Abstract
Genome-wide association studies (GWAS) have identified polymorphic loci associated with coronary artery disease (CAD) risk factors (i. e. serum lipids) in adult populations (42–69 yrs). We hypothesized that younger populations would show a greater relative genetic component due to fewer confounding variables. We examined the influence of 20 GWAS loci associated with serum lipids and insulin metabolism, in a university student cohort (n=548; mean age= 24 yrs), and replicated statistically associated results in a second study cohort of primary school students (n=810, mean age= 11.5 yrs). 19 loci showed no relationship with studied risk factors in young adults. However, the ancestral allele of the rs646776 (SORT1) locus was strongly associated with increased low density lipoprotein cholesterol {LDL (C)} in young adults (TT: 97.6 ± 1.0 mg/dL {n=345}, vs. CT/CC: 87.3 ± 1.0 mg/dL {n=203}; p = 3 × 10−6) and children (TT: 94.0 ± 1.3 mg/dL {n=551}, vs. CT/CC: 84.7 ± 1.4 mg/dL {n=259}; p = 4 × 10−6). This locus is responsible for 3.6% of population variance in young adults and 2.5% of population variance in children. The effect size of the SORT1 locus is considerably higher in young populations (2.5%–4.1%) compared to older subjects (1%).
INTRODUCTION
Coronary artery disease is a major health problem of industrialized countries. Some risk factors for CAD such as insulin resistance and type II diabetes mellitus (T2DM) are also rising even amongpediatric populations primarily because of changes in life-style including a high caloric intake and low physical activity.
Many GWA studies have been used to identify genetic risk factors for markers associated with selected CAD risk factors (1, 2). Typically, GWAS has used dichotomous traits (affected vs. non-affected controls) to identify new loci for such CAD risk factors as glucose levels (insulin resistance) (1, 2), T2DM (1), obesity (BMI) (3), blood pressure (4), and serum lipids (5).
A consistent finding of all of these GWAS studies of individual CAD risk factors and associated phenotypes has been the relatively weak contribution of any specific locus to the relative risk. Dichotomous studies of disease risk for T2DM, for example, have typically found only a 1.1-fold increase in relative risk for any specific genetic marker. This low effect size shows a marked difference from twin studies for each of the biochemical, physiological, and anthropomorphic measures associated with the development of such CAD risk factors as T2DM (6). Moreover, the poor predictive power of genetic loci, either singly or in isolation, have led some to conclude that clinical measures are in fact more prognostic than genetic predispositions (7). Likewise, quantitative (continuous) trait studies typically report that specific genetic components have an effect size of 1% or less. This has led to the hypothesis that traits associated with CAD risk factors are highly polygenic, with many loci each contributing a very small proportion of variation.
We hypothesized that study of younger populations could prove more valuable in identifying genetic contributions to CAD risk for two reasons: genetic markers associated with CAD risk factors could be highly valuable tools to identify individuals who might benefit most from early interventions and young adults lack many of the co-morbidities that complicate gene/phenotype associations. The co-variance of the co-morbidities in older subjects would serve to lessen the effect size of any specific genetic locus. Thus, young adults may show much larger effect sizes. Finally, the study of young adults should uncover the genetic loci associated with onset of the disease, rather than later disease progression. There are also disadvantages to studying young adults. The variance of values is considerably less in young adults, with fewer subjects showing abnormal values of the marker, and this might be expected to reduce statistical power. However, we hypothesized that the reduced statistical power would be offset by larger effect sizes of strong genetic components.
In this study, we analyzed 20 GWAS-identified single nucleotide polymorphisms (SNP) in younger populations. The loci studied have been replicated in numerous studies, with seven loci associated with serum lipids (5, 8, 9), 10 associated with T2DM (1), and three associated with development of CAD (8, 10). The initial cohort studied was college-aged individuals (Functional Polymorphism Associated with Human Muscle Size and Strength; FAMUSS), and gene associations queried for LDL (C), high-density lipoprotein cholesterol {HDL (C)}, triglycerides, glucose, homoeostasis model assessment (HOMA), and insulin. Validation of positive associations was performed in a cohort of 6TH grade volunteers (Cardiovascular Health Intervention Program; CHIP).
We establish that the 1p13.3 LDL (C) locus was strongly associated with a CAD risk factor in both of these young populations. LDL (C) is strong predictor of CAD (11), and this locus could be used to identify individuals who could benefit from early interventions to prevent CAD.
Methods
Genotyping of the 20 GWAS-associated SNPs was performed in FAMUSS to identify loci associated with the following CAD risk factors: LDL (C), HDL (C), triglycerides (TG), glucose, HOMA, and insulin with and without adjustment for age and BMI. Significant loci were validated in the CHIP cohort.
FAMUSS cohort
The FAMUSS study design is a population-based study of individuals (average age = 24 yrs old) designed to identify genetic factors associated with skeletal muscle changes as a result of resistance training (12). We collected selected baseline CAD risk factors such as serum lipids {LDL (C), HDL (C), and TG}, glucose, and insulin following an overnight fast (Quest Diagnostics, Chantilly, VA, 20153) in 548 White participants. The HOMA values were obtained using this calculation: (insulin × fasting glucose)/22.5 (13). The study was approved by the Children's National Medical Center Institutional Review Board (protocol #2449) in compliance with the Helsinki Declaration. Informed consent was obtained from all subject participants.
CHIP cohort
CHIP is a population-based study of 6th graders (11.5 yrs of age) in Michigan that includes a screening component and health education program (manuscript in preparation). The screenings have included the following CAD risk factor assessments: physical activity, cardiovascular fitness, body composition, blood pressure, family history, fasted blood lipids {LDL (C), HDL (C), and TG}, glucose, dietary assessments, grip strength, and a blood spot sample. Serum lipids and markers of glucose metabolism were available for 810 participants. The study was approved by the Central Michigan University in compliance with the Helsinki Declaration. Informed consent was obtained from all mothers and/or fathers of participants in the CHIP study. De-identified blood spots were sent to Children’s National Medical Center for genotyping, and genotyping was approved by the CNMC IRB.
