Abstract
Cytokinin is an essential phytohormone controlling various biological processes, including environmental stress responses. In Arabidopsis, although the cytokinin (CK)-related phosphorelay—consisting of three histidine kinases, five histidine phosphotransfer proteins (AHPs), and a number of response regulators—has been known to be important for stress responses, the AHPs required for CK signaling during drought stress remain elusive. Here, we report that three Arabidopsis AHPs, namely AHP2, AHP3, and AHP5, control responses to drought stress in negative and redundant manner. Loss of function of these three AHP genes resulted in a strong drought-tolerant phenotype that was associated with the stimulation of protective mechanisms. Specifically, cell membrane integrity was improved as well as an increased sensitivity to abscisic acid (ABA) was observed rather than an alteration in ABA-mediated stomatal closure and density. Consistent with their negative regulatory functions, all three AHP genes’ expression was down-regulated by dehydration, which most likely resulted from a stress-induced reduction of endogenous CK levels. Furthermore, global transcriptional analysis of ahp2,3,5 leaves revealed down-regulation of many well-known stress- and/or ABA-responsive genes, suggesting that these three AHPs may control drought response in both ABA-dependent and ABA-independent manners. The discovery of mechanisms of activation and the targets of the downstream components of CK signaling involved in stress responses is an important and challenging goal for the study of plant stress regulatory network responses and plant growth. The knowledge gained from this study also has broad potential for biotechnological applications to increase abiotic stress tolerance in plants.
Keywords: microarray analysis, plant adaptation, stress mitigation, two-component system
A wide range of environmental factors, including drought and salinity stress, adversely affect plant growth, productivity, and seed quality. During the process of evolution, plants have acquired elaborate and sensitive protection systems that enable them to rapidly sense, respond, and properly adapt to these stresses (1–3). Phosphorylation modulated by two-component systems (TCSs), is one of the key mechanisms for intracellular signal transduction in both eukaryotic and prokaryotic cells. In plants, TCSs form a complex His-to-Asp phosphorelay that makes use of a “hybrid” sensor histidine kinase (HK), which contains both a His-kinase domain and a receiver domain. The TCSs also include His-containing phosphotransfer (HPt) proteins that function as signaling modules that connect to the final response regulators (RRs). This three-step phosphorelay implies an advantageous mechanism that provides multiple regulatory checkpoints for signal cross-talk or negative regulation by specific phosphatases (4–7).
In Arabidopsis thaliana (At), cytokinin (CK) was reported to play a negative role in the adaptation of plants to stresses (8, 9). Within the CK signaling cascade, all three CK receptor Arabidopsis HKs—AHK2, AHK3, and AHK4/cytokinin response 1 (CRE1)—are known to be involved in the response to cold, drought, and salt stresses (10, 11). Analyses of ahk2, ahk3, and ahk4 single, double, and triple mutants suggest that AHK2, AHK3, and AHK4 function as negative regulators in abscisic acid (ABA), salt, and drought signaling pathways (10, 12). However, the specific functions of the downstream At HPts (AHPs) and most Arabidopsis response regulators (ARRs) within CK signaling during abiotic stress responses remain to be elucidated.
In the CK-related phosphorelay, all five AHPs (AHP1–AHP5) mediate the transfer of the phosphoryl group from the AHKs to the ARRs (5, 13). Although the subcellular localization of these AHPs is independent of CK and CK-receptor AHKs (14), AHP gene expression is induced by CK (15, 16). All five AHPs, as well as those in rice and soybean, contain a consensus sequence of HQXKGSSXS with a highly conserved His residue (17, 18). Analyses of ahp multiple mutants indicated that with the exception of AHP4, for which consistent evidence is lacking, all remaining AHPs function as redundant positive regulators of CK signaling and affect many aspects of plant development (13). Among the authentic AHP proteins, at least AHP1, 2, 3, and 5 are capable of interacting with AHK2, AHK3, and AHK4 (19, 20).
Using a loss-of-function approach, we characterized the in planta functions of AHP2, AHP3, and AHP5 in response to drought stress. Various combinations of ahp double and triple mutants were investigated to study the mechanism by which these AHPs modulate drought responses. We also analyzed the transcriptional regulation of AHP genes in WT and CK-deficient plants to examine how plants counteract the negative regulatory role of CK signaling to mitigate the effects of abiotic stress. Using microarray analyses, we identified the downstream components of CK signaling that are potentially implicated in the regulation of drought stress response.
Results
ahp2,3,5 Triple Mutant Is Strongly Drought-Tolerant.
We previously published that the ahk2,3 double mutant, which has a distinguishable shoot growth retardation phenotype in comparison with other single and double combinations of ahk2, ahk3, and ahk4 genes, possessed the strongest drought-tolerant phenotype (10). To further characterize the functional role of the five Arabidopsis AHP-encoding genes in drought stress response, we collected various combinations of ahp mutants from available public resources (13). During early stages of development, ahp2-1,3,5-2 (hereafter referred to as ahp2,3,5) triple mutant seedlings, but not any single mutants or double-mutant combinations for AHP2, AHP3, and AHP5, were found to have smaller leaves relative to WT. These data were similar to those observed for the ahk2,3 double mutant (Fig. S1). Furthermore, all of the ahp mutant combinations, which contains the ahp2-1, ahp3, and ahp5-2 alleles, exhibited a similar dwarf shoot phenotype, as does the ahp2,3,5 (13). Thus, in this study, we focused on the functional analyses of these three AHPs, which are the most closely homologous to each other (13).
