-wise Bland-Altman Plots comparing manual versus SBHV automatically derived manual labels for the Sunnybrook LOOCV. An optimized protocol was used for SBHV segmentation, which fused the 15 best matching label sets in target image space.
2.2.2 ADNI-1 cross-validation dataset
The range of right hippocmapal volumes for the ADNI-1 validation was greater than the Sunnybrook LOOCV, which also included bilateral hippocampal labels. When all groups were pooled, the ADNI-1 NVD values were similar to the LOOCV dataset. For P1 and P4 the NVD values were lower in AD versus MCI and NC. However, volume differences were consistently greater for P2 across all groups. Further, the ADNI-1 SBHV segmentations demonstrated a similar pattern of protocol-wise bias to the LOOCV, as Figure 3 shows that SBHV underestimated ADNI-1 volumes in comparison to manual tracings (Table 8).
Figure 3.
Protocol-wise Bland-Altman plots comparing manual versus SBHV automatically derived manual labels of the right hippocampus for the ADNI-1 cross-validation study. An optimized protocol was used for SBHV segmentation, which propagated to and fused the 15 best matching template library label sets in target (query) image space.
Table 8.
Alzheimer’s Disease Neuroimaging Initiative-1 group-wise cross-validation experiment median and interquartile range (IQR) uncorrected right hippocampal volumes derived from manual (Man) and automated (Auto) labeling methods based on 3 anatomically unique segmentation protocols (P1= (Haller et al., 1997) P2=(Killiany et al., 1993) and P4= (Pruessner et al., 2000)) (n=30). Percent normalized volume differences (NVDs) are based on the difference between right hippocampal manual and automatic labels and are reported as percentages with a bootstrap 95% confidence intervals (CI) and interquartile ranges (IQR). Manual and automated labels were generated across protocols for a random sample of normal controls (NC), persons with mild cognitive impairment (MCI) and Alzheimer disease (AD).
| Dx Group | Atlas Protocol |
Method | Volume (mm3) | % Normalized Volume Difference | ||
|---|---|---|---|---|---|---|
| Median | IQR | Median | IQR | |||
| NC (n=10) | P1 | Auto | 2319..72 | 723.37 | 8.12 | 4.81 |
| Man | 2346.69 | 894.89 | ||||
| P2 | Auto | 1686.64 | 627.85 | 7.63 | 11.1 | |
| Man | 1813.89 | 557.87 | ||||
| P4 | Auto | 2533.29 | 789.69 | 9.28 | 6.51 | |
| Man | 2578.03 | 935.98 | ||||
| MCI (n=10) | P1 | Auto | 2100.76 | 438.24 | 8.50 | 4.78 |
| Man | 2297.82 | 457.91 | ||||
| P2 | Auto | 1663.79 | 382.55 | 9.01 | 12.55 | |
| Man | 1938.60 | 381.28 | ||||
| P4 | Auto | 2285.13 | 479.33 | 6.33 | 5.06 | |
| Man | 2457.76 | 452.20 | ||||
| AD (n=10) | P1 | Auto | 1559.38 | 542.48 | 4.27 | 4.84 |
| Man | 1697.74 | 696.87 | ||||
| P2 | Auto | 1188.74 | 520.27 | 9.91 | 7.74 | |
| Man | 1360.42 | 682.75 | ||||
| P4 | Auto | 1732.02 | 548.51 | 5.85 | 9.09 | |
| Man | 1890.68 | 792.54 | ||||
| All Groups (n=30) | P1 | Auto | 2026.19 | 820.31 | 7.95 | 5.59 |
| Man | 2198.50 | 636.57 | ||||
| P2 | Auto | 1532.41 | 572.95 | 8.73 | 8.29 | |
| Man | 1760.89 | 589.61 | ||||
| P4 | Auto | 2227.76 | 894.41 | 7.42 | 7.22 | |
| Man | 2392.86 | 686.24 | ||||
2.3 Qualitative analysis using error distribution maps
2.3.1 LOOCV error maps
For the LOOCV study there was a greater proportion of FN (volume underestimation) versus FP (volume overestimation) labels.
Caudal region: The most pronounced segmentation errors were localized to the caudal/posterior hippocampus. Caudal underestimation was most conspicuous for P2, which used the crus of the fornix as a boundary landmark (Figure 4), P1 also suffered from caudal underestimation in comparison to definitions which segmented the first ovoid mass of posterior grey matter (i.e. P3–P5). There was some overestimation of the superior-posterior hippocampus across protocols, which labeled portions of the crus of the fornix (Supplementary Figure 2). For P4, the posterior medial compartment demonstrated greater FP and FN errors compared to P3 and P5. This error was attributable to the manually placed vertical line used to separate the gyrus fasciolaris and Andreas-Retzius gyrus. Moreover, this may partially explain the high variance in NVD realized for the P4 LOOCV analysis.
Anterior region: The anterior hippocampus suffered from moderate underestimation and overestimation equally across protocols.
Dorsal border: Protocols 1 and P2 excised the alveus and fimbria located on the dorsal hippocampal surface. The grey-white matter interface for this compartment suffered from low contrast resolution and partial volume effects for both datasets at the acquired 1.5 Tesla in-plane resolutions. SBHV slightly overestimated the medial anterior-superior white matter compartment for P1 and P2, resulting in a modest volume underestimation (Figure 5); whereas, SBHV overestimated this compartment for P1 and P2 throughout the body and tail to a greater extent than P3–P5 (Figure 6).
Inferior border: All protocols showed some background labeling of the parahippocampal white matter, resulting in a slight overestimation of the inferior hippocampus (Figure 7).
Medial region: In addition, the medial compartment was occasionally mislabeled by SBHV. Automatic segmentation modestly underestimated the posterior medial subiculum (i.e. excluding the presubiculum) among more medially inclusive protocols (i.e. P1 and P5). However, protocols that used an oblique line to separate the parahippocampal gyrus from the subiculum (e.g. P4), tended to label more inferomedial parahippocampal white matter along the hippocampal body.
Right versus left volumes: Visually, there was slightly less voxel-wise error in the posterior and superior compartments on the right hippocampus versus the left (Figure 4 and 5).
Figure 4.
False negative (FN) coronal distribution maps for SBHV segmentation of the posterior hippocampal region from the results of the LOOCV. The color masks represent voxel-wise FN counts (underestimation) across the five different protocols overlaid on the Sunnybrook average elderly 100-brain template. Each row of panels represent 4 serial slices from posterior (right) to anterior (left) for a given protocol. Panel row A represents the posterior border region for P1 and P3–P5 with no overlay, whereas row B shows the border region for P2 with no overlay, which is located more anterior to the other protocols. SBHV often underestimated the caudal hippocampal region across all protocols; however, the more inclusive protocols P3–P5 demonstrated less FN errors than P1 and P2, which excluded portions of the hippocampal tail.
Figure 5.
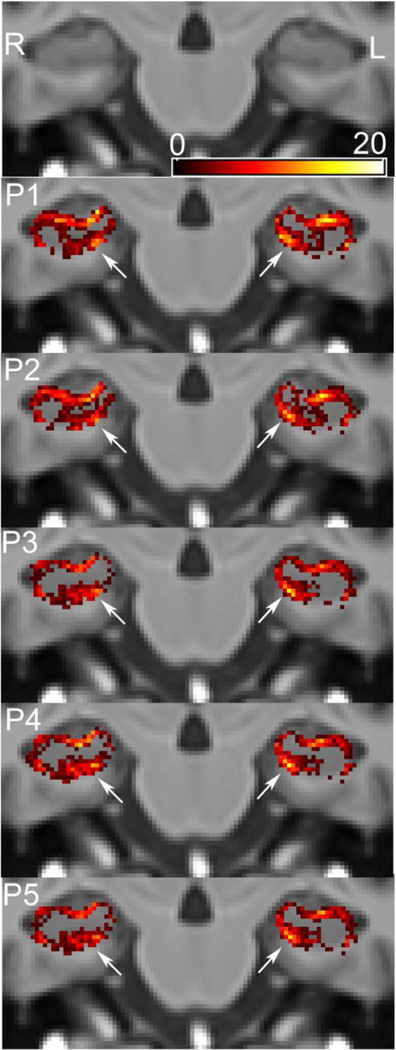
Protocol-wise coronal false negative (FN) distribution maps for SBHV segmentation of the medial anterior-superior alveus from the results of the LOOCV. The color masks represent voxel-wise FN counts (underestimation) across the five different protocols overlaid on the Sunnybrook average elderly 100-brain template. The most offending regions are highlighted with white arrows. SBHV tended to overestimate the anterior-superior medial white matter compartment (white arrows) to a greater extent in protocols, which excluded the alveus and fimbria (i.e. P1 and P2).
Figure 6.
Protocol-wise coronal false positive (FP) distribution maps for SBHV segmentation of the superior white matter compartment across the hippocampal body (i.e. alveus/fimbria) from the results of the LOOCV. The color masks represent voxel-wise FP counts (overestimation) across all five protocols projected onto the Sunnybrook average elderly 100-brain template. The most offending regions/protocols are highlighted with white arrows. P1 and P2, which excluded the alveus and fimbria tended to overestimate the superior white matter compartment, and this may be partially explained by the poor contrast realized between grey and white matter within this region.
Figure 7.
Protocol-wise coronal false positive (FP) distribution maps for SBHV segmentation from the results of the LOOCV. The color masks represent voxel-wise FP counts across the five different surveyed protocols projected onto the Sunnybrook average elderly 100- brain template. White arrows highlight marked overestimation (FP errors) of the inferior hippocampal compartment, which includes background regions of parahippocampal white matter. This FP error similarly affected all protocols in the LOOCV.
2.3.2 ADNI-1 cross validation error maps
The ADNI-1 cross validation study revealed a consistent topographical distribution of errors when compared to the LOOCV.