The Study of Targeted Risk Reduction Intervention through Defined Exercise study (STRRIDE)
This study examines defined modes, doses and intensities of exercise in a structured exercise program, with the goal to define the relationship between mode, amount and intensities of exercise and improvements in risk parameters of cardiovascular health. The STRRIDE encompassed 555 middle-aged subjects with features of metabolic syndrome (25<BMI<35; fasting hyperinsulinemia {>10 IU/ml}, and dyslipidemia with LDL cholesterol between 130–190, and/or HDL < 45 for women, < 35 for men). Subjects were assigned to one of six groups addressing exercise mode, intensity and duration. STRRIDE participants signed informed consent associated with IRB approval by Duke University Medical Center.
Genotyping Methods
Genomic DNA was isolated using Puregene kits (Gentra Systems, MN) from peripheral blood (FAMUSS), or from blood spots using Qiagen kits (CHIP). Genotyping was done using TaqMan allele discrimination assays (14). Both alleles were detected simultaneously and automatically called (see Table 1).
Table 1.
Assay ID#s for SNPs genotyped in FAMUSS Study.
| Region | SNP | ABI ASSAY ID# |
|---|---|---|
| 1p31.3 | rs12130333 | C__31145189_10 |
| 1p13.3 | rs646776 | C___3160062_10 |
| 1q42.13 | rs4846914 | C__27968379_10 |
| 3p25.2 | rs1801282 | C___1129864_10 |
| 3q22.3 | rs9818870 | C__26070086_10 |
| 3q27.2 | rs4402960 | C___2165199_10 |
| 6p22.3 | rs7756992 | C___2504058_20 |
| 6p22.3 | rs10946398 | C__31635230_10 |
| 7q11.23 | rs17145738 | C___2632544_10 |
| 8q24.11 | rs13266634 | C____357888_10 |
| 8q24.13 | rs17321515 | C__33068431_10 |
| 9p21.3 | rs1333049 | C___1754666_10 |
| 9p21.3 | rs10811661 | C__31288917_10 |
| 10q23.33 | rs1111875 | C__11214581_10 |
| 10q23.33 | rs7923837 | C__31982553_10 |
| 10q25.2 | rs7901695 | C____384583_10 |
| 11p15.1 | rs5219 | C__11654065_10 |
| 12q24.11 | rs2338104 | C___2877290_10 |
| 12q24.31 | rs2259816 | C__11753816_10 |
| 19p13.11 | rs16996148 | C__33703643_10 |
Statistical Analyses
All genotype data sets were tested for Hardy-Weinberg equilibrium by using a χ2 tests to compare the observed genotype frequencies to those expected under equilibrium. Normality of each quantitative trait was tested using the Shapiro-Wilk normality test and any trait not meeting the criteria was appropriately transformed.
Bivariate correlation analyses of each quantitative measurement showed several significant correlations with age and baseline weight; therefore, associations between each SNP and quantitative phenotype were assessed using analysis of covariance methods using both dominant genetic models and quantitative trait models.
Linear regression analysis, including likelihood ratio tests between full (containing genotype and covariates) and constrained (containing covariates only) models, were performed to estimate the proportion of variance in volumetric measurements attributable to each SNP’s genotype. All statistical analyses were performed using Stata V.8 (StataCorp, College Park, TX).
The CHIP cohort was used as a validation data set. Thus, only statistically significant associations surviving multiple testing thresholds seen in FAMUSS were subsequently tested in the CHIP cohort. Statistical significance threshold was established as p< 0.05 as this was a validation cohort with no multiple testing.
RESULTS
Analysis of Selected CAD Risk Factor GWAS loci in young adults
The FAMUSS cohort was utilized as the test cohort. Traits used for association studies were fasting glucose (mmol/L), HOMA, insulin (µU/ml), LDL (C) (mg/dl), HDL (C) (mg/dl), triglycerides (mg/dL), with and without adjustment for BMI (kg/m2). Average values for all subjects and stratified by sex are provided in Table 2.
Table 2.
Sample Characteristics for FAMUSS and CHIP Population-Based Cohorts.