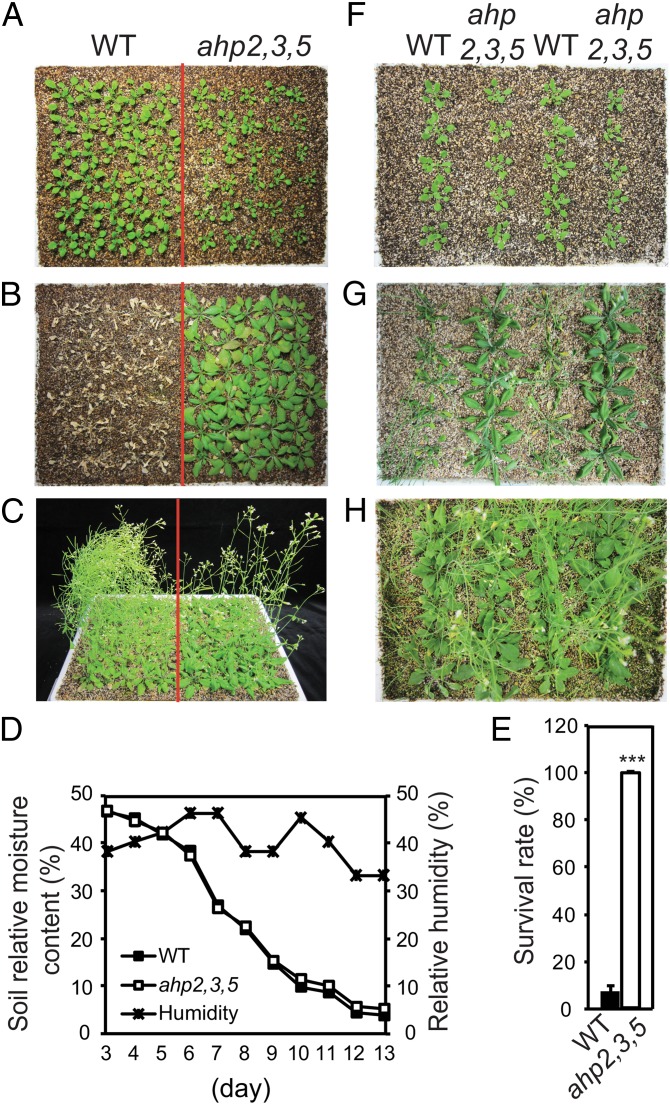
To determine whether the loss of function of AHP2, AHP3, and AHP5 alters drought response, ahp2,3,5 seedlings were subjected to a drought-tolerance assay using the same-tray method that ensures a valid comparison of genotypes with different growth rates (21) (Fig. 1 A–C). Only minimal differences in quantified soil moisture content were observed between the WT (Left) and ahp2,3,5 (Right) sides of the trays used for the drought tolerance test (Fig. 1D), indicating that ahp2,3,5 is strongly drought tolerant (Fig. 1 B and E). This finding was confirmed by a more stringent approach in which ahp2,3,5 and WT plants were grown in an alternate order within the same tray and subjected to drought stress (Fig. 1 F–H). Under this condition, the WT plants exhibited severely wilted or dead leaves, whereas the leaves of ahp2,3,5 plants remained turgid (Fig. 1G). Taken together, our results clearly demonstrate that ahp2,3,5 plants have enhanced drought tolerance and suggest that these AHP proteins function as important negative regulators for adaptation to drought stress.
Fig. 1.
Loss-of-function of AHP2, AHP3, and AHP5 results in enhanced drought tolerance. (A) Two-week-old WT and ahp2,3,5 plants were transferred from germination medium (GM) plates to soil and grown for an additional week. (B) Three-week-old plants were exposed to drought stress for 13 d and photographed 3 d subsequent to rewatering and after the removal of inflorescences. (C) For control purposes, 2-wk-old WT and ahp2,3,5 plants were transferred from GM plates to trays and grown under well-watered conditions in parallel with the drought test as shown in (B). (D) Soil relative moisture contents and relative humidity were monitored during the drought tolerance test (A and B). (E) Survival rates and SEs (error bars) were calculated from the results of three independent experiments (n = 30 plants/genotype). Asterisks indicate significantly higher survival rates than WT as determined by a Student’s t test analysis (***P < 0.001). (F) Two-week-old WT and ahp2,3,5 plants were transferred to soil in an alternate order and grown for an additional week. (G) Three-week-old plants were exposed to drought stress; plants were photographed 20 d after the withholding of water. (H) For control purposes, WT and ahp2,3,5 plants were grown in an alternate order in parallel under well-watered conditions.
AHP2, AHP3, and AHP5 Function as Redundant Negative Regulators of Drought Stress Response.
To determine the effect of ahp2, ahp3, and ahp5 mutations on drought stress response, we investigated the drought-tolerant phenotype of the lower-order mutants, namely the ahp2,3, ahp2,5, and ahp3,5 double mutants. These double mutant plants were subjected to a long-term drought stress treatment similar to the treatment applied to ahp2,3,5 (Fig. S2 A–C). Although all of the double mutants exhibited enhanced drought tolerance relative to WT, their level of drought tolerance was less than that of ahp2,3,5 (Fig. S2). The data obtained also suggested that AHP2 and AHP3 might play a more prominent role than AHP5 in regulation of drought response. These findings indicate that AHP2, AHP3, and AHP5 have redundant and negative regulatory roles in drought stress response. In other words, AHP2, AHP3, and AHP5 function in combination to effectively regulate drought stress signaling.
AHP2, AHP3, and AHP5 Gene Expression Is Repressed by Abiotic Stress in CK-Dependent Manners.
