Abstract
Endoplasmic reticulum (ER) stress sensors use a related luminal domain to monitor the unfolded protein load and convey the signal to downstream effectors, signaling an unfolded protein response (UPR) that maintains compartment-specific protein folding homeostasis. Surprisingly, perturbation of cellular lipid composition also activates the UPR, with important consequences in obesity and diabetes. However, it is unclear if direct sensing of the lipid perturbation contributes to UPR activation. We found that mutant mammalian ER stress sensors, IRE1α and PERK, lacking their luminal unfolded protein stress-sensing domain, nonetheless retained responsiveness to increased lipid saturation. Lipid saturation-mediated activation in cells required an ER-spanning transmembrane domain and was positively regulated in vitro by acyl-chain saturation in reconstituted liposomes. These observations suggest that direct sensing of the lipid composition of the ER membrane contributes to the UPR.
Keywords: lipid bilayer, membrane fluidity, integral membrane protein, palmitic acid
Protein folding homeostasis in the lumen of the endoplasmic reticulum (ER) requires matching the flux of unfolded proteins entering the organelle to the capacity of the machinery for processing this biosynthetic load. To achieve this, eukaryotes have evolved signaling pathways that couple ER perturbation (or stress) to a rectifying gene expression program referred to as the unfolded protein response (UPR). Upstream in these pathways are ER localized transmembrane proteins, such as the product of the Inositol REquiring 1 gene (IRE1) (1, 2) and Protein kinase r-like Endoplasmic Reticulum Kinase (PERK) (3), that sense a luminal stress signal(s) and convey it to downstream effectors via their enzymatic activities.
The unfolded protein load in the ER regulates IRE1 and PERK activation by two complementary processes: One involves the luminal chaperone immunoglobulin heavy chain binding protein (BiP), which complexes with the luminal domain of IRE1 and PERK, maintaining the enzymes in an inactive, monomeric form. Increases in unfolded protein correlate with disruption of this repressive complex, oligomerization, and activation of the sensor (4–6). The other process involves unfolded proteins that may also engage the stress-sensing luminal domain directly and favor its dimerization and thereby downstream signaling (7, 8).
Perturbation of cellular lipid composition also activates the UPR. Enhanced UPR signaling has been observed in cholesterol-loaded macrophages (9), in pancreatic beta cells exposed to saturated fatty acids (10), and in the liver of mice fed a high-fat diet (11) and is believed to play a role in the lipotoxicity associated with obesity and type II diabetes mellitus. In yeast, too, lipid perturbation activates the UPR (12, 13), but the mechanisms remain obscure.
In mammals, altered membrane lipid composition leads to ER calcium depletion (11, 14). This is proposed to promote unfolded protein stress by interfering with calcium-dependent chaperones and enzymes required for protein folding, leading to activation of the UPR by conventional sensing mechanisms. However, a recent study reported that the luminal stress-sensing domain of yeast IRE1 is dispensable to activation by lipid perturbation (15), suggesting that UPR signaling by altered lipid composition may also proceed independently of conventional unfolded protein stress sensing in the yeast ER lumen. It is presently unknown if a similar process prevails in mammals, but recent studies hint at possible mechanisms: The cytosolic, catalytic, effector domain of yeast IRE1 can be directly activated by flavonols, which engage a hydrophobic pocket at the dimer interface stabilizing the active form of yeast IRE1 (16). It is unclear, however, if other hydrophobic ligands, such as lipids, can regulate IRE1’s effector domain by a similar mechanism; and the kinase domain of mammalian PKR binds palmitate directly (17); however, it is unknown if the highly related kinase effector domain of PERK is also regulated by lipids.
Here we report on a dissection of lipid perturbation-mediated UPR activation in mammalian cells. Our findings uncover a luminal-independent component, which nonetheless requires an ER-spanning transmembrane domain, thus revealing the potential of the lipid bilayer to directly modulate UPR signaling, independently of changes to protein folding homeostasis in the ER lumen.
Results
Luminal ER Stress-Sensing Domain of IRE1 and PERK Is Dispensable to Their Activation by Lipid Perturbation.
