Abstract
ARID4A and ARID4B are homologous members of the ARID (AT-rich interaction domain) gene family. ARID4A and ARID4B physically interact with each other. ARID4A is a retinoblastoma (RB)-binding protein. Biological function of these interactions is still unknown. Here, we report that mice with complete deficiency of Arid4a, combined with haploinsufficiency of Arid4b (Arid4a−/−Arid4b+/−), showed progressive loss of male fertility, accompanied by hypogonadism and seminal vesicle agenesis/hypodysplasia. Arid4a and Arid4b are expressed mainly in Sertoli cells of testes, which implies that their roles in Sertoli cell function are to support spermatogenesis and create the impermeable blood–testis barrier. In fact, evaluation of germ cell development in the Arid4a−/−Arid4b+/− mice showed spermatogenic arrest at the stages of meiotic spermatocytes and postmeiotic haploid spermatids. Analysis of the integrity of the blood–testis barrier showed increased permeability of seminiferous tubules in the Arid4a−/−Arid4b+/− testes. Interestingly, phenotypic Sertoli cell dysfunction in the Arid4a−/−Arid4b+/− mice, including spermatogenic failures and the impaired blood–testis barrier, recapitulated the defects found in the Sertoli cell-specific androgen receptor (AR) knockout mice and the Sertoli cell-specific RB knockout mice. Investigation of the molecular mechanism identified several AR- and RB-responsive genes as downstream targets of ARID4A and ARID4B. Our results thus indicate that ARID4A and ARID4B function as transcriptional coactivators for AR and RB and play an integral part in the AR and RB regulatory pathways involved in the regulation of Sertoli cell function and male fertility.
Keywords: male reproductive function, androgen receptor coactivator
Male fertility is a complex process that involves growth and formation of male reproductive organs as well as initiation and maintenance of spermatogenesis. Spermatogenesis is a developmental process in which mature spermatozoa are generated within seminiferous tubules of testes (reviewed in ref. 1). In addition to the germ cells, testes contain three somatic cell types including Sertoli cells, peritubular myoid cells, and Leydig cells. Sertoli cells are the epithelium of the seminiferous tubules and provide both physical and nutritional supports for the developing germ cells (2). Between adjacent Sertoli cells, tight junctions are formed to create the blood–testis barrier, which segregates the developing germ cells into the immunologically privileged adluminal compartment and prevents passage of cytotoxic substances into the seminiferous tubules (2, 3).
Androgen receptor (AR), a member of the steroid hormone receptor superfamily, mediates the androgen action and plays an important role in male fertility (4). Global AR knockout male mice exhibited infertility with strict defects in male reproductive organs and severe disruption of germ cell development (5). The cell-specific AR knockout mouse models further clarified the functions of AR in each specific type of testicular cells (6). Sertoli cell-specific AR knockout affected the Sertoli cell functions of nourishing and supporting the developing germ cells, resulting in spermatogenesis arrest before first meiotic division and during the transition from round to elongated spermatids (7–9). In addition, lack of AR in Sertoli cells causes impairment of the blood–testis barrier (10). As expected, the Sertoli cell-specific AR conditional knockout male mice were infertile (7–9).
AR functions as a ligand-dependent transcription factor, and its transcriptional activity is known to require the interactions with various coregulators that act in a sequential and combinatorial manner to regulate expression of the AR-responsive genes. Retinoblastoma (RB) has been reported to interact with AR and functions as a coactivator to induce AR transcriptional activity (11). The Sertoli cell-specific Rb conditional knockout mice displayed progressive infertility in males (12). These infertile male mice exhibited Sertoli cell dysfunction, leading to loss of elongating spermatids and spermatozoa, as well as loss of the integrity of the blood–testis barrier (12).
ARID4A/Arid4a and ARID4B/Arid4b are homologous members of the ARID (AT-rich interaction domain) gene family. Previously, ARID4A and ARID4B were known as RB-binding protein 1 (RBBP1, RBP1) (13) and RBBP1-like protein 1 (RBBP1L1) (14), respectively. Biochemical analyses have suggested that ARID4A and ARID4B are members of the chromatin-remodeling complex and function as transcriptional repressors upon recruitment by RB (15, 16). Recently, we generated the Arid4a and Arid4b knockout mouse models and demonstrated that ARID4A and ARID4B physically and functionally interact with each other (17, 18). Studies of these mouse models provided in vivo evidence that Arid4a and Arid4b play a role in genomic imprinting and function as leukemia suppressor genes through regulation of epigenetic modifications (17, 18).
Although ARID4A and ARID4B are part of the RB complex, the biological relationship of ARID4A and ARID4B to RB has not been defined. In this study, using the Arid4a and Arid4b knockout mouse model, we found that Arid4a and Arid4b play important roles in Sertoli cell function. Investigation of the transcriptional consequences after the Arid4a and Arid4b knockout identified their downstream targets overlapping the AR- and RB-responsive genes. We further demonstrated that ARID4A and ARID4B function as coactivators to enhance the AR and RB transcriptional activity. Our studies indicate that ARID4A and ARID4B not only physically interact with AR and RB, but also functionally link to the AR and RB pathways in regulation of male fertility.
Results
Mice Deficiency for Arid4a and Haploinsufficiency for Arid4b Displayed Progressive Loss of Fertility in Males.
