Abstract
Genetic variants influencing the transcriptome have been extensively studied. However, the impact of the genetic factors on the human proteome is largely unexplored, mainly due to lack of suitable high-throughput methods. Here we present unique and comprehensive identification of genetic variants affecting the human plasma protein profile by combining high-throughput and high-resolution mass spectrometry (MS) with genome-wide SNP data. We identified and quantified the abundance of 1,056 tryptic-digested peptides, representing 163 proteins in the plasma of 1,060 individuals from two population-based cohorts. The abundance level of almost one-fifth (19%) of the peptides was found to be heritable, with heritability ranging from 0.08 to 0.43. The levels of 60 peptides from 25 proteins, 15% of the proteins studied, were influenced by cis-acting SNPs. We identified and replicated individual cis-acting SNPs (combined P value ranging from 3.1 × 10−52 to 2.9 × 10−12) influencing 11 peptides from 5 individual proteins. These SNPs represent both regulatory SNPs and nonsynonymous changes defining well-studied disease alleles such as the ɛ4 allele of apolipoprotein E (APOE), which has been shown to increase risk of Alzheimer's disease. Our results show that high-throughput mass spectrometry represents a promising method for large-scale characterization of the human proteome, allowing for both quantification and sequencing of individual proteins. Abundance and peptide composition of a protein plays an important role in the etiology, diagnosis, and treatment of a number of diseases. A better understanding of the genetic impact on the plasma proteome is therefore important for evaluating potential biomarkers and therapeutic agents for common diseases.
Keywords: protein quantitative trait loci, population proteomics
Our understanding of the impact of genetic variation on human traits has been greatly advanced using high-throughput SNP genotyping and massively parallel sequencing. The large number of genome-wide association studies (GWAS) performed have resulted in the identification of hundreds of SNPs that are associated with human traits and diseases (1, 2). The functional impact of most of the SNPs influencing human traits has not been well characterized. Whereas nonsynonymous SNPs affect the amino acid sequence directly and could alter the function of the resulting protein, other SNPs may have an impact on splice sites (3) or influence amount or stability of the mRNA (4). GWAS studies relating the genetic variability to the transcript profile have identified a number of cis-regulatory SNPs affecting expression quantitative traits (eQTs) (5, 6). Evidently, the expression of many genes is influenced by a nearby SNP, with cis-regulatory SNPs being overrepresented among SNPs associated with human phenotypes (1). Studies have also addressed the impact of genetic variability on levels of endogenous metabolites, such as sugars, biogenic amines, acylcarnitines, and glycerophospho- and sphingolipids, which can be measured in either human urine or plasma. These studies have identified a series of metabolic quantitative trait loci (mQTL) affecting the level of these biomolecules (7, 8).
By contrast, genome-wide analyses of the impact of genetic variability on the proteome profile have been missing, due to methodological limitations. A recent study in mouse fibroblasts (9) estimates that mRNA levels explain around 40% of the variability in protein levels, underscoring the need for studies directly addressing the correlation between the genetic and the proteome profiles. Most studies of the effect of genetic variation on the proteome profile have focused on single proteins or a limited set of proteins. Studies of selected proteins, e.g., those affecting the immune response or biomarkers for a specific disease, have demonstrated the presence of protein quantitative trait loci (pQTL) (10–12), i.e., genetic variants impacting protein expression. The main limitation in the identification of pQTL has been access to high-throughput methods in proteomics to study the abundance of individual proteins in human clinical samples, which can be applied to the analysis of large cohorts.
In this paper we have used high-throughput high-resolution mass spectrometry (MS) to identify and quantify peptides in plasma from more than 1,000 individuals from a population-based study. We have identified genetic determinants of the peptide profile using genome-wide SNP data. To our knowledge, this is the largest study to date using MS to quantify peptides and assess the genetic determinants of the human plasma proteome. Our results show that abundance of a large number of the plasma proteins is heritable and affected by genetic variants.
Results
Heritability of the Plasma Proteome.