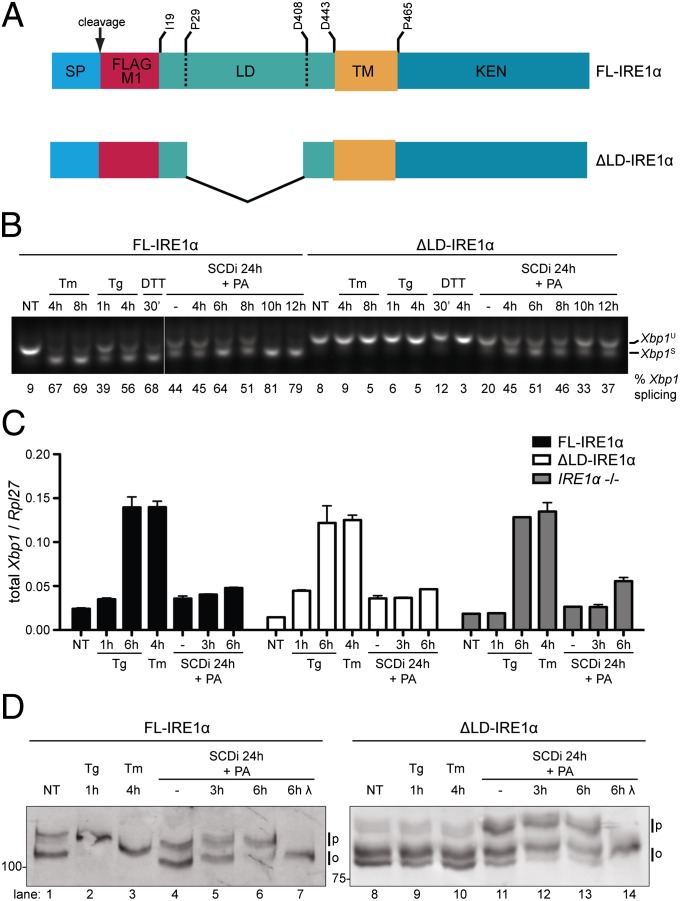
To explore mechanisms of lipid perturbation-induced UPR signaling, mouse cells homozygous for a mutant null allele of Ern1, encoding IRE1α, were transduced with retroviruses expressing Flag-M1–tagged full-length IRE1α (FL-IRE1α) or a Flag-M1–tagged ER-targeted mutant IRE1α lacking the luminal stress-sensing domain (ΔLD-IRE1α) (Fig. 1A and Fig. S1). The mutant cells have no endogenous IRE1 activity and express no detectable IRE1 protein and thus report on the activity of the transduced IRE1 with no interference by the endogenous protein (18). Stable clones, expressing roughly comparable levels of IRE1α effector (Fig. S2A) were exposed to the glycosylation inhibitor, tunicamycin; the ER calcium pump inhibitor, thapsigargin; or the reducing agent DTT, all known inducers of ER unfolded protein stress. IRE1 activity was measured by the splicing of its substrate, the X-box binding protein 1 encoding Xbp1 mRNA and was readily detected in FL-IRE1α–expressing cells subjected to unfolded protein stress. By contrast, no increase in Xbp1 mRNA splicing was detected in similarly treated ΔLD-IRE1α–expressing cells (Fig. 1B and Fig. S2B). Total Xbp1 mRNA levels increase in ER stressed cells, independently of IRE1 (18). Therefore, the comparable increase in total Xbp1 mRNA in tunicamycin and thapsigargin-treated ΔLD-IRE1α and FL-IRE1α–expressing cells reported on similar levels of unfolded protein stress (Fig. 1C) and confirmed the unresponsiveness of ΔLD-IRE1α to luminal signals, suggested by Fig. 1B.
Fig. 1.
Luminal unfolded protein stress-sensing domain is dispensable to IRE1 activation by lipid saturation. (A) Schema of ER-targeted full-length (FL) and luminal domain-deleted (ΔLD) IRE1α proteins. The signal peptide (SP) whose cleavage exposes the Flag M1 tag, luminal (LD), transmembrane (TM), and kinase-endonuclease (KEN) domains are noted. Residue numbering is based on UniProt O75460. (B) RT-PCR of Xbp1 mRNA from IRE1α−/− cells transduced with FL-IRE1α and ΔLD-IRE1α. Cells were exposed to tunicamycin (Tm; 5 µg/mL), thapsigargin (Tg; 1 µM), DTT (2 mM), or treated for 24 h with an SCD1 inhibitor, followed, where indicated, by palmitic acid (PA, 0.5 mM). The unspliced (Xbp1U) and spliced (Xbp1S) products are noted and the fraction of spliced Xbp1 mRNA in each sample is indicated below the photograph. (C) Quantitative PCR analysis of total Xbp1 mRNA from the cells described in B (shown is the mean ± SEM, n = 3 of values normalized to Rpl27 mRNA, a housekeeping gene). (D) Immunoblot of IRE1α proteins immunopurified from IRE1α−/− cells transduced with FL-IRE1α or ΔLD-IRE1α and treated as in B. Samples were resolved on a SDS/PAGE gel containing Phos-tag. The position of hypophosphorylated (0) and phosphorylated (P) IRE1α is indicated. In lanes 7 and 14, lambda phosphatase (λ) was applied in vitro to the purified IRE1α.
Increased membrane lipid saturation by genetic or pharmacological inhibition of stearoyl CoA desaturase 1 (SCD1) (19) and by loading of cultured cells with the saturated fatty acid palmitate (10) has been reported to promote UPR signaling. In keeping with these observations, exposure to an SCD1 inhibitor resulted in a fourfold increase in Xbp1 mRNA splicing in FL-IRE1α–expressing cells, which increased to eightfold when compounded with palmitate loading. Interestingly, SCD1 inhibition and palmitate loading also activated Xbp1 mRNA splicing in ΔLD-IRE1α–expressing cells (Fig. 1B), but not in the parental IRE1α knockout cells (Fig. S2B).
Activation of IRE1’s RNase activity is preceded by oligomerization-mediated transautophosphorylation (20), which can be detected by the change in mobility of the protein on SDS/PAGE gels impregnated with a phosphate ligand (Phos-tag). FL-IRE1α extracted from untreated cells migrated as two species on Phos-tag gels. Exposure to thapsigargin or tunicamycin led to disappearance of the faster-migrating species and its replacement by slower migrating species, unique to each manipulation (compare lanes 1–3 in Fig. 1D). SCD1 inhibition and palmitate loading were also associated with a shift toward slower-migrating species that collapsed to a faster-migrating one upon exposure of the sample to phosphatase in vitro (Fig. 1D, lanes 6 and 7). Thus, both luminal unfolded protein stress and lipid perturbation are associated with activating phosphorylation of FL-IRE1α.
ΔLD-IRE1α extracted from untreated cells gave rise to two prominent fast migrating immunoreactive species and a fainter slower-migrating one. This Phos-tag gel pattern was unaltered by exposure of cells to thapsigargin or tunicamycin (compare lanes 8–10, Fig. 1D). By contrast, SCD1 inhibition and palmitate loading led to redistribution of the ΔLD-IRE1α to lower-mobility species that also collapsed to a single faster migrating species upon dephosphorylation in vitro (lanes 11–14, Fig. 1D). Lipid-driven activation of ΔLD-IRE1α likely proceeds by conventional transautophosphorylation, as it is blocked by the K599A (21) mutation in the ATP-binding pocket of the kinase domain (Fig. S3). Thus, ΔLD-IRE1α activation is selectively responsive to lipid perturbation.