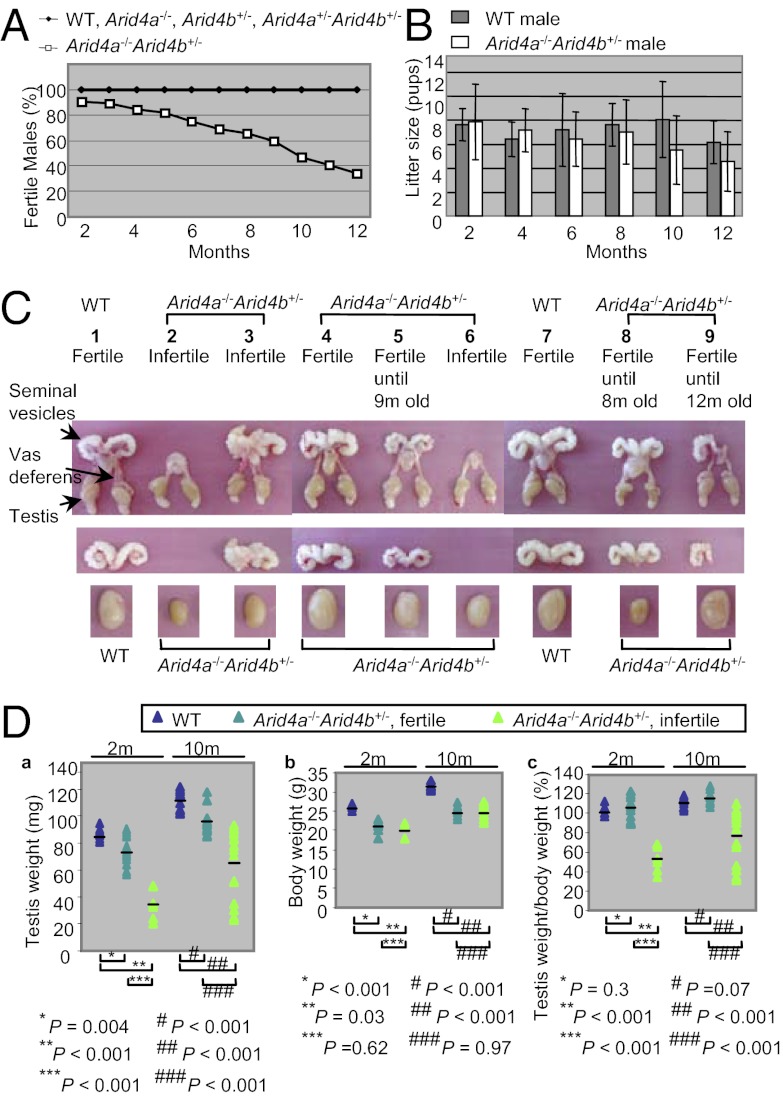
To determine the involvement of Arid4a and Arid4b in male fertility, male mice with different Arid4a and Arid4b genotypes, including Arid4a−/− (n = 25), Arid4b+/− (n = 21), Arid4a+/−Arid4b+/− (n = 19), Arid4a−/−Arid4b+/− (n = 32), and wild type (n = 15), were mated with adult wild-type females. We monitored fertility of male mice from 2 to 12 mo of age. Adult Arid4a−/−, Arid4b+/−, Arid4a+/−Arid4b+/−, and wild-type males were fully fertile (Fig. 1A). In contrast, adult Arid4a−/−Arid4b+/− males displayed age-related reduced fertility. We found that 29 of 32 (∼90%) Arid4a−/−Arid4b+/− males were fertile at 2 mo of age. The fertility decreased from 3 mo of age onward, with less than 40% of the Arid4a−/−Arid4b+/− males remaining fertile at 1 y of age (Fig. 1A). These results showed that complete deficiency of Arid4a in combination with haploinsufficiency of Arid4b leads to progressive infertility in male mice, suggesting that Arid4a collaborates with Arid4b to regulate male fertility. Despite the progressive loss of fertility, no significant difference in the litter size between the fertile Arid4a−/−Arid4b+/− males and wild-type males was noted (Fig. 1B). Mice homozygous for Arid4b deficiency die early in embryogenesis between embryonic day 3 (E3) and E7 (17); therefore, conventional knockout mice with all four alleles mutated (Arid4a−/−Arid4b−/−) are not available for the fertility assay.
Fig. 1.
Arid4a−/−Arid4b+/− male mice displayed defects in male reproductive function. (A and B) Progressive loss of fertility in the Arid4a−/−Arid4b+/− male mice. Percentages of fertile males (A) and litter size (number of pups) (B) were compared between wild-type and the Arid4a−/−Arid4b+/− male mice from 2 mo of age through 1 y of age. (C) Hypogonadism and seminal vesicle agenesis/hypodysplasia in the Arid4a−/−Arid4b+/− male mice. Morphology of the whole male reproductive system (Top), seminal vesicles (Middle), and testes (Bottom) was compared between wild-type and Arid4a−/−Arid4b+/− mice. (D) Testis weight (a), body weight (b), and testis weight normalized against body weight (c) were shown from wild type and the Arid4a−/−Arid4b+/− mice at 2 and 10 mo of age. Horizontal lines indicate the means. Wild type, 2 mo (“2m”) old—4 males and 10 mo old—6 males; fertile Arid4a−/−Arid4b+/−, 2 mo old—6 males and 10 mo old—6 males; infertile Arid4a−/−Arid4b+/−, 2 mo old—3 males and 10 mo old—8 males.
We next examined the male reproductive organs for any morphological defect that may account for reduced fertility. Of the 32 Arid4a−/−Arid4b+/− males monitored from 2 mo of age, 3 mice were infertile for their entire lives (Fig. 1A). These three infertile males showed reduced size of both testes compared with that of wild-type mice (Fig. 1C, the Arid4a−/−Arid4b+/− mice 2, 3, and 6; wild-type mice 1 and 7). In addition, two of the three mice displayed bilateral absence of seminal vesicles (Fig. 1C, mice 2 and 6), and the third infertile male showed abnormal fusion of seminal vesicles (Fig. 1C, mouse 3). The majority of the Arid4a−/−Arid4b+/− male mice were fertile at a younger age but lost their fertility at an older age (from 3 to 12 mo of age) (Fig. 1A). These mice showed decreased size of testes with or without fused or hypotrophic seminal vesicles (Fig. 1C, mice 5 and 8). We also found that one male showed unilateral agenesis of seminal vesicles (Fig. 1C, mouse 9), but remained fertile up to 12 mo old.
Measurements of testes revealed reduced testicular weights in the Arid4a−/−Arid4b+/− males compared with wild-type males at 2 and 10 mo of age (Fig. 1 D, a). Testes of the infertile Arid4a−/−Arid4b+/− mice weighed even less than the fertile Arid4a−/−Arid4b+/− mice (Fig. 1 D, a). The body weights of the Arid4a−/−Arid4b+/− mice were also less than that of wild-type mice (17) (Fig. 1 D, b). It should be noted that only the infertile Arid4a−/−Arid4b+/− males, but not the fertile Arid4a−/−Arid4b+/− males, showed a significant decrease in the ratios of testicular weight vs. body weight at both 2 and 10 mo of age (Fig. 1 D, c). On the other hand, both fertile and infertile Arid4a−/−Arid4b+/− males showed no significant difference in reduced body weight (Fig. 1 D, b), suggesting that the fertility defects were not a result of reduced body weight.
Arid4a and Arid4b Express in Sertoli Cells of Testes.