Caudal region: Figure 8 shows a similar underestimation of the posterior hippocampus across protocols in relation to the LOOCV. However, qualitative assessment of ADNI-1 group-wise FN map differences shown in Figure 8 revealed greater heterogeneity of caudal error distributions between protocols in AD compared to MCI and NC, and this finding is congruous with our ADNI-1 protocol-wise DSC comparisons, which found significant differences between all protocols in AD. There was also some overestimation within the posterior-medial compartment for P1 and P4.
Anterior region: Similar to the LOOCV, the anterior hippocampus suffered from volume underestimation and some overestimation medially.
Dorsal border: SBHV tended to underestimate the anterior superior white matter compartment for the NC group to a greater extent than the MCI and AD group. In contrast, there was comparatively less overestimation of the dorsal white matter compartment (i.e. alveus and fimbria) for the body of the hippocampus in comparison to the LOOCV, and these observations to some extent support the higher DSC values reported in the ADNI-1 validation compared to the LOOCV.
Inferomedial region: The medial hippocampus was also overestimated across protocols and diagnostic groups. Specifically, there were fewer inferomedial errors observed for the AD group versus the MCI and NC group (Figure 9). Indeed, this finding partially explains the poorer segmentation accuracy of the SBHV tool in NC and MCI in comparison to the AD group.
Figure 8.
False negative (FN) coronal distribution maps for SBHV segmentation of the posterior hippocampal region from the results of the ADNI-1 cross-validation study. The color masks represent voxel-wise FN counts (underestimation) across P1, P2 and P4 (rows) and NC/MCI/AD ADNI-1 groups (columns), projected onto the Sunnybrook average elderly 100-brain template. Note, the P2 posterior border started more anterior to P1 and P4. Caudal FN error distributions for the ADNI-1 validation are similar to those observed in the LOOCV. Qualitatively, caudal FN distributions between protocols varied the most in AD, and within the AD sample all protocols showed significantly different median Dice similarity measures (P4>P1>P2). Further, P2 demonstrated the greatest caudal error as a result of the landmark-based definition used to demarcate the posterior border. For within protocol comparisons, only P4 demonstrated significantly different voxel-wise accuracy measurements between groups (AD>NC).
Figure 9.

ADNI-1 group-wise coronal false positive (FP) distribution maps for SBHV-P4 segmentation from the results of the ADNI-1 cross-validation study. White arrows highlight overestimation (FP errors) of the inferior hippocampal compartment. The color masks represent voxel-wise FP counts (overestimation) projected onto the Sunnybrook average elderly 100-brain template. Note that the FP error count was greater along the inferomedial hippocampus in NC and MCI than AD, which may partially explain the lower Dice similarity results in NC and MCI versus AD.
3.0 Protocol-wise hippocampal biomarker performance applied to the entire ADNI-1 dataset
3.1 ADNI-1 cross-sectional group-wise volumetric comparisons
Three subjects had inconsistent clinical conversions over the 24-month study window and were included in the sMCI group. Specifically, subject 127_S_0112 (MCI) reverted → converted → reverted, subject 136_S_0429 (MCI) converted → reverted, and subject 137_S_0669 (MCI) reverted → converted → reverted. Protocol-by-diagnostic mean (SD) unadjusted total (right + left) baseline volumes are reported in Supplementary Table 1 and are shown juxtaposed in Figure 10. All baseline comparisons were corrected for ICV, age and gender. All protocols showed significantly larger mean baseline hippocampi in the NC group in comparison to both the MCI and AD groups (p<0.001). The MCI group had significantly smaller total adjusted hippocampal volumes than NC but more volume than the AD group (p<0.001). The cMCI group had significantly smaller adjusted hippocampal volumes than sMCI (p<0.001) and rMCI (p<0.001), but significantly greater volumes than AD (p<0.05) across protocols. The rMCI group was not significantly different than the sMCI group. For all protocols, the sMCI and cMCI hippocampal volumes were significantly smaller than NC (p<0.001); however, the rMCI group was not significantly different from NC.
Figure 10.
Protocol-specific comparisons of baseline total hippocampal volume (right + left) including ADNI-1 principal groups: Normal controls (NC), Mild Cognitive Impairment (total group) (tMCI) and Alzheimer’s Disease (AD) in addition to ADNI-1 MCI subgroups: MCI converters after 24-months (cMCI), MCI subjects who reverted back to normal elders (rMCI) and MCI subjects who remained stable after 24-months (sMCI). The whiskers represent the 10th and 90th percentiles, and all data beyond these values are plotted. P1=(Haller et al., 1997), P2= (Killiany et al., 1993), P3= (Malykhin et al., 2007), P4= (Pruessner et al., 2000) and P5 = (Pantel et al., 2000).
3.2 ADNI-1 longitudinal group-wise volumetric comparisons
Protocol-by-diagnostic mean (SD) baseline and scan interval normalized total (right + left) rates of change are reported in Supplementary Table 1 and are shown juxtaposed in Figure 11. All of the group-wise longitudinal comparisons were adjusted for age and gender. For all protocols, both the AD and total MCI (tMCI) groups had significantly greater rates of 24-month hippocampal atrophy in comparison to the NC group (p<0.001), and the AD group had a greater rate of atrophy than the tMCI and sMCI groups (p<0.001). The tMCI, sMCI, cMCI and AD groups had significantly greater adjusted hippocampal atrophy than NC (p<0.001), while the rMCI group was not significantly different from normal elders. In addition, the cMCI subgroup had significantly less atrophy than the AD group (p<0.001), and only P1 and P4 demonstrated significant differences between rMCI and AD (p<0.01), although the rMCI sample size was small (N=10). An important finding was that the most inclusive protocols (i.e. P1 and P3–P5) demonstrated greater adjusted rates of atrophy for cMCI compared to sMCI (p<0.01); however, P2 rates were not significantly different (p=0.065).
Figure 11.
Protocol-specific comparisons of hippocampal 24-month rates of change normalized to baseline volume and serial scan window including ADNI-1 principal groups: Normal controls (NC), Mild Cognitive Impairment (total group) (tMCI) and Alzheimer’s Disease (AD) in addition to ADNI-1 MCI subgroups: MCI converters after 24-months (cMCI), MCI subjects who reverted back to normal elders (rMCI) and MCI subjects who remained stable after 24-months (sMCI). The whiskers represent the 10th and 90th percentiles, and all data beyond these values are plotted. P1=(Haller et al., 1997), P2= (Killiany et al., 1993), P3= (Malykhin et al., 2007), P4= (Pruessner et al., 2000) and P5 = (Pantel et al., 2000).
3.3 ADNI-1 hippocampal volumetry and episodic memory associations
3.3.1 MCI group correlations
Adjusted baseline hippocampal volumes were significantly associated with baseline AVLT scores within the MCI group for all protocols (Table 9). There were no marked differences among hippocampal protocols for associations with baseline AVLT scores in the MCI group. Similarly, Table 9 shows that the MCI baseline LM scores were significantly associated with adjusted hippocampal volumes across protocols, and there were no marked differences between associations. For the MCI group, there were no significant associations between the rate of hippocampal change and 24-month change in memory performance on the AVLT. In contrast, all protocols with the exception of P2 were significantly correlated with 24-month change on the LM (Table 9).
Table 9.
Protocol-specific standardized β coefficients grouped by diagnosis for ADNI-1 participants’ cognitive test performance versus hippocampal volumetry. The standardized β represents the number of standard deviations the cognitive measure increases/decreases with a one standard deviation increase of the hippocampal measurement. Note, significant standardized β values (p<0.05) are indicated in bold. Both the logical memory 1 (LM) immediate recall test and the Auditory Verbal Learning Test (AVLT) (sum of Trials I–IV) baseline and 24-month change in test scores were associated with corresponding absolute total (right + left) hippocampal volumes and 24-month percent hippocampal atrophy. All cross-sectional models included age, gender and intra-cranial volume whereas all longitudinal linear models controlled for age and gender. MCI = Mild Cognitive Impairment and AD = Alzheimer’s disease, P1= (Haller et al., 1997), P2= (Killiany et al., 1993), P3= (Malykhin et al., 2007), P4= (Pruessner et al., 2000) and P5= (Pantel et al., 2000).
| P1 | P2 | P3 | P4 | P5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β | p-value | β | p-value | β | p-value | β | p-value | β | p-value | |
| Baseline Hippocampal Volume & AVLT Score | ||||||||||
| MCI | 0.22 | <0.001 | 0.19 | 0.001 | 0.22 | 0.001 | 0.23 | <0.001 | 0.22 | <0.001 |
| AD | 0.22 | 0.021 | 0.21 | 0.025 | 0.22 | 0.017 | 0.27 | 0.003 | 0.22 | 0.016 |
| Baseline Hippocampal Volume & LM Score | ||||||||||
| MCI | 0.14 | 0.019 | 0.11 | 0.05 | 0.14 | 0.013 | 0.16 | 0.005 | 0.14 | 0.013 |
| AD | 0.24 | 0.005 | 0.22 | 0.011 | 0.25 | 0.004 | 0.28 | 0.001 | 0.26 | 0.003 |
| 24 month atrophy & AVLT change | ||||||||||
| MCI | −0.07 | 0.25 | −0.03 | 0.629 | −0.08 | 0.247 | −0.09 | 0.153 | −0.08 | 0.247 |
| AD | −0.01 | 0.912 | −0.02 | 0.832 | −0.01 | 0.894 | −0.01 | 0.914 | −0.01 | 0.930 |
| 24 month atrophy & LM change | ||||||||||
| MCI | −0.17 | 0.008 | −0.10 | 0.11 | −0.18 | 0.005 | −0.22 | <0.001 | −0.18 | 0.004 |
| AD | −0.16 | 0.106 | −0.15 | 0.134 | −0.14 | 0.159 | −0.126 | 0.205 | −0.137 | 0.171 |
3.3.2 AD group correlations
Only baseline hippocampal and neurocognitive scores were significantly associated for the AD group (Table 9). Baseline AVLT score was significantly associated with adjusted baseline hippocampal volume across protocols. Moreover, baseline LM scores were significantly associated with adjusted baseline hippocampal volumes (p<0.01). P4 was slightly more associated with baseline memory performance in comparison to the other protocols.