| Characteristic | FAMUSS (Test Cohort) | CHIP (Replication Cohort) | ||||
|---|---|---|---|---|---|---|
| Mean ± SD | Mean ± SD | |||||
| All | Females | Males | All | Females | Males | |
| N | 548 | 348 | 200 | 810 | 428 | 382 |
| Mean Age (years) | 23.86 ± 5.71 | 23.67 ± 5.71 | 24.19 ± 5.71 | 11.52 ± 0.81 | 11.44 ± 0.78 | 11.61 ± 0.83 |
| BMI (kg/m2)* | 24.48 ± 4.84 | 23.89 ± 4.64 | 25.49 ± 5.03 | 21.56 ± 5.13 | 21.67 ± 5.45 | 21.44 ± 4.73 |
| Total cholesterol (mg/dl) | 166.49 ± 31.35 | 166.62 ± 32.24 | 166.26 ± 29.81 | 165.39 ± 31.89 | 163.88 ± 30.58 | 167.08 ± 33.26 |
| LDL (C) (mg/dl) | 97.54 ± 27.85 | 94.99 ± 28.02 | 101.97 ± 27.04 | 95.21 ± 28.95 | 93.17 ± 27.01 | 97.49 ± 30.85 |
| HDL (C) (mg/dl) | 48.26 ± 12.82 | 52.32 ± 11.81 | 41.21 ± 11.40 | 51.26 ± 14.43 | 51.52 ± 14.53 | 50.97 ± 14.33 |
| Triglycerides (mg/dL) | 103.48 ± 52.23 | 96.62 ± 45.76 | 115.41 ± 60.17 | 96.70 ± 51.92 | 99.17 ± 53.84 | 93.93 ± 49.61 |
| Insulin (µU/mL) | 5.58 ± 4.65 | 5.26 ± 3.70 | 6.13 ± 5.92 | N/A | N/A | N/A |
| Fasting Glucose (mmol/L) | 86.84 ± 7.43 | 85.43 ± 7.11 | 89.30 ± 7.37 | 95.81 ± 10.25 | 94.67 ± 10.71 | 97.09 ± 9.56 |
| HOMA | 1.21 ± 1.11 | 1.11 ± 0.82 | 1.38 ± 1.48 | N/A | N/A | N/A |
| Mean Blood Pressure (mmHg)**, § | 86.71 ± 9.19 | 85.07 ± 8.55 | 89.55 ± 9.58 | 82.40 ± 8.12 | 81.81 ± 8.51 | 83.07 ± 7.61 |
| Systolic blood pressure (mmHg)§ | 115.41 ± 12.07 | 112.59 ± 11.38 | 120.30 ± 11.69 | 108.81 ± 10.66 | 108.30 ± 10.49 | 109.37 ± 10.83 |
| Diastolic blood pressure (mmHg)§ | 72.36 ± 9.94 | 71.31 ± 9.21 | 74.18 ± 10.88 | 69.20 ± 8.90 | 68.56 ± 9.33 | 69.92 ± 8.34 |
Mean Blood Pressure defined as (Systolic blood pressure + Diastolic blood pressure × 2)/3
FAMUSS Female N=347 for BMI due to missing weight
FAMUSS Female N=341 and Male N=197 due to missing BP measurement
The percentage of subjects showing abnormal levels of each CAD risk factor is provided in Table 3. BMI was >30 in 12.0% of all subjects (9.5% of females; 16.5% of males). A relatively large proportion of volunteers showed abnormal HDL (C) levels (81% of males; 29% of females). Females showed an overall lower risk for mean blood pressure (mmHg) ≥ 100 (2.6% abnormal values in females; 14.5% in males), and for the diagnosis of metabolic syndrome (4.6% of females; 18.0% of males).
Table 3.
Incidence of abnormal values for FAMUSS and CHIP.
| FAMUSS (Ave. Age 24 yrs) | |||
|---|---|---|---|
| All | Females | Males | |
| N | 548 | 348 | 200 |
| BMI (kg/m2) ≥ 25 | 34.1% | 28.4% | 44.0% |
| BMI (kg/m2) ≥ 30 | 12.0% | 9.5% | 16.5% |
| Total cholesterol (mg/dl) ≥ 240 | 2.7% | 3.5% | 1.5% |
| LDL (C) (mg/dl) ≥ 130 | 12.2% | 10.3% | 15.5% |
| HDL (C) (mg/dl) ≤ 40 (for males) ≤ 50 (for females) | 48.0% | 29.0% | 81.0% |
| Triglycerides (mg/dL) ≥150 | 15.1% | 12.1% | 20.5% |
| Insulin resistance | 6.9% | 5.5% | 9.5% |
| Mean BP (mmHg) ≥ 100 | 6.9% | 2.6% | 14.5% |
| Metabolic Syndrome | 9.5% | 4.6% | 18.0% |
| CHIP (Ave. Age 11.5 yrs) | |||
| All | Females | Males | |
| N | 810 | 428 | 382 |
| BMI (kg/m2)* | 44.6% | 41.8% | 47.6% |
| Total cholesterol (mg/dl) ≥ 170 | 41.2% | 40.7% | 42.0% |
| LDL (C) (mg/dl) | 27.3% | 25.9% | 28.8% |
| HDL (C) (mg/dl) ≤ 40 | 23.5% | 23.4% | 23.6% |
| Triglycerides (mg/dL) ≥110 | 27.8% | 29.4% | 25.9% |
| Insulin resistance | ND** | ND** | ND** |
| Mean BP (mmHg) > 90th percentile, based on CDC growth charts | 11.4% | 10.7% | 12.0% |
| Metabolic Syndrome | 7.6% | 7.7% | 7.6% |
Thresholds for abnormal BMI were based on age and sex: Males 10 yrs ≥ 19; 11 yrs ≥ 20; 12 yrs ≥ 21. Females 10 yrs ≥ 20; 11 yrs ≥ 21; 12 yrs ≥ 22.
ND: Not Determined
BP: Blood Pressure
Loci previously reported to be associated with CAD, CAD risk factors, or glucose metabolism phenotypes were genotyped in FAMUSS (Table 1; Table 4). Resulting genotypes were tested for Hardy-Weinberg equilibrium; two SNPs showed divergence from expected allele frequencies (rs4846914 and rs1333049). Twenty loci were tested against six phenotypes {fasting glucose, triglycerides, HDL (C), LDL (C), fasting insulin, and HOMA} in FAMUSS. This required adjustment for multiple testing with a p-value threshold of < 0.00042 for significance (adjusted for the testing of 20 SNPs and six phenotypes).
Table 4.
SNPs genotyped in FAMUSS Study.