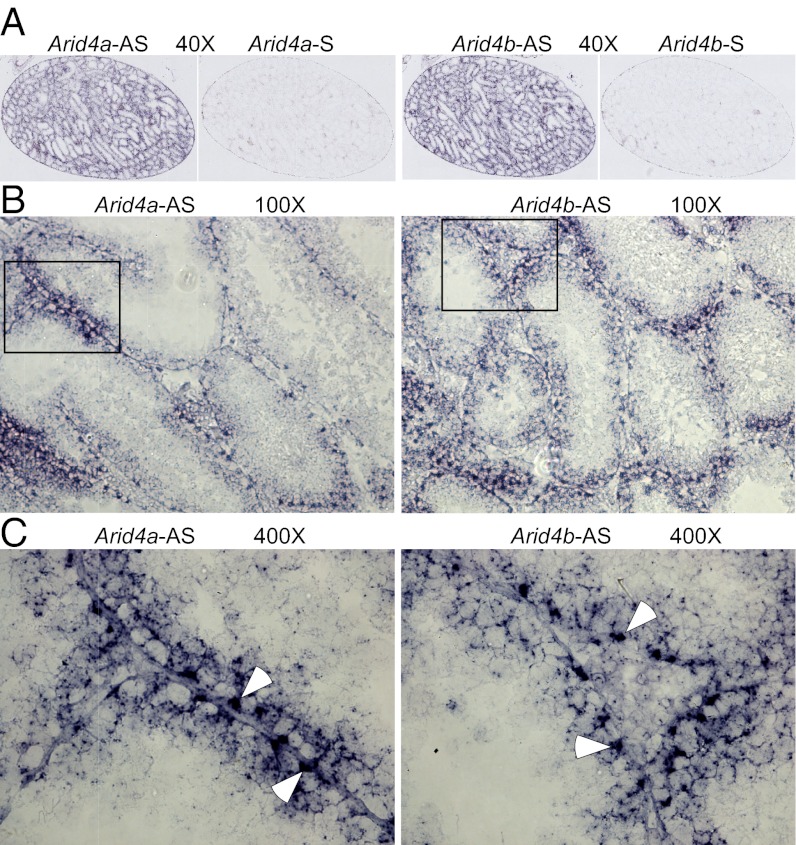
A testis contains four primary cell types: germ cells, Sertoli cells, peritubular myoid cells, and Leydig cells. Within the seminiferous tubules of testes, germ cells develop from spermatogonia to spermatocytes to spermatids to spermatozoa; Sertoli cells are the epithelium of seminiferous tubules. Peritubular myoid cells are located on the outer border of the seminiferous tubules, and Leydig cells are located between seminiferous tubules. Each type of cell plays different roles in male reproductive function. To define the function of Arid4a and Arid4b in male reproduction, it is necessary to identify their expression in testicular cells. In situ hybridization analysis was performed using probes encompassing Arid4a and Arid4b antisense RNA to detect the Arid4a and Arid4b transcripts, respectively, whereas probes containing sense RNA were used as negative controls. Our results showed that both Arid4a and Arid4b are expressed in testes (Fig. 2A). Specifically, microscopic analysis under higher magnification revealed that the majorities of the Arid4a and Arid4b transcripts were detected in Sertoli cells (Fig. 2 B and C). Sertoli cells in testes are critical for supporting spermatogenesis and creating the blood–testis barrier. Abundant expression of Arid4a and Arid4b in Sertoli cells implies their role in Sertoli cell function.
Fig. 2.
In situ hybridization analyses of Arid4a and Arid4b expression in testes from wild-type mice. (A–C) Sections of wild-type testes were hybridized with a digoxigenin-labeled Arid4a and Arid4b antisense (Arid4a-AS and Arid4b-AS) or sense (Arid4a-S and Arid4b-S) probes. Original magnifications were 40× (A), 100× (B), and 400× (C). White arrowheads indicate Sertoli cells (C).
Arid4a−/−Arid4b+/− Male Mice Displayed Spermatogenic Failure at the Meiosis II and the Postmeiotic Spermatid Stages.
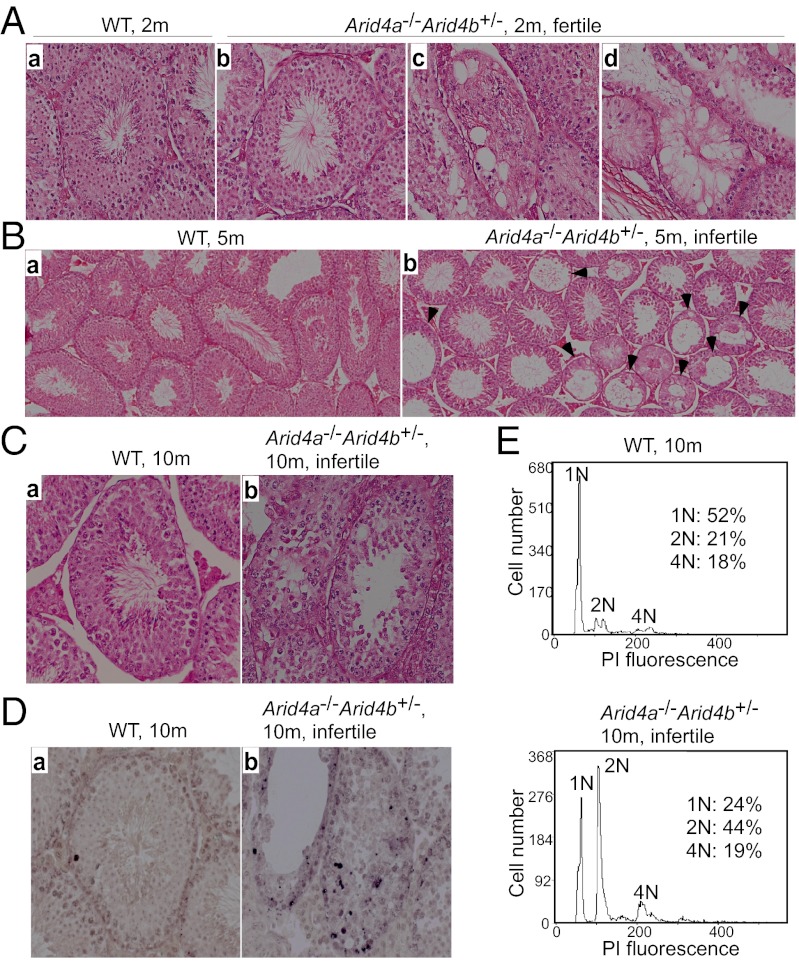
Next, we investigated spermatogenesis for potential causes of the observed reduced fertility in the Arid4a−/−Arid4b+/− male mice. Histological analysis of testes from the fertile Arid4a−/−Arid4b+/− and wild-type males at 2 mo of age showed spermatogenic cells with different stages of maturation ranging from spermatogonia spermatocyte, spermatid, and finally to spermatozoa in seminiferous tubules (Fig. 3 A, a and b). However, we also found that several seminiferous tubules underwent vacuolar degeneration in the fertile Arid4a−/−Arid4b+/− mice at 2 mo of age (Fig. 3 A, c and d). In the infertile Arid4a−/−Arid4b+/− males at 5 mo of age, seminiferous tubules revealed decreased widths with many showing vacuolization and a complete loss of architecture (Fig. 3 B, b). Moreover, in the Arid4a−/−Arid4b+/− males that became infertile at 10 mo of age, spermatogenesis was impaired as evidenced by an apparent differentiation arrest at the transition between early to late spermatocytes (Fig. 3 C, a and b). TUNEL assay further revealed that the seminiferous tubules from these infertile 10-mo-old Arid4a−/−Arid4b+/− mice contained an increased number of apoptotic cells compared with that from wild-type controls (Fig. 3 D, a and b).
