Abstract
Discovering novel treatments for Autism Spectrum Disorders (ASD) is a challenge. Its etiology and pathology remain largely unknown, the condition shows wide clinical diversity, and case identification is still solely based on symptomatology. Hence clinical trials typically include samples of biologically and clinically heterogeneous individuals. ‘Core deficits', that is, deficits common to all individuals with ASD, are thus inherently difficult to find. Nevertheless, recent reports suggest that new opportunities are emerging, which may help develop new treatments and biomarkers for the condition. Most important, several risk gene variants have now been identified that significantly contribute to ASD susceptibility, many linked to synaptic functioning, excitation–inhibition balance, and brain connectivity. Second, neuroimaging studies have advanced our understanding of the ‘wider' neural systems underlying ASD; and significantly contributed to our knowledge of the complex neurobiology associated with the condition. Last, the recent development of powerful multivariate analytical techniques now enable us to use multi-modal information in order to develop complex ‘biomarker systems', which may in the future be used to assist the behavioral diagnosis, aid patient stratification and predict response to treatment/intervention. The aim of this review is, therefore, to summarize some of these important new findings and highlight their potential significant translational value to the future of ASD research.
Keywords: Autism spectrum disorder, biomarkers, drug development, genetics, neuroimaging
Introduction
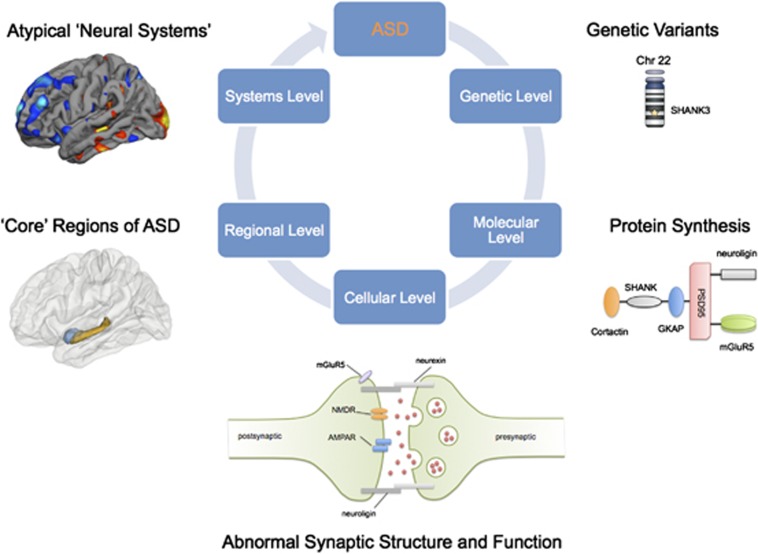
Autism Spectrum Disorders (ASD) are a group of conditions in which heterogeneity, both causative and phenotypic, is emerging as a dominant theme.1 This poses a complex challenge to developing new treatments, and biomarkers. Also, case identification is still solely based on symptomatology (that is, impaired social communication, social reciprocity and repetitive/stereotypic behavior2). Hence, clinical studies typically include samples of biologically heterogeneous cases, and this has hampered studies of clinical outcomes as well as research into the specific molecular and genetic underpinnings of the condition. However, several lines of evidence are now emerging that may unite findings from separate disciplines (for example, genetics, neuropathology and neuroimaging). These seem to converge in suggesting a common etiological pathway for ASD, namely: defective synaptic functioning, excitation–inhibition balance and brain connectivity. Further, they present new (and tractable) opportunities to identify biomarkers that can be used to fractionate the disorder and underpin the discovery of new drug targets. Here, we examine the genetic and neurobiological ‘building blocks' of ASD, and suggest a pathway from abnormalities in the synapse to the formation of large-scale cortical systems (Figure 1).
Figure 1.
Synaptic functioning and brain connectivity in Autism Spectrum Disorders (ASD)—from the molecular level to the neural systems level.
Genetic models of ASD and their importance for drug development
It has long been known that ASD is among the most genetically determined neuro-psychiatric conditions, with concordance rates between identical twins reported at nearly 90% in some studies.3, 4 The high heritability of the condition has been linked to several risk gene variants that significantly contribute to the genetic architecture of ASD (reviewed in Abrahams et al.1). These include common as well as rare genetic variants, which may plausibly help guide the development of new pharmacological targets and interventions in the future.
Despite the high heritability of the condition, the number of common genetic variants that have been reliably associated with ASD is surprisingly small. Such common variants are generally identified by genome-wide association studies (GWAS), which examine the association of a large number of common polymorphisms with a trait or groups of individuals (for example, ASD vs ‘neurotypicals'). For example, Wang et al.5 found a significant association between ASD and cadherin 10 (CDH10 and CDH9) genes on region 5p14.1, which code for neuronal cell-adhesion molecules. Another genome-wide linkage study has also identified a potential role for semophorin 5A (SEMA5A), a gene implicated in axonal guidance and reported to be downregulated in ASD.6 It has generally been assumed that the relative paucity of significant genome-wide results may be a consequence of the small effect sizes attributable to any particular gene, and conservative statistical thresholds resulting from multiple comparison corrections.1 More recent studies, however, also highlight the importance of a number of distinct and individually rare genetic variants for ASD, which significantly contribute to the genetic architecture of the condition.
Rare copy number variants (CNVs), which are insertions or deletions of relatively large DNA fragments, are often not transmitted from the parent but instead occur de novo. Such rare CNVs are currently thought to account for ∼10% of cases with idiopathic autism, that is, those with no known cause (unlike ASD occurring in Fragile X or Rett syndrome). To date, the rare mutations have predominantly been observed in genes encoding for synaptic cell adhesion molecules, which have a crucial role in synaptogenesis and neuronal differentiation. For example, well-replicated findings implicate CNVs in genes encoding for a family of proteins known as neuroligins, neurexins and SHANK3.
Neuroligins and neurexins
One of the first CNVs groups found to be associated with ASD included neuroligin (NLGN) genes such as neuroligin 3 (NLGN3) and neuroligin 4 X-linked (NLGN4X).7 Neuroligins are a family of cell adhesion proteins whose members are localized postsynaptically, and are believed to be involved in the formation and consolidation of synaptic contacts.8 Neuroligins form a transsynaptic contact with presynaptic neurexins, which is thought to trigger synaptogenesis, and abnormalities in this process have been linked to ASD.9 Neuroligins (and neurexins) can induce the formation of both excitatory and inhibitory synapses, depending on their specific subtypes (reviewed in Dalva et al.10). Furthermore, it has been demonstrated in neuroligin-knockout animals that depletion of the gene leads to a shift in the balance between excitation and inhibition.11 This is of importance because increasing evidence points to a role for disturbances to the excitation–inhibition balance being implicated in ASD (Rubenstein and Merzenich12).