3.4 Protocol-wise ADNI-1 sample size estimates
Table 10 demonstrates that with the exception of P2 (which consistently underperformed compared to all other protocols) the ranking of labeling methods in MCI with respect to sample size changed when adjusted for normal aging. Sample sizes for P3, P4 and P5 were smaller than P1 and P2, and P1 was smaller than P2. Within the AD group, all protocols demonstrated smaller sample sizes than P2, and this relationship remained after correction for normal aging.
Table 10.
Protocol-wise (P1–P5) sample sizes (N) and bootstrap derived 95% confidence intervals (CI) required for a hypothetical therapeutic trial in Alzheimer’s disease (AD) or in mild cognitive impairment (MCI,) designed to detect a 25 percent change in the 24-month rate of hippocampal atrophy in comparison to a placebo group (with significance set at 0.05 and power at 0.8). Sample size calculations are reported as unadjusted or adjusted based on the mean 24-month protocol-specific rate of hippocampal atrophy observed in ADNI-1 normal elders. All rates of atrophy were entered as percentage change from baseline and normalized for each participant’s ADNI scan interval, which often was greater than 24-months.
| Dx Group | P1 | P2 | P3 | P4 | P5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | 95% CI | n | 95% CI | n | 95% CI | n | 95% CI | n | 95% CI | |
| MCI Unadjusted | 260 | (202, 353) | 339 | 254, 479) | 225 | (175, 306) | 214 | (165, 292) | 217 | (169, 293) |
| MCI Adjusted | 891 | (670, 1841) | 1109 | (834, 2661) | 870 | (594,1585) | 902 | (640, 1844) | 893 | (510, 1258) |
| AD Unadjusted | 152 | (100, 263) | 207 | (132, 381) | 145 | (97, 247) | 135 | (92, 229) | 143 | (95, 243) |
| AD Adjusted | 287 | (194, 601) | 379 | (250, 867) | 297 | (192, 585) | 294 | (198, 600) | 302 | (179, 534) |
Discussion
Based on the original multi-template work of (Aljabar et al., 2009; Barnes et al., 2008a; Heckemann et al., 2006) in conjunction with the pipelines of (Collins and Pruessner 2010; Leung et al., 2010; Wang et al., 2011b), we developed a fully automated multi-atlas hippocampal segmentation tool, SBHV, that spatially maps labels using the SyN diffeomorphic registration algorithm. What distinguishes this study from previous atlas comparisons in AD is the large number of unique hippocampal atlas libraries that were generated by the first author – 5 hippocampal libraries totaling 500 manual hippocampal tracings. Moreover, this was the first atlas library in AD to include a more heterogeneous sample of AD pathologies including patients with VaD and SVD, which is important if an automated tool is to be applied clinically or to large cohort studies in AD. This automatic framework coupled with the entire ADNI-1 baseline and 24-month dataset, enabled the largest automated head-to-head comparison of hippocampal atlas protocols in AD to-date and the first of its kind to assess voxel-wise accuracy and voxel-wise error distributions among anatomically distinct multi-atlas libraries. Here we report a number of important biomarker performance-based findings among the 5 atlas protocols tested including: group-wise discrimination, differential accuracy association with cognition. We also describe how both multi-template segmentation accuracy and inclusivity among the protocols sampled may influence hippocampal biomarker performance in MCI and AD.
DSC Manual Vs. automatic segmentation accuracy
Inter- and intra- rater agreement were excellent (intra-ICC: 0.95–0.97, DSC = 0.92–0.93: and inter-ICC: 0.92–0.96, DSC = 0.90–0.91) and within the range of previous studies (Barnes et al., 2009; Boccardi et al., 2011; Collins and Pruessner 2010; Leung et al., 2010). All protocols demonstrated high accuracy in comparison to manual tracings across both the LOOCV and ADNI-1 datasets. One of the principal findings of this study was that the accuracy of automated hippocampal segmentation appears to be modulated by morphological definition. The most inclusive protocols (P3–P5) demonstrated the highest accuracies in the LOOCV (DSC = 0.88, 0.88 and 0.89 respectively), and were not significantly different. The DSC values were modestly improved across all protocols in the ADNI-1 validation over the LOOCV; however, there were differences between datasets, which may explain these findings. First, the LOOCV contained greater heterogeneity among disease profiles (i.e. VaD, AD, mixed AD, AD with SVD and NC) when compared to the ADNI-1 NC, MCI and AD participants. A recent study by (Scher et al., 2011) reported differential patterns of hippocampal atrophy across dementia subgroups (AD versus VaD), and this morphological heterogeneity may have contributed to the greater protocol-wise DSC measures observed in the ADNI-1 sample versus the LOOCV. Moreover, diagnostic group was shown to modulate both DSC and NVD in our ADNI-1 validation. Second, the ADNI-1 validation only compared n = 30 right hippocampal volumes per protocol versus n = 70 right + left volumes for the LOOCV. Although not reported in the results, right hippocampal segmentations demonstrated a ½ percentage point improvement in median DSC over left labels for the LOOCV. Finally, the median volumes were slightly larger for the ADNI-1 NC and MCI validation groups compared to the LOOCV dataset, which may have biased the ADNI-1 DSC results. Despite these experimental differences, the ADNI-1 validation reflected the same rank order of protocol-wise DSC accuracy, comparable DSC variation within protocols and similar FP/FN error distributions to those observed in the Sunnybrook LOOCV.
Definition of the posterior border was most variable between protocols, and upon visual inspection of the FN/FP error maps, was frequently underestimated by SBHV, with occasional inclusion (overestimation) of the fornix. Certain hippocampal protocols use landmarks to determine the posterior border (Bartzokis et al., 1998; deToledo-Morrell et al., 2004; Jack 1994; Killiany et al., 1993; Watson et al., 1992). And although manually identified landmarks were designed to improve inter- and intra- labeler precision, automated reproducibility of these rule-based boundaries appears to be less accurate in heterogeneous cohorts (e.g. elders and AD). For example, P4 used an orthogonal pair of lines to manually define the hippocampal tail from the surrounding parenchyma, which SBHV failed to consistently reproduce. Moreover, protocols that truncated caudal regions of the hippocampus (P1 and P2) tended to suffer from greater posterior volume underestimation. The most extreme example was P2 (Killiany et al., 1993), which excluded significant portions of the hippocampal tail posterior to where the crus of the fornix was visible in full profile. In dementia, there is frequently thalamic atrophy that can modulate where an operator defines the posterior boundary, often more rostral when atrophy is present, and a registration algorithm may not appropriately capture this boundary shift.
Strengthening these observations, (Carmichael et al., 2005) found marked label error along the posterior hippocampus, and this finding was also observed using a multi-atlas based method in MCI and AD (Leung et al., 2010). In a recent multi-atlas hippocampal segmentation study, which examined spatial bias in voting-based label fusion, the greatest error (automatic label underestimation) was realized for convex regions of the hippocampus, particularly in the posterior and anterior regions (Wang and Yushkevich 2012). However, the small bias towards the mean observed for atrophic hippocampi in the LOOCV and ADNI-1 study suggests that volume underestimation may be less pronounced in smaller hippocampi. We did not explicitly test the effect of registering small versus large templates to an atrophic hippocampus. Inclusion of a weighted voting strategy may also reduce averaging bias. Alternatively, a greater number of atrophic hippocampi may also reduce the affect of heterogeneous template fusion.
The next most variable region among protocols was the dorsal hippocampal border (i.e. alveus and fimbria white matter compartment). Protocols that excluded the alveus and fimbria (P1 and P2) were significantly less accurate and had greater DSC variation than structures including these dorsal white matter compartments (P3–P5). Visual inspection of error distribution maps revealed a pattern of greater SBHV FP errors along the superior hippocampal body and FN errors at the anterior-superior pole for both P1 and P2, which were generated from the low contrast realized at this grey-white matter interface
Further visual inspection of FN/FP error maps revealed that certain hippocampi were mislabeled along the inferomedial border, which has been previously reported in (van der Lijn et al., 2008). A thin layer of white matter separates the hippocampus from the parahippocampl parenchyma, and occasionally, the SBHV tool segmented these background structures across protocols. However, intensity based thresholding reduced FP labels, and comparable accuracy was achieved among protocols that predominantly varied by medial definition (e.g. P3 versus P5).
For our ADNI-1 voxel-overlap comparisons, there were significant protocol-wise DSC differences within groups, but only the AD group showed significant differences among all protocols sampled. More inclusive protocols always outperformed less inclusive definitions in AD (P4>P1>P2). However, in MCI only the most inclusive protocol (P4) outperformed the least inclusive P2, while in NC, P2 was less accurate than the more inclusive P1 and P4. This suggests that more inclusive protocols (i.e. that include the alveus/fimbria and >hippocampal tail) provide superior accuracy across groups, and protocol accuracy as a function of structural assembly appears to matter most in AD. Indeed, our findings shown in Figure 8 strengthen this notion, demonstrating greater voxel-wise error differences between protocols in AD versus MCI and NC.