Fig. 3.
Deterioration of seminiferous tubules and arrest of spermatogenesis in the Arid4a−/−Arid4b+/− testes. (A) H&E-stained sections of seminiferous tubules of testes from wild-type (a) and fertile Arid4a−/−Arid4b+/− (b–d) mice at 2 mo of age. Original magnification was 400× . (B and C) H&E-stained sections of wild-type (a) and infertile Arid4a−/−Arid4b+/− (b) testis from mice at 5 mo of age (B) and at 10 mo of age (C). Original magnifications were 200× (B) and 400× (C). Black arrowheads indicate loss of architecture of the seminiferous tubules in the Arid4a−/−Arid4b+/− testes (B, b). (D) In situ TUNEL assay of testis sections from wild-type (a) and infertile Arid4a−/−Arid4b+/− (b) male mice at 10 mo of age. Apoptotic cells were stained brown. Original magnification was 400× . (E) Flow cytometric analysis of propidium iodide fluorescence-stained DNA of testicular cells from wild-type (Upper) and infertile Arid4a−/−Arid4b+/− (Lower) male mice at 10 mo of age.
In adult testes, the process of spermatogenesis begins from a diploid spermatogonia (2N DNA content), which undergoes mitotic division to produce two diploid primary spermatocytes (2N DNA content). Each primary spermatocyte duplicates its chromosomes (4N DNA content), followed by two meiotic divisions, meiosis I and meiosis II. In meiosis I, primary spermatocytes divide into secondary spermatocytes (2N DNA content). In meiosis II, secondary spermatocytes divide to produce haploid round spermatids (1N DNA content) (19). To analyze sperm development in the Arid4a−/−Arid4b+/− mice, DNA contents of testicular cells from the infertile 10-mo-old male mutants were analyzed by flow cytometry with propidium iodide staining. We found that the percentage of primary spermatocytes (4N) in the Arid4a−/−Arid4b+/− mice was not significantly different from that in wild-type mice (Fig. 3E). However, the Arid4a−/−Arid4b+/− testes revealed a marked increase of the 2N DNA peak concomitant with a decrease of the 1N peak (Fig. 3E). These results suggest that germ cells are able to complete meiosis I to form diploid secondary spermatocytes (2N DNA content), but had delayed entry into meiosis II to become haploid round spermatids (1N DNA content). This result suggests a maturation defect at meiosis II stages of spermatogenesis in the infertile Arid4a−/−Arid4b+/− males at 10 mo of age.
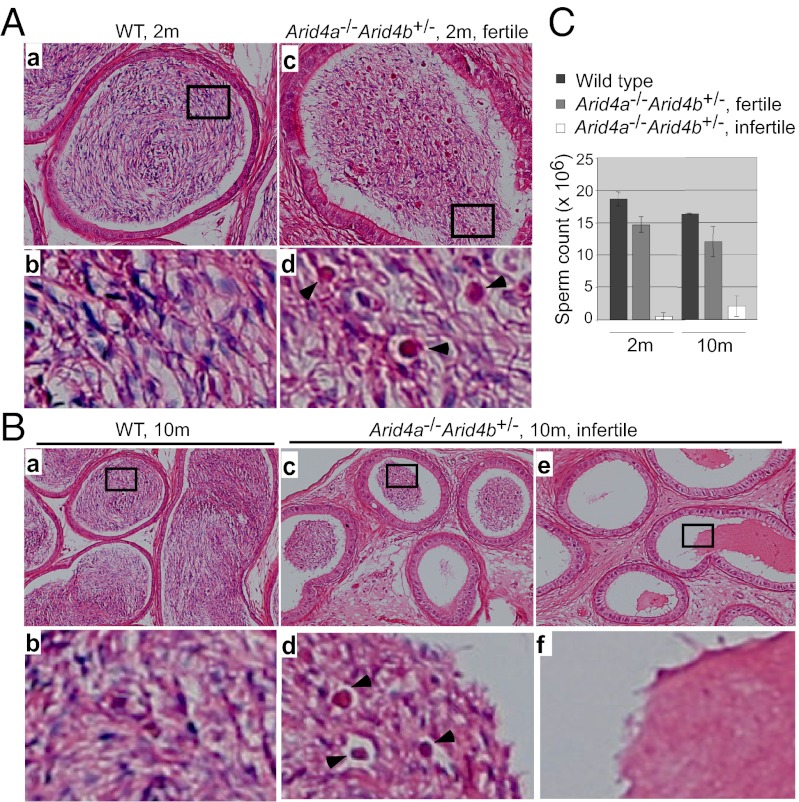
Round spermatids differentiate into elongated spermatids and undergo a series of morphological changes leading to the formation of mature spermatozoa, which then pass from testes into the epididymis (20). Evaluation of epididymal lumens in wild-type mice at 2 mo of age displayed numerous fully mature spermatozoa (Fig. 4 A, a and b), whereas the fertile Arid4a−/−Arid4b+/− males showed the presence of round spermatids (Fig. 4 A, c and d). These results revealed a partial maturation block at the spermatid stage with round spermatids sloughing off into the epididymal lumens, suggesting that Arid4a and Arid4b have effects on postmeiotic events for adhesion of round and elongated spermatids in testes. Moreover, the infertile Arid4a−/−Arid4b+/− males at 10 mo of age revealed a significant decrease or a complete absence of germ cells in epididymal lumens (Fig. 4 B, c–f) compared with wild-type controls (Fig. 4 B, a and b). To further characterize the basis for infertility, spermatozoa were isolated from epididymis. Consistent with the histological findings, the spermatozoa number decreased dramatically in the infertile Arid4a−/−Arid4b+/− males at 2 and 10 mo of age (Fig. 4C).