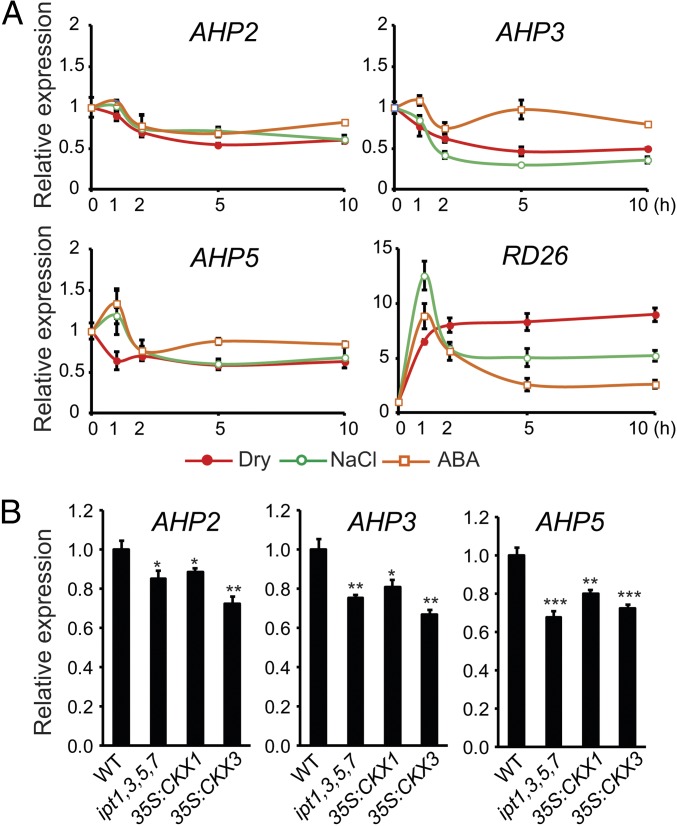
To further examine the in planta function of the AHP2, AHP3, and AHP5 genes under abiotic stress, we analyzed their expression patterns after exposure to dehydration and high salinity stress. Because plants can respond to drought stress in an ABA-dependent or ABA-independent manner (22), expression of the AHP2, AHP3, and AHP5 genes was also assessed under ABA treatment. All three AHP genes were shown to be down-regulated by dehydration and high salinity, with the strongest repression observed for the AHP3 gene (Fig. 2A). The expression of the three AHP genes examined was reduced within 1 h and reached the lowest level at 5 h after dehydration. During salt stress, transcription of AHP3 started to reduce after 1 h and reached the minimum after 5 h of treatment, whereas that of AHP2 and AHP5 decreased by high salinity only after 2 h with the lowest expression level observed at 10 h and 5 h for AHP2 and AHP5, respectively. ABA treatment also influenced expression of the examined AHP genes. Although expression of the AHP2 and AHP3 genes was reduced after 2 h of treatment, the AHP5 transcript seemed to slightly increase at 1 h, but rapidly decreased below level of control after 2 h of ABA treatment. The dynamic changes in transcript levels identified in the time-course experiments show the different responses of AHP2, AHP3, and AHP5 to various stresses at transcriptional level and further confirm the involvement of AHP2, AHP3, and AHP5 in the regulation of stress responses, perhaps in both ABA-dependent and ABA-independent manners.
Fig. 2.
CK-dependent repression of AHP2, AHP3, and AHP5 genes under stress conditions. (A) Expression of AHP2, AHP3, and AHP5 genes in 2-wk-old WT plants after exposure to dehydration, salt stress (250 mM), and ABA (100 µM) treatments. Relative expression levels were normalized to a value of 1 in the respective mock control plants. Data represent the means and SEs of three independent biological replicates. The stress-inducible RD26 gene was used as ABA, NaCl, and dehydration stress markers to confirm the efficacy of our stress treatments. (B) Down-regulated expression of AHP2, AHP3, and AHP5 genes in 10-d-old WT and CK-deficient plants. Relative expression levels were normalized to a value of 1 in WT plants. Data represent the means and SE of four independent biological replicates.
Previous studies indicated that the CK level decreases as a result of plant adaptive responses to drought and salt stresses (8, 23). Thus, we hypothesized that reductions in CK levels upon stress exposure might contribute to the reduced steady-state levels of AHP transcripts observed during dehydration and high salinity treatments (Fig. 2A). With this aim, we examined the expression of the AHP2, AHP3, and AHP5 genes in ipt1,3,5,7, 35S:CKX1, and 35S:CKX3 CK-deficient plants. AHP expression levels were reduced by ∼20–30% in the CK-deficient plants compared with the WT plants (Fig. 2B). This relatively low reduction rate in expression of each AHP gene might further indicate the functional redundancy among the three AHP genes and that corepression of multiple AHP genes is required for an efficient adaptation to stress. These results suggest that the stress-mediated repression of AHP genes is, at least in part, dependent upon endogenous CK levels.
Drought Tolerance Mediated by Loss of Function of AHP2, AHP3, and AHP5 Is Associated with Maintenance of Cell Membrane Integrity.