ASD has also been linked to another member of the neurexin family, contactin associated protein-line 2 (CNTNAP2). CNTNAP2 has been shown to modulate language function in ASD (and other conditions), and may modulate long-range brain connectivity. CNTNAP2 messenger RNA is significantly enriched in the developing human brain in the frontal and temporal lobes, as well as in striatal circuits and in the frontal cortex of the adult brain.13 In humans, these regions support speech and language learning, which are frequently impaired in ASD. On the molecular level, CNTNAP2 is thought to have a major role in assisting interactions important for cellular migration and subsequent laminar organization, indicating a role for this CNVs being the construction of neural circuits.14 The cortical pattern of CNTNAP2 expression particularly implicates frontal lobe connectivity, which has been reported to be atypical not only in ASD but also in a variety of ASD-related conditions, such as attention deficit-hyperactivity disorder15 (also discussed below). This finding is of particular importance as the genetic mediation of disconnection across multiple phenotypes suggests that certain genetic variants, and their associated brain phenotypes, may not be specific to ASD. Instead, they may increase the overall risk of ASD by affecting isolated neural systems that mediate specific autistic symptoms.
SHANK3, glutamate and GABAergic signaling in ASD
Another promising gene, which has been linked to ASD, is SHANK3.16 SHANK3 codes for a key postsynaptic density protein at glutamatergic synapses, which is believed to function as a scaffold for the assembly of the postsynaptic signaling complex. Mice with a deletion of the SHANK3 gene have been reported to exhibit autistic-like symptoms such as deficits in social interaction and repetitive behavior.17 Further, SHANK3 modulates striatal size (larger in knockout mouse) as well as the anatomy of cortico-striatal circuits known to be affected in ASD18—and for which there is preliminary evidence to suggest that glutamate metabolism is abnormal in individuals with ASD.19 In humans, it has also recently been demonstrated that mutations in SHANK3 strongly affects the development and morphology of dendritic spines and reduces synaptic transmission in mature neurons.20 Hence it has been suggested21 that variation in SHANK3 may directly modulate some aspects of brain phenotype in ASD.
In addition, the direct effects of SHANK3 may also be combined with other genetically (and/or environmentally) determined risk factors affecting the balance of excitation–inhibition—for example, the balance of glutamate and γ-aminobutyric acid (GABA)).22 GABA is an inhibitory amino acid neurotransmitter synthesized by decarboxylation of glutamate by the enzyme glutamic acid decarboxylase (GAD). In the adult brain, GAD exists in two major isoforms, GAD65 and GAD67, which are products of two independently regulated genes located on chromosomes 2 and 10, respectively.23 In ASD, chromosome 10 has been shown to reach genome-wide significance in linkage studies, and particularly in the region 10p12.1, which also encodes GAD65.24 It has further been shown that individuals with ASD have a ∼50% reduction in protein levels of the enzymes GAD65 and GAD67 in parietal (BA40) and cerebellar cortices,20 which are both regions where individuals with ASD show differences in brain anatomy from controls. During development, GABA can also act as a trophic factor and influence neuronal proliferation, migration, differentiation, synapse maturation and cell death.21 Hence, SHANK3 (alone or in combination with other factors) may affect synaptic functioning and brain connectivity, and increase the risk for ASD.
Implications for research and drug development
These findings coming from genetic investigations have important implications for ASD. First, it seems that very few genetic variants are ‘causal' for ASD. Instead, they affect a wide variety of phenotypes with variable symptom expressivity, which may or may not meet the clinical cutoffs for ASD (that is, the ‘broader autism phenotype').25 Thus, there is a strong need for detailed phenotypic studies not only of patients with autism but also of unaffected individuals with more or less autistic traits who harbor such rare potentially causal mutations. For example, a recent study has also shown that a distinct autism-related CNTNAP2 ‘risk allele' is associated with reduced functional connectivity in frontal lobe networks, regardless of whether participants were autistic or ‘neurotypical'.26 The findings of this study are important as they imply that some genetic variants may predispose individuals to ASD (that is, increase the risk of ASD) without necessarily being ‘causal' for the condition (that is, lead to clinical diagnosis of ASD). Such noncausal genetic variants may also affect the functional and structural makeup of particular brain regions (or neural systems) that contribute to the wider brain phenotype of ASD. Investigations of a similar study design, which investigate the association between genetic variants and autistic traits across various conditions—and in neurotypicals—are hence of high importance for the future, as this will allow us to link specific aspects of brain structure, function and connectivity to specific CNVs that increase autism susceptibility.
The existence of specific ASD genotypes also implies that these could be rescued (that is, ‘manipulated') by specific, targeted molecular treatment. There is, therefore, now an opportunity to exploit these new findings and make progress on the development of new therapies for ASD, including both children and adults. For example, by specifically targeting CNVs of known developmental genes, efforts can be focused onto precisely those elements where these genes would be predicted to make the most impact. So far, drug discovery in ASD has also been hampered by the availability of valid human cellular assays that recapitulate normal and diseased neural function in vitro. Emerging data from both genetic and clinical studies in addition to rodent models of disease might, however, soon be used to configure cellular assays, which model the etiology of ASD. One approach being pioneered as part of a large-scale EU Innovative Medicines Initiative on Autism (EU-AIMS; http://www.eu-aims.eu) is to use induced human pluripotential stem cells from specific cohorts of patients to generate (in a culture dish) human neurons carrying the specific genotype of ASD individuals, and to examine the effect of these in mouse models. If successful, this approach will allow us to target specific aspects of neurobiology that are most atypical in ASD, rapidly ‘assay' potential new treatments, and translate the findings from ‘bench to bedside' (and back). Also, this approach may be used to find clusters of genotypes with similar neuronal phenotypes, which could then be used for case stratification in clinical studies and to evaluate the effect(s) of specific, targeted molecular treatment.
ASD—a multi-etiological disorder of defective synaptic structure and functioning
Although several CNVs are now known to mediate the risk of ASD, it is likely that each CNVs does not act in isolation. Instead, several significant ‘gene clusters' have now been identified, which can be grouped together according to their functional involvement in ASD. For example, Gilman et al.27 identified several large functional network of genes affected by rare de novo CNVs in ASD using a network-based analysis of genetic associations (NETBAG). These networks included many of the CNVs discussed above, and primarily include genes coding for cell adhesion (for example, NRXN1, NLGN3) and scaffolding proteins (for example, SHANK2/3). Atypical cell adhesion and scaffolding affect the formation and consolidation of synaptic contacts, and hence have an impact on the formation of local neuronal connections on the microscopic level. Synaptogenesis has therefore become a ‘common theme'— linking CNVs associated with ASD functionally—and directly impacts on the structural architecture of synaptic terminal on the microscropic level. The analysis of the functional impact across genes and/or CNVs supports the hypothesis that perturbed synaptogenesis may be a ‘core deficit' of ASD, that is, common to all individuals with autistic symptoms and traits (see also28). In addition, the findings of genetic studies imply that synaptic structure and functioning—mediated by synaptic transmission—is affected in ASD.