Fig. 4.
Characteristics of sperms in the Arid4a−/−Arid4b+/− epididymis. (A) Sections of epididymis from wild type (a and b) and fertile Arid4a−/−Arid4b+/− (c and d) males at 2 mo of age were stained with H&E. High magnification is shown in b and d. Arrowheads indicate round spermatids inside epididymal lumens of Arid4a−/−Arid4b+/− mice (d). (B) H&E-stained epididymis sections from a wild-type male (a and b) and two infertile Arid4a−/−Arid4b+/− males (c–f) at 10 mo of age. High magnification is shown in b, d, and f. Arrowheads indicate round spermatids (d). (C) Sperm count from wild-type males, fertile Arid4a−/−Arid4b+/− males, and infertile Arid4a−/−Arid4b+/− males at 2 and 10 mo of age. Sperms were collected from the epididymides of each mouse. Spermatozoa containing both a head and a tail were counted. Counts were obtained from at least three mice per genotype at the indicated age.
To determine whether these effects were hormone related, we examined the levels of serum testosterone, luteinizing hormone (LH), and follicle-stimulating hormone (FSH) in the Arid4a−/−Arid4b+/− and wild-types males at 2 and 10 mo of ages. No significant differences were found between the fertile mutants, infertile mutants, and wild-type controls from either group of age (Fig. S1).
Taken together, Arid4a and Arid4b are important for normal sperm development. Complete deficiency of Arid4a combined with partial deficiency of Arid4b leads to spermatogenic failure at the meiosis II and the postmeiotic spermatid stages, even though the hormonal levels are maintained.
Dysregulation of AR-Responsive Genes in the Arid4a−/−Arid4b+/− Male Mice.
To investigate the mechanism leading to spermatogenic failure in the Arid4a−/−Arid4b+/− mice, we evaluated the effects of the Arid4a and Arid4b mutations on gene expression in the testes. Microarray analysis was performed to examine genome-wide gene expression patterns using RNAs purified from testes of three infertile Arid4a−/−Arid4b+/− mice and three wild-type mice at 10 mo of age. Compared with wild-type mice, 42 genes were down-regulated (Table S1) and 65 genes were up-regulated (Table S2) in the Arid4a−/−Arid4b+/− mice. It is of note that microarray analysis showed decreased expression of both Arid4a and Arid4b in the Arid4a−/−Arid4b+/− testes (Table S1).
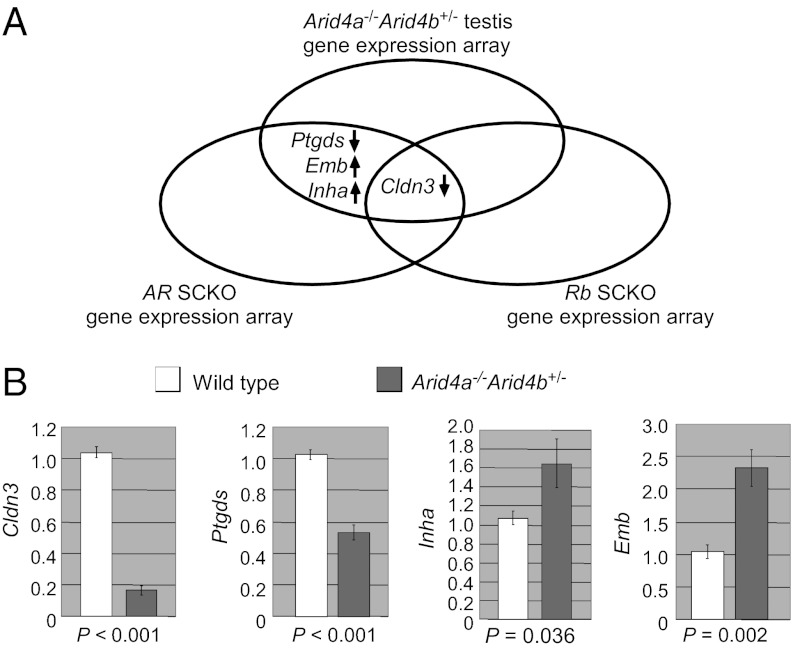
ARID4A and ARID4B are two RB-binding related proteins (13, 14). RB is a known coregulator of AR (11). Both AR and RB are indispensable for Sertoli cell function in supporting germ cell development (7–9, 12). We hypothesized that ARID4A and ARID4B are involved in the AR and RB pathways to control spermatogenesis, and therefore spermatogenic failure in the Arid4a−/−Arid4b+/− mice might be due, in part, to dysregulation of AR-responsive and/or Rb-responsive genes. To test this hypothesis, we compared our microarray data to the gene lists that contained putative AR- or RB-regulated genes identified by microarray gene expression analyses using the Sertoli cell-specific AR knockout mice (21) or the Sertoli cell-specific Rb knockout mice (12). Interestingly, claudin 3 (Cldn3) was down-regulated in all three kinds of knockout mice (Fig. 5A), suggesting that Cldn3 is a common target gene regulated by AR, Rb, Arid4a, and Arid4b. In addition, three more putative AR-regulated genes, including prostaglandin D2 synthase (Ptgds), inhibin alpha (Inha), and embigin (Emb), were also identified in our microarray analysis. In both Arid4a−/−Arid4b+/− testes and Sertoli cell-specific AR knockout testes, Ptgds was down-regulated, whereas Inha and Emb were up-regulated (Fig. 5A).
Fig. 5.
Changes of gene expression in the Arid4a−/−Arid4b+/− testes. (A) Genes identified by microarray analyses of the Arid4a−/−Arid4b+/− testes, AR conditional knockout Sertoli cells, and Rb conditional knockout Sertoli cells show the overlaps. (B) qRT-PCR analyses confirmed decreased expression of Cldn3 and Ptgds and increased expression of Inha and Emb in testes from the Arid4a−/−Arid4b+/− mice at 2, 5, and 10 mo of age. Three mice for each genotype at each age were analyzed. The level of gene expression from one of wild-type mice at each age was set as 1.