A total of 1,056 tryptic digested peptides were identified and quantified by high-throughput high-resolution mass spectrometry in the plasma samples from 1,060 individuals. A total of 87.3% of the peptides were unique to one protein (using the September 2011 release of the International Protein Index database) (13). This is similar to what was found for peptides in the Human Plasma Proteome Project (88.3%, SI Methods). Our peptides mapped to 163 plasma proteins, with an average of 6.4 peptides per protein and a range from 1 to 173 peptides per protein (Table S1). A total of 1,029 individuals passed the genotyping quality control, had peptide values measured, and represent the basis of the analyses. More than 93% of the peptides (n = 989) were detected in at least 400 individuals and used in the association analyses. We first tested the correlation between the peptide values and covariates such as sex, age, body mass index (BMI), allergy, lipid lowering treatment, and antihypertensive treatment. Of the 989 peptides tested, 226 were significantly influenced by age, 29 by BMI, and 24 by sex [false discovery rate (FDR) q value <0.05]. None of the peptides was influenced by allergy status, lipid lowering, or antihypertensive treatment (FDR q value >0.05 for all peptides). In a further analysis we used sex, age, and BMI as covariates.
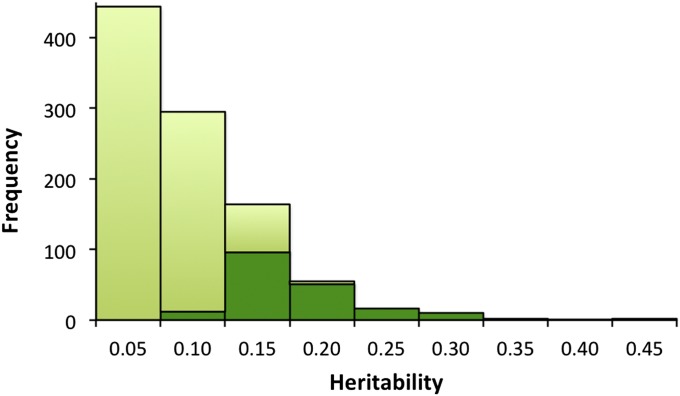
In the total dataset (both cohort KA06 and KA09), 190 (19%) peptides (FDR q value <0.05) showed a significant heritability (Fig. 1), with heritability estimates ranging from 0.08 to 0.43 (Table S2). The 190 heritable peptides were derived from 57 proteins, and consequently 35% of the proteins identified had at least one peptide showing a significant heritability. The highest heritability was found for a peptide from the complement 3 (CO3) protein (h2 = 0.43), encoded by the C3 gene, followed by a peptide from the Alpha-2-macroglobulin (FETUA) protein (h2 = 0.42).
Fig. 1.
Heritability of peptide abundance levels. Histogram of estimated heritabilities for all peptides. The dark green indicates significant observations (FDR P value <0.05).
Identification of cis-pQTL.
To identify cis-pQTL we performed an association analysis using the discovery cohort (KA06) and found 1,815 genome-wide significant SNPs distributed over 32 peptides from 16 proteins (Table S3). The inflation factor lambda for the association analyses was 1.0054 in the discovery cohort confirming that the results are not due to internal population structuring. The top SNP for 11 of these 32 peptides showed a significant association also in the replication cohort (KA09) (Table 1). About half (5/11) of the associated SNPs either themselves result in an altered amino acid sequence of the peptide or are in high linkage disequilibrium (LD) with a nonsynonymous SNP (nsSNP) located in the same peptide region. This number is substantially higher than the genome-wide fraction (0.0022%) of SNPs that are either themselves a nonsynonymous SNP or in LD with a nonsynonymous SNP (SI Methods). Among the associations identified, that of rs2230203 (reference SNP number) with one peptide in CO3, and rs429358 with one peptide in apolipoprotein E (APOE), both represent nonsynonymous SNPs changing an amino acid.
Table 1.