| Region | SNP | Risk Allele |
Risk allele Frequency |
Nearest Gene (s) | Phenotype associated with SNP |
|---|---|---|---|---|---|
| 1p31.3 | rs12130333 | C | 0.77 | ANGPTL3, DOCK, ATG4C | TG |
| 1p13.3 | rs646776 | T | 0.83 | CELSR2, PSCR1, SORT1 | LDL |
| 1q42.13 | rs4846914 | G | 0.43 | GALNT2 | HDL, TG |
| 3p25.2 | rs1801282 | C | 0.87 | PPARG2 | T2DM |
| 3q22.3 | rs9818870 | T | 0.16 | MRAS | CAD |
| 3q27.2 | rs4402960 | T | 0.35 | IGF2BP2 | T2DM |
| 6p22.3 | rs7756992 | G | 0.26 | CDKAL1 | T2DM |
| 6p22.3 | rs10946398 | C | 0.31 | CDKAL1 | T2DM |
| 7q11.23 | rs17145738 | G | 0.92 | BCL7B, TBL2, MLXIPL | TG |
| 8q24.11 | rs13266634 | C | 0.71 | SCL30A8 | T2DM |
| 8q24.13 | rs17321515 | A | 0.55 | TRIB1 | TG |
| 9p21.3 | rs1333049 | C | 0.51 | ANRIL | CAD |
| 9p21.3 | rs10811661 | T | 0.83 | CDKN2A/2B | T2DM |
| 10q23.33 | rs1111875 | G | 0.39 | HHEX | T2DM |
| 10q23.33 | rs7923837 | G | 0.61 | HHEX | T2DM |
| 10q25.2 | rs7901695 | T | 0.72 | TCF7L2 | T2DM |
| 11p15.1 | rs5219 | A | 0.35 | KCNJ11 | T2DM |
| 12q24.11 | rs2338104 | G | 0.46 | KCTD10 | HDL |
| 12q24.31 | rs2259816 | T | 0.35 | HNF1A | CAD |
| 19p13.11 | rs16996148 | G | 0.07 | CILP2, PBX4 | LDL, TG |
TG: Triglycerides: LDL (C); Low-density lipoprotein cholesterol; HDL (C): High-density lipoprotein; T2DM: Type 2 Diabetes Mellitus; CAD: Cornary Artery Disase
Nineteen of the 20 loci tested did not reach the adjusted p-value threshold of 0.00042 for any T2DM-related or lipid trait in the FAMUSS group. Given the lower average serum values in young adults to compared to older adults, and lower variance, our data does not preclude the other 19 loci from having a phenotype effect in a larger population. We are underpowered to detect a <1% effect size published in larger adult studies.
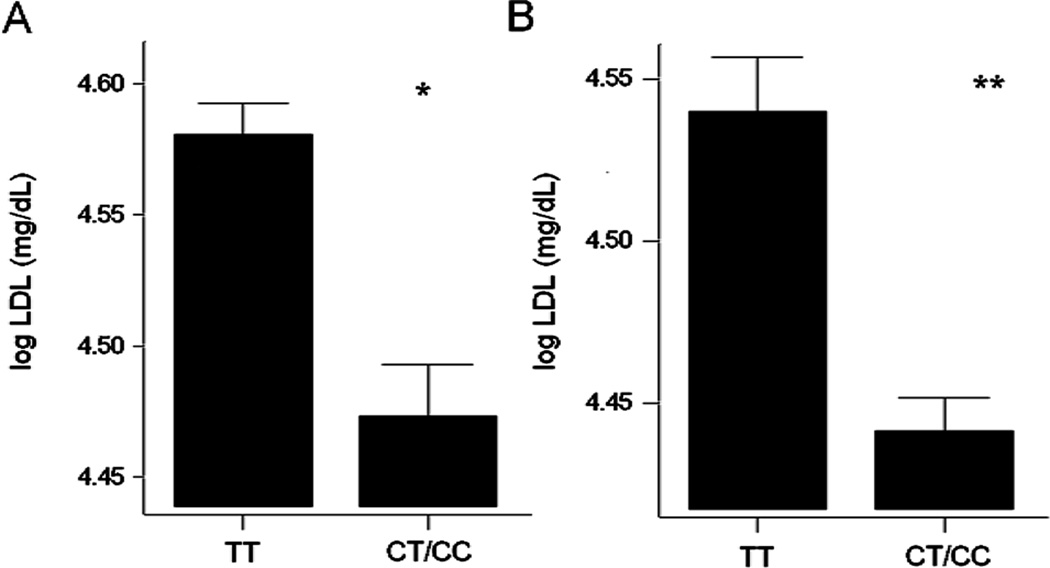
A single locus met our threshold for significance, and this was the rs646776 locus at 1p13.3, containing the SORT1 gene with association for circulating LDL (C) levels (Table 5; Figure 1A). There was a dose effect of the polymorphism, suggesting a quantitative or additive effect of the alleles, however, the numbers of homozygotes for the rare allele were relatively low (Figure 1A). The association between rs646776 and LDL (C) was significant for the entire cohort (p = 3.0×10−6), and significant in both male (p = 0.009) and female (p = 8.8×10−5) sub-cohorts. Inclusion of age and/or BMI as covariates in the statistical model showed little change in statistical support for the association of rs646776 with LDL (C). The amount of variation in serum LDL (C) explained by the rs646776 genotype was 3.6% for the entire cohort (4.1% in females; 3.0% in males; Table 5).
Table 5.
1p13.3 variant (rs646776) is associated with LDL (C) in FAMUSS cohort (24 yr olds).
| Phenotype | N: unadjusted mean ± SEM | p-value | % variation attributable to genotype |
|---|---|---|---|
| LDL (C) (log) | TT (N=345; 4.58 ± 0.0) CT/CC (N=203; 4.47 ± 0.02) | 0.000003 | 3.6% |
| LDL (C) (log) in females | TT (N=212; 4.56 ± 0.02) CT/CC (N=136; 4.44 ± 0.02) | 0.000088 | 4.1% |
| LDL (C) (log) in males | TT (N=133; 4.622 ± 0.022) CT/CC (N=67; 4.522 ± 0.031) | 0.0094 | 3.0% |
Figure 1. 1p13.3 variant (rs646776) shows an effect on LDL (C) in both FAMUSS and CHIP cohorts.
Both sexes are shown for the FAMUSS study (1A) and the CHIP study (1B). In both population-based studies, individuals with the TT genotype had significantly higher LDL (C) values than the CT/CC genotypes. Figure 1A: *p=0.000003; N=345 for TT, N=203 for CT/CC. Figure 1A: **p=0.000008; N=551 for TT, N=259 for CT/CC.