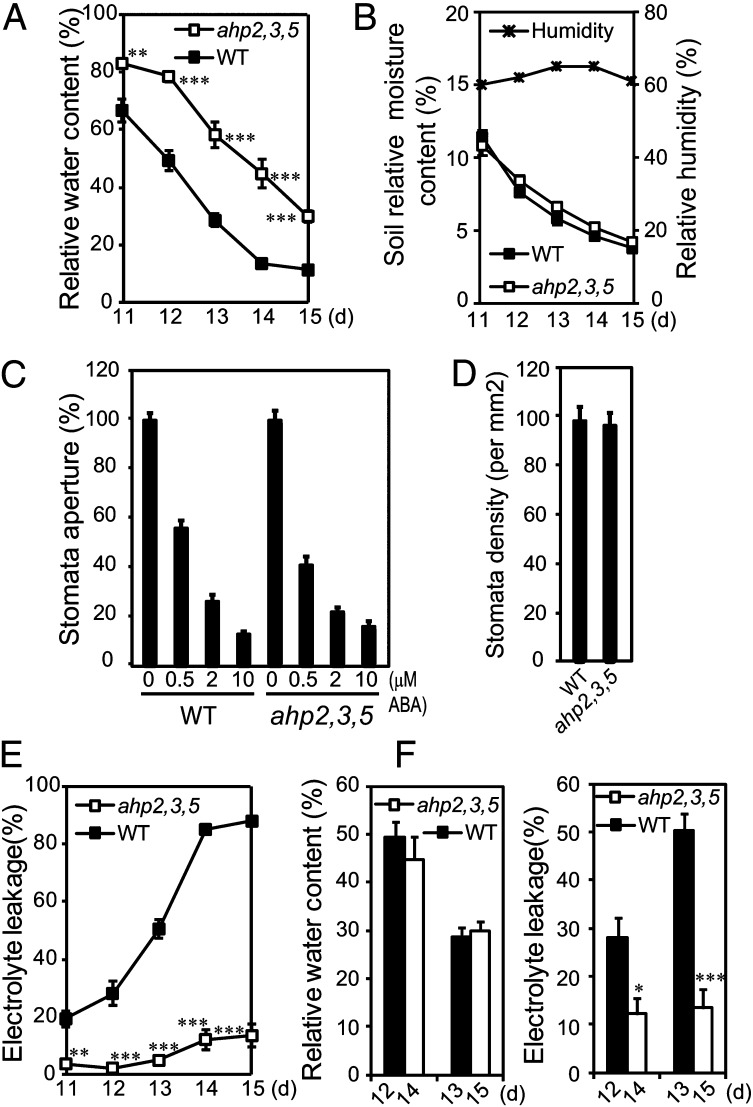
We next sought to identify the mechanisms mediating drought stress tolerance resulting from a disruption of AHP2, AHP3, and AHP5. Under drought stress, relative water content (RWC) was more rapidly reduced in WT plants relative to the ahp2,3,5 triple mutant (Fig. 3A) at similar soil moisture content (Fig. 3B). In addition, we measured stomatal apertures in the WT and ahp2,3,5 plants to determine whether alteration in ABA-mediated stomatal closure contributes to the enhanced maintenance of RWC in the ahp2,3,5 mutant. Both WT and the ahp2,3,5 plants maintained similar stomatal apertures at various concentrations of ABA (Fig. 3C). Stomatal density is another important factor in the regulation of leaf RWC. However, no significant differences of stomatal density were observed between WT and ahp2,3,5 plants (Fig. 3D). Taken together, these data suggest that neither ABA-mediated stomatal closure nor stomatal density plays a critical role in regulating the lower rates of water loss observed with the ahp2,3,5 mutant.
Fig. 3.
Comparison of RWC, electrolyte leakage, stomatal aperture, and density of WT and ahp2,3,5 plants. (A) The WT and ahp2,3,5 plants were grown and exposed to drought stress as described in Fig. 1 A and B. At the indicated time points, plants were harvested for measurement of RWC. Error bars represent SEs (n = 5). (B) Soil relative moisture contents and relative humidity were recorded just before sample collection for measurements of RWC and electrolyte leakage. (C) Average stomatal aperture of rosette leaves from 4-wk-old WT and ahp2,3,5 plants in the presence or absence of ABA. Error bars represent SEs (n > 28). (D) Average stomatal density of rosette leaves from 4-wk-old WT and ahp2,3,5 plants. Error bars represent SE (n = 25). (E) Electrolyte leakage of the WT and ahp2,3,5 plants exposed to drought stress as described in (A). Error bars represent SEs (n = 5). (F) Comparison of electrolyte leakage levels between WT and ahp2,3,5 plants collected at a similar RWC during drought stress. Error bars represent SE (n = 5). Asterisks indicate significant differences as determined by a Student’s t- test analysis (*P < 0.05, **P < 0.01, ***P < 0.001).
The maintenance of stability and integrity of cell membranes under water stress is an important component of drought tolerance in plants. Electrolyte leakage analysis was performed to quantify the differences of cell membrane integrity after exposure to drought stress in WT and ahp2,3,5 plants. The ahp2,3,5 plants exhibited significantly lower electrolyte leakage than WT at all time points examined during drought stress (Fig. 3E). At two time points at which WT and ahp2,3,5 plants exhibited a similar RWC, electrolyte leakage was significantly lower in ahp2,3,5 relative to WT (Fig. 3F). Thus, the enhanced stress tolerance of ahp2,3,5 can be attributed, at least in part, to an increased cell membrane integrity and stability rather than to differences in stomatal density and ABA-mediated stomatal closure.
Loss of Function of AHP2, AHP3, and AHP5 Increases Sensitivity to ABA.