SNPs with a genome-wide significant association in the discovery cohort (KA06) and replicated in the second cohort (KA09)
| UniProt ID (gene symbol) Peptide | SNP | Chr:position | Freq | KA06 beta* (SE)† | P value | KA09 beta* (SE)† | P value | Pooled P value‡ |
| HPT (HP) | ||||||||
| AVGDKLPECEAVCGKPK | rs217184§ | 16:72105965 | 0.18 | 0.56 (0.08) | 1.7 × 10−12 | 0.46 (0.11) | 1.5 × 10−5 | 8.0 × 10−17 |
| GDKLPECEAVCGKPK | rs77303550§ | 16:72079657 | 0.18 | 0.47 (0.077) | 1.6 × 10−9 | 0.39 (0.11) | 2.4 × 10−4 | 1.5 × 10−12 |
| TEGDGVYTLNDKK | rs77303550§ | 16:72079657 | 0.18 | 0.71 (0.08) | 8.7 × 10−19 | 0.68 (0.11) | 1.0 × 10−10 | 4.4 × 10−27 |
| TEGDGVYTLNNEK | rs217184§ | 16:72105965 | 0.18 | 0.86 (0.08) | 2.8 × 10−27 | 0.58 (0.13) | 3.7 × 10−6 | 2.9 × 10−31 |
| VDSGNDVTDIADDGCPKPPEIAHGYVEHSVR | rs77303550§ | 16:72079657 | 0.18 | 0.4 (0.078) | 2.7 × 10−7 | 0.39 (0.11) | 1.9 × 10−4 | 4.3 × 10−10 |
| A1AT (SERPINA1) | ||||||||
| DTEEEDFHVDQVTTVK¶ | rs1243165 | 14:94844305 | 0.21 | −0.9 (0.075) | 3.6 × 10−33 | −0.6 (0.089) | 1.5 × 10−11 | 3.9 × 10−41 |
| IVDLVKELDRDTVFALVNYIFFK | rs17090693 | 14:94841331 | 0.21 | −0.88 (0.069) | 5.9 × 10−37 | −0.81 (0.096) | 3.0 × 10−17 | 3.1 × 10−52 |
| TLNQPDSQLQLTTGNGLFLSEGLK|| | rs709932 | 14:94849201 | 0.13 | −0.48 (0.08) | 3.0 × 10−9 | −0.57 (0.15) | 1.7 × 10−4 | 2.9 × 10−12 |
| CO3 (C3) | ||||||||
| LLDGVQNPR** | rs2230203 | 19:6710782 | 0.10 | −0.85 (0.12) | 4.2 × 10−13 | −0.44 (0.12) | 1.7 × 10−4 | 1.9 × 10−14 |
| APOE (APOE) | ||||||||
| LGADMEDVCGR†† | rs429358 | 19:45411941 | 0.18 | −0.52 (0.083) | 6.3 × 10−10 | −0.34 (0.10) | 7.8 × 10−4 | 3.9 × 10−12 |
| FETUA (AHSG) | ||||||||
| HTFMGVVSLGSPSGEVSHPR‡‡ | rs2070635 | 3:186336176 | 0.48 | 0.53 (0.057) | 7.0 × 10−21 | 0.36 (0.081) | 9.9 × 10−6 | 1.7 × 10−24 |
Beta: The additive effect of the minor allele on the abundance level of the peptide.
SE: SE of the effect (beta).
Pooled: KA06 and KA09 analyzed together.
rs217184 and rs77303550 are in LD (R2 = 0.95).
DTEEEDFHVDQVTTVK is located at position 226–241 of the protein. Position 237 has a natural variant, coded by the SNP rs6647, which is in LD with rs1243165.
TLNQPDSQLQLTTGNGLFLSEGLK is located at position 126–149 of the protein. Position 125 has a natural variant, coded by our top SNP rs709932.
LLDGVQNPR is located at position 307–315 of the protein. Position 314 has a natural variant coded by the SNP rs1047286, which is in LD with our top SNP rs2230203.
LGADMEDVCGR is located at position 122–132 of the protein. Position 130 has a natural variant, coded by our top SNP rs429358.
HTFMGVVSLGSPSGEVSHPR is located at position 318–337 of the protein. Position 317 has a natural variant, coded by the SNP rs35457250, which is not in strong LD with our top SNP rs2070635. The P value for rs35457250 (P value = 5.1E-14 in the pooled analyses) is not as significant as our top SNP.