Evidence for perturbed synaptic functioning come from several proton magnetic resonance spectroscopy studies. For example, individuals with ASD show differences in N-acetyl-aspartate (a measure of neuronal density and mitochondrial function) concentration,29 and in choline-containing compounds (Cho)—a measure of membrane turnover. However, recent studies have also highlighted the importance of abnormalities in glutamatergic neurotransmission in ASD, which can now be measured using proton magnetic resonance spectroscopy. For instance, in adults with ASD, glutamate concentrations were found to be increased in the amygdala–hippocampal regions in adults,19 but to be decreased in many brain regions in children with ASD.30 Thus, synaptic defects in ASD may not be confined to their structural architecture but also to synaptic inhibition and excitation mediated by glutamate and GABA. The balanced interaction between glutamate and GABA transmission is essential for regulating cognition (for example, learning and memory) and emotional behaviors, and an imbalance between gluatamate excitation and GABA inhibition, leading to hyperexcitation, has been linked to ASD.31, 32, 33, 34 There is recent in vivo evidence from a Positron Emission Tomography study demonstrating proof of concept that individuals with ASD have significant differences from controls in GABA α-5 receptor binding.35 Last, there is preliminary evidence implicating abnormalities in serotonergic system to ASD. For example, a significant proportion of ASD individuals may have hyperserotonemia (Hranilovic et al.36); and there are significant associations between ASD and genetic polymorphisms for serotonin synthesis,37 transporters38 and receptors.39 Additionally, neuroimaging studies report that individuals with ASD have significant differences in serotonin synthesis and reductions in serotonin2A receptor binding, and the 5HT transporter, in brain regions involved in social communication (for example, cingulate cortices).40, 41, 42 Furthermore, very recent evidence shows that abnormal brain activity during facial emotion processing in individuals with ASD is modulated by cortical serotonin levels, which may underpin some of the impairments in social functioning.43 Although it has previously been suggested that selective serotonin reuptake inhibitors may not be effective in the treatment of all individuals with ASD,44 it remains to be established if such small effect sizes are due to the large degree of phenotypic heterogeneity within the ASD population. Thus, individual's response to modulation of 5HT may potentially be used as a possible stratification tool in the future and to elucidate the various biological phenotypes of the condition.
Cortical neuropathology of ASD—from synapses to local circuits
Synaptic defects mediated by genetic variation in ASD not only affect their structural architecture, but also affect the formation of local neuronal circuitry. These, in turn, constitute the cyto-architectural and microstructural makeup of neocortical regions. For example, GABA—mediated by genes encoding for enzymes GAD65/67, which have both been associated with ASD (discussed above)—has a crucial role as ‘trophic' factor during neurodevelopment.45 However, GABA not only affects synaptic structure and functioning during neurodevelopment but also has an important role in the formation and functioning of local microcircuitry in the mature brain. Here, GABAergic interneurons are an integral part of cortical ‘minicolumns', the basic architectonic and physiological elements of the neocortex,46 and are located mostly in the so-called peripheral neuropil space surrounding the column ‘core'. In ASD, minicolumns have been reported to be more numerous and narrower than in neurotypicals, which is also associated with reduced peripheral neuropile space.47, 48 Thus, ASD may be linked to a reduction in GABAergic inhibitory activity and an imbalance of inhibition and excitation, which in turn may explain some clinical symptoms of ASD (for example, increased incidence of seizures and hypersensitivity to visual/auditory stimulation).12 This example demonstrates how genetic variation associated with ASD can cause atypical development and morphology of synaptic terminals on the microstructural level, which then leads to altered neuronal organization and synaptic transmission in local neuronal circuits (for example, within minicolumns). In turn, altered neuronal organization will affect the cytoarchitectural makeup, gross anatomy and function of individual brain regions, which has been investigated in ASD using various neuroimaging techniques.
The anatomy of the brain in ASD
It is well established that ASD is accompanied by differences in brain anatomy (also reviewed in49). During early childhood, these differences are most prominent on the global level and include differences in overall brain growth trajectory of total gray- orwhite-matter volume (Courchesne et al.50), head circumferences (Lainhart et al.51) and/or surface area (Hazlett et al.52). During later childhood, adolescence and adulthood, however, ASD are associated with abnormalities in specific spatially distributed neural systems that may mediate specific symptoms and traits.49 During early postnatal life, the brain in ASD may undergo a period of precocious accelerated growth, which may then be followed by an atypically slow or arrested growth during childhood, so that no global differences are generally observed in adulthood.53 In childhood, increased brain size in ASD affects both gray and white matter,54 and may be mainly driven by an increase in surface area—rather than cortical thickness.52 Normal intelligence adults with ASD, however, do not differ significantly from ‘neurotypicals' in whole brain gray/white matter volume, but do in spatially distributed patterns of significant gray- or white-matter differences, indicating the wider neural systems implicated in ASD.55 It has also been suggested that the overgrowth is idiosyncratic for different lobes of the brain, with frontal and temporal lobes being more affected than occipital and parietal lobes (Carper et al.56). Both the temporal and the frontal lobe are also brain areas that mature relatively late during normal development,57 which further strengthens the hypothesis that the altered time course of brain maturation may lead to neuroanatomical differences in multiple neural systems rather than isolated regions. Across studies, the most replicated structural differences associated with ASD have been described in the cerebellum,58 the amygdala–hippocampal complex,59, 60, 61 fronto-temporal regions59, 60 and the caudate nuclei.18, 62 It has also been suggested that neuroanatomical differences in specific brain structures mediate specific clinical symptoms. For instance, abnormalities in; (1) Broca's and Wernicke's area have been associated with social communication and language deficits;63 (2) fronto-temporal regions and the amygdala have been related to abnormalities in socio-emotional processing;64, 65 and 3) the orbitofrontal cortex and the caudate nucleus (that is, fronto-striatal system) may mediate repetitive and stereotyped behaviors.66 These studies were important first steps that add weight to the suggestion that differences in brain anatomy observed in ASD underpin specific clinical symptoms and traits.