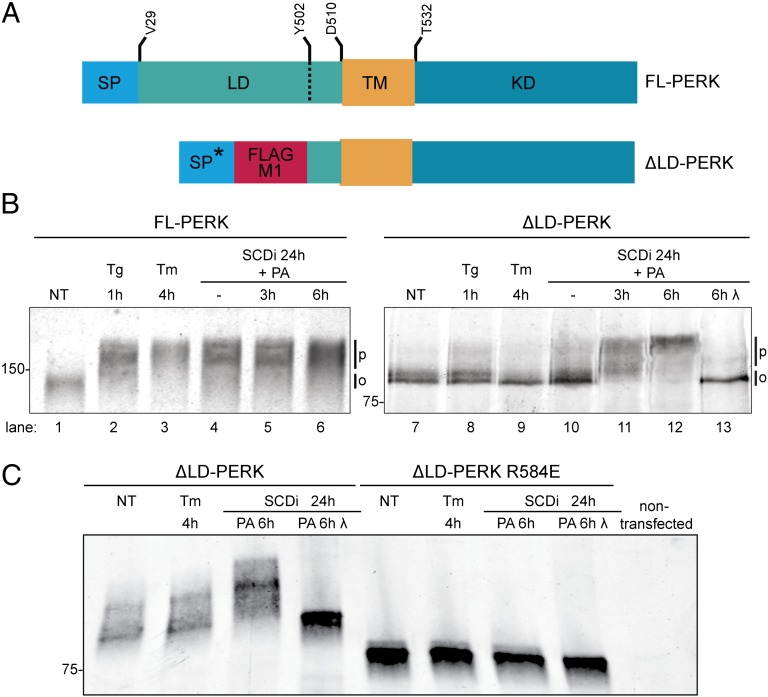
PERK is also activated by membrane lipid saturation (10, 19). To test if luminal-independent signals are involved in that process too, an ER-targeted PERK lacking its luminal stress-sensing domain (ΔLD-PERK, Fig. 2A) was expressed in mouse fibroblasts deleted of endogenous Eif2ak3/PERK. PERK undergoes extensive stress-induced autophosphorylation, which is readily detected by a shift to slower migrating forms on SDS/PAGE (3). As expected, phosphorylation of endogenous PERK was significantly increased in wild-type cells exposed to thapsigargin or tunicamycin (Fig. 2B, lanes 1–3). PERK phosphorylation was increased in cells treated with an SCD1 inhibitor, alone or in combination with palmitate loading (Fig. 2B, lanes 4–6). Only a minimal decrease in ΔLD-PERK mobility on SDS/PAGE was observed in cells exposed to thapsigargin and none in response to tunicamycin (Fig. 2B, lanes 7–9), indicating that the phosphorylation of ΔLD-PERK does not increase substantially in response to luminal unfolded protein stress. By contrast, a modest increase in ΔLD-PERK phosphorylation was observed in cells treated with the SCD1 inhibitor, and phosphorylation was markedly enhanced by palmitate loading (Fig. 2B, lanes 10–12). In vitro treatment with lambda phosphatase collapsed the heterogenous slower-migrating bands of ΔLD-PERK into a faster-migrating species, confirming that the former represent hyperphosphorylated PERK (Fig. 2B, lane 13). Lipid activation of ∆LD-PERK likely proceeds by conventional downstream mechanisms as it was disrupted by the arginine 584 to glutamate mutation (R584E), which interferes with the formation of a back-to-back active PERK kinase domain dimer (22, 23) (Fig. 2C). These observations indicate that both PERK and IRE1α can be activated by luminal-independent signals in cells with enhanced lipid saturation.
Fig. 2.
Luminal unfolded protein stress-sensing domain is dispensable to PERK activation by lipid saturation. (A) Schema of the PERK (FL) and the Flag-M1–tagged ER-targeted luminal domain-deleted (ΔLD) PERK proteins. The heterologous signal peptide of the ∆LD-PERK (SP*) whose cleavage exposes the Flag M1 tag, luminal (LD), transmembrane (TM), and kinase (KD) domains are noted. Residue numbering is based on UniProt Q9Z2B5. (B) Immunoblot of PERK immunopurified from wild-type mouse fibroblasts (FL-PERK) or PERK−/− cells transduced with ΔLD-PERK. Cells were exposed to thapsigargin (Tg; 1 µM), tunicamycin (Tm; 5 µg/mL) for the indicated times, or an SCD1 inhibitor for 24 h followed, where indicated, by palmitic acid (PA, 0.5 mM). The position of hypophosphorylated (0) and phosphorylated (P) PERK is indicated. λ-Phosphatase was applied in vitro to the purified protein (lane 13). (C) Immunoblot of wild-type and R584E dimerization mutant ∆LD-PERK immunopurified from transfected HEK293T cells treated as in B.
Membrane Tethering Mediates Sensitivity to Lipid Saturation.