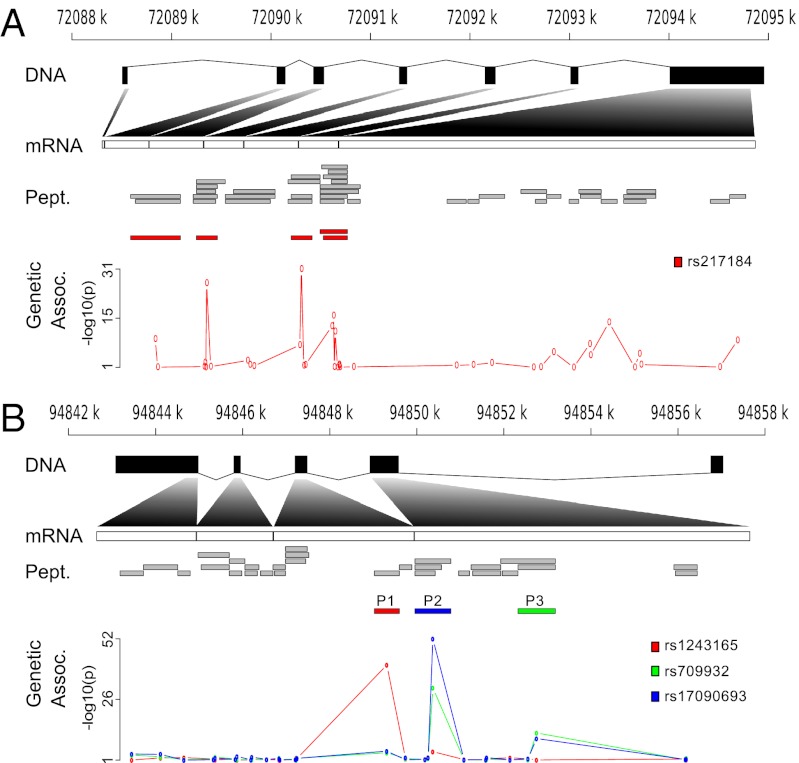
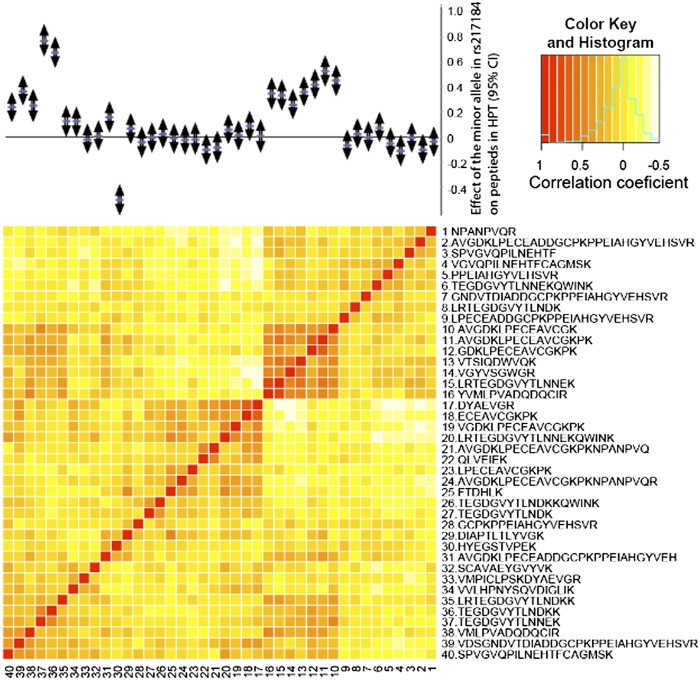
A second group of SNPs is associated with the abundance of multiple, nonoverlapping, peptides from the same protein (Fig. 2A). For instance, five different peptides from the Haptoglobin protein (HPT) were strongly associated with three SNPs in high LD with each other (rs217181, rs217184, and rs77303550, pairwise R2 > 0.95). Using a more relaxed threshold for genome-wide significance in the KA06 cohort, we found that each of these SNPs was associated with 16 of the 40 HPT peptides quantified (Fig. 3). It should be noted that the minor allele in rs217184 has a positive effect on all but one (HYEGSTVPEK) peptide. This exception is most likely due to an association with another SNP (not present in our dataset) that is negatively correlated with rs217184. Although many of the HPT peptides are associated with this SNP, the pairwise correlation between peptide measurements is quite modest (Fig. 3), with a maximum correlation coefficient of 0.71. However, it is obvious that a set of peptides (10 to 15 in Fig. 3), which are highly correlated, also show a similar association with the top SNP (rs217184). The Manhattan plot of P values across the ∼200-kb region surrounding the haptoglobin (HP) gene encoding the HPT protein for the different peptides, shows two peaks with SNPs located on both sides of the gene (rs77303550 and rs217184/rs217181) (Fig. S1A). When studying the distribution of the association signal relative to the location of the peptides, the highest signal was found for SNPs located in or near exons 3 and 5 of the HP gene. The strong LD between the three top SNPs indicates that any of these three, a combination of all three, or other SNPs (which are not present on the SNP array, not imputed with high enough certainty, or not present in the reference panels used for the imputations) is affecting the expression of the HP gene. A similar situation is found for FETUA, encoded by the alpha-2-HS-glycoprotein (AHSG) gene (Table 1), where the minor allele of rs2070635 is nominally associated with abundance of 5 of the 16 peptides quantified, suggesting that this might represent a regulatory SNP influencing the protein level.