However, despite growing evidence for the involvement of specific brain regions in ASD, many previous findings are nonreplicated. For example, cerebellum and amygdala have been variously reported to be normal, smaller and increased in size. For a long time, it was believed that this variability arose because most studies were of relatively small samples that differed in several key aspects within and across subject groups (for example, diagnostic criteria, intelligence quotient and age). However, recent work demonstrates that the effect sizes for neuroanatomical differences between individuals with ASD and neurotypicals remain moderate even when samples are large and well-characterized. For instance, Ecker et al.55 investigated neuroanatomical differences in a large sample of male adults with ASD and matched ‘neurotypicals' (N=176) using a multi-center acquisition paradigm. Despite the large sample size, however, relatively few clusters survived correction for multiple comparisons (for example, significant increases in volume of the anterior temporal and frontal lobe in ASD relative to controls). This may indicate that (1) the clinical diversity of the behaviorally defined autistic phenotype is also reflected on the level of brain anatomy (that is, multiple brain phenotypes) so that increasing sample sizes will not necessarily improve effect sizes. Alternatively, it may suggest that (2) individuals with ASD have differences in brain anatomy that are difficult to describe using conventional analytical approaches.
More recent theoretical models suggest the need to consider ASD as a disorder of several large-scale neurocognitive networks. Regional or voxel-level analytical methods, which rely on conservative statistical thresholds mandated by the large number of voxels compared between groups, may not be optimal for detecting differences that are theoretically expected to be subtle and spatially distributed (that is, standard voxel-level approaches powerfully detect regional differences, but they are not are not suitable for systems-level questions). Multivariate or multi-voxel approaches, which are statistically more powerful and hence offer higher exploratory power, are therefore becoming increasingly popular to examine the brain in ASD. For example, the recent introduction of pattern classification techniques (discussed below) has proven invaluable in detecting brain regions that distinguish individuals with ASD from ‘neurotypicals' on the basis of several neuroanatomical features (for example, cortical thickness, regional brain volumes and so on.). Taken together, these findings suggest that instead of asking the question ‘what can ASD tell us about its neuroanatomy?', we ought to be examining what neuroanatomy can tell us about ASD. In this undertaking, the development of novel analytical approaches to identify the potentially multiple brain phenotypes of ASD as well as their genetic and molecular underpinnings will be essential. It is imperative that future investigations do not only target the genetic underpinnings of differences in brain volume in ASD, but also associate genetic markers with measures of brain connectivity in several large-scale ‘neural systems'.
ASD—a ‘neural systems' condition
It has been noted that the altered neurodevelopmental trajectory is likely to interfere with brain connectivity in ASD (recently reviewed in Vissers et al.67), as the time window of overgrowth coincides with the period when synaptogenesis, dendritic growth and myelination are at their peak (reviewed in Courchesne et al.68 and Rippon et al.69). Moreover, the maturation of higher-order cortical systems, such as the frontal and temporal lobes, rely on the earlier maturation of lower-order and phylogenetically older cortical systems (for example, somatosensory and visual cortices57) so that any developmental dysregulation(s) during this critical time will not only affect the neural architecture of isolated brain regions (and their local connectivity), but also the formation of their global circuitry.68 Thus, ASD is most likely a ‘neural systems' condition that is mediated by abnormalities in spatially distributed, large-scale cortical networks rather than isolated brain regions. ASD has therefore also been referred to as a ‘developmental disconnection syndrome'.70
Evidence for atypical structural connectivity in ASD comes from an increasing number of neuroimaging studies measuring white-matter anatomy. For example, prior studies found that individuals with ASD have significant differences in white-matter volume,60, 65 and microstructural integrity—as measured by diffusion tensor magnetic resonance imaging.71, 72, 73, 74, 75 Furthermore, it has been reported that individuals with ASD undergo abnormal postnatal white-matter development. Such prior reports mostly highlight significant increases in white matter during early childhood, which may precede the abnormal pattern of growth in gray matter.54 In adults, however, ASD is associated with a pattern of regional reductions in white matter, which suggests that white-matter differences may also result from differences in neurodevelopmental trajectories. Several core deficits seen in ASD have also been associated with atypical connectivity of specific white-matter fiber tracts. For example, (1) diffusivity measures of the corpus callosum are significantly correlated with reduced processing speed in performance intelligence quotient tests in ASD;74 (2) the severity of social impairments in ASD are related to abnormal diffusion anisotropy in fibers of the superior cerebellar peduncle73 or (3) that individuals with ASD have significant differences in the anatomy and maturation of limbic tracts, which predominantly relay information underpinning socio-emotional processing.72 Thus, although it is difficult to link specific cognitive functions to differences in white-matter anatomy, altered brain connectivity in addition to structural gray-matter differences may explain some of the behavioral features typically observed in ASD.
Altered connectivity in ASD has also been reported by functional magnetic resonance imaging (MRI) studies, which have led to one of the dominant theories regarding brain connectivity in ASD; namely, that there is long distance under-connectivity and local over-connectivity of the frontal cortex.68, 76 Reduced long-range cortical functional connectivity has been reported predominantly in frontal regions using a variety of fMRI paradigms including executive functioning,77 working memory for faces78 and facial-affect processing.79 Less evidence has been provided for the concept of local under-connectivity, which may be mainly owing to the fact that it is difficult to measure. However, these MRI findings complement genetic investigations suggesting that atypical connectivity on the cellular level (that is, defect synaptic functioning) may affect interregional connectivity on the ‘systems level' in ASD.
Autism biomarkers
Owing to the new insights described above, there is now a search for biomarkers that can be used to assist the behavioral diagnosis, aid patient stratification and predict response to treatment/intervention.
So far, the discovery of biomarkers has been hindered by the complexity of the condition, as ASD has multiple causes (see above), comorbid conditions and varies in the type and severity of symptoms expressed. In addition, ASD is a neurodevelopmental condition and phenotypes are likely to vary with age. Therefore, it is unlikely that ASD can be linked to a single biomarker (that is, a single gene or brain region) across the neurodevelopmental time course. Instead, ASD biomarkers are most likely to be multivariate and complex, encompassing data from different aspects of biology as well as genetics. However, no single analytical framework has so far been powerful enough to establish such complex ‘biomarker systems'.
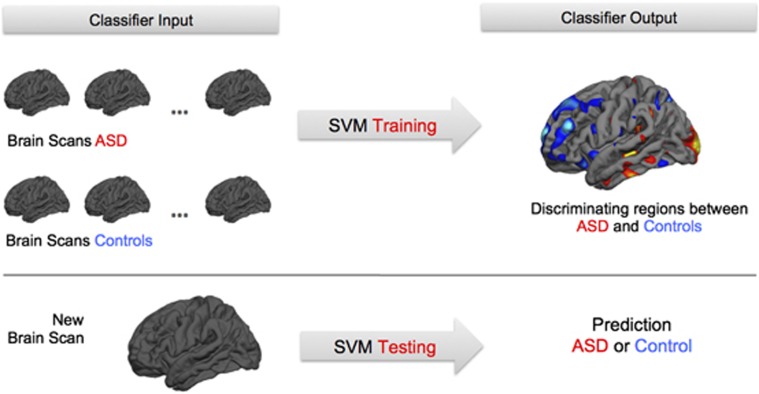
Recent advances in analytical techniques now make it possible to utilize such complex, multivariate data in order to make a prediction. In the context of brain imaging, these techniques have been described as ‘brain-reading' or ‘brain-decoding' methods,80 and belong to a broad group of techniques collectively known as ‘machine learning'. The basic idea of machine learning is to train a computer algorithm to identify a complex pattern of data that can then be applied in new individuals to make a prediction. Training usually occurs in a well-characterized sample by finding a boundary or ‘hyperplane' that best discriminates between different classes (for example, patients and controls). Once the classifier is ‘trained', it can subsequently be used to predict group membership of a new test example (for example, new individual with unknown group membership) (see Figure 2). A key feature of pattern classification is their potential to detect global, complex and potentially multimodal patterns of abnormalities that cannot be efficiently identified with univariate methods (for example, general linear model). Machine-learning approaches are therefore particularly suited to explore biomarkers for ASD.