Importantly, quantitative RT-PCR (qRT-PCR) analyses confirmed decreased expression of Cldn3 and Ptgds, as well as increased expression of Inha and Emb, in the Arid4a−/−Arid4b+/− testes (Fig. 5B). In all cases, the expression levels of AR and Rb from the Arid4a−/−Arid4b+/− testes were comparable to that from the wild-type controls (Fig. S2, D and E), suggesting that the effects are not due to insufficiency of AR or RB.
ARID4A and ARID4B Function as Coactivators of AR and RB.
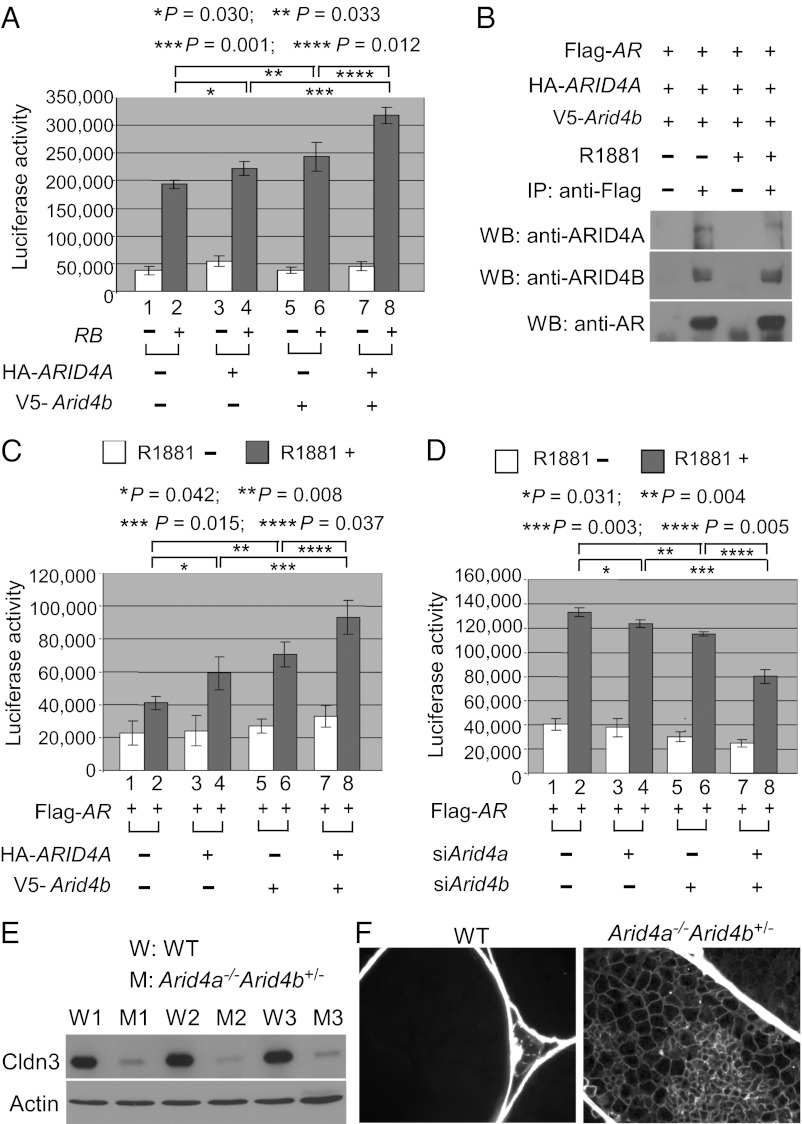
Given that Cldn3 is commonly regulated by Arid4a, Arid4b, Rb, and AR (Fig. 5A), we investigated whether ARID4A and ARID4B collaborate with RB or AR to regulate expression of Cldn3. For this purpose, a reporter plasmid containing the promoter fragment of Cldn3 (from −972 to +229 bp) was used for luciferase reporter gene assays. We performed transient transfection experiments combined with luciferase activity analyses in the testis Sertoli cell line TM4. Whereas transfection of RB activated the Cldn3 promoter (Fig. 6A, columns 1 and 2), transfection of ARID4A or ARID4B alone or ARID4A and ARID4B combined without cotransfection of RB did not have significant effects on the Cldn3 promoter (Fig. 6A, columns 1, 3, 5, and 7). Interesting, cotransfection of ARID4A or ARID4B alone with RB slightly increased the RB-induced activation of Cldn3 (Fig. 6A, columns 2, 4, and 6), and both ARID4A and ARID4B combined enhanced the up-regulation (Fig. 6A, columns 2, 4, 6, and 8). These results suggest that ARID4A and ARID4B regulate expression of Cldn3 by acting as positive coregulators for RB.
Fig. 6.
Arid4a and Arid4b regulate Cldn3. (A) ARID4A and Arid4b induce RB activity on the Cldn3 promoter. TM4 cells were transfected with the Cldn3 promoter-regulated luciferase reporter along with the expression vectors for RB, HA-ARID4A, or V5-Arid4b as indicated. Luciferase activity was measured 48 h after transfection. Shown are means ± SD from three experiments performed in triplicate. (B) ARID4A and ARID4B were associated with AR in a ligand-independent manner. TM4 cells were transfected with the expression vectors for Flag-AR, HA-ARID4A, and V5-Arid4b. After the transfected cells were treated with or without R1881 (100 nM), immunoprecipitation experiments were performed using the anti-Flag Ab, and then Western blot analyses were performed by using antibodies against ARID4A, ARID4B, or AR. (C) ARID4A and ARID4B additively induce AR activity on the Cldn3 promoter. TM4 cells were transfected with the Cldn3 promoter-regulated luciferase reporter along with the expression vectors for Flag-AR, HA-ARID4A, or V5-Arid4b as indicated. The transfected cells were treated with or without R1881, and luciferase activity was measured thereafter. Shown are means ± SD from three experiments performed in triplicate. (D) Knockdown of Arid4a and Arid4b synergistically reduced the AR activity on the Cldn3 promoter. TM4 cells were first transfected with siArid4a and/or siArid4b. The next day, these cells were transfected with the Cldn3 promoter-regulated luciferase reporter and the expression vector for AR. After the cells were treated with or without R1881, luciferase activity was determined. Shown are means ± SD from three experiments performed in triplicate. (E) Decreased expression of Cldn3 in the testes of the Arid4a−/−Arid4b+/− mice was validated at the protein level by Western blot analysis. Actin was used as a loading control. Cldn3, 23 kDa; actin, 47 kDa. (F) Increased permeability of the blood–testis barrier in the Arid4a−/−Arid4b+/− testes. The permeability of the blood–testis barrier was assessed by injection of a biotin tracer into testes of the Arid4a−/−Arid4b+/− and wild-type mice at 3 mo of age. The biotin tracer was detected by Alexa Fluor 488 streptavidin. Biotin is present in the adluminal space of the Arid4a−/−Arid4b+/− testes.