LDL (C) levels have been shown to have a weak correlation with for BMI in children and adolescents (ages 3–18 yrs old) (15). Oxidized LDL (C) has a strong relationship to waist circumference and has been shown to have a strong relationship to LDL (C) levels (16–19). Our association data in young adults has shown that more of the variation of LDL (C) in 24 yr olds was due to rs646776 than to an increase BMI which is supported by the findings in the Lee et al. 2009 paper (15). To test this, regression models of LDL (C) were done with and without rs646766 genotype and BMI to determine the percent variation attributable to each factor. In both the FAMUSS and CHIP cohorts, more of the variation in LDL (C) can be attributed to rs646766 than BMI (3.6% versus 3.0% in FAMUSS; 2.5% versus 1.4% in CHIP). While there are significant associations between LDL (C) and BMI in both cohorts (p=0.03 in FAMUSS; p=0.02 in CHIP), the associations between LDL (C) and rs646766 are statistically much more significant (p=0.000003 in FAMUSS; p=0.00008 in CHIP).
The association between rs646776 and LDL (C) is replicated in the 11.5 yr old CHIP cohort
The CHIP cohort is comprised of 810 predominantly White 6th grade students attending public school in Michigan. Average values for selected CAD risk factors in the 11.5 yr old CHIP volunteers were healthier compared to the FAMUSS, as expected given the well-accepted increase in risk factors with age (Table 2).
We then looked at the abnormal threshold levels for each phenotype, using age-adjusted criteria (Table 3) (20). In this population-based study, 44.6% of children met the criteria for an abnormally high BMI (41.8% for girls, 47.6% for boys). 40% showed abnormal total cholesterol, while about 25% of the children showed abnormal values for HDL (C), LDL (C), and TG as individual laboratory phenotypes.
As this was a replication cohort, we tested only the association of rs646776 with serum LDL (C). There was a strong statistical association between rs646776 and LDL (C), similar to that observed in FAMUSS (Figure 1B; Table 6). There was a similar dosage effect of the genotype, with homozygotes for the ancestral common allele showing the highest LDL (C) levels. The percentage of variation explained by genotype was 2.5% for the entire cohort (2.7% for females; 2.0% for males).
Table 6.
Replication data for 1p13.3 variant (rs646776) shows association with LDL (C) in the CHIP cohort (11.5 yr olds).
| Phenotype | N: unadjusted mean ± SEM | p-value | % variation attributable to genotype |
|---|---|---|---|
| LDL (C) (log) | TT (N=551; 4.54 ± 0.02) CT/CC (N=259; 4.44 ± 0.01) | 0.0000076 | 2.5% |
| LDL (C) (log) in females | TT (N=280; 4.53 ± 0.03) CT/CC (N=148; 4.42 ± 0.02) | 0.0006 | 2.7% |
| LDL (C) (log) in males | TT (N=271; 4.56 ± 0.03) CT/TT (N=; 111; 4.46 ± 0.02) | 0.0061 | 2.0% |
The rs646776 locus at 1p13.3 1 shows a greater effect in females than males
In both populations, females showed a stronger percent variation attributable to SORT1 genotype (see Tables 4 and 5). In the FAMUSS study, the SORT1 locus contributed 4.1% to to LDL (C) values in females while contributing 3.0% to males. We see a similar trend in the validation population, CHIP, where SORT1 contributed 2.7% to LDL (C) values in females and 2.0% in males.
The rs646776 locus at 1p13.3 1 shows a greater effect in young populations, compared to older, sicker populations
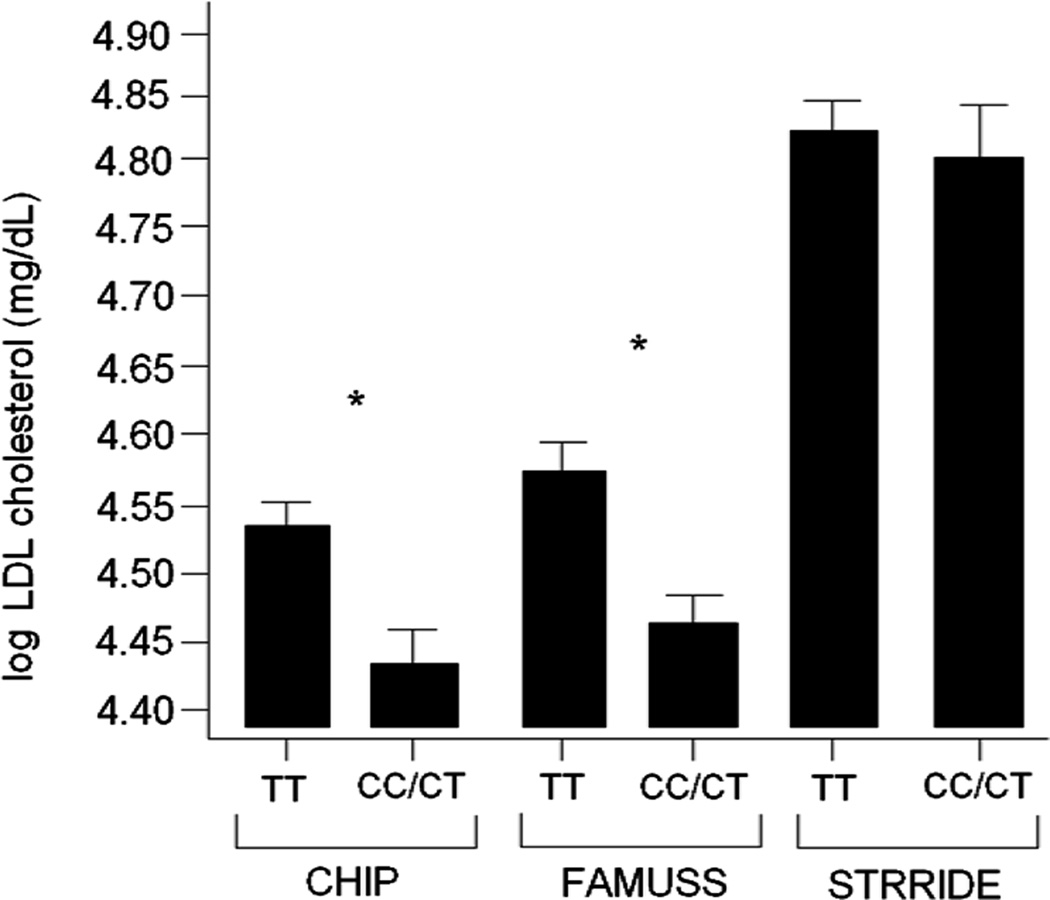
The association between rs646776 locus at 1p13.3 and LDL (C) has been studied in many older populations (5), some with overt T2DM (9). While each study has shown a similar effect of ancestral (common) allele associated with higher LDL (C), the effect size (proportion of variation explained by genotype) has been consistently quite small (~1%). We hypothesized that the larger effect size seen in the younger populations studied here (e.g. 4.1% in 24 yr old females; 2.7% in 11.5 yr old females) could be explained by the relative absence of co-morbidities that serve to obscure underlying genetic associations. To test this, we compared the association data from FAMUSS and CHIP to STRRIDE. STRRIDE includes older (age = 54 yrs; Table 7) subjects with features of metabolic syndrome, but without a T2DM diagnosis (21). As expected, STRRIDE participants showed much higher average serum LDL (C) values (Figure 2) but not a significant association of LDL (C) with rs646776 genotype. This contrasts to the 11.5 yr old CHIP children, and 24 yr old FAMUSS young adults that showed a significant genotype correlation. However, the 24 yr olds showed a higher genotype effect than the 11.5 yr olds (3.6% vs. 2.5%, respectively).