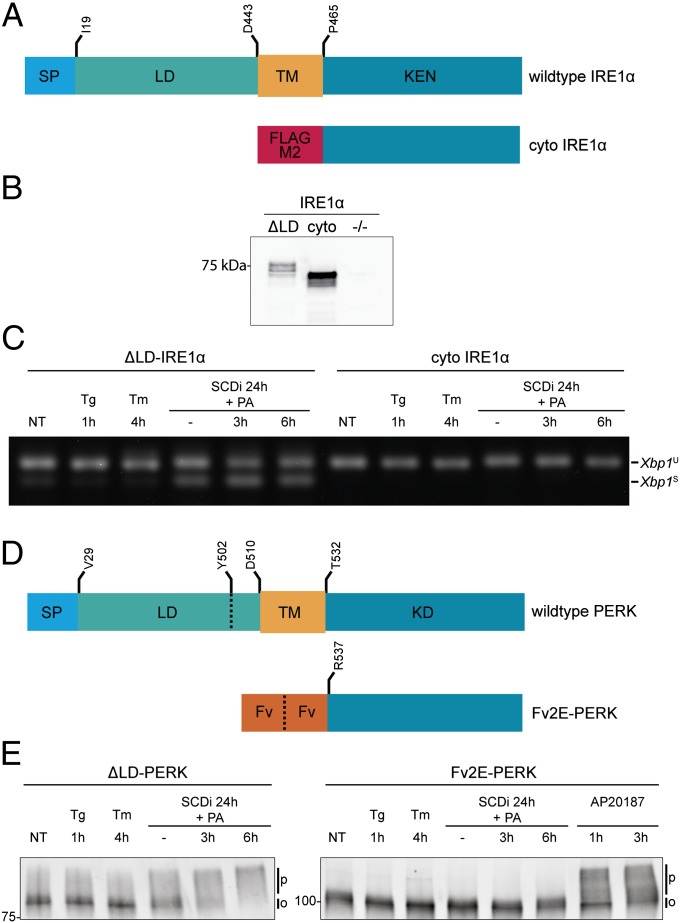
The isolated cytosolic domains of IRE1α and PERK are both enzymatically active, in vitro. Therefore, to further define the minimal requirements for IRE1α and PERK activation by lipid saturation in cells, we tested whether their isolated cytosolic effector domains could be activated in the absence of a membrane-spanning segment. The cytosolic effector domain of human IRE1α (residues 464–977, Fig. 3A) was readily expressed as a Flag-tagged fusion protein in the cytoplasm of cells lacking endogenous IRE1 (Fig. 3B). However, it failed to splice endogenous Xbp1 mRNA, unlike the transmembrane domain-containing ΔLD-IRE1α, which responded to lipids (Fig. 3C).
Fig. 3.
Membrane tethering is required for sensitivity to lipid perturbation. (A) Schema of Flag-M2–tagged cyto-IRE1α (residues 464–977), lacking the luminal and membrane-spanning domains. Schema of wild-type human IRE1α is provided as a reference. Signal peptide (SP) luminal (LD), transmembrane (TM), and kinase-endonuclease (KEN) domains are noted. Residue numbering is based on UniProt O75460. (B) Flag-M2 immunoblot of IRE1α immunopurified from IRE1α−/− cells transduced with Flag-M1–tagged ΔLD-IRE1α (∆LD), the Flag-M2–tagged cytosolic portion lacking the membrane-spanning domain (cyto), and the untransduced cells (−/−). (C) RT-PCR analysis of Xbp1 mRNA purified from IRE1α−/− cells transduced with Flag-M1–tagged ΔLD-IRE1α or the Flag-M2–tagged cytosolic portion (cyto IRE1α) and exposed to the indicated manipulations. (D) Schema of Fv2E-PERK, a fusion of a modified FK506 binding domain (Fv) to PERK's cytosolic effector domain (residues R537–1113). A scheme of endogenous mouse PERK (UniProt Q9Z2B5) is provided as a reference with the signal peptide (SP) luminal (LD), transmembrane (TM), and kinase (KD) domains noted. (E) Immunoblot of PERK immunopurified from PERK−/− cells expressing ΔLD-PERK or a cytosolic, ligand-activated PERK kinase (Fv2E-PERK). Cells were exposed to the indicated manipulations, which included the activating ligand of Fv2E-PERK, AP20187 (1 nM).
The specific residues constituting the transmembrane domain seemed less important, as both FL-IRE1α and ΔLD-IRE1α remained responsive to lipid saturation when the sequence of their transmembrane domain was scrambled or replaced by that of the unrelated ER localized transmembrane protein calnexin (Fig. S4 A and B). Nonetheless, not all membrane-spanning protein kinases are activated by lipid perturbations that activate the membrane-tethered UPR transducers. The epidermal growth-factor (EGF) receptor undergoes activating autophosphorylation in cells exposed to EGF, but is not activated by SCD1 inhibition and palmitate loading (Fig S4 C and D).
To study the role of membrane tethering in PERK activation by lipid saturation, we made use of a previously characterized fusion of a modified FK506 binding domain (Fv2E) to PERK's cytosolic effector domain (Fv2E-PERK) that undergoes ligand-induced dimerization (24) (Fig. 3D). As expected, Fv2E-PERK was strongly responsive to its ligand, the compound AP20187, reflected in a shift in its mobility on SDS/PAGE; however, unlike ΔLD-PERK the cytosolic Fv2E-PERK was unresponsive to lipid perturbation (Fig. 3E).
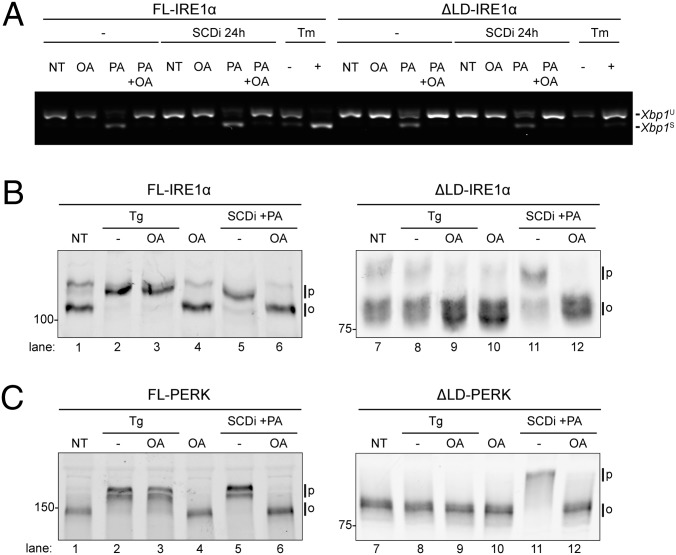
These observations demonstrate an essential role for membrane tethering in lipid saturation-induced activation of IRE1 and PERK. The ratio of unsaturated and saturated acyl chains affects membrane fluidity and thickness (25). IRE1 and PERK responded to this ratio, as UPR activation by SCD1 inhibition and palmitate loading was antagonized by coexposing cells to the monounsaturated oleic acid that restores the balance between saturated and unsaturated acyl chains in the membrane (Fig. 4 A–C). These observations suggest that IRE1 and PERK could directly sense changes in the biophysical properties of membranes, via their transmembrane domains.