It is important to note that FP/FN at the boundary of smaller volumes can have a larger impact on voxel-wise similarity metrics in comparison to more inclusive structures, and may partially explain the lower DSC values for P1 and P2 in comparison to P3–P5. Moreover, the ADNI-1 MCI and AD validation samples had a greater median absolute volume in comparison to the median volume of the Sunnybrook LOOCV, which may have partially contributed to the higher DSC ADNI-1 validation results. Indeed, (Patenaude et al., 2011) reported lower DSC values for small structures such as the amygdala, nucleus accumbens and hippocampus in comparison to larger subcortical compartments including the putamen, caudate and thalamus. However, the FP and FN error maps in the current study revealed greater absolute posterior and superior labeling errors for P1 and P2 than the more inclusive P3–P5, suggesting that lower DSC values are driven to a larger extent by protocol definition versus the well-known bias of measuring smaller structures.
Interestingly, in the current study, persons with AD despite having significantly smaller hippocampi and greater surface-area-to-volume ratios than NC demonstrated comparable segmentation results to normal elders. One explanation for this inconsistent finding may be that the enlarged CSF compartment (cistern, choroid fissure and inferior horns) surrounding atrophic hippocampi improved delineation along the complete lateral extent, superior parenchymal surface, and amygdalar hippocampal interface, facilitating more robust registration outcomes. Additionally, SBHV segmentation accuracy along the inferior border was improved in AD versus NC, which may also support these findings.
ADNI provides a platform for direct quantitative comparisons among segmentation methods in AD. Since its inception, there have been numerous head-to-head comparisons of hippocampal segmentation algorithms (see (Weiner et al., 2012) for a detailed review), and across these studies, much attention has been focused on algorithmic differences. As manual “ground-truth” labels are not widely available, definitive performance evaluation among atlas protocols and automated methods in general, is complicated without consistent manual labels for each protocol on the same MRI dataset. Thus, comparisons across previous validation studies should be interpreted cautiously. Here we compare our voxel-overlap results to multi-template studies based on similar manual protocols. In particular, we replicated DSC measures of a validation study in healthy adults by (Collins and Pruessner 2010), using a smaller atlas library with greater pathological heterogeneity. Moreover, this result was achieved with a modified version of the labeling criteria developed by (Pruessner et al., 2000) (P4), as our protocol defined the hippocampus along the AC-PC line without normalization to Talairach space. Additionally, we used the SyN nonlinear registration whereas Collins and Pruessner used an elastic registration method (Collins et al., 1995).
We also demonstrated comparable results to previous studies of multi-atlas based segmentation that employ other anatomical definitions: (Barnes et al., 2008b; Hammers et al., 2007; Kim et al., 2012; van der Lijn et al., 2008; Wang et al., 2011b; Wolz et al., 2009) (Table 11). After semantic harmonization of current multi-atlas protocols in Table 11, there are only a few widely used protocols for multi-atlas segmentation to-date. All methods include the alveus/fimbria and similarly define the anterior border. Protocols principally differ by inclusion/exclusion of the caudal pole and medial compartment. Unfortunately, there is no clear relationship between structural definition and voxel-overlap in the literature, partly owing to algorithmic and validation sample differences. Among state-of-the-art methods, DSCs ranged between 0.83–0.91, and (Wang et al., 2011b) demonstrated the highest accuracies to-date, also using the SyN registration tool. However, the investigators derived their gold standard labels from manually corrected automatic volumes and adjusted automated labels using a learning based wrapper. In summary, SBHV with the P3, P4 or P5 library achieves high voxel-overlap compared to manual labels both within a large multi-centre study and a diverse memory clinic cohort that is consistent with recent work in dementia.
Table 11.
Comparison of SBHV-P1, -P2 and -P4 atlas composition and accuracy with recent automated hippocampal segmentation techniques based on multi-atlas registration and label fusion.
| Method | Segmentation protocol | Anatomical Definition | DSC | JI | Validation Sample | Template Library Composition |
||
|---|---|---|---|---|---|---|---|---|
| Posterior | Alveus/Fi mbria |
Medial | ||||||
| (Hammers et al., 2007) | (Niemann et al., 2000) | − | + | + | 0.76 0.83 |
TLE Unilateral Sclerotic TLE Contralateral Side |
Healthy Young Adults |
|
| (van der Lijn et al., 2008) | Based on modified protocol similar to (Jack 1994)but including the entire hippocampal tail | + | + | + | 0.852 (left side) 0.864 (right side) |
Healthy Elderly | 20 Healthy Elderly Subjects |
|
| (Barnes et al., 2008a) | (Watson et al., 1992) | − | + | + | 0.87 0.86 |
NC AD |
NC and AD | |
| (Wolz et al., 2009) | N/Aα | N/A | N/A | N/A | 0.860 | NC, MCI and AD | NC, MCI and AD | |
| (Lotjonen et al., 2010) | N/Aα | N/A | N/A | N/A | 0.885β | NC, MCI and AD | NC, MCI and AD | |
| (Collins and Pruessner 2010) | (Pruessner et al., 2000) | + | + | − | 0.887 | 0.796 | Healthy Young Adults |
Healthy Young Adults |
| (Leung et al., 2010) | (Watson et al., 1992) | − | + | + | 0.80 0.81 0.79 |
NC MCI AD |
NC and AD | |
| (Wang et al., 2011b) | (Hasboun et al., 1996) | − | + | − | 0.887 0.908 0.872 0.893 |
0.798 0.833 0.774 0.808 |
NC NC (error corrected) MCI MCI (error corrected) |
NC and MCI |
| (Kim et al., 2012) | (Watson et al., 1992) | − | + | + | 0.835 0.854 0.807 0.841 0.824 0.823 |
NC (left side) NC (right side) LTLE (left side) LTLE (right side) RTLE (right side) RTLE (left side) |
NC, LTLE and RTLE |
|
| SBHC-P1 | (Haller et al., 1997) | + | − | + | 0.88 0.89 0.90 |
NC MCI AD |
NC, AD, Mixed Dementia, AD+SVD and VaD |
|
| SBHC- P 2 | (Killiany et al., 1993) | − | − | − | 0.86 0.88 0.88 |
NC MCI AD |
NC, AD, Mixed Dementia, AD+SVD and VaD |
|
| SBHC- P4 | (Pruessner et al., 2000) | + | + | − | 0.89 0.90 0.91 |
NC MCI AD |
NC, AD, Mixed Dementia, AD+SVD and VaD |
|
The authors reported that manual labels were provided by ADNI consortium”,
Best result was achieved using atlas selection and an expectation maximization algorithm.
DSC= Dice Similarity Coefficient, JI = Jaccard Index = (M ∩ A)/(M ∪ A), Where M is the manually traced label set, A is the SBHV automatically derived label set and ∩ is the intersection operator, (R or L) TLE= Right/Left Temporal Lobe Epilepsy, + = more inclusive protocol, − = less inclusive protocol
Manual Vs. automatic segmentation volume differences
We also tested volumetric differences between manual and automated labels, and our data show that all 5 automated protocols tended to underestimate absolute volumes for both the Sunnybrook LOOCV and the ADNI-1 derived manual volumes. Additionally, the error distribution maps revealed a similar pattern of FN labels compared to the LOOCV dataset. The higher variation in NVD for P4 in the LOOCV and ADNI-1 AD validation may be partially explained by the orthogonal lines used to demarcate the posterior compartment. In certain subjects, this compartment was inconstantly labeled by SBHV, generating greater NVDs.
Over all, automatic-manual volumetric similarity amongst the assessed protocols was congruous with previous multi-atlas work in healthy adults (Collins and Pruessner 2010) (NVD = 4.9%), right temporal lobe epilepsy (TLE) (Kim et al., 2012) (absolute volume = 3134 mm3 manual vs. 3301 mm3 automatic) and AD (Leung et al., 2010; Wang et al., 2011b) (absolute volume difference = 56–81 mm3). When volumetric accuracies are taken together with voxel-wise overlap outcomes, these results suggest that more inclusive protocols furnish superior accuracy versus conservative atlas definitions, especially in AD and diagnostically heterogeneous samples. Our results also support the notion that SBHV voxel-wise segmentation accuracy is lower in more diagnostically heterogeneous samples (i.e. Sunnybrook versus ADNI-1). Finally, SBHV + a more anatomically inclusive template library provides high fidelity segmentation accuracy when compared to expert tracings and so is a suitable method to replace manual segmentation for both the analysis of large multi-centre AD studies and for use in a general memory clinic cohort, with the caveat that image quality must be sufficient to perform unbiased registration.
Cross-sectional and longitudinal group-wise volumetric comparisons for the entire ADNI-1 dataset
Although an automated hippocampal volumetric technique may demonstrate high technical accuracy, it is equally important to assess its utility as a biomarker to measure disease progression, discriminate amongst clinical cohorts, prognosticate decline and serve as a useful indirect marker for clinical trials. Here we report on the biomarker performance outcomes of the 5 different SBHV atlas-protocols. Baseline volumes ranged considerably between methods. In fact, mean P2 NC volumes were 32% smaller than corresponding P5 volumes. Nevertheless, all methods compare well with previous ADNI-1 cross-sectional studies (Leung et al., 2010; Morra et al., 2009; Mouiha, Duchesne, Alzheimer's Disease Neuroimaging Initiative 2011; Schuff et al., 2009; Wolz et al., 2010b), distinguishing principal groups by baseline volume (NC>MCI>AD) and similarly discriminated across principal groups for 24-month atrophy measures (AD>MCI>NC). Consistent with previous studies, cMCI showed reduced baseline volume versus sMCI. However, the least inclusive protocol, P2, did not significantly differentiate cMCI from sMCI, whereas all other protocols showed cMCI rates to be greater than sMCI. This may suggest that definitions approaching the minimal hippocampal assembly are not sufficient to capture atrophic changes automatically in multi-centre studies of prodromal AD. Moreover, the absolute caudal segmentation error realized for P2 may reduce the signal-to-noise ratio necessary to detect group-wise separation.