Figure 2.
Pattern classification using support vector machine based on structural magnetic resonance images in Autism Spectrum Disorders (ASD). The classifier is initially ‘trained' on a well-characterized sample of individuals with ASD and controls. Training results in a ‘discrimination map' indicating regions that can be used to discriminate between ASD and controls. This map can then be used to predict group membership of a new test example (for example, ASD or control).
A growing number of recent publications have therefore started to explore the diagnostic (that is, predictive) value of various measures of brain anatomy, functioning and connectivity for ASD. Most of the initial studies were based on measures of brain anatomy. Using a common variant of machine-learning, the so-called support vector machine, Ecker et al.81 explored the diagnostic value of whole-brain structural MRI scans measuring regional gray- and white-matter volume. In this sample, support vector machine correctly identified individuals with ASD based on their brain anatomy with ∼90% specificity and sensitivity. In addition to the overall binary classification (that is, being autistic or not), support vector machine provided a ‘test margin' for each subject indicating how ‘prototypical' the test example is of each class, which is related to the confidence with which a new participant can be classified. These test margins were positively correlated with the severity of current autistic symptoms—suggesting that support vector machine may be able to measure ASD along a continuum based on neuroanatomy. The development of a quantitative ‘dimensional' approach (rather than a categorical classification) is of importance, as any usable biomarker needs to provide a quantitative measure of a pathogenic process rather than simply testing for the existence of a particular pathological phenotype. These original reports, which provided a proof of concept, are now supported by several other studies using different imaging modalities; and which also examined different age groups. Using structural MRI data, high classification accuracies have been replicated in children and adolescents with ASD,82, 83 as well as females with the condition.84 It has also been demonstrated that ASD may be detected using measures of brain functioning,85 as well as using functional and structural connectivity indices.86, 87, 88 Notably, a recent study, which also incorporated genetic information into the classifier, proved highly accurate in distinguishing individuals with Asperger syndrome from high-functioning autism.89 In summary, initial work on the ability of biomarkers to classify people having autism looks promising, but several crucial questions need to be addressed first before these novel methods find their way into clinical practice. One of these crucial issues is the clinical specificity.
Although the established methods seem highly successful at distinguishing individuals with ASD from ‘neurotypicals', it is currently unknown how well the biomarkers are able to separate ASD from related comorbid conditions (for example, deficit-hyperactivity disorder, social anxiety or obsessive–compulsive disorders). Preliminary evidence suggest that the proposed methods are indeed specific to ASD rather than neurodevelopmental conditions in general,90 but conclusive evidence is still required. Also, it is currently unknown if the proposed ASD biomarkers will be able to deal with the clinical heterogeneity of the condition. So far, only one study has looked at a specific subtype of ASD—low-functioning children with ASD—and demonstrated that these can be differentiated from children with a general intellectual disability using perfusion (Positron Emission Tomography) data.91 This study was an important step forward, as classifier accuracy was investigated across different autism subtypes.2
Thus, although the search for ASD biomarkers is still in its infancy, the availability of new analytical techniques with high exploratory power and predictive value offers promising new avenues into finding a biomarker whose complexity equals the etiology and phenotype of the condition. If successful, such a biomarker (or a set of biomarkers) might one day prove invaluable in helping diagnose, treat and characterize ASD.
Conclusions
Over the last decade, the behavioral diagnosis of ASD has been invaluable in the clinical setting, as it can accommodate all variations of the ASD spectrum, regardless of their etiology. However, for any biologist (geneticist or neuroscientist) trying to solve what is often referred to as the ‘puzzle of neurobiology' in ASD, starting with the behavioral phenotype, represents a heuristic challenge, which can only be compared with solving an inverse problem. However, new approaches to genotype clustering and the identification of their common functional pathways may enable us in the future to find comparable (that is, homogenous) groups of individuals with ASD, which can be combined in order to elucidate their common underlying neurobiology. Given the genetic and phenotypic heterogeneity of ASD, neither ‘top-down' clinical and translational studies, nor ‘bottom-up' model system analysis are therefore likely to impact on ASD alone. Rather, we need to integrate proven technologies around animal models, cellular assays, together with new analytical approaches in clinical populations to elucidate the complex etiology and phenotype of the condition. Recent animal and postmortem studies are important steps forward in this quest. For example, recent work17 demonstrate how causal interactions between geno- and phenotype can be established by means of active genetic manipulation. Furthermore, the discovery of multiple, specific ASD genotypes implies that these may be rescued (that is, ‘manipulated') by specific, targeted molecular treatment. There is therefore now an opportunity to exploit these new findings and make progress on the development of new therapies for ASD, including both children and adults. There is no doubt that advances in genetic and imaging technologies offer promising new ways of finding biomarkers and/or treatment targets for ASD. If successful, these new approaches may one day prove invaluable in diagnosis, treating and characterizing ASD.
Acknowledgments
This work was supported by the AIMS Consortium (Autism Imaging Multicentre Study) funded by the Medical Research Council UK (G0400061), IMI funded EU AIMS (http://www.eu-aims.eu), and the NIHR Biomedical Research Centre for Mental Health at King's College London, Institute of Psychiatry and South London & Maudsley NHS Foundation Trust.
WS is employed by F. Hoffmann-La Roche. None of the remaining authors have declared any conflict of interest of financial interest, which may arise from being names as an author on the manuscript.