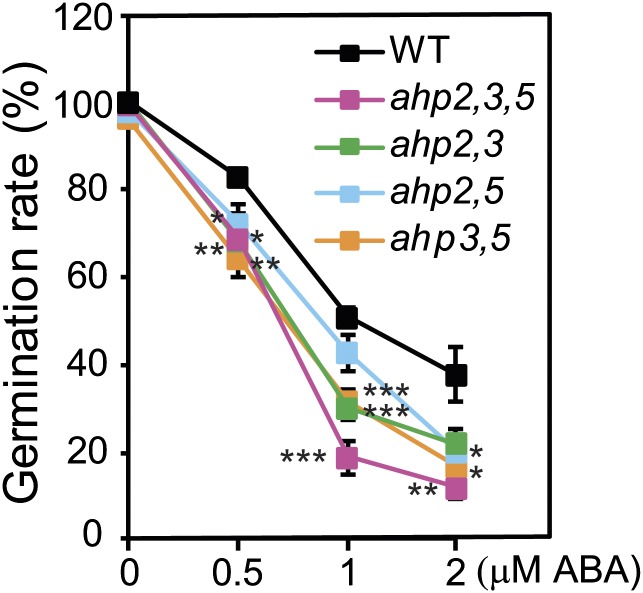
Because expression analyses of the AHP2, AHP3, and AHP5 genes indicated that they might control drought stress response in an ABA-dependent manner, we examined the responsiveness of the ahp triple and double mutants to ABA to determine whether loss of function of AHP2, AHP3, and AHP5 alters ABA responses. In comparison to WT, all of the ahp mutants exhibited reduced germination rates in the presence of exogenous ABA (Fig. 4 and Fig. S3). These data indicate increased sensitivity of the ahp mutants to ABA. In addition, we observed that the ahp triple mutant was more sensitive to ABA than ahp double mutants, suggesting that the three AHPs function to negatively regulate ABA response in a redundant manner.
Fig. 4.
Response of the ahp double and triple mutants to treatment with exogenous ABA. Seeds were sown on germination medium-1% sucrose containing the indicated ABA concentrations; germination rates were quantified after 4 d of incubation by counting the number of open cotyledons. Error bars indicate SEs that were calculated from the results of four independent experiments. Asterisks indicate significant differences as determined by a Student’s t test analysis (*P < 0.05, **P < 0.01, ***P < 0.001).
Comparative Transcriptome Analysis of WT and ahp2,3,5 Leaves Under Well-Watered and Drought Stress Conditions.
To elucidate the molecular mechanisms associated with drought tolerance and identify downstream genes involved in drought response that are regulated by AHP2, AHP3, and AHP5, we conducted a genomewide transcriptional analysis to compare the leaf transcriptomes of the WT and ahp2,3,5 plants under well-watered and drought stress conditions using whole genome Arabidopsis microarrays (see Fig. S4 A–C for experimental design). The results of the microarray analyses are available in Dataset S1 or can be accessed through the Gene Expression Omnibus (GSE42290). Using the criteria of the ratio change ≥ 2, and a false discovery rate–corrected P value (q-value) <0.05, 190 and 83 genes were up- and down-regulated in ahp2,3,5, respectively, in comparison with WT under well-watered conditions (Fig. S4D, M-C/W-C, and Datasets S2A and S3A). A number of the up-regulated genes were also induced by drought stress (Fig. S5A, compare M-C/W-C and W-D/W-C; Dataset S4A). We hypothesized that the expression of more stress-inducible genes would increase in ahp2,3,5 during drought stress, which would render this mutant more tolerant to drought stress than WT. Indeed, when leaf transcriptomes of ahp2,3,5 and WT plants were compared during drought stress, we identified 4,977 up-regulated and 3,431 down-regulated genes with a ratio of ≥ 2 in ahp2,3,5 plants subjected to drought in relation to the expression of the same genes in drought-stressed WT plants (Fig. S4D, M-D/W-D, and Datasets S2B and S3B). At least 68 genes up-regulated in this comparison were identified as drought stress–inducible (Fig. S5A, compare M-D/W-D and W-D/W-C, and Dataset S4B). Furthermore, the majority of the up-regulated genes identified in well-watered ahp2,3,5 versus well-watered WT were also identified among the up-regulated genes of drought-stressed ahp2,3,5 plants compared with WT plants subjected to the same treatment (Fig. S5A, compare M-C/W-C and M-D/W-D, and Dataset S4C). Several genes were chosen to validate our microarray data by quantitative RT-PCR analysis (Fig. S5B and Dataset S2B).
Among the genes with increased transcript abundance identified in comparative analysis of drought-stressed ahp2,3,5 and WT plants, we identified many regulatory genes, including those encoding APETALA2/ethylene-responsive element binding proteins (AP2/EREBP), basic leucine zipper motif (bZIP), myeloblastosis (MYB), homeobox, NAC (NAM, no apical meristem; ATAF, Arabidopsis transcription activation factor; and CUC, cup-shaped cotyledon), nuclear factor Y (NF-Y), WRKY (trytophan, arginine, lysine, and tyrosine), and zinc finger-type transcription factors; receptor-like and salt overly sensitive (SOS) 4 kinases; calcium sensor (SOS3); and sodium transporter proteins (HKT1). In addition, genes encoding heat shock proteins were also found among up-regulated genes. Several key transcription factor encoding genes that were up-regulated, such as DREB1A/CBF3, DREB1B/CBF1, and NF-YA10/At5G06510, are worth mentioning because their overexpression is well known to significantly enhance drought tolerance in transgenic plants (24, 25). Because we observed the up-regulation of several key salt stress-responsive genes in ahp2,3,5 during drought stress (compare M-D/W-D), such as HKT1, SOS3, and SOS4, we were interested to characterize the response of ahp2,3,5 to salt stress. Increased survival of ahp2,3,5 was observed relative to WT under salt stress, indicating a clear salt-tolerant phenotype for ahp2,3,5 (Fig. S6 A and B).