Our annualized mean longitudinal measures in AD, P1 = 4.12 %, P2 = 4.07%, P3 = 4.13%, P4 = 4.52% and P5 =4.16% and in NC, P1 = 1.11%, P2 = 1.04%, P3 = 1.22%, P4 = 1.43% and P5 = 1.27% compare favorably with a metaanalysis by (Barnes et al., 2009) of manually derived volumes in NC = 1.41% and AD = 4.66%. Our annualized rates based on independent measures are consistent with a 1-year ADNI-1 study evaluating automatically derived volumes: NC = 1.40%, and AD = 4.57% (Leung et al., 2011). While our 24-month mean rates of change and variances are greater than an ADNI-1 study using multi-atlas segmentation with 4D graph cuts by (Wolz et al., 2010b), who report markedly lower 24-month mean rates of change (± SD) for NC / MCI / AD of: 1.66% ± 2.07 (n=114), 4.50% ± 3.12 (n=157) and 6.74% ± 2.89 (n=81) when compared to our results in Table 10. These differences may in part be explained by differing sample sizes that do not represent the more complete 24-month ADNI-1 sample. Indeed, 12-month and 24-month annualized measures reported in (Wolz et al., 2010b), notably underestimate the expected meta-analytically computed manual rate of change reported in (Barnes et al., 2009). However, there are several sources of bias between serial acquisitions including intensity differences, Interpolation asymmetries, software upgrades and hardware drift (Fox, Ridgway, Schott 2011), which can introduce morphometric variability. Thus, simultaneous hippocampal segmentation of aligned scan pairs (i.e. comparative analysis) using, for example, 4D graph cuts (Wolz et al., 2010b) or the Boundary Shift Integral (BSI) with bias field correction (Leung et al., 2011) has been shown to lower variance and improve sensitivity. Nevertheless, our goal was to directly compare different atlas protocols, not algorithms, and the direct nature of this study design ensured all protocols experienced identical algorithmic biases and very similar intensity inhomogeneity and resampling biases depending on subregions included/excluded. Finally, our ADNI-1 results contrast with a recent study by (Mouiha, Duchesne, Alzheimer's Disease Neuroimaging Initiative 2011), who reported that both a semi-automated atlas-based registration method called SNT, Medtronic Surgical Navigation Technologies (Louisville, CO) (Haller et al., 1997) and FreeSurfer (Fischl et al., 2002) annualized automatic measurements appear to significantly overestimate the rate of meta-analytically measured mean atrophy in AD (7.75% and 10.09%) and in NC (2.95% and 1.67%). However, multi-atlas methods have previously demonstrated greater accuracy than both FreeSurfer (Wang et al., 2011b) and SNT (Leung et al., 2011).
Protocol-wise associations with episodic memory
To be a valid surrogate marker of AD progression, hippocampal volumetry should relate to cognitive phenotype. The hippocampus measured in vivo has been strongly implicated in memory networks, and has been associated with episodic memory impairment in AD. However, anatomical variability among atlas-protocols may attenuate hippocampal-neurocognitive correlations. Therefore we directly tested whether differences existed between template ROI configuration and episodic memory performance. Our findings showed that all protocols demonstrated comparable associations with AVLT and LM derived episodic memory performance. Concordant with previous ADNI-1 hippocampal studies (Apostolova et al., 2010; Leung et al., 2010) baseline cognitive score was modestly associated with unadjusted total baseline hippocampal volume in both MCI and AD. Protocol 4 consistently demonstrated the highest association with memory measures among techniques whilst P2 (the least comprehensive definition) showed slightly lower associations with cognitive measures. A few studies have examined hippocampus sub-regional associations with episodic memory in AD (Costafreda et al., 2011; Lim et al., 2012; Shen et al., 2010). H.K. Lim and colleagues used the FMRIB’s Integrated Registration and Segmentation Tool (FIRST) (Patenaude et al., 2011), a shape based method, and demonstrated correlations in drug naïve patients with AD. The authors report associations with verbal memory within the lateral subiculum and CA1 extending from the hippocampal head to tail. These results are strengthened by the findings of (Costafreda et al., 2011) who demonstrated more lateral and anterior localized hippocampal associations with memory performance in AD compared to persons with MCI. As all protocols (P1–P5) included similar anterior-lateral hippocampal anatomy, it is unsurprising that we found only minor variation between methods in relation to cognitive performance. Moreover, the majority of voxel-wise segmentation error differences among protocols were localized to the posterior and superior borders and not the lateral/anterior compartments, The small variation that did exist between protocols for MCI and AD associations may reflect the differential error realized at the posterior border shown in Figure 8 of the ADNI-1 validation study.
Protocol-wise sample sizes in MCI and AD
Another important application of automated hippocampal volumetry is towards quantitative assessment of macroscopic brain changes to evaluate drug efficacy in MCI and the early stages of AD. Hippocampal imaging markers may have the potential to lower sample sizes, which can expedite trials of putative disease modifying therapies. While, we acknowledge that using comparative serial measurement methods such as the BSI would likely reduce sample sizes across protocols than to our independent serial measures (baseline - 24-months), we showed comparable sample sizes to previous ADNI-1 multi-atlas studies (Leung et al., 2010) and other techniques reviewed in (Weiner et al., 2012). Interestingly, our SBHV-P1 24-month MCI sample estimates (when adjusted for 90 percent power) based on the protocol of (Haller et al., 1997), were 31% lower, N=489, than 1-year sample sizes reported in (Schuff et al., 2009), N=698, using SNT, two time-points and the same atlas protocol. A possible explanation for our superior results based on two time points, is that hippocampal atrophy in MCI may accelerate and provide a larger effect size at 2-years than 1-year serial measures. Indeed, Jack and colleagues found an accelerated trajectory of brain atrophy in amnestic MCI subjects (Jack et al., 2008a), and (Schuff et al., 2009) also showed evidence for accelerated hippocampal change in ADNI-1 MCI over 1-year. However, as previously mentioned, bias within different techniques and between scan pairs can modulate longitudinal measures. While the trajectory of MCI atrophy is not entirely clear over 24-months, the superior performance of multi-atlas segmentation compared to SNT reported in (Leung et al., 2010) and our results, suggests that algorithmic differences and susceptibility to bias rather than pathogenesis, may be importantly implicated.
Sample size estimates globally increased after adjusting for the rate of change in normal aging, which detects the maximum potential treatment effect. Interestingly, adjusting for atrophy in normal aging appears to significantly improve the relative performance of P1 to detect changes in MCI, while no significant changes were realized among tracing methods in AD. The smaller adjusted sample size derived from P1 atlases is due to the lower rate of atrophy measured in the NC group. While P3 and P4 by definition excluded portions of the medial body including part of the medial subiculum, sample sizes were not remarkably different when compared to P1 and P5, which integrated more of this region. This may be ascribed to either registration errors along the medial boundary, which have been observed in the current study and by (van der Lijn et al., 2008), or that atrophy of both the subiculum and adjacent parahippocmapal gyrus in AD, reduces medial volumetric differences amongst protocols. Thus, medial body definition across the protocols sampled does not appear to markedly alter sensitivity of multi-atlas segmentation to detect changes in disease progression in either MCI or AD. Another striking observation was that more inclusive protocols (P1 and P3–P5) consistently generated smaller sample sizes than the least inclusive protocol (P2) in both MCI and AD. It is tempting to speculate that the differential sensitivity to detect disease progression between the protocols surveyed is largely driven by atrophy within the hippocampal tail. Although we did not explicitly test this hypothesis, such findings make intuitive sense given emerging shape-based analyses in AD, which suggest there are significant atrophic changes present in the posterior hippocampus (Gerardin et al., 2009; Shen et al., 2012). Specifically, (Shen et al., 2012) recently demonstrated in a subset of ADNI-1 participants that hippocampal atrophy in AD appears to involve the CA1, subiculum and regions of the hippocampal tail. More inclusive protocols would capture these putative changes and gain signal to detect volumetric differences between pathological and healthy hippocampi. However, the proportionally higher error distribution at the caudal and dorsal boundary of P2 and to a lesser extent P1 compared to P3–5 suggests that automatic label accuracy may partially modulate sample size differences among hippocampal protocols.
Protocols Excluded from Comparison
The widespread use of several hippocampal methods in the literature has generated interest to harmonize hippocampal protocols in AD, and The EADC-ADNI hippocampal initiative is currently working towards a unified protocol for the manual delineation of the hippocampus from 3D MRI. Briefly, the principal objectives of this initiative include reviewing the literature, generating a robust in-vivo definition of the hippocampus, and finally validating and qualifying a single consensus protocol on pathologically confirmed samples. Although the current study compared a variety of different hippocampal assemblies, we did not include all 12 published morphological variations assessed by the harmonization initiative. To reduce the number of manual tracings by the first author and to facilitate manual labeling in a common orientation, we selected 5/12 protocols for comparison. There were, however, a few notable protocol exclusions that were based on the long axis orientation of the hippocampus. First, the protocol of (Bartzokis et al., 1998) was the most conservative anatomical definition among all 12 protocols, excluding the entire tail of the hippocampus and dorsal WM compartment. Second, (Convit et al., 1997) used the most restrictive definition at the level of the hippocampal body, which excluded the parahippocampal gyrus and large portions of the subiculum. Finally, we did not include the protocols of (Jack 1994), (deToledo-Morrell et al., 2004) and (Watson et al., 1992), which excluded the tail of the hippocampus similar to (Killiany et al., 1993) (P2), but included notably more anatomy along the medial body similar to P5.