References
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing L, Gould J, Gillberg C. Autism spectrum disorders in the DSM-V: better or worse than the DSM-IV. Res Dev Disabil. 2011;32:768–773. doi: 10.1016/j.ridd.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Muhle R, Trentacoste SV, Rapin I. The genetics of autism. Pediatrics. 2004;113:e472–e486. doi: 10.1542/peds.113.5.e472. [DOI] [PubMed] [Google Scholar]
- Rosenberg RE, Law JK, Yenokyan G, McGready J, Kaufmann WE, Law PA. Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch Pediatr Adolesc Med. 2009;163:907–914. doi: 10.1001/archpediatrics.2009.98. [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Arking DE, Daly MJ, Chakravarti A. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Råstam M, Colineaux C, Gillberg IC, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Autism Genome Project Consortium Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: signalling functions at the synapse. Nat Rev Neurosci. 2007;8:206–220. doi: 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, et al. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Rubenstein JLR, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams BS, Tentler D, Perederiy JV, Oldham MC, Coppola G, Geschwind DH. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc Natl Acad Sci USA. 2007;104:17849–17854. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- Casey BJ, Nigg JT, Durston S. New potential leads in the biology and treatment of attention deficit-hyperactivity disorder. Curr Opin Neurol. 2007;20:119–124. doi: 10.1097/WCO.0b013e3280a02f78. [DOI] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peça J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen M, Durston S, Staal WG, Palmen SJMC, van Engeland H. Caudate nucleus is enlarged in high-functioning medication-naive subjects with autism. Biol Psychiatry. 2007;62:262–266. doi: 10.1016/j.biopsych.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Page LA, Daly E, Schmitz N, Simmons A, Toal F, Deeley Q, et al. In vivo 1H-magnetic resonance spectroscopy study of amygdala-hippocampal and parietal regions in autism. Am J Psychiatry. 2006;163:2189–2192. doi: 10.1176/appi.ajp.163.12.2189. [DOI] [PubMed] [Google Scholar]
- Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T, et al. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry. 2012;17:71–84. doi: 10.1038/mp.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert MR. SHANK3, the synapse, and autism. N Engl J Med. 2011;365:173–175. doi: 10.1056/NEJMcibr1104261. [DOI] [PubMed] [Google Scholar]
- Pizzarelli R, Cherubini E. Alterations of GABAergic signaling in autism spectrum disorders. Neural Plast. 2011;2011:297153. doi: 10.1155/2011/297153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlander MG, Tillakaratne NJ, Feldblum S, Patel N, Tobin AJ. Two genes encode distinct glutamate decarboxylases. Neuron. 1991;7:91–100. doi: 10.1016/0896-6273(91)90077-d. [DOI] [PubMed] [Google Scholar]
- Trikalinos TA, Karvouni A, Zintzaras E, Ylisaukko-oja T, Peltonen L, Järvelä I, et al. A heterogeneity-based genome search meta-analysis for autism-spectrum disorders. Mol Psychiatry. 2006;11:29–36. doi: 10.1038/sj.mp.4001750. [DOI] [PubMed] [Google Scholar]
- Geschwind DH. Genetics of autism spectrum disorders. Trends Cogn Sci. 2011;15:409–416. doi: 10.1016/j.tics.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Van Zeeland AA, Abrahams BS, Alvarez-Retuerto AI, Sonnenblick LI, Rudie JD, Ghahremani D, et al. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci Transl Med. 2010;2:56ra80. doi: 10.1126/scitranslmed.3001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DGM, Critchley HD, Schmitz N, McAlonan G, van Amelsvoort T, Robertson D, et al. Asperger syndrome: a proton magnetic resonance spectroscopy study of brain. Arch Gen Psychiatry. 2002;59:885–891. doi: 10.1001/archpsyc.59.10.885. [DOI] [PubMed] [Google Scholar]
- DeVito TJ, Drost DJ, Neufeld RWJ, Rajakumar N, Pavlosky W, Williamson P, et al. Evidence for cortical dysfunction in autism: a proton magnetic resonance spectroscopic imaging study. Biol Psychiatry. 2007;61:465–473. doi: 10.1016/j.biopsych.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Chao H-T, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahanty RJ, Kang JQ, Brune CW, Kistner EO, Courchesne E, Cox NJ, et al. Maternal transmission of a rare GABRB3 signal peptide variant is associated with autism. Mol Psychiatry. 2011;16:86–96. doi: 10.1038/mp.2009.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogolla N, Leblanc JJ, Quast KB, Südhof TC, Fagiolini M, Hensch TK. Common circuit defect of excitatory-inhibitory balance in mouse models of autism. J Neurodev Disord. 2009;1:172–181. doi: 10.1007/s11689-009-9023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson DR, Adusei DC, Pacey LKK. The neurochemical basis for the treatment of autism spectrum disorders and Fragile X Syndrome. Biochem Pharmacol. 2011;81:1078–1086. doi: 10.1016/j.bcp.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Mendez MA, Horder J, Myers J, Coghlan S, Stokes P, Erritzoe D, et al. The brain GABA-benzodiazepine receptor alpha-5 subtype in autism spectrum disorder: A pilot [(11)C]Ro15-4513 positron emission tomography study Neuropharmacology 2012. PMID: 22546616 (in press). [DOI] [PMC free article] [PubMed]
- Hranilovic D, Bujas-Petkovic Z, Vragovic R, Vuk T, Hock K, Jernej B. Hyperserotonemia in adults with autistic disorder. J Autism Dev Disord. 2007;37:1934–1940. doi: 10.1007/s10803-006-0324-6. [DOI] [PubMed] [Google Scholar]
- Nabi R, Serajee FJ, Chugani DC, Zhong H, Huq AHMM. Association of tryptophan 2,3 dioxygenase gene polymorphism with autism. Am J Med Genet B Neuropsychiatr Genet. 2004;125B:63–68. doi: 10.1002/ajmg.b.20147. [DOI] [PubMed] [Google Scholar]
- Devlin B, Cook EH, Coon H, Dawson G, Grigorenko EL, McMahon W, et al. Autism and the serotonin transporter: the long and short of it. Mol Psychiatry. 2005;10:1110–1116. doi: 10.1038/sj.mp.4001724. [DOI] [PubMed] [Google Scholar]