Many cell wall- and lipid metabolism-related genes were up-regulated in both well-watered and drought-stressed ahp2,3,5 plants compared with the corresponding WT control. These genes included those involved in the synthesis of cuticular wax, such as the AP2/EREBP-type SHN2 and SHN3, and those encoding fatty acid desaturases (FADs), such as FAD3 and FAD8 (Fig. S7 and Dataset S5). Fatty acids (FAs) are crucial components of cellular membranes and cuticular waxes, whose formation is tightly regulated in response to both developmental and environmental cues (26). Overexpression of Brassica napus FAD3 and Arabidopsis FAD8 increased the tolerance of transgenic tobacco to drought stress by increasing the levels of unsaturated FAs to adjust the fluidity of membrane lipids (27). On the other hand, overexpression of Arabidopsis SHN2 and SHN3 enhanced the drought tolerance of transgenic Arabidopsis by increasing wax production and modulating cuticular permeability (28). In addition, the majority of these lipid metabolism-related genes were also induced by drought stress independently of ABA (Dataset S5).
Discussion
The CK receptors AHK2 and AHK3 were previously reported to act as negative regulators in drought response (10, 12). However, the physiological and molecular mechanisms by which the CK signaling acts, the stress-related specific functions of other TCS components, and the identification of downstream components of CK phosphorelay involved in the regulation of drought response were not completely understood and required further in-depth characterization (7). In this study, we designed a series of experiments that aimed to analyze the specific functions of AHP2, AHP3, and AHP5 in drought stress responses by investigating the consequences of loss of function of these AHP proteins on the adaptation of Arabidopsis plants to drought stress. We found that all combinations of the ahp double mutants displayed tolerance against drought stress. Furthermore, in comparison with all versions of the ahp double mutants, the ahp2,3,5 exhibited the strongest drought-tolerant phenotype (Fig. 1 and Fig. S2). These results indicate that all three AHP proteins act as negative and redundant regulators in drought stress response.
Our examinations of several physiological aspects have demonstrated that the improved performance of the ahp2,3,5 mutant under drought stress was attributed to its ability to maintain a higher water status (Fig. 3A). This trait was associated with improved protection of membrane structures, as evidenced by lower ion leakage (Fig. 3 E and F), rather than alteration in ABA-mediated stomatal closure or stomatal density (Fig. 3 C and D). In good accordance with our result, the repression of CK signaling via a reduction of endogenous CK levels also resulted in enhanced protection of membrane structures, and thereby drought tolerance of CK-deficient plants, whereas the ABA-mediated stomatal closure and stomatal density remained relatively unchanged (8). Our data are also supported by a study of the ABA-responsive AREB1 gene that reported that constitutive overexpression of the active form of AREB1 (AREB1ΔQT) gene in Arabidopsis enhanced drought tolerance independent from the status of stomatal closure. The improved performance of the AREB1ΔQT transgenic plants under limiting water conditions was associated with their increased ABA sensitivity, leading to the induction of a number of downstream ABA-responsive genes (29).
Similar to the AREB1 study, all ahp double and triple mutants were also found to be hypersensitive to ABA (Fig. 4 and Fig. S3). Therefore, the elevated drought tolerance of the ahp mutant plants might also be attributed to their ability to react faster to ABA upon exposure to drought stress. As a result, this would lead to an up-regulation of ABA-signaling genes; a result that would show firm molecular evidence for the existence of an active but antagonistic interaction between CK and ABA signalings in controlling drought responses. Several lines of evidence have demonstrated that the impairment of CK signaling by disruption of various members of CK signaling, namely the CK receptor encoding AHKs, resulted in enhanced tolerance to drought stresses, which also coincided with their ABA-hypersensitive phenotype (10, 12). In an attempt to explain this phenomenon at a molecular level, we performed a comparative microarray analysis of ahp2,3,5 and WT plants under both well-watered and drought stress conditions to identify the downstream genes of CK signaling that function in drought response. Many ABA- and/or stress-responsive genes, such as DREB1A, DREB1B, AtMYC2, and NF-YA10 (Dataset S2B), whose overexpression enhanced drought tolerance (24, 25, 30, 31), were up-regulated in either well-watered or drought-stressed ahp2,3,5 relative to WT. These data indicate that both ABA-dependent and ABA-independent pathways cross-talk with the CK pathway to regulate drought response. In addition, we also found that many genes involved in the regulation of cuticular wax biosynthesis and FA desaturation were significantly induced in ahp2,3,5 relative to WT with or without drought treatment (Dataset S5). It is possible that this observed difference in gene expression could result in increased levels of cuticular wax and unsaturated FAs, which in turn may function to enhance protection from abiotic stress (27, 28). This finding also suggested that the drought-tolerant and reduced electrolyte leakage phenotypes observed in ahp2,3,5 could be related to enhanced cuticular wax accumulation and modification of membrane fluidity mediated by changes in unsaturated FA levels. Furthermore, these results suggested that the FA unsaturation, mobilization, and regulation as well as the synthesis of cuticular wax might be regulated by CK with an important function contributed by AHPs. Additionally, we noticed that a significantly lower number of genes were up-regulated in ahp2,3,5 relative to WT during growth under sufficient rather than water-limiting conditions (Fig. S4 and Dataset S2). Consistently, only a few up-regulated genes were found in a transcriptomic analysis of 3-wk-old ahk2,3 seedlings versus WT under normal growth conditions (10). This result also indicates that the defense mechanism of CK signaling is activated in ahp2,3,5 during stress rather than being maintained in a ready state under sufficient water as a preventive measure.