Fig. 4.
Lipid-mediated UPR signaling depends on the balance between saturated and monounsaturated phospholipids. (A) RT-PCR analysis of Xbp1 mRNA purified from IRE1α−/− cells transduced with FL-IRE1α and ΔLD-IRE1α. Cells were exposed for 6 h to the monounsaturated oleic acid (OA, 0.5 mM), palmitic acid (PA, 0.5 mM), or both, in the absence or presence of an SCD1 inhibitor. (B) Immunoblot of a Phos-tag gel of FL and ∆LD-IRE1α that had been exposed to an SCD1 inhibitor, oleic acid, palmitic acid, thapsigargin (Tg), or combinations thereof. (C) Immunoblot of FL and ∆LD-PERK from PERK−/− cells treated as in B.
Modulation of UPR Signaling by Lipid Composition in Proteoliposomes.
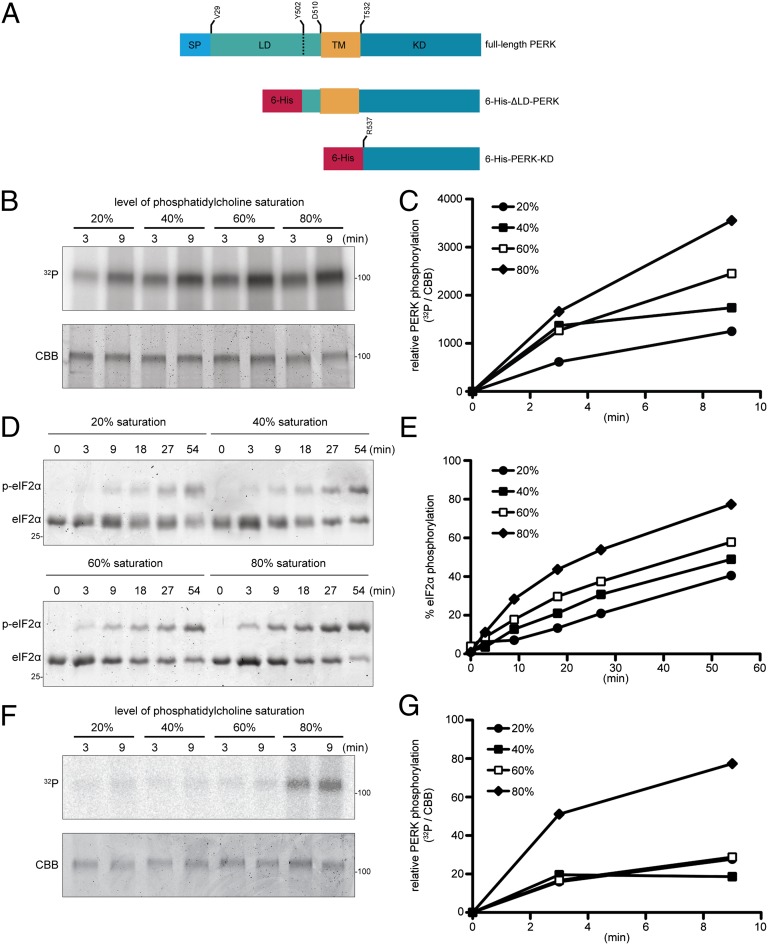
To further isolate the effects of lipids on UPR activity from its effects on luminal unfolded protein stress we reconstituted aspects of PERK activity in proteoliposomes that are devoid of unfolded ER client proteins. The transmembrane and kinase domain of PERK was expressed in Escherichia coli as a hexahistidine-tagged fusion protein (6-His-ΔLD-PERK, Fig. 5A), purified and reconstituted into liposomes containing variable amounts of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), a monounsaturated phosphatidylcholine, and 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), a saturated phosphatidylcholine (PC) (similar attempts to express ΔLD-IRE1 in bacteria were unsuccessful). PERK-containing proteoliposomes were isolated by flotation. A histidine-tagged PERK kinase domain lacking the transmembrane domain (6-His-PERK-KD, Fig. 5A) did not associate with liposomes after flotation, indicating that 6-His-ΔLD-PERK integrated into the liposomes via its transmembrane domain (Fig. S5). In 6-His-ΔLD-PERK proteoliposomes, both PERK autokinase activity (Fig. 5 B and C) and phosphorylation of its substrate, the purified alpha subunit of translation initiation factor 2 (eIF2α) (Fig. 5 D and E), were greater in liposomes containing more saturated acyl chains. Importantly, the positive effect of lipid saturation on PERK enzymatic activity was also observed across the 40% saturation mark, which corresponds to the saturation levels measured in ER membranes (26) and in response to changes in saturation observed in disease models (27).
Fig. 5.