Table 7.
Sample Characteristics for STRRIDE Study.
| Phenotype | Females (N=114) | Males (N=117) |
|---|---|---|
| Mean ± SD | Mean ± SD | |
| Age (years) | 53.65 ± 5.61 | 50.64 ± 6.64 |
| Triglycerides (mg/dL) | 125.02 ± 59.77 | 152.60 ± 78.59 |
| Total cholesterol (mg/dL) | 206.61 ± 22.05 | 184.39 ± 28.96 |
| Fasting glucose (mg/dL)* | 89.96 ± 11.32 | 94.77 ± 10.17 |
| HDL (C) (mg/dL) | 54.99 ± 14.15 | 39.22 ± 8.19 |
| LDL (C) (mg/dL) | 130.78 ± 24.00 | 121.17 ± 23.83 |
| Height (m) | 1.63 ± 0.07 | 1.78 ± 0.07 |
| Weight (kg) | 79.71 ± 10.56 | 95.37 ± 12.58 |
| BMI | 29.90 ± 3.20 | 30.12 ± 3.11 |
Glucose values available for only 111 females and 113 males
Figure 2. LDL (C) cholesterol as a function of 1p genotype shows higher effect size in young population-based cohorts compared to older subjects with features of metabolic syndrome.
For the CHIP cohort, LDL (C) was significantly different between genotypes (*p=<0.001) with a mean ΔLDL (C) = 0.103 and with genotype explaining 2.5% of the variability in LDL (C). For the FAMUSS cohort, LDL (C) was significantly different between genotypes (*p=<0.001) with a mean ΔLDL (C) = 0.116 and with genotype explaining 3.6% of the variability in LDL (C). For the STRRIDE cohort, LDL (C) was not significantly different between genotypes with a mean ΔLDL (C) = 0.014 and with genotype explaining <0.1% of the variability in LDL (C).
DISCUSSION
GWAS studies have enabled the identification of key polymorphisms linked to complex disease, including T2DM (1) and related phenotypes (see reviews from Rideerstrale and Groop 2009) (22). This approach is frequently cited as holding potential to transform the practice of medicine. Most frequently, the integration of genetic risk factors into clinical practice is envisioned through preventive medicine. For example, those at highest risk for T2DM and/or CAD can be identified prior to onset of clinical symptoms, with interventions targeted towards those at greatest risk, perhaps personalizing interventions for specific genotypes. There are some shortcomings to this rationale. First, clinical trials will be needed to provide the evidence-based data to bring such pre-symptomatic interventions into standard of care. Since T2DM and CAD take decades to develop, the progression to overt disease is unlikely to be a practical endpoint in such trials. Thus, alternative sensitive and reliable endpoints are needed that can be assessed over a reasonable amount of time (e.g. 1 yr). Second, many CAD risk factor loci such as those for T2DM have been identified with case/control cohorts, with advanced or end-stage disease. Thus, it is possible, if not likely, that existing GWAS CAD risk factor loci identified in adults are associated with advanced development of the risk factor. We hypothesized that the identification of T2DM- and dyslipidemia-related loci showing association with related biomarkers in young population-based cohorts would overcome many of these limitations.
We tested 20 CAD risk factors and T2DM GWAS loci (Table 4), and found that the large majority (19/20) showed no evidence of association with related phenotypes as quantitative traits in a young adult (24 yr old) population. The phenotypes tested were serum lipids (LDL (C), HDL (C), TG), glucose, HOMA, and insulin resistance with and without adjustment for age and BMI. One locus, rs646776 at 1p13.3, showed strong associations with serum LDL (C) in our 24 yr old FAMUSS cohort and this association (Table 5; Figure 1A) remained significant after adjustment for multiple testing in the entire cohort (p = 3.0 × 10−6), males (p = 0.009) and females (p = 8.8 × 10−5). The association between rs646776 and LDL (C) was validated in a population-based 11.5 yr old replication cohort, and found to show similar significance (Table 6; Figure 1B).