Given that ARID4A and ARID4B are two RB-binding related proteins (13, 14) and that RB has been reported to specifically bind to AR (11), we examined whether ARID4A and ARID4B interact with AR. Coimmunoprecipitation combined with Western blotting analyses in the TM4 cells clearly showed that both ARID4A and ARID4B can be coimmunoprecipitated with AR even in the absence of AR agonist R1881, and treatment of R1881 did not enhance their interactions (Fig. 6B). These ligand-independent interactions of ARID4A and ARID4B with AR are similar to the reported ligand-independent interaction between RB and AR (11).
Because expression of Cldn3 is also regulated by AR (10, 21, 22), the interaction of ARID4A and ARID4B with AR led us to examine whether ARID4A and ARID4B are involved in the AR-induced transcriptional activation of Cldn3. Our results showed that the Cldn3 promoter was activated by AR in a ligand R1881-dependent manner in the TM4 cells (Fig. 6C, columns 1 and 2), and cotransfection of either Arid4a or Arid4b alone with AR increased the activation of the Cldn3 promoter (Fig. 6C, columns 2, 4, and 6). In addition, cotransfection of both Arid4a and Arid4b combined with AR further enhanced this up-regulation (Fig. 6C, columns 2, 4, 6, and 8). These results suggest that both ARID4A and ARID4B function as coactivators for AR in activation of Cldn3.
Next, we determined whether knockdown of endogenous Arid4a and Arid4b by siRNA affects the ability of AR to activate the Cldn3 promoter. Our results showed that, whereas knockdown of either Arid4a or Arid4b slightly reduced the AR activity on the Cldn3 promoter (Fig. 6D, columns 2, 4, and 6), knockdown of both combined further inhibited the AR-induced activation of Cldn3 (Fig. 6D, columns 2, 4, 6, and 8). In all cases, knockdown of Arid4a and Arid4b by siRNA was verified by qRT-PCR analyses (Fig. S3). Together, our results suggest that ARID4A and ARID4B contribute to the optimal expression of Cldn3 by functioning as positive coregulators in the context of the AR and RB complex.
ARID4A and ARID4B Regulate the Permeability of the Blood–Testis Barrier.
Study of the mouse models carrying the Arid4a and/or Arid4b knockout mutations showed that expression of Cldn3 was partially decreased in the Arid4a−/− testes (Fig. S4), but was markedly reduced in the Arid4a−/−Arid4b+/− testes (Fig. 5B and Fig. S4), suggesting that Arid4a and Arid4b collaborate to regulate expression of Cldn3. The severe down-regulation of Cldn3 in the Arid4a−/−Arid4b+/− testes was further verified at the protein level by Western blotting assay (Fig. 6E).
Cldn3 encodes a tight junction component of the blood–testis barrier (10). Down-regulation of Cldn3 has been reported to be related to increased permeability of the blood–testis barrier in the Sertoli cell-specific AR knockout mice (10) and the Sertoli cell-specific Rb knockout mice (12). To examine whether the integrity of the blood–testis barrier was compromised in the Arid4a−/−Arid4b+/− mice, we injected a biotin tracer into the testis interstitium of 3-mo-old mice and monitored the location of the biotin tracer. In wild-type mice, the biotin tracer remained in the basal compartment and was excluded from the adluminal compartment of seminiferous tubules (Fig. 6F). In contrast, the biotin tracer was present in the adluminal compartment and pervaded the tubules of the Arid4a−/−Arid4b+/− testes (Fig. 6F), indicating increased permeability of the blood–testis barrier. This result suggests that the integrity of the blood–testis barrier was impaired in the Arid4a−/−Arid4b+/− testes. Together, the Arid4a−/−Arid4b+/− mice recapitulated Sertoli cell dysfunction, including spermatogenic failures (Figs. 3 and 4) and the impaired blood–testis barrier (Fig. 6F).
Discussion
In this study, we characterized the Arid4a and Arid4b knockout mouse models and identified their previously unrecognized roles in male reproduction. We demonstrated that haploinsufficiency of Arid4b in combination with complete deficiency for Arid4a leads to progressive loss of fertility in the Arid4a−/−Arid4b+/− males, but the presence of two functional copies of Arid4a and/or Arid4b is sufficient to maintain normal fertility in the Arid4a−/− or Arid4a+/−Arid4b+/− males. This genetic interaction study suggests that these two homologous Arid4 genes are functionally redundant in the control of male fertility. Because mice homozygous for Arid4b deficiency die at early embryonic stages (17), we analyzed the available conventional knockout mutants with triple allelic mutations (Arid4a−/−Arid4b+/−). If the gene dosage effect of Arid4a and Arid4b plays a role in progressive loss of male fertility, the complete loss-of-function phenotypes could be revealed only when the final Arid4b wild-type allele is removed. To circumvent embryonic lethality and to completely remove Arid4b function, we are working on generation of a conditional Arid4b knockout mice line. We expect that mice with all four Arid4 alleles mutated (Arid4a−/−Arid4b−/−) might display an early and complete loss of male fertility.
Infertility in the Sertoli cell-specific Rb mutant males is also progressive (12). RB is not essential for initial maturation of Sertoli cells, but is important in maintaining mature Sertoli cell function, which includes maintenance of maturing germ cells and the blood–testis barrier (12). ARID4A and ARID4B are two RB-binding related proteins (13, 14). In this study, our results suggested that ARID4A and ARID4B are involved in the RB pathways to control male fertility. Complete deficiency of Arid4a combined with haploinsufficiency deficiency of Arid4b might affect RB function in Sertoli cells, leading to progressive loss of male fertility.
ARID4A and ARID4B are critical for physiological function of Sertoli cells, which supports spermatogenesis and maintains the impermeable blood–testis barrier, leading to normal male fertility. The phenotypes of the Arid4a−/−Arid4b+/− mice, including spermatogenic failures and increased permeability of the blood–testis barrier, are strikingly similar to that found in the Sertoli cell-specific AR knockout mice and the Sertoli cell-specific RB knockout mice. Investigation into the molecular mechanism identified several genes as common targets of AR, RB, ARID4A, and ARID4B. As two RB-binding related proteins, ARID4A and ARID4B also interact with AR and function as transcriptional coactivators for AR and RB. Our results link ARID4A and ARID4B to the AR and RB pathways, suggesting a mechanism by which ARID4A and ARID4B control male fertility.