Based on these findings, we were interested to investigate how plants could mitigate the negative effect of CK signaling during exposure to stress conditions. The results of the expression study shown in Fig. 2A suggest that plants may attempt to survive exposure to abiotic stress by triggering a mechanism that represses the CK signaling through suppression of AHP gene expression upon their exposure to adverse environmental conditions. Furthermore, the stress-triggered down-regulation of AHP genes was CK-dependent (Fig. 2B). Because the endogenous CK levels are known to decrease under stress conditions (8, 23) as an adaptive mechanism to tolerate adverse environments (8, 9), we hypothesize that during stress, plants activate a yet-unknown mechanism to reduce endogenous CK levels. This in turn leads to a repression of CK signaling, at least by down-regulation of the AHP2, AHP3, and AHP5 genes (Fig. 2B) and a reduction in HK activities (20, 32). Because of the reduction of the negative effect of CK signaling, the expression of downstream genes is up-regulated, leading to an optimized adaptation of plants to stresses (Fig. S8). Our findings also open the possibility to engineer transgenic crops with enhanced drought tolerance by reducing AHP levels using a drought-regulated gene-silencing system.
Materials and Methods
Plant Materials and Stress Treatments.
Arabidopsis plants (Columbia ecotype) were grown on germination medium agar plates for 2 wk (22 °C, 16 h light/8 h dark cycle, 60 µmol⋅m–2⋅s–1 photon flux density) and treated with water (hydroponic control), 100 µM ABA, or 250 mM NaCl for the indicated periods. For dehydration treatment, 2-wk-old plants were gently removed from plates and allowed to dry on parafilm. The ahp mutants (different combinations of ahp2-1, ahp3, and ahp5-2) used in this study were obtained from the Arabidopsis Biological Resource Center. Characterization of ahp mutants has been previously described (13). The ahk2,3 and CK-deficient ipt1,3,5,7 mutants were obtained from Higuchi et al. (33) and Miyawaki et al. (34), respectively, whereas the 35S:CKX1 and 35S:CKX3 overexpressors were obtained from Werner et al. (35).
Assessment of Drought Tolerance.
Drought tolerance was experimentally evaluated as previously described using the same tray method and “Dio Propagation Mix No.2 for Professional” soil (Dio Chemicals Ltd.) (8). Rewatering was done when the lethal effect of water withholding was observed on almost all control plants (ahp triple vs. WT comparison) or the best possible difference could be identified between control and mutant plants (ahp double vs. WT comparisons). Soil relative moisture was monitored using a HydroSense soil moisture probe (Campbell Scientific Inc.) to simultaneously monitor soil conditions during the stress treatment (8). A 50% volumetric soil moisture content was used to set the upper reference level (100% soil relative moisture content), whereas the soil that had been made for drought survival test experiment and dried for 20 d was used to set the lower reference level (0% soil relative moisture content).
Measurements of RWC and Electrolyte Leakage from Drought-Stressed Plants.
RWC and electrolyte leakage of the detached aerial parts of plants during exposure to long-term drought stress were measured according to the previously described methods (8).
Stomatal Movement Assays.
Stomatal movement assays were conducted as previously described (8).
Germination Assay.
A germination assay was performed on germination medium containing 1% sucrose and various concentrations of ABA as previously described (10). Germination was recorded as the number of seeds that had opened cotyledons under the various assay conditions.
Expression Analyses.
Total RNA was extracted with TRIzol Reagent according to the supplier’s instructions (Invitrogen). cDNA synthesis and quantitative RT-PCR were performed as previously described (36). Used primer pairs are listed in Dataset S6. UBQ10 was used as an internal control for expression analysis.
Plant Materials for Microarray Analysis.
WT and ahp2,3,5 plants (30 plants each) were grown on the same trays as in the drought tolerance test. After 10 d of water withholding, rosette leaves were collected from both well-watered and drought-stressed plants in three biological replicates, frozen in liquid nitrogen, and stored at −80 °C until used for RNA extraction. Leaf RWCs of WT and ahp2,3,5 plants were 30.9% ± 2.58% and 56.1% ± 2.66% (SE), respectively.
Microarray Analysis.
Total RNA from three biological replicates was extracted using an RNeasy Plant Mini Kit (Qiagen). Hybridization to the Arabidopsis Oligo 44K DNA microarray (Version 4.0, Agilent Technology), data analyses, and data mining were performed as previously described (37). The raw microarray data and the detailed protocol were deposited in the Gene Expression Omnibus database. MAPMAN classification of the transcripts was performed using the Web-tool Classification Superviewer (http://bar.utoronto.ca). When necessary, the ABA and stress-responsive gene expression was examined in the Arabidopsis eFP browser (http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi).
Supplementary Material
Acknowledgments
We thank Dr. T. Kakimoto for providing the ahk2,3 and ipt1,3,5,7 mutants, Dr. T. Schmulling for the 35:CKX1 and 35S:CKX3 overexpressors, and Dr. J. J. Kieber and the Arabidopsis Biological Resource Center for the ahp mutants. This research was supported by Start-up Support Grant M36-57000 from the RIKEN Yokohama Institute Director Discretionary Funds (to L.-S.P.T.) and Grant 55007646 from Howard Hughes Medical Institute (to L.H.-E.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus database, www.ncbi.nlm.nih.gov/geo (accession no. GSE42290).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302265110/-/DCSupplemental.