We found a considerably larger effect size of the 1p13.3 LDL (C)-associated locus in young populations, compared to the previously published older cohorts. All previous studies have shown that rs646776 explains approximately 1% of the population variation in LDL (C) levels (5, 8). For comparison, a recent GWAS identified 30 loci for dyslipidemia (including the 1p13.3 locus) where the cumulative effect size of all 30 loci was approximately 5% of the population variance in lipoprotein levels (5). In contrast, our data in 24 yr olds showed the rs646776 SNP explained 3.6% of population variation – a considerably higher effect size than seen in most other loci (Table 5). This was despite lower average serum LDL (C) levels in our younger population (Figure 2), and lower population variance in LDL (C) values. Our data in 6th graders (Table 6) also showed larger effect sizes than previously published (2.5% vs. 1%); however, the effect size in our young population (11.5 yrs old) was not significantly higher than our 24 yr olds (FAMUSS; 3.6% effect size).
The specific functional variant at the 1p13.3 LDL (C)-associated locus has recently been shown to be a non-coding SNP in the gene promoter of the SORT1 gene, a multi-ligand sorting receptor directly involved in lipid trafficking in the liver (23). This locus showed results consistent with our hypothesis that younger populations would show a stronger genetic component in population variation in serum markers. However, the other 19 loci tested did not show significance in the 24 yr olds. This could be due to poor statistical power in younger subjects due to lower average serum marker levels and lower variance in blood levels for the serum markers. Also, some genetic loci significant in older subjects may require the onset of age-related co-variates before showing the published genetic associations. For example, the well-studied TCF7L2 T2DM locus has been associated with renal failure in diabetics and non-diabetics (24); a phenotype not expected to be expressed in younger subjects.
The rs646776 locus may provide the first opportunity to conduct clinical trials aimed at genotype-stratified prevention of dyslipidemia. Specifically, in our 11.5 yr old 6th grader cohort, 27.3% of children showed levels of LDL (C) above the threshold considered abnormal (Table 3). Increasing LDL (C) levels increase the risk of CAD (11). Interventions can be designed to test the efficacy of treatment strategies such as dietary interventions and low dose, lifelong lipid-lowering medication to decrease LDL (C) in children identified at risk by rs646776 genotype.
Our findings were consistent with previous reports showing that the ancestral common allele is associated with higher LDL (C) levels. Thus, the larger population is at high risk for elevated LDL (C), whereas the more recent derived allele imparts protection from high LDL (C) levels to the subset of rare allele carriers. The relatively low relative risk associated with rs646776 and T2DM (1.1 fold), and small effects sizes (1%) in older sicker populations is likely a consequence of the majority (2/3rd) of individuals carrying that high risk genotype for increased LDL (C) levels (homozygous for the common rs646776 genotype), and that multiple environmental and polygenic confounders obscure some of the stronger effects seen earlier in young adults (Figure 2).
Acknowledgments
Statement of financial support: This work was supported by NIAMS—5R01AR055100-07 (FAMUSS), NIDDK—1R01DK081559-01A1 (STRRIDE) and Owosso Memorial Health Care System (CHIP).
P.D.T is supported by a research grants from GlaxoSmithKline, Merck, Roche, Pfizer, AstraZeneca, B. Braun, Genomas, and Hoffman-LaRoche, served as a consultant for Astra Zenica, Merck, Schering-Plough, Takeda, Roche, Genomas, Abbott and Runners World; received speaker’s fees from Merck, Pfizer, Abbott, Astra Zenica, Schering-Plough, and is a stock holder of Zoll, General Electric, JA Wiley Publishing, Zimmer, Medtronic, Merck, Isis and J&J.
ABBREVIATIONS
- CAD
Coronary Artery Disease
- CHIP
Cardiovascular Health Intervention Program
- FAMUSS
Functional Polymorphism Associated with Human Muscle Size and Strength
- GWAS
Genome-Wide Association Study
- HOMA
Homoeostasis Model Assessment
- T2DM
Type II Diabetes Mellitus
- SNP
Single Nucleotide Polymosphim
- STRRIDE
Studies of a Targeted Risk Reduction Intervention through Defined Exercise
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.McCarthy MI, Zeggini E. Genome-wide association studies in type 2 diabetes. Curr Diab Rep. 2009;9:164–171. doi: 10.1007/s11892-009-0027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ioannidis JP. Prediction of cardiovascular disease outcomes and established cardiovascular risk factors by genome-wide association markers. Circ Cardiovasc Genet. 2009;2:7–15. doi: 10.1161/CIRCGENETICS.108.833392. [DOI] [PubMed] [Google Scholar]
- 3.Walley AJ, Asher JE, Froguel P. The genetic contribution to non-syndromic human obesity. Nat Rev Genet. 2009;10:431–442. doi: 10.1038/nrg2594. [DOI] [PubMed] [Google Scholar]
- 4.Ehret GB. Genome-wide association studies: contribution of genomics to understanding blood pressure and essential hypertension. Curr Hypertens Rep. 2010;12:17–25. doi: 10.1007/s11906-009-0086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PI, O'Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck-Nielsen H, Vaag A, Poulsen P, Gaster M. Metabolic and genetic influence on glucose metabolism in type 2 diabetic subjects--experiences from relatives and twin studies. Best Pract Res Clin Endocrinol Metab. 2003;17:445–467. doi: 10.1016/s1521-690x(03)00041-1. [DOI] [PubMed] [Google Scholar]