Lipid bilayer acyl-chain saturation enhances activity of ∆LD-PERK in proteoliposomes. (A) Schema of bacterially expressed hexahistidine-tagged PERK proteins used to constitute proteoliposomes. Full-length mouse PERK (UniProt Q9Z2B5) is provided as a reference with the signal peptide (SP), luminal (LD), transmembrane (TM), and kinase (KD) domains noted. (B) Autoradiogram (32P, Upper) and Coomassie Brilliant Blue stain (CBB, Lower) of purified bacterially expressed His-tagged ∆LD-PERK that had been incorporated into liposomes of the indicated phosphatidylcholine saturation, purified by flotation, reacted with 32P γ-labeled ATP for the indicated time, and resolved by SDS/PAGE. (C) Level of PERK phosphorylation as determined by quantitation of the signals in B. Shown is a representative experiment reproduced four times. (D) Coomassie Blue-stained Phos-tag gel of eIF2α N-terminal lobe purified from bacteria and reacted for the indicated time with His-tagged ∆LD-PERK reconstituted in liposomes of the indicated saturation in the presence of ATP (0.2 mM). The phosphorylated and nonphosphorylated forms of eIF2α are indicated. (E) Fraction of eIF2α phosphorylated as determined by quantitation of the signals in D. Shown is a representative experiment reproduced three times. (F) Autoradiogram (32P, Upper) and Coomassie Brilliant Blue stain (CBB, Lower) of purified bacterially expressed His-tagged PERK-KD that had been incorporated into Ni-NTA–tagged liposomes of the indicated phosphatidylcholine saturation, purified by flotation, reacted with 32P γ-labeled ATP for the indicated time, and resolved by SDS/PAGE. (G) Level of PERK phosphorylation as determined by quantitation of the signals in F. Shown is a representative experiment reproduced three times.
To gauge the role of the transmembrane domain in the sensitivity of PERK kinase activity to saturation levels, the histidine-tagged PERK kinase domain lacking the transmembrane domain (6-His-PERK-KD, Fig. 5A) was bound to liposomes containing 1% lipid with a nickel-nitrilotriacetic acid (NTA)–bearing head group (28). The enzymatic activity of nickel-NTA–bound PERK was weak by comparison with the 6-His-ΔLD-PERK–containing proteoliposomes (Fig. 5, compare B and F) and unresponsive to lipid saturation in the physiological range (Fig. 5 F and G). These observations suggest that the transmembrane domain can mediate aspects of the regulation of PERK kinase activity by lipids, independently of unfolded protein signaling.
Discussion
In mammalian cells, IRE1 and PERK lacking the luminal unfolded protein-stress sensing domain retained the ability to detect perturbation of membrane lipid composition. Activation required ER retention via a transmembrane domain and the positive relationship between acyl chain saturation and enzymatic activity of PERK was reconstituted in vitro, in isolation from luminal unfolded protein stress. Together these observations call attention to the potential role of IRE1 and PERK as direct lipid sensors.
Dimerization of the cytosolic effector domains is a proximate driver of PERK (4, 23, 24) and IRE1 activation (20). In the intact protein, this is initiated by dimerization of the stress-sensing luminal domain incorporating the entire protein, including its transmembrane domain into the oligomer. Our findings suggest that perturbation of membrane lipid composition could promote IRE1 and PERK activation by enhanced dimerization via their transmembrane domain. Dimerization requires disruption of interactions between the phospholipid acyl chains and amino acids at the dimer interface. Saturated acyl chains are less flexible than unsaturated acyl chains and interact less strongly with transmembrane domains. Thus, the barrier to delipidation of membrane-embedded proteins is weaker in a saturated than unsaturated lipid bilayer, favoring protein–protein over protein–lipid interactions (29, 30). This biophysical principle could contribute to the sensitivity of the UPR transducers to their lipid environment if, as seems likely, approximation of the transmembrane segments in the active oligomeric state entails disruption of some protein–lipid interactions.
Acyl chain saturation also decreases lipid bilayer fluidity, favoring a gel phase over a liquid phase. Transmembrane peptides are preferentially excluded from the gel phase and as a consequence, their local density increases in the liquid phase, favoring dimerization (25). Finally, the extended conformation of saturated acyl chains increases lipid bilayer thickness. An expanded lipid bilayer may promote dimer formation by altering the tilting angle of the transmembrane domains and by favoring interactions among polar residues that had been forced into the expanded lipid bilayer (31).
∆LD-IRE1’s responsiveness to acyl chain saturation survived both swapping of the transmembrane domain with that of an unrelated ER protein and sequence scrambling. These findings support the primacy of the aforementioned generic mechanisms and suggest that specific interactions among side chains of the transmembrane domain may be less important to recognition of changes in lipid composition. However, it is worth noting that activation of IRE1 and PERK by this lipid-based mechanism is dependent on special features of their soluble effector domains, such as the ability to form back-to-back dimers, in the case of PERK, or autophosphorylation, in the case of IRE1. Furthermore not all membrane-spanning signal transducers are activated by the lipid perturbation; the EGF receptor that we tested here is not activated. Unfortunately, the resolution of our assays of IRE1 or PERK activity is inadequate for careful quantitative assessment of the role of the transmembrane domain sequence. Thus, the significance of the hint that scrambling of the transmembrane domain sequence reproducibly weakened the response of ∆LD-IRE1α to lipids is difficult to explore, at present.
In cells subjected to an identical perturbation of the lipid environment, both the full-length IRE1 and the full-length PERK have a stronger response than their comparably expressed ∆LD versions. This may reflect the contribution of the lipid perturbation to unfolded protein stress that is sensed through the luminal domain. Thus, the direct and indirect effects of altered lipid saturation on IRE1 and PERK activity may be additive. The relatively feeble activity of the ∆LD transducers may also reflect cooperativity between the luminal and transmembrane domains in promoting dimerization independently of changes in luminal unfolded protein stress. The crystal structure of both the yeast and mammalian IRE1 luminal domains is that of a dimer (7, 32), which may also engage in higher order oligomer formation. It is thus possible that the luminal domain stabilizes a dimer that is favored by lipid saturation, amounting to lipid sensing by IRE1 and PERK that can be independent of changes in luminal signals.