References
- 1.Hadiarto T, Tran LS. Progress studies of drought-responsive genes in rice. Plant Cell Rep. 2011;30(3):297–310. doi: 10.1007/s00299-010-0956-z. [DOI] [PubMed] [Google Scholar]
- 2.Choudhary SP, Yu JQ, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. Benefits of brassinosteroid crosstalk. Trends Plant Sci. 2012;17(10):594–605. doi: 10.1016/j.tplants.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Jogaiah S, Ramsandra Govind S, Tran LS. System biology-based approaches towards understanding drought tolerance in food crops. Crit Rev Biotechnol. 2013;33(1):23–39. doi: 10.3109/07388551.2012.659174. [DOI] [PubMed] [Google Scholar]
- 4.Mizuno T. Two-component phosphorelay signal transduction systems in plants: From hormone responses to circadian rhythms. Biosci Biotechnol Biochem. 2005;69(12):2263–2276. doi: 10.1271/bbb.69.2263. [DOI] [PubMed] [Google Scholar]
- 5.Mähönen AP, et al. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311(5757):94–98. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 6.Schaller GE, Shiu SH, Armitage JP. Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol. 2011;21(9):R320–R330. doi: 10.1016/j.cub.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 7.Ha S, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. Cytokinins: Metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012;17(3):172–179. doi: 10.1016/j.tplants.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Nishiyama R, et al. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell. 2011;23(6):2169–2183. doi: 10.1105/tpc.111.087395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Werner T, et al. Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell. 2010;22(12):3905–3920. doi: 10.1105/tpc.109.072694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tran LS, et al. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA. 2007;104(51):20623–20628. doi: 10.1073/pnas.0706547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeon J, et al. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J Biol Chem. 2010;285(30):23371–23386. doi: 10.1074/jbc.M109.096644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang NY, Cho C, Kim NY, Kim J. Cytokinin receptor-dependent and receptor-independent pathways in the dehydration response of Arabidopsis thaliana. J Plant Physiol. 2012;169(14):1382–1391. doi: 10.1016/j.jplph.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Hutchison CE, et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell. 2006;18(11):3073–3087. doi: 10.1105/tpc.106.045674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Punwani JA, Hutchison CE, Schaller GE, Kieber JJ. The subcellular distribution of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin signaling. Plant J. 2010;62(3):473–482. doi: 10.1111/j.1365-313X.2010.04165.x. [DOI] [PubMed] [Google Scholar]
- 15.Hradilova J, Malbeck J, Brzobohaty B. Cytokinin regulation of gene expression in the AHP gene family in Arabidopsis thaliana. J Plant Growth Regul. 2007;26(3):229–244. [Google Scholar]
- 16.Hoth S, et al. Monitoring genome-wide changes in gene expression in response to endogenous cytokinin reveals targets in Arabidopsis thaliana. FEBS Lett. 2003;554(3):373–380. doi: 10.1016/s0014-5793(03)01194-3. [DOI] [PubMed] [Google Scholar]
- 17.Pareek A, et al. Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 2006;142(2):380–397. doi: 10.1104/pp.106.086371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mochida K, et al. Genome-wide analysis of two-component systems and prediction of stress-responsive two-component system members in soybean. DNA Res. 2010;17(5):303–324. doi: 10.1093/dnares/dsq021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dortay H, Mehnert N, Bürkle L, Schmülling T, Heyl A. Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J. 2006;273(20):4631–4644. doi: 10.1111/j.1742-4658.2006.05467.x. [DOI] [PubMed] [Google Scholar]
- 20.Mähönen AP, et al. Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr Biol. 2006;16(11):1116–1122. doi: 10.1016/j.cub.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 21.Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006;45(4):523–539. doi: 10.1111/j.1365-313X.2005.02593.x. [DOI] [PubMed] [Google Scholar]
- 22.Tran LS, Nakashima K, Shinozaki K, Yamaguchi-Shinozaki K. Plant gene networks in osmotic stress response: From genes to regulatory networks. Methods Enzymol. 2007;428:109–128. doi: 10.1016/S0076-6879(07)28006-1. [DOI] [PubMed] [Google Scholar]
- 23.Ghanem ME, et al. Hormonal changes during salinity-induced leaf senescence in tomato (Solanum lycopersicum L.) J Exp Bot. 2008;59(11):3039–3050. doi: 10.1093/jxb/ern153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol. 1999;17(3):287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- 25.Leyva-González MA, Ibarra-Laclette E, Cruz-Ramírez A, Herrera-Estrella L. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS ONE. 2012;7(10):e48138. doi: 10.1371/journal.pone.0048138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Upchurch RG. Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol Lett. 2008;30(6):967–977. doi: 10.1007/s10529-008-9639-z. [DOI] [PubMed] [Google Scholar]
- 27.Zhang M, et al. Modulated fatty acid desaturation via overexpression of two distinct omega-3 desaturases differentially alters tolerance to various abiotic stresses in transgenic tobacco cells and plants. Plant J. 2005;44(3):361–371. doi: 10.1111/j.1365-313X.2005.02536.x. [DOI] [PubMed] [Google Scholar]
- 28.Aharoni A, et al. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16(9):2463–2480. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujita Y, et al. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17(12):3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abe H, et al. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9(10):1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knight H, Zarka DG, Okamoto H, Thomashow MF, Knight MR. Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Physiol. 2004;135(3):1710–1717. doi: 10.1104/pp.104.043562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reiser V, Raitt DC, Saito H. Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J Cell Biol. 2003;161(6):1035–1040. doi: 10.1083/jcb.200301099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higuchi M, et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA. 2004;101(23):8821–8826. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyawaki K, et al. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA. 2006;103(44):16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Werner T, et al. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15(11):2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le DT, et al. Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res. 2011;18(1):17–29. doi: 10.1093/dnares/dsq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishiyama R, et al. Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS ONE. 2012;7(2):e32124. doi: 10.1371/journal.pone.0032124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.