Materials and Methods
Mouse Lines.
Arid4a null (Arid4a−/−) mice, mice heterozygous for the Arid4b deletion (Arid4b+/−), and mice null for Arid4a and heterozygous for the Arid4b deletion (Arid4a−/−Arid4b+/−) have been described previously (17).
Male Fertility Analysis.
Two-month-old males with different Arid4a and Arid4b genotypes were set up for mating with two adult wild-type females and were housed together. Percentages of fertile males and litter size (number of pups) were recorded over a 10-mo period.
Supplementary Material
Acknowledgments
We thank Dr. Sophia Y. Tsai for her helpful comments and critical reading of the manuscript, Dr. Alexander Agoulnik for advice, Xiaodong Zhai and Silvia Briones for technical assistance, and Minnie Freeman for help maintaining the mouse colonies. This work was supported by a startup fund from The George Washington University (to R.-C.W.) and by Grant HD037283 from the National Institutes of Health (to A.L.B.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218318110/-/DCSupplemental.
References
- 1.Jan SZ, et al. Molecular control of rodent spermatogenesis. Biochim Biophys Acta. 2012;1822(12):1838–1850. doi: 10.1016/j.bbadis.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Mruk DD, Cheng CY. Sertoli-Sertoli and Sertoli-germ cell interactions and their significance in germ cell movement in the seminiferous epithelium during spermatogenesis. Endocr Rev. 2004;25(5):747–806. doi: 10.1210/er.2003-0022. [DOI] [PubMed] [Google Scholar]
- 3.Vogl AW, Pfeiffer DC, Mulholland D, Kimel G, Guttman J. Unique and multifunctional adhesion junctions in the testis: Ectoplasmic specializations. Arch Histol Cytol. 2000;63(1):1–15. doi: 10.1679/aohc.63.1. [DOI] [PubMed] [Google Scholar]
- 4.Verhoeven G, Willems A, Denolet E, Swinnen JV, De Gendt K. Androgens and spermatogenesis: Lessons from transgenic mouse models. Philos Trans R Soc Lond B Biol Sci. 2010;365(1546):1537–1556. doi: 10.1098/rstb.2009.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeh S, et al. Generation and characterization of androgen receptor knockout (ARKO) mice: An in vivo model for the study of androgen functions in selective tissues. Proc Natl Acad Sci USA. 2002;99(21):13498–13503. doi: 10.1073/pnas.212474399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai MY, et al. Differential effects of spermatogenesis and fertility in mice lacking androgen receptor in individual testis cells. Proc Natl Acad Sci USA. 2006;103(50):18975–18980. doi: 10.1073/pnas.0608565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang C, et al. Infertility with defective spermatogenesis and hypotestosteronemia in male mice lacking the androgen receptor in Sertoli cells. Proc Natl Acad Sci USA. 2004;101(18):6876–6881. doi: 10.1073/pnas.0307306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Gendt K, et al. A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc Natl Acad Sci USA. 2004;101(5):1327–1332. doi: 10.1073/pnas.0308114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holdcraft RW, Braun RE. Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development. 2004;131(2):459–467. doi: 10.1242/dev.00957. [DOI] [PubMed] [Google Scholar]
- 10.Meng J, Holdcraft RW, Shima JE, Griswold MD, Braun RE. Androgens regulate the permeability of the blood-testis barrier. Proc Natl Acad Sci USA. 2005;102(46):16696–16700. doi: 10.1073/pnas.0506084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh S, et al. Retinoblastoma, a tumor suppressor, is a coactivator for the androgen receptor in human prostate cancer DU145 cells. Biochem Biophys Res Commun. 1998;248(2):361–367. doi: 10.1006/bbrc.1998.8974. [DOI] [PubMed] [Google Scholar]
- 12.Nalam RL, Andreu-Vieyra C, Braun RE, Akiyama H, Matzuk MM. Retinoblastoma protein plays multiple essential roles in the terminal differentiation of Sertoli cells. Mol Endocrinol. 2009;23(11):1900–1913. doi: 10.1210/me.2009-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Defeo-Jones D, et al. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature. 1991;352(6332):251–254. doi: 10.1038/352251a0. [DOI] [PubMed] [Google Scholar]
- 14.Cao J, Gao T, Stanbridge EJ, Irie R. RBP1L1, a retinoblastoma-binding protein-related gene encoding an antigenic epitope abundantly expressed in human carcinomas and normal testis. J Natl Cancer Inst. 2001;93(15):1159–1165. doi: 10.1093/jnci/93.15.1159. [DOI] [PubMed] [Google Scholar]
- 15.Lai A, et al. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol Cell Biol. 2001;21(8):2918–2932. doi: 10.1128/MCB.21.8.2918-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleischer TC, Yun UJ, Ayer DE. Identification and characterization of three new components of the mSin3A corepressor complex. Mol Cell Biol. 2003;23(10):3456–3467. doi: 10.1128/MCB.23.10.3456-3467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu MY, Tsai TF, Beaudet AL. Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain. Genes Dev. 2006;20(20):2859–2870. doi: 10.1101/gad.1452206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu MY, Eldin KW, Beaudet AL. Identification of chromatin remodeling genes Arid4a and Arid4b as leukemia suppressor genes. J Natl Cancer Inst. 2008;100(17):1247–1259. doi: 10.1093/jnci/djn253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handel MA, Schimenti JC. Genetics of mammalian meiosis: Regulation, dynamics and impact on fertility. Nat Rev Genet. 2010;11(2):124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- 20.O’Donnell L, Nicholls PK, O’Bryan MK, McLachlan RI, Stanton PG. Spermiation: The process of sperm release. Spermatogenesis. 2011;1(1):14–35. doi: 10.4161/spmg.1.1.14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denolet E, et al. The effect of a Sertoli cell-selective knockout of the androgen receptor on testicular gene expression in prepubertal mice. Mol Endocrinol. 2006;20(2):321–334. doi: 10.1210/me.2005-0113. [DOI] [PubMed] [Google Scholar]
- 22.Willems A, et al. Selective ablation of the androgen receptor in mouse Sertoli cells affects Sertoli cell maturation, barrier formation and cytoskeletal development. PLoS ONE. 2010;5(11):e14168. doi: 10.1371/journal.pone.0014168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.