- Anderson BM, Schnetz-Boutaud NC, Bartlett J, Wotawa AM, Wright HH, Abramson RK, et al. Examination of association of genes in the serotonin system to autism. Neurogenetics. 2009;10:209–216. doi: 10.1007/s10048-009-0171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugani DC, Muzik O, Behen M, Rothermel R, Janisse JJ, Lee J, et al. Developmental changes in brain serotonin synthesis capacity in autistic and nonautistic children. Ann Neurol. 1999;45:287–295. doi: 10.1002/1531-8249(199903)45:3<287::aid-ana3>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Murphy DGM, Daly E, Schmitz N, Toal F, Murphy K, Curran S, et al. Cortical serotonin 5-HT2A receptor binding and social communication in adults with Asperger's syndrome: an in vivo SPECT study. Am J Psychiatry. 2006;163:934–936. doi: 10.1176/ajp.2006.163.5.934. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Sekine Y, Ouchi Y, Tsujii M, Yoshikawa E, Futatsubashi M, et al. Brain serotonin and dopamine transporter bindings in adults with high-functioning autism. Arch Gen Psychiatry. 2010;67:59–68. doi: 10.1001/archgenpsychiatry.2009.137. [DOI] [PubMed] [Google Scholar]
- Daly E, Deeley Q, Ecker C, Craig M, Hallahan B, Murphy C, et al. Serotonin and the neural processing of facial emotions in autism; an fMRI study using acute tryptophan depletion. Arch Gen Psychiatry. 2012;4:1–11. doi: 10.1001/archgenpsychiatry.2012.513. [DOI] [PubMed] [Google Scholar]
- Williams K, Wheeler DM, Silove N, Hazell P.Selective serotonin reuptake inhibitors for the treatment of autism spectrum disorders. Cochrane database of systematic reviews. Advance online publication, 8 September 2010. [DOI] [PubMed]
- Represa A, Ben-Ari Y. Trophic actions of GABA on neuronal development. Trends Neurosci. 2005;28:278–283. doi: 10.1016/j.tins.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Buxhoeveden DP, Casanova MF. The minicolumn and evolution of the brain. Brain Behav Evol. 2002;60:125–151. doi: 10.1159/000065935. [DOI] [PubMed] [Google Scholar]
- Casanova MF, Buxhoeveden DP, Brown C. Clinical and macroscopic correlates of minicolumnar pathology in autism. J Child Neurol. 2002;17:692–695. doi: 10.1177/088307380201700908. [DOI] [PubMed] [Google Scholar]
- Casanova MF, Buxhoeveden DP, Switala AE, Roy E. Neuronal density and architecture (Gray Level Index) in the brains of autistic patients. J Child Neurol. 2002;17:515–521. doi: 10.1177/088307380201700708. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Karns CM, Davis HR, Ziccardi R, Carper RA, Tigue ZD, et al. Unusual brain growth patterns in early life in patients with autistic disorder: an MRI study. Neurology. 2001;57:245–254. doi: 10.1212/wnl.57.2.245. [DOI] [PubMed] [Google Scholar]
- Lainhart JE, Bigler ED, Bocian M, Coon H, Dinh E, Dawson G, et al. Head circumference and height in autism: a study by the Collaborative Program of Excellence in Autism. Am J Med Genet A. 2006;140:2257–2274. doi: 10.1002/ajmg.a.31465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett HC, Poe MD, Gerig G, Styner M, Chappell C, Smith RG, et al. Early brain overgrowth in autism associated with an increase in cortical surface area before age 2 years. Arch Gen Psychiatry. 2011;68:467–476. doi: 10.1001/archgenpsychiatry.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E. Abnormal early brain development in autism. Mol Psychiatry. 2002;7 (Suppl 2:S21–S23. doi: 10.1038/sj.mp.4001169. [DOI] [PubMed] [Google Scholar]
- Herbert MR, Ziegler DA, Deutsch CK, O'Brien LM, Lange N, Bakardjiev A, et al. Dissociations of cerebral cortex, subcortical and cerebral white matter volumes in autistic boys. Brain. 2003;126:1182–1192. doi: 10.1093/brain/awg110. [DOI] [PubMed] [Google Scholar]
- Ecker C, Suckling J, Deoni SC, Lombardo MV, Bullmore ET, Baron-Cohen S, et al. Brain anatomy and its relationship to behavior in adults with autism spectrum disorder: a multicenter magnetic resonance imaging study. Arch Gen Psychiatry. 2012;69:195–209. doi: 10.1001/archgenpsychiatry.2011.1251. [DOI] [PubMed] [Google Scholar]
- Carper RA, Moses P, Tigue ZD, Courchesne E. Cerebral lobes in autism: early hyperplasia and abnormal age effects. Neuroimage. 2002;16:1038–1051. doi: 10.1006/nimg.2002.1099. [DOI] [PubMed] [Google Scholar]
- Gogtay N, Thompson PM. Mapping gray matter development: implications for typical development and vulnerability to psychopathology. Brain Cogn. 2010;72:6–15. doi: 10.1016/j.bandc.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Yeung-Courchesne R, Press GA, Hesselink JR, Jernigan TL. Hypoplasia of cerebellar vermal lobules VI and VII in autism. N Engl J Med. 1988;318:1349–1354. doi: 10.1056/NEJM198805263182102. [DOI] [PubMed] [Google Scholar]
- Abell F, Krams M, Ashburner J, Passingham R, Friston K, Frackowiak R, et al. The neuroanatomy of autism: a voxel-based whole brain analysis of structural scans. Neuroreport. 1999;10:1647–1651. doi: 10.1097/00001756-199906030-00005. [DOI] [PubMed] [Google Scholar]
- McAlonan GM, Cheung V, Cheung C, Suckling J, Lam GY, Tai KS, et al. Mapping the brain in autism. A voxel-based MRI study of volumetric differences and intercorrelations in autism. Brain. 2005;128:268–276. doi: 10.1093/brain/awh332. [DOI] [PubMed] [Google Scholar]
- Saitoh O, Karns CM, Courchesne E. Development of the hippocampal formation from 2 to 42 years: MRI evidence of smaller area dentata in autism. Brain. 2001;124:1317–1324. doi: 10.1093/brain/124.7.1317. [DOI] [PubMed] [Google Scholar]
- Sears LL, Vest C, Mohamed S, Bailey J, Ranson BJ, Piven J. An MRI study of the basal ganglia in autism. Prog Neuropsychopharmacol Biol Psychiatry. 1999;23:613–624. doi: 10.1016/s0278-5846(99)00020-2. [DOI] [PubMed] [Google Scholar]
- Redcay E, Courchesne E. Deviant functional magnetic resonance imaging patterns of brain activity to speech in 2-3-year-old children with autism spectrum disorder. Biol Psychiatry. 2008;64:589–598. doi: 10.1016/j.biopsych.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas DC, Peterson E, Winterrowd E, Reite ML, Rogers SJ, Tregellas JR. Regional gray matter volumetric changes in autism associated with social and repetitive behavior symptoms. BMC Psychiatry. 2006;6:56. doi: 10.1186/1471-244X-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddaert N, Chabane N, Gervais H, Good CD, Bourgeois M, Plumet M-H, et al. Superior temporal sulcus anatomical abnormalities in childhood autism: a voxel-based morphometry MRI study. Neuroimage. 2004;23:364–369. doi: 10.1016/j.neuroimage.2004.06.016. [DOI] [PubMed] [Google Scholar]
- McAlonan GM, Daly E, Kumari V, Critchley HD, van Amelsvoort T, Suckling J, et al. Brain anatomy and sensorimotor gating in Asperger's syndrome. Brain. 2002;125:1594–1606. doi: 10.1093/brain/awf150. [DOI] [PubMed] [Google Scholar]