Algorithm-based limitations
There have been a number of recent technical developments that aim to advance multi-template driven segmentation and may improve the results reported here. Nevertheless, the spirit of the current study was to develop a common platform inspired by previously published work to directly evaluate the performance of template design for multiatlas segmentation. With this in mind, there were some limitations to the present study. Emerging evidence suggests that regional nonlinear registration may offer superior registration outcomes in comparison to whole brain approaches (Yousefi, Kehtarnavaz, Gholipour 2011). While the current study used global versus regionally specific nonlinear registration, we endeavored to optimize a multi-atlas scheme that could accommodate several other discrete subcortical structures implicated in AD, including the inferior horn, thalamus and also the cingulate gyrus. Moreover, we compared intensity similarity between template and target images using a localized VOI over the hippocampal region.
The current method used only the SyN nonlinear registration algorithm, which is computationally expensive. However, this method outperformed several nonlinear registration methods in a large head-to-head comparison (Klein et al., 2009). Moreover, computationally intensive registration algorithms are increasingly accessible for most investigators given rapid improvements in multiplex computing, processing speed and memory.
We cannot fully exclude the possibility that a larger template library (>100 templates) might improve segmentation results in AD and NC, particularly with outliers. Nevertheless, the SBHV library was selected based on a priori criteria to ensure sufficient template variation in a heterogeneous AD sample and normal aging.
Additionally, we used a non-weighted voting strategy to compare protocol libraries. However, more sophisticated weighted priors and label fusion strategies may furnish even greater accuracy (Robitaille and Duchesne 2012; Wang et al., 2011a). We also affine registered templates to the MNI-152 template brain to compare similarity over the hippocampal region, since this has been previously shown to identify similarity among MTL ROIs centered over the hippocampus (Collins and Pruessner 2010; Kim et al., 2012). However, this linear fitting to a healthy average brain may lower variability to assess similarity among atrophic brains (e.g. in AD); so future work should evaluate accuracy using an average elderly or AD-specific brain template.
Recently, (Wang et al., 2011b) developed a wrapper based correction method, which detects systematic bias across an input dataset and accordingly adjusts final label volumes. This method improved multi-atlas segmentation accuracy by 1% (DSC =0.0887 → 0.908) for multi-template based segmentations and may significantly improve segmentation outcomes for less inclusive protocols.
Conclusions
Accurate and precise automated hippocampal volumetry is ever more important for ROI demarcation of functional imaging measurements, supporting a diagnosis of AD and determining therapeutic efficacy in putative disease modifying MCI and AD trials. Although exceptional efforts are underway to manually harmonize the hippocampus in AD, several automated methods are based on a variety of labeling protocols, are widely used in other diseases besides AD and a direct comparison among atlas protocols has not been previously conducted to determine optimal hippocampal definitions for multi-atlas methods. The SBHV fully automated method uses a template library derived from a representative memory clinic cohort and demonstrates comparable to better voxel-overlap outcomes (DSC=0.85–0.88) compared to previous approaches in NC and AD. Although a consensus definition is ongoing for the hippocampus, it remains integral to determine how automatic segmentation performance (accuracy and sensitivity) is impacted by atlas composition. Given our findings, the most accurate results were for protocols that included the majority of the hippocampal tail, alveus and fimbria: P3 (DSC=0.88), P4 (0.88) and P5 (0.88). Anatomical differences for the medial hippocampal body did not markedly affect accuracy among the most inclusive atlas protocols (P3–P5). In contrast, voxel-wise error differences among protocols were principally distributed around the alveus/fimbria grey-white matter border and the posterior hippocampus. Moreover, errors affecting the caudal hippocampus were more pronounced and desperate in ADNI-1 AD subjects when the posterior definition varied among protocols. Voxel-wise segmentation accuracy was lower across protocols for the more pathologically heterogeneous Sunnybrook sample (NC, AD+VaD, VaD, AD+SVD and AD) versus the ADNI-1 validation dataset, which included only NC, AD and amnestic MCI subjects. All protocols discriminated between NC, MCI and AD in the expected directions and showed similar associations with episodic memory performance and decline in both MCI and AD. At the same time, our findings confirm manually derived rates of change in the literature. Finally, more inclusive protocols appear to furnish slightly better group-wise separation between MCI subgroups modestly better associations with episodic memory measures. A broad interpretation of our results suggests that on the whole, more inclusive hippocampal definitions that include the alveus, fimbria and >hippocampal tail capture slightly more pathological change and offer more robust segmentation outcomes, which together may explain the improved biomarker performance in MCI and AD when compared to less inclusive definitions. Given that the majority of automated techniques rely on prior structural information, our ADNI-1 and Sunnybrook Dementia Study performance findings have application to other automatic hippocampal segmentation techniques. Moreover, these results extend beyond AD to studies in healthy aging and may be relevant to other neurodegenerative diseases. It is the investigators’ hope that these performance findings ultimately advance the selection, design and interpretation of atlases used for automatic hippocampal segmentation in AD.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge financial support from the following sources. The development and testing of the hippocampal segmentation algorithm was supported by several grants, most notably from the Canadian Institutes of Health Research (MT#13129), the Alzheimer Society of Canada and Alzheimer Association (US), the Heart and Stroke Foundation Centre for Stroke Recovery, and the LC Campbell Foundation. SN receives financial support from a Canadian Institutes of Health Research Fredrick Banting and Charles Best Canada Doctoral Scholarship, and SEB from the Departments of Medicine at Sunnybrook and U of T, including the Brill Chair in Neurology. EG, FQG, and SEB also receive salary support from the HSF Centre for Stroke Recovery. We would like to thank Gregory Szilagyi for coding a multiplexing batch script in Python and the anonymous reviewers for their helpful comments. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for NeuroImaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wpcontent/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
References
- Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D. Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. NeuroImage. 2009;46(3):726–738. doi: 10.1016/j.neuroimage.2009.02.018. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Morra JH, Green AE, Hwang KS, Avedissian C, Woo E, Cummings JL, Toga AW, Jack CR, Jr, Weiner MW, et al. Automated 3D mapping of baseline and 12-month associations between three verbal memory measures and hippocampal atrophy in 490 ADNI subjects. NeuroImage. 2010;51(1):488–499. doi: 10.1016/j.neuroimage.2009.12.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J, Bartlett JW, van de Pol LA, Loy CT, Scahill RI, Frost C, Thompson P, Fox NC. A meta-analysis of hippocampal atrophy rates in alzheimer's disease. Neurobiology of Aging. 2009;30(11):1711–1723. doi: 10.1016/j.neurobiolaging.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J, Foster J, Boyes RG, Pepple T, Moore EK, Schott JM, Frost C, Scahill RI, Fox NC. A comparison of methods for the automated calculation of volumes and atrophy rates in the hippocampus. NeuroImage. 2008a;40(4):1655–1671. doi: 10.1016/j.neuroimage.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Barnes J, Foster J, Boyes RG, Pepple T, Moore EK, Schott JM, Frost C, Scahill RI, Fox NC. A comparison of methods for the automated calculation of volumes and atrophy rates in the hippocampus. NeuroImage. 2008b;40(4):1655–1671. doi: 10.1016/j.neuroimage.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Bartzokis G, Altshuler LL, Greider T, Curran J, Keen B, Dixon WJ. Reliability of medial temporal lobe volume measurements using reformatted 3D images. Psychiatry Research. 1998;82(1):11–24. doi: 10.1016/s0925-4927(98)00007-9. [DOI] [PubMed] [Google Scholar]
- Boccardi M, Ganzola R, Bocchetta M, Pievani M, Redolfi A, Bartzokis G, Camicioli R, Csernansky JG, de Leon MJ, deToledo-Morrell L, et al. Survey of protocols for the manual segmentation of the hippocampus: Preparatory steps towards a joint EADC-ADNI harmonized protocol. Journal of Alzheimer's Disease : JAD. 2011;26(Suppl 3):61–75. doi: 10.3233/JAD-2011-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael O, Schwarz C, Drucker D, Fletcher E, Harvey D, Beckett L, Jack CR, Jr, Weiner M, DeCarli C Alzheimer's Disease Neuroimaging Initiative. Longitudinal changes in white matter disease and cognition in the first year of the alzheimer disease neuroimaging initiative. Archives of Neurology. 2010;67(11):1370–1378. doi: 10.1001/archneurol.2010.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael OT, Aizenstein HA, Davis SW, Becker JT, Thompson PM, Meltzer CC, Liu Y. Atlas-based hippocampus segmentation in alzheimer's disease and mild cognitive impairment. NeuroImage. 2005;27(4):979–990. doi: 10.1016/j.neuroimage.2005.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins DL, Holmes CJ, Peters TM, Evans AC. Automatic 3-D model-based neuroanatomical segmentation. Human Brain Mapping. 1995;3(3):190–208. [Google Scholar]