Direct modulation of IRE1α and PERK’s activity by the lipid environment and luminal stress-induced activation are two parallel pathways that likely intertwine. By stabilizing the dimer, an increase in the acyl chain saturation could lower the threshold of luminal stress required for unfolded protein-dependent activation of IRE1α and PERK, and levels of unfolded protein stress could modulate the response to lipids by a similar cooperative mechanism. Lipid sensing by IRE1 and PERK may account for instances, such as B-cell development, marked by an apparent discrepancy between a modest level of unfolded protein load and substantial UPR signaling (33–35). It may therefore be interesting to examine the potential role of lipids as instigators of what has been termed an anticipatory UPR (36, 37).
The UPR effectors modulate important aspects of lipid metabolism. For example, spliced XBP1 contributes to membrane expansion by promoting phospholipid synthesis (38), and interfering with PERK signaling by overexpressing an eIF2α phosphatase in the liver attenuates hepatosteatosis in mice (39). Elevated UPR markers have been observed in the liver and adipose tissue of obese humans (40). Obesity, diet-induced ER stress, and UPR activation have been linked to insulin resistance (11, 41) and pancreatic β-cell failure (10) and may therefore have a role in the development of type 2 diabetes. Activation of IRE1α and PERK by changes in lipid composition suggests a mechanism by which UPR signaling may contribute to the aforementioned pathophysiological processes directly, as sensors of a primary perturbation.
Materials and Methods
Cell Manipulation.
Knockout cells were rescued with IRE1 and PERK derivatives by retroviral transduction, as described in detail in SI Materials and Methods.
ER stress was induced by exposure of cells to tunicamycin, thapsigargin, or DTT. SCD1 inhibitors CVT-11127 (42) (also known as GS-456332) (Gilead Sciences) and MF-438 (43) (Merck Sharp & Dohme) were dissolved in DMSO and were used interchangeably at 1 µM, as they provided equivalent Xbp1 mRNA splicing. Palmitic acid (Sigma) and oleic acid (Sigma) were dissolved in 90% (vol/vol) ethanol. For pretreatment with the SCD1 inhibitors, cells were cultured in DMEM containing 10% FCS, nonessential amino acids, penicillin/streptomycin, glutamine, 50 µM beta-mercaptoethanol, and 14 µg/mL gentamicin. Solubilized oleate and palmitate were added at 0.5 mM in DMEM containing 1% FCS and 1% BSA.
Analysis of protein expression by immunoprecipiation and immunoblotting and the measurement of mRNA expression and Xbp1 mRNA splicing are described in detail in SI Materials and Methods.
Liposome Formation.
The liposome mixture contained PC/phosphatidylethanolamine (PE)/rhodamine-PE at a mass ratio of 8/1.9/0.1. PC was composed of a mix of the monounsaturated DOPC and of the saturated DSPC. We adjusted the degree of PC saturation by varying the DOPC/DSPC mass ratio (r), as follows: 20% saturation (r = 6.4/1.6), 40% (r = 4.8/3.2), 60% (r = 3.2/4.8), and 80% (r = 1.6/6.4). Where indicated, liposomes also contained a phospholipid with a nickel-NTA–bearing head group (1,2-dioleoyl-sn-glycero-3-{[N(5-amino-1-carboxypentyl)iminodi-acetic acid]succinyl} nickel salt (DOGS-NTA-Ni)). Nickel-bearing liposomes contained a mixture of PC/PE/rhodamine-PE/DOGS-NTA-Ni at a mass ratio of 8/1.8/0.1/0.1. All of the phospholipids were from Avanti Polar Lipids. Subsequent steps in the liposomes preparation were carried out as described previously (44).
Proteoliposome Formation.
Methods of protein expression in bacteria and kinase assays are described in detail in SI Materials and Methods.
Liposomes were partially solubilized in kinase buffer (20 mM Tris⋅HCl, pH = 7.5, 150 mM NaCl, 2 mM MgCl2, 1 mM DTT) containing 0.25% deoxy-Big CHAP (DBC) (Merck) and then incubated with bacterially expressed 6×-His-ΔLD-PERK in a lipid:protein mass ratio of 80:1. This mixture containing 800 µg of lipids and 10 µg of protein was added to 30 mg of Biobeads SM2 (Bio-Rad) and incubated overnight at 4 °C. The fluid phase was recovered and floated in a 50–30–5% (wt/vol) Optiprep (Sigma) gradient (250,000 × g, 3 h, 4 °C) to remove unincorporated protein. Proteoliposomes were recovered at the 30–5% interface, diluted in kinase buffer, and subsequently subjected to an ultracentrifugation (250,000 × g, 45 min, 4 °C) to concentrate proteoliposomes and dilute the Optiprep medium. Proteoliposomes were snap frozen in liquid nitrogen and stored at −80 °C until further use. The protein content of proteoliposomes was determined by running the preparations on SDS/PAGE followed by Coomassie stain using Instant Blue (Expedion). Coomassie stain was visualized on an Odyssey scanner (Li-Cor). Protein content was normalized for each lipid mixture and the lipid content was quantified by measuring the fluorescence of rhodamine-PE.
Supplementary Material
Acknowledgments
We are indebted to Ramanujan Hegde for expert advice on liposome reconstitution and Gilead Sciences and Merck Sharp & Dohme for the kind gift of SCD1 inhibitors. This work was supported by a Wellcome Trust Intermediate Clinical Fellowship (to R.V.), a Wellcome Trust Principal Research Fellowship (to D.R.), and European Union Seventh Framework Programme Grant (Beta Bat, 277713).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217611110/-/DCSupplemental.