- Vissers ME, Cohen MX, Geurts HM. Brain connectivity and high functioning autism: a promising path of research that needs refined models, methodological convergence, and stronger behavioral links. Neurosci Biobehav Rev. 2012;36:604–625. doi: 10.1016/j.neubiorev.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Pierce K. Brain overgrowth in autism during a critical time in development: implications for frontal pyramidal neuron and interneuron development and connectivity. Int J Dev Neurosci. 2005;23:153–170. doi: 10.1016/j.ijdevneu.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Rippon G, Brock J, Brown C, Boucher J. Disordered connectivity in the autistic brain: challenges for the ‘new psychophysiology. Int J Psychophysiol. 2007;63:164–172. doi: 10.1016/j.ijpsycho.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Cheung C, Chua SE, Cheung V, Khong PL, Tai KS, Wong TKW, et al. White matter fractional anisotrophy differences and correlates of diagnostic symptoms in autism. J Child Psychol Psychiatry. 2009;50:1102–1112. doi: 10.1111/j.1469-7610.2009.02086.x. [DOI] [PubMed] [Google Scholar]
- Pugliese L, Catani M, Ameis S, Dell'acqua F, Thiebaut de Schotten M, Murphy C, et al. The anatomy of extended limbic pathways in Asperger syndrome: a preliminary diffusion tensor imaging tractography study. Neuroimage. 2009;47:427–434. doi: 10.1016/j.neuroimage.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Catani M, Jones DK, Daly E, Embiricos N, Deeley Q, Pugliese L, et al. Altered cerebellar feedback projections in Asperger syndrome. Neuroimage. 2008;41:1184–1191. doi: 10.1016/j.neuroimage.2008.03.041. [DOI] [PubMed] [Google Scholar]
- Alexander AL, Lee JE, Lazar M, Boudos R, Dubray MB, Oakes TR, et al. Diffusion tensor imaging of the corpus callosum in Autism. Neuroimage. 2007;34:61–73. doi: 10.1016/j.neuroimage.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Bloemen OJN, Deeley Q, Sundram F, Daly EM, Barker GJ, Jones DK, et al. White matter integrity in Asperger syndrome: a preliminary diffusion tensor magnetic resonance imaging study in adults. Autism Res. 2010;3:203–213. doi: 10.1002/aur.146. [DOI] [PubMed] [Google Scholar]
- Belmonte MK, Allen G, Beckel-Mitchener A, Boulanger LM, Carper RA, Webb SJ. Autism and abnormal development of brain connectivity. J Neurosci. 2004;24:9228–9231. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just MA, Cherkassky VL, Keller TA, Kana RK, Minshew NJ. Functional and anatomical cortical underconnectivity in autism: evidence from an FMRI study of an executive function task and corpus callosum morphometry. Cereb Cortex. 2007;17:951–961. doi: 10.1093/cercor/bhl006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshino H, Kana RK, Keller TA, Cherkassky VL, Minshew NJ, Just MA. fMRI investigation of working memory for faces in autism: visual coding and underconnectivity with frontal areas. Cereb Cortex. 2008;18:289–300. doi: 10.1093/cercor/bhm054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchew DE, Ashwin C, Berkouk K, Salvador R, Suckling J, Baron-Cohen S, et al. Functional disconnectivity of the medial temporal lobe in Asperger's syndrome. Biol Psychiatry. 2005;57:991–998. doi: 10.1016/j.biopsych.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Cox DD, Savoy RL. Functional magnetic resonance imaging (fMRI) ‘brain reading': detecting and classifying distributed patterns of fMRI activity in human visual cortex. Neuroimage. 2003;19:261–270. doi: 10.1016/s1053-8119(03)00049-1. [DOI] [PubMed] [Google Scholar]
- Ecker C, Rocha-Rego V, Johnston P, Mourão-Miranda J, Marquand A, Daly EM, et al. Investigating the predictive value of whole-brain structural MR scans in autism: a pattern classification approach. Neuroimage. 2010;49:44–56. doi: 10.1016/j.neuroimage.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Chen R, Ke X, Chu K, Lu Z, Herskovits EH. Predictive models of autism spectrum disorder based on brain regional cortical thickness. Neuroimage. 2010;50:589–599. doi: 10.1016/j.neuroimage.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Menon V, Young CB, Ryali S, Chen T, Khouzam A, et al. Multivariate searchlight classification of structural magnetic resonance imaging in children and adolescents with autism. Biol Psychiatry. 2011;70:833–841. doi: 10.1016/j.biopsych.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderoni S, Retico A, Biagi L, Tancredi R, Muratori F, Tosetti M. Female children with autism spectrum disorder: an insight from mass-univariate and pattern classification analyses. Neuroimage. 2012;59:1013–1022. doi: 10.1016/j.neuroimage.2011.08.070. [DOI] [PubMed] [Google Scholar]
- Coutanche MN, Thompson-Schill SL, Schultz RT. Multi-voxel pattern analysis of fMRI data predicts clinical symptom severity. Neuroimage. 2011;57:113–123. doi: 10.1016/j.neuroimage.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange N, Dubray MB, Lee JE, Froimowitz MP, Froehlich A, Adluru N, et al. Atypical diffusion tensor hemispheric asymmetry in autism. Autism Res. 2010;3:350–358. doi: 10.1002/aur.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalhalikar M, Parker D, Bloy L, Roberts TPL, Verma R. Diffusion based abnormality markers of pathology: toward learned diagnostic prediction of ASD. Neuroimage. 2011;57:918–927. doi: 10.1016/j.neuroimage.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JS, Nielsen JA, Froehlich AL, Dubray MB, Druzgal TJ, Cariello AN, et al. Functional connectivity magnetic resonance imaging classification of autism. Brain. 2011;134:3739–3751. doi: 10.1093/brain/awr263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Chen R, Ke X, Cheng L, Chu K, Lu Z, et al. Predictive models for subtypes of autism spectrum disorder based on single-nucleotide polymorphisms and magnetic resonance imaging. Adv Med Sci. 2011;56:334–342. doi: 10.2478/v10039-011-0042-y. [DOI] [PubMed] [Google Scholar]
- Ecker C, Marquand A, Mourão-Miranda J, Johnston P, Daly EM, Brammer MJ, et al. Describing the brain in autism in five dimensions—magnetic resonance imaging-assisted diagnosis of autism spectrum disorder using a multiparameter classification approach. J Neurosci. 2010;30:10612–10623. doi: 10.1523/JNEUROSCI.5413-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchesnay E, Cachia A, Boddaert N, Chabane N, Mangin J-F, Martinot J-L, et al. Feature selection and classification of imbalanced datasets: application to PET images of children with autistic spectrum disorders. Neuroimage. 2011;57:1003–1014. doi: 10.1016/j.neuroimage.2011.05.011. [DOI] [PubMed] [Google Scholar]