- 7.Jakobsdottir J, Gorin MB, Conley YP, Ferrell RE, Weeks DE. Interpretation of genetic association studies: markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genet. 2009;5:e1000337. doi: 10.1371/journal.pgen.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, Altshuler DM, Newton-Cheh C, Orho-Melander M. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 9.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erdmann J, Grosshennig A, Braund PS, Konig IR, Hengstenberg C, Hall AS, Linsel-Nitschke P, Kathiresan S, Wright B, Tregouet DA, Cambien F, Bruse P, Aherrahrou Z, Wagner AK, Stark K, Schwartz SM, Salomaa V, Elosua R, Melander O, Voight BF, O'Donnell CJ, Peltonen L, Siscovick DS, Altshuler D, Merlini PA, Peyvandi F, Bernardinelli L, Ardissino D, Schillert A, Blankenberg S, Zeller T, Wild P, Schwarz DF, Tiret L, Perret C, Schreiber S, El Mokhtari NE, Schafer A, Marz W, Renner W, Bugert P, Kluter H, Schrezenmeir J, Rubin D, Ball SG, Balmforth AJ, Wichmann HE, Meitinger T, Fischer M, Meisinger C, Baumert J, Peters A, Ouwehand WH, Deloukas P, Thompson JR, Ziegler A, Samani NJ, Schunkert H. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingelsson E, Schaefer EJ, Contois JH, McNamara JR, Sullivan L, Keyes MJ, Pencina MJ, Schoonmaker C, Wilson PW, D'Agostino RB, Vasan RS. Clinical utility of different lipid measures for prediction of coronary heart disease in men and women. JAMA. 2007;298:776–785. doi: 10.1001/jama.298.7.776. [DOI] [PubMed] [Google Scholar]
- 12.Thompson PD, Moyna N, Seip R, Price T, Clarkson P, Angelopoulos T, Gordon P, Pescatello L, Visich P, Zoeller R, Devaney JM, Gordish H, Bilbie S, Hoffman EP. Functional polymorphisms associated with human muscle size and strength. Med Sci Sports Exerc. 2004;36:1132–1139. doi: 10.1249/01.mss.0000132274.26612.23. [DOI] [PubMed] [Google Scholar]
- 13.Keskin M, Kurtoglu S, Kendirci M, Atabek ME, Yazici C. Homeostasis model assessment is more reliable than the fasting glucose/insulin ratio and quantitative insulin sensitivity check index for assessing insulin resistance among obese children and adolescents. Pediatrics. 2005;115:e500–e503. doi: 10.1542/peds.2004-1921. [DOI] [PubMed] [Google Scholar]
- 14.Orkunoglu-Suer FE, Gordish-Dressman H, Clarkson PM, Thompson PD, Angelopoulos TJ, Gordon PM, Moyna NM, Pescatello LS, Visich PS, Zoeller RF, Harmon B, Seip RL, Hoffman EP, Devaney JM. INSIG2 gene polymorphism is associated with increased subcutaneous fat in women and poor response to resistance training in men. BMC Med Genet. 2008;9:117. doi: 10.1186/1471-2350-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JM, Gebremariam A, Card-Higginson P, Shaw JL, Thompson JW, Davis MM. Poor performance of body mass index as a marker for hypercholesterolemia in children and adolescents. Arch Pediatr Adolesc Med. 2009;163:716–723. doi: 10.1001/archpediatrics.2009.109. [DOI] [PubMed] [Google Scholar]
- 16.Holvoet P, Kritchevsky SB, Tracy RP, Mertens A, Rubin SM, Butler J, Goodpaster B, Harris TB. The metabolic syndrome, circulating oxidized LDL, and risk of myocardial infarction in well-functioning elderly people in the health, aging, and body composition cohort. Diabetes. 2004;53:1068–1073. doi: 10.2337/diabetes.53.4.1068. [DOI] [PubMed] [Google Scholar]
- 17.Holvoet P, Lee DH, Steffes M, Gross M, Jacobs DR., Jr Association between circulating oxidized low-density lipoprotein and incidence of the metabolic syndrome. JAMA. 2008;299:2287–2293. doi: 10.1001/jama.299.19.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopprasch S, Pietzsch J, Kuhlisch E, Fuecker K, Temelkova-Kurktschiev T, Hanefeld M, Kuhne H, Julius U, Graessler J. In vivo evidence for increased oxidation of circulating LDL in impaired glucose tolerance. Diabetes. 2002;51:3102–3106. doi: 10.2337/diabetes.51.10.3102. [DOI] [PubMed] [Google Scholar]
- 19.Lapointe A, Couillard C, Piche ME, Weisnagel SJ, Bergeron J, Nadeau A, Lemieux S. Circulating oxidized LDL is associated with parameters of the metabolic syndrome in postmenopausal women. Atherosclerosis. 2007;191:362–368. doi: 10.1016/j.atherosclerosis.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 20.Kavey RE, Daniels SR, Lauer RM, Atkins DL, Hayman LL, Taubert K. American Heart Association guidelines for primary prevention of atherosclerotic cardiovascular disease beginning in childhood. Circulation. 2003;107:1562–1566. doi: 10.1161/01.cir.0000061521.15730.6e. [DOI] [PubMed] [Google Scholar]
- 21.Slentz CA, Aiken LB, Houmard JA, Bales CW, Johnson JL, Tanner CJ, Duscha BD, Kraus WE. Inactivity, exercise, and visceral fat. STRRIDE: a randomized, controlled study of exercise intensity and amount. J Appl Physiol. 2005;99:1613–1618. doi: 10.1152/japplphysiol.00124.2005. [DOI] [PubMed] [Google Scholar]
- 22.Ridderstråle M, Groop L. Genetic dissection of type 2 diabetes. Mol Cell Endocrinol. 2009;297:10–17. doi: 10.1016/j.mce.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, Pirruccello JP, Muchmore B, Prokunina-Olsson L, Hall JL, Schadt EE, Morales CR, Lund-Katz S, Phillips MC, Wong J, Cantley W, Racie T, Ejebe KG, Orho-Melander M, Melander O, Koteliansky V, Fitzgerald K, Krauss RM, Cowan CA, Kathiresan S, Rader DJ. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Köttgen A, Hwang SJ, Rampersaud E, Coresh J, North KE, Pankow JS, Meigs JB, Florez JC, Parsa A, Levy D, Boerwinkle E, Shuldiner AR, Fox CS, Kao WH. TCF7L2 variants associate with CKD progression and renal function in population-based cohorts. J Am Soc Nephrol. 2008;19:1989–1999. doi: 10.1681/ASN.2007121291. [DOI] [PMC free article] [PubMed] [Google Scholar]