References
- 1.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73(6):1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 2.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74(4):743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 3.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397(6716):271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 4.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2(6):326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 5.Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem Biophys Res Commun. 2000;279(2):445–450. doi: 10.1006/bbrc.2000.3987. [DOI] [PubMed] [Google Scholar]
- 6.Pincus D, et al. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010;8(7):e1000415. doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2005;102(52):18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 2011;333(6051):1891–1894. doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng B, et al. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5(9):781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- 10.Cunha DA, et al. Initiation and execution of lipotoxic ER stress in pancreatic beta-cells. J Cell Sci. 2008;121(Pt 14):2308–2318. doi: 10.1242/jcs.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu S, et al. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473(7348):528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox JS, Chapman RE, Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell. 1997;8(9):1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pineau L, et al. Lipid-induced ER stress: Synergistic effects of sterols and saturated fatty acids. Traffic. 2009;10(6):673–690. doi: 10.1111/j.1600-0854.2009.00903.x. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, et al. Enrichment of endoplasmic reticulum with cholesterol inhibits sarcoplasmic-endoplasmic reticulum calcium ATPase-2b activity in parallel with increased order of membrane lipids: Implications for depletion of endoplasmic reticulum calcium stores and apoptosis in cholesterol-loaded macrophages. J Biol Chem. 2004;279(35):37030–37039. doi: 10.1074/jbc.M405195200. [DOI] [PubMed] [Google Scholar]
- 15.Promlek T, et al. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol Biol Cell. 2011;22(18):3520–3532. doi: 10.1091/mbc.E11-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiseman RL, et al. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol Cell. 2010;38(2):291–304. doi: 10.1016/j.molcel.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho H, et al. Molecular mechanism by which palmitate inhibits PKR autophosphorylation. Biochemistry. 2011;50(6):1110–1119. doi: 10.1021/bi101923r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 19.Ariyama H, Kono N, Matsuda S, Inoue T, Arai H. Decrease in membrane phospholipid unsaturation induces unfolded protein response. J Biol Chem. 2010;285(29):22027–22035. doi: 10.1074/jbc.M110.126870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15(12):3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 21.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12(12):1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui W, Li J, Ron D, Sha B. The structure of the PERK kinase domain suggests the mechanism for its activation. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 5):423–428. doi: 10.1107/S0907444911006445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dey M, Cao C, Sicheri F, Dever TE. Conserved intermolecular salt bridge required for activation of protein kinases PKR, GCN2, and PERK. J Biol Chem. 2007;282(9):6653–6660. doi: 10.1074/jbc.M607897200. [DOI] [PubMed] [Google Scholar]
- 24.Lu PD, et al. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23(1):169–179. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9(2):112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keenan TW, Morré DJ. Phospholipid class and fatty acid composition of golgi apparatus isolated from rat liver and comparison with other cell fractions. Biochemistry. 1970;9(1):19–25. doi: 10.1021/bi00803a003. [DOI] [PubMed] [Google Scholar]
- 27.Borradaile NM, et al. Disruption of endoplasmic reticulum structure and integrity in lipotoxic cell death. J Lipid Res. 2006;47(12):2726–2737. doi: 10.1194/jlr.M600299-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125(6):1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 29.White SH, Wimley WC. Membrane protein folding and stability: Physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 30.Anbazhagan V, Schneider D. The membrane environment modulates self-association of the human GpA TM domain—implications for membrane protein folding and transmembrane signaling. Biochim Biophys Acta. 2010;1798(10):1899–1907. doi: 10.1016/j.bbamem.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 31.Sparr E, et al. Self-association of transmembrane alpha-helices in model membranes: Importance of helix orientation and role of hydrophobic mismatch. J Biol Chem. 2005;280(47):39324–39331. doi: 10.1074/jbc.M502810200. [DOI] [PubMed] [Google Scholar]
- 32.Zhou J, et al. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci USA. 2006;103(39):14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Anken E, et al. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 2003;18(2):243–253. doi: 10.1016/s1074-7613(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 34.Gass JN, Gifford NM, Brewer JW. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem. 2002;277(50):49047–49054. doi: 10.1074/jbc.M205011200. [DOI] [PubMed] [Google Scholar]
- 35.Hu CC, Dougan SK, McGehee AM, Love JC, Ploegh HL. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. EMBO J. 2009;28(11):1624–1636. doi: 10.1038/emboj.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vitale A, Boston RS. Endoplasmic reticulum quality control and the unfolded protein response: Insights from plants. Traffic. 2008;9(10):1581–1588. doi: 10.1111/j.1600-0854.2008.00780.x. [DOI] [PubMed] [Google Scholar]
- 37.Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol. 2010;189(5):783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: A link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167(1):35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7(6):520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregor MF, et al. Endoplasmic reticulum stress is reduced in tissues of obese subjects after weight loss. Diabetes. 2009;58(3):693–700. doi: 10.2337/db08-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozcan U, et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313(5790):1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koltun DO, et al. Novel, potent, selective, and metabolically stable stearoyl-CoA desaturase (SCD) inhibitors. Bioorg Med Chem Lett. 2009;19(7):2048–2052. doi: 10.1016/j.bmcl.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 43.Léger S, et al. Synthesis and biological activity of a potent and orally bioavailable SCD inhibitor (MF-438) Bioorg Med Chem Lett. 2010;20(2):499–502. doi: 10.1016/j.bmcl.2009.11.111. [DOI] [PubMed] [Google Scholar]
- 44.Mariappan M, et al. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature. 2011;477(7362):61–66. doi: 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.