- Collins DL, Pruessner JC. Towards accurate, automatic segmentation of the hippocampus and amygdala from MRI by augmenting ANIMAL with a template library and label fusion. NeuroImage. 2010;52(4):1355–1366. doi: 10.1016/j.neuroimage.2010.04.193. [DOI] [PubMed] [Google Scholar]
- Convit A, De Leon MJ, Tarshish C, De Santi S, Tsui W, Rusinek H, George A. Specific hippocampal volume reductions in individuals at risk for alzheimer's disease. Neurobiology of Aging. 1997;18(2):131–138. doi: 10.1016/s0197-4580(97)00001-8. [DOI] [PubMed] [Google Scholar]
- Costafreda SG, Dinov ID, Tu Z, Shi Y, Liu CY, Kloszewska I, Mecocci P, Soininen H, Tsolaki M, Vellas B, et al. Automated hippocampal shape analysis predicts the onset of dementia in mild cognitive impairment. NeuroImage. 2011;56(1):212–219. doi: 10.1016/j.neuroimage.2011.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Heijer T, van der Lijn F, Koudstaal PJ, Hofman A, van der Lugt A, Krestin GP, Niessen WJ, Breteler MM. A 10-year follow-up of hippocampal volume on magnetic resonance imaging in early dementia and cognitive decline. Brain : A Journal of Neurology. 2010;133(Pt 4):1163–1172. doi: 10.1093/brain/awq048. [DOI] [PubMed] [Google Scholar]
- deToledo-Morrell L, Stoub TR, Bulgakova M, Wilson RS, Bennett DA, Leurgans S, Wuu J, Turner DA. MRI-derived entorhinal volume is a good predictor of conversion from MCI to AD. Neurobiology of Aging. 2004;25(9):1197–1203. doi: 10.1016/j.neurobiolaging.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Dubois B, Feldman HH, Jacova C, Dekosky ST, Barberger-Gateau P, Cummings J, Delacourte A, Galasko D, Gauthier S, Jicha G, et al. Research criteria for the diagnosis of alzheimer's disease: Revising the NINCDS-ADRDA criteria. Lancet Neurology. 2007;6(8):734–746. doi: 10.1016/S1474-4422(07)70178-3. [see comment] [DOI] [PubMed] [Google Scholar]
- Duvernoy HM. The human hippocampus. Heidelberg: Springer-Verlag; 1998. [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, et al. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fleiss JL. Design and analysis of clinical experiments. New York: John Wiley & Sons, Inc; 1986. [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. "Mini-mental state". A practical method for grading the cognitive state of patients for the clinician. Journal of Psychiatric Research. 1975;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Fonov VS, Evans AC, McKinstry RC, Almli CR, Collins DL. Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. NeuroImage. 2009;47:S102. [Google Scholar]
- Fonov V, Evans AC, Botteron K, Almli CR, McKinstry RC, Collins DL Brain Development Cooperative Group. Unbiased average age-appropriate atlases for pediatric studies. NeuroImage. 2011;54(1):313–327. doi: 10.1016/j.neuroimage.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox NC, Ridgway GR, Schott JM. Algorithms, atrophy and alzheimer's disease: Cautionary tales for clinical trials. NeuroImage. 2011;57(1):15–18. doi: 10.1016/j.neuroimage.2011.01.077. [DOI] [PubMed] [Google Scholar]
- Fox NC, Cousens S, Scahill R, Harvey RJ, Rossor MN. Using serial registered brain magnetic resonance imaging to measure disease progression in alzheimer disease: Power calculations and estimates of sample size to detect treatment effects. Archives of Neurology. 2000;57(3):339–344. doi: 10.1001/archneur.57.3.339. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Jack CR. Harmonization of magnetic resonance-based manual hippocampal segmentation: A mandatory step for wide clinical use. Alzheimer's & Dementia : The Journal of the Alzheimer's Association. 2011;7(2):171–174. doi: 10.1016/j.jalz.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Gerardin E, Chetelat G, Chupin M, Cuingnet R, Desgranges B, Kim HS, Niethammer M, Dubois B, Lehericy S, Garnero L, et al. Multidimensional classification of hippocampal shape features discriminates alzheimer's disease and mild cognitive impairment from normal aging. NeuroImage. 2009;47(4):1476–1486. doi: 10.1016/j.neuroimage.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachinski V, Iadecola C, Petersen RC, Breteler MM, Nyenhuis DL, Black SE, Powers WJ, DeCarli C, Merino JG, Kalaria RN, et al. National institute of neurological disorders and stroke-canadian stroke network vascular cognitive impairment harmonization standards. Stroke; a Journal of Cerebral Circulation. 2006;37(9):2220–2241. doi: 10.1161/01.STR.0000237236.88823.47. [DOI] [PubMed] [Google Scholar]
- Haller JW, Banerjee A, Christensen GE, Gado M, Joshi S, Miller MI, Sheline Y, Vannier MW, Csernansky JG. Three-dimensional hippocampal MR morphometry with high-dimensional transformation of a neuroanatomic atlas. Radiology. 1997;202(2):504–510. doi: 10.1148/radiology.202.2.9015081. [DOI] [PubMed] [Google Scholar]
- Hammers A, Heckemann R, Koepp MJ, Duncan JS, Hajnal JV, Rueckert D, Aljabar P. Automatic detection and quantification of hippocampal atrophy on MRI in temporal lobe epilepsy: A proof-of-principle study. NeuroImage. 2007;36(1):38–47. doi: 10.1016/j.neuroimage.2007.02.031. [DOI] [PubMed] [Google Scholar]
- Hampel H, Frank R, Broich K, Teipel SJ, Katz RG, Hardy J, Herholz K, Bokde AL, Jessen F, Hoessler YC, et al. Biomarkers for alzheimer's disease: Academic, industry and regulatory perspectives. Nature Reviews.Drug Discovery. 2010;9(7):560–574. doi: 10.1038/nrd3115. [DOI] [PubMed] [Google Scholar]
- Hasboun D, Chantome M, Zouaoui A, Sahel M, Deladoeuille M, Sourour N, Duyme M, Baulac M, Marsault C, Dormont D. MR determination of hippocampal volume: Comparison of three methods. AJNR.American Journal of Neuroradiology. 1996;17(6):1091–1098. [PMC free article] [PubMed] [Google Scholar]
- Heckemann RA, Hajnal JV, Aljabar P, Rueckert D, Hammers A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage. 2006;33(1):115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- Holland D, McEvoy LK, Dale AM the Alzheimer's Disease Neuroimaging Initiative. Unbiased comparison of sample size estimates from longitudinal structural measures in ADNI. Human Brain Mapping. 2011;33(11):2586–2602. doi: 10.1002/hbm.21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Coupe P, Pruessner JC, Collins DL. Appearance-based modeling for segmentation of hippocampus and amygdala using multi-contrast MR imaging. NeuroImage. 2011;58(2):549–559. doi: 10.1016/j.neuroimage.2011.06.054. [DOI] [PubMed] [Google Scholar]
- Jack CR., Jr MRI-based hippocampal volume measurements in epilepsy. Epilepsia. 1994;35(Suppl 6):S21–S29. doi: 10.1111/j.1528-1157.1994.tb05986.x. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Weigand SD, Shiung MM, Przybelski SA, O'Brien PC, Gunter JL, Knopman DS, Boeve BF, Smith GE, Petersen RC. Atrophy rates accelerate in amnestic mild cognitive impairment. Neurology. 2008a;70(19 Pt 2):1740–1752. doi: 10.1212/01.wnl.0000281688.77598.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Lowe VJ, Weigand SD, Wiste HJ, Senjem ML, Knopman DS, Shiung MM, Gunter JL, Boeve BF, Kemp BJ, et al. Serial PIB and MRI in normal, mild cognitive impairment and alzheimer's disease: Implications for sequence of pathological events in alzheimer's disease. Brain : A Journal of Neurology. 2009;132(Pt 5):1355–1365. doi: 10.1093/brain/awp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Barkhof F, Bernstein MA, Cantillon M, Cole PE, Decarli C, Dubois B, Duchesne S, Fox NC, Frisoni GB, et al. Steps to standardization and validation of hippocampal volumetry as a biomarker in clinical trials and diagnostic criterion for alzheimer's disease. Alzheimer's & Dementia : The Journal of the Alzheimer's Association. 2011;7(4):474–485. e4. doi: 10.1016/j.jalz.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski BJ, Britson PJ, Whitwell J, Ward C, et al. The alzheimer's disease neuroimaging initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging : JMRI. 2008b;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, et al. Reliability in multi-site structural MRI studies: Effects of gradient non-linearity correction on phantom and human data. NeuroImage. 2006;30(2):436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Killiany RJ, Moss MB, Albert MS, Sandor T, Tieman J, Jolesz F. Temporal lobe regions on magnetic resonance imaging identify patients with early alzheimer's disease. Archives of Neurology. 1993;50(9):949–954. doi: 10.1001/archneur.1993.00540090052010. [DOI] [PubMed] [Google Scholar]
- Kim H, Chupin M, Colliot O, Bernhardt BC, Bernasconi N, Bernasconi A. Automatic hippocampal segmentation in temporal lobe epilepsy: Impact of developmental abnormalities. NeuroImage. 2012;59(4):3178–3186. doi: 10.1016/j.neuroimage.2011.11.040. [DOI] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage. 2009;46(3):786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad C, Ukas T, Nebel C, Arolt V, Toga AW, Narr KL. Defining the human hippocampus in cerebral magnetic resonance images--an overview of current segmentation protocols. NeuroImage. 2009;47(4):1185–1195. doi: 10.1016/j.neuroimage.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KK, Barnes J, Modat M, Ridgway GR, Bartlett JW, Fox NC, Ourselin S Alzheimer's Disease Neuroimaging Initiative. Brain MAPS: An automated, accurate and robust brain extraction technique using a template library. NeuroImage. 2011;55(3):1091–1108. doi: 10.1016/j.neuroimage.2010.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KK, Barnes J, Ridgway GR, Bartlett JW, Clarkson MJ, Macdonald K, Schuff N, Fox NC, Ourselin S Alzheimer's Disease Neuroimaging Initiative. Automated cross-sectional and longitudinal hippocampal volume measurement in mild cognitive impairment and alzheimer's disease. NeuroImage. 2010;51(4):1345–1359. doi: 10.1016/j.neuroimage.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HK, Jung WS, Ahn KJ, Won WY, Hahn C, Lee SY, Kim I, Lee CU. Relationships between hippocampal shape and cognitive performances in drug-naive patients with alzheimer's disease. Neuroscience Letters. 2012;516(1):124–129. doi: 10.1016/j.neulet.2012.03.072. [DOI] [PubMed] [Google Scholar]
- Lotjonen J, Wolz R, Koikkalainen J, Julkunen V, Thurfjell L, Lundqvist R, Waldemar G, Soininen H, Rueckert D Alzheimer's Disease Neuroimaging Initiative. Fast and robust extraction of hippocampus from MR images for diagnostics of alzheimer's disease. NeuroImage. 2011;56(1):185–196. doi: 10.1016/j.neuroimage.2011.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotjonen JM, Wolz R, Koikkalainen JR, Thurfjell L, Waldemar G, Soininen H, Rueckert D Alzheimer's Disease Neuroimaging Initiative. Fast and robust multi-atlas segmentation of brain magnetic resonance images. NeuroImage. 2010;49(3):2352–2365. doi: 10.1016/j.neuroimage.2009.10.026. [DOI] [PubMed] [Google Scholar]
- Malykhin NV, Bouchard TP, Ogilvie CJ, Coupland NJ, Seres P, Camicioli R. Three-dimensional volumetric analysis and reconstruction of amygdala and hippocampal head, body and tail. Psychiatry Research. 2007;155(2):155–165. doi: 10.1016/j.pscychresns.2006.11.011. [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of alzheimer's disease: Report of the NINCDS-ADRDA work group under the auspices of department of health and human services task force on alzheimer's disease. Neurology. 1984;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr, Kawas CH, Klunk WE, Koroshetz WJ, Manly JJ, Mayeux R, et al. The diagnosis of dementia due to alzheimer's disease: Recommendations from the national institute on aging-alzheimer's association workgroups on diagnostic guidelines for alzheimer's disease. Alzheimer's & Dementia : The Journal of the Alzheimer's Association. 2011;7(3):263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Toga AW, Jack CR, Jr, Schuff N, et al. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with alzheimer's disease, mild cognitive impairment, and elderly controls. NeuroImage. 2009;45(1 Suppl):S3–S15. doi: 10.1016/j.neuroimage.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouiha A, Duchesne S Alzheimer's Disease Neuroimaging Initiative. Hippocampal atrophy rates in alzheimer's disease: Automated segmentation variability analysis. Neuroscience Letters. 2011;495(1):6–10. doi: 10.1016/j.neulet.2011.02.065. [DOI] [PubMed] [Google Scholar]
- Niemann K, Hammers A, Coenen VA, Thron A, Klosterkotter J. Evidence of a smaller left hippocampus and left temporal horn in both patients with first episode schizophrenia and normal control subjects. Psychiatry Research. 2000;99(2):93–110. doi: 10.1016/s0925-4927(00)00059-7. [DOI] [PubMed] [Google Scholar]
- Pantel J, O'Leary DS, Cretsinger K, Bockholt HJ, Keefe H, Magnotta VA, Andreasen NC. A new method for the in vivo volumetric measurement of the human hippocampus with high neuroanatomical accuracy. Hippocampus. 2000;10(6):752–758. doi: 10.1002/1098-1063(2000)10:6<752::AID-HIPO1012>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage. 2011;56(3):907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen JA, Sathiyamoorthy G, Gao FQ, Szilagyi G, Nadkarni NK, St George-Hyslop P, Rogaeva E, Black SE. Microbleed topography, leukoaraiosis, and cognition in probable alzheimer disease from the sunnybrook dementia study. Archives of Neurology. 2008;65(6):790–795. doi: 10.1001/archneur.65.6.790. [DOI] [PubMed] [Google Scholar]
- Pruessner JC, Li LM, Serles W, Pruessner M, Collins DL, Kabani N, Lupien S, Evans AC. Volumetry of hippocampus and amygdala with high-resolution MRI and three-dimensional analysis software: Minimizing the discrepancies between laboratories. Cerebral Cortex (New York NY 1991. 2000;10(4):433–442. doi: 10.1093/cercor/10.4.433. [DOI] [PubMed] [Google Scholar]
- Ramirez J, Gibson E, Quddus A, Lobaugh NJ, Feinstein A, Levine B, Scott CJ, Levy-Cooperman N, Gao FQ, Black SE. Lesion explorer: A comprehensive segmentation and parcellation package to obtain regional volumetrics for subcortical hyperintensities and intracranial tissue. NeuroImage. 2011;54(2):963–973. doi: 10.1016/j.neuroimage.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Rey A. L'examen clinique en psychologie. France: Presses Universitaires de France; 1964. [Google Scholar]
- Robitaille N, Duchesne S. Label fusion strategy selection. International Journal of Biomedical Imaging. 2012;2012 doi: 10.1155/2012/431095. 431095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfing T, Maurer CR., Jr Shape-based averaging. IEEE Transactions on Image Processing : A Publication of the IEEE Signal Processing Society. 2007;16(1):153–161. doi: 10.1109/tip.2006.884936. [DOI] [PubMed] [Google Scholar]
- Scher AI, Xu Y, Korf ES, Hartley SW, Witter MP, Scheltens P, White LR, Thompson PM, Toga AW, Valentino DJ, et al. Hippocampal morphometry in population-based incident alzheimer's disease and vascular dementia: The HAAS. Journal of Neurology, Neurosurgery, and Psychiatry. 2011;82(4):373–377. doi: 10.1136/jnnp.2008.165902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JA, Arvanitakis Z, Bang W, Bennett DA. Mixed brain pathologies account for most dementia cases in community-dwelling older persons. Neurology. 2007;69(24):2197–2204. doi: 10.1212/01.wnl.0000271090.28148.24. [DOI] [PubMed] [Google Scholar]
- Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, Thompson PM, Jack CR, Jr, Weiner MW Alzheimer's Disease Neuroimaging I. MRI of hippocampal volume loss in early alzheimer's disease in relation to ApoE genotype and biomarkers. Brain. 2009;132(Pt 4):1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen KK, Fripp J, Meriaudeau F, Chetelat G, Salvado O, Bourgeat P Alzheimer's Disease Neuroimaging Initiative. Detecting global and local hippocampal shape changes in alzheimer's disease using statistical shape models. NeuroImage. 2012;59(3):2155–2166. doi: 10.1016/j.neuroimage.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Shen L, Saykin AJ, Kim S, Firpi HA, West JD, Risacher SL, McDonald BC, McHugh TL, Wishart HA, Flashman LA. Comparison of manual and automated determination of hippocampal volumes in MCI and early AD. Brain Imaging and Behavior. 2010;4(1):86–95. doi: 10.1007/s11682-010-9088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Transactions on Medical Imaging. 1998;17(1):87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lijn F, den Heijer T, Breteler MM, Niessen WJ. Hippocampus segmentation in MR images using atlas registration, voxel classification, and graph cuts. NeuroImage. 2008;43(4):708–720. doi: 10.1016/j.neuroimage.2008.07.058. [DOI] [PubMed] [Google Scholar]
- Wang H, Yushkevich PA. Spatial bias in multi-atlas based segmentation. Computer Vision and Pattern Recognition. 2012:909–916. doi: 10.1109/CVPR.2012.6247765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Suh JW, Pluta J, Altinay M, Yushkevich P. Optimal weights for multi-atlas label fusion. Information Processing in Medical Imaging : Proceedings of the …Conference. 2011a;22:73–84. doi: 10.1007/978-3-642-22092-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Das SR, Suh JW, Altinay M, Pluta J, Craige C, Avants B, Yushkevich PA Alzheimer's Disease Neuroimaging Initiative. A learning-based wrapper method to correct systematic errors in automatic image segmentation: Consistently improved performance in hippocampus, cortex and brain segmentation. NeuroImage. 2011b;55(3):968–985. doi: 10.1016/j.neuroimage.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Transactions on Medical Imaging. 2004;23(7):903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson C, Andermann F, Gloor P, Jones-Gotman M, Peters T, Evans A, Olivier A, Melanson D, Leroux G. Anatomic basis of amygdaloid and hippocampal volume measurement by magnetic resonance imaging. Neurology. 1992;42(9):1743–1750. doi: 10.1212/wnl.42.9.1743. [DOI] [PubMed] [Google Scholar]
- Wechsler D. Manual for the wechsler adult intelligence scale revised. New York: Psychological Corporation; 1981. [Google Scholar]
- Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Harvey D, Jack CR, Jagust W, Liu E, et al. The alzheimer's disease neuroimaging initiative: A review of papers published since its inception. Alzheimer's & Dementia. 2012;8(1 Suppl):S1–S68. doi: 10.1016/j.jalz.2011.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MW, Aisen PS, Jack CR, Jr, Jagust WJ, Trojanowski JQ, Shaw L, Saykin AJ, Morris JC, Cairns N, Beckett LA, et al. The alzheimer's disease neuroimaging initiative: Progress report and future plans. Alzheimer's & Dementia : The Journal of the Alzheimer's Association. 2010;6(3):202–11. e7. doi: 10.1016/j.jalz.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolz R, Aljabar P, Heckemann RA, Hammers A, Rueckert D. Segmentation of subcortical structures and the hippocampus in brain MRI using graph-cuts and subject-specific a-priori information; IEEE International Symposium on Biomedical Imaging — ISBI; 2009. pp. 470–473. [Google Scholar]
- Wolz R, Aljabar P, Hajnal JV, Hammers A, Rueckert D Alzheimer's Disease Neuroimaging Initiative. LEAP: Learning embeddings for atlas propagation. NeuroImage. 2010a;49(2):1316–1325. doi: 10.1016/j.neuroimage.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolz R, Heckemann RA, Aljabar P, Hajnal JV, Hammers A, Lotjonen J, Rueckert D Alzheimer's Disease Neuroimaging Initiative. Measurement of hippocampal atrophy using 4D graph-cut segmentation: Application to ADNI. NeuroImage. 2010b;52(1):109–118. doi: 10.1016/j.neuroimage.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Yousefi S, Kehtarnavaz N, Gholipour A. Improved labeling of subcortical brain structures in atlas-based segmentation of magnetic resonance images. IEEE Transactions on Bio-Medical Engineering. 2011;59(7):1808–1817. doi: 10.1109/TBME.2011.2122306. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.