Fig. 2.
Gene structure and location of the identified and quantified peptides. (A) Peptides in HPT and the association between rs217184 and each peptide. (B) Peptides in ATA1 and the association between three different SNPs (rs1243165, rs709932, and rs17090693) and each peptide.
Fig. 3.
Heatmap of pairwise correlation coefficients between HPT peptides. Each point in the heatmap represents the correlation coefficient between two peptides ranging from R = −0.5 (white) to R = 1 (red). (Upper) Effect of rs217184 on the abundance level of each peptide is illustrated. Each peptide is represented by the effect (beta) and the 95% confidence interval (CI) of the effect. The minor allele in rs217184 is associated with increased abundance levels for 15 peptides (P value nominal <0.05) and with decreased abundance levels for one peptide. Observations where the CI does not include zero represent nominally significant observations (P < 0.05). The color histogram (Upper Right) shows the distribution of correlation coefficients.
For a third group of proteins, several SNPs not in LD with each other, showed an association with the abundance of different peptides from the same protein. For example, a number of SNPs were associated with the abundance of different (nonoverlapping) peptides of the Alpha-1-antitrypsin (A1AT) protein encoded by the serpin peptidase inhibitor, clade A, member 1(SERPINA1) gene (Fig. S2). Of the three SNPs that showed the strongest association with A1AT peptides (Table 1), two (rs709932 and rs17090693) showed a high genetic correlation (R = 0.73), whereas the third (rs1243165) had a lower, but still significant, correlation (R = −0.31 between rs1243165 and rs17090693 and R = −0.21 between rs1243165 and rs709932) (P < 4.7 × 10−12 for all correlations). The Manhattan plot of P values across the SERPINA1 region (Fig. S1B) for the different peptides shows that the association pattern differs between SNPs. Both rs709932 and rs17090693 showed an effect on the same two peptides: IVDLVKELDRDTVFALVNYIFFK (Fig. 2B, P2) and TLNQPDSQLQLTTGNGLFLSEGLK (Fig. 2B, P3). When adjusting for the strongest association for each of these two peptides, no association was seen for the other SNP (P > 0.05). The third SNP, rs1243165, showed the strongest association to one peptide: DTEEEDFHVDQVTTVK (Fig. 2B, P1). This variation in association pattern between SNPs in SERPINA1 is not surprising because rs709932 and rs1243165 either directly change the amino acid of a peptide or disrupt the trypsin cleavage site (Table 1). rs17090693 on the other hand is not located in the region coding for the IVDLVKELDRDTVFALVNYIFFK peptide (or known to be in LD with). However, there is a variant form of the A1AT protein, which has the amino acid sequence QGKIVDLVK instead of GFQNAILVR at positions 190–198 (14). Because the IVDLVKELDRDTVFALVNYIFFK peptide is located at position 193–215 of the protein, it is possible that rs17090693 tags the underlying genetic variant that results in the aberrant A1AT protein. Similar to the pattern for HPT, the peptides of the proteins A1AT, FETUA, APOE, and CO3 also showed limited correlation within each protein (Fig. S3). Of the 5 proteins with a significant association, 4 overlap with an eQTL previously identified in monocytes (15). The only pQTL not overlapping with an eQTL is the AHSG gene (FETUA protein). This gene is predominantly expressed in liver and the transcripts might not be measurable in blood cells. Because our genetic data has been imputed using the 1000 Genomes reference panels, we have information on most of the SNPs reported as eQTL by Zeller et al. (15). We also tested if known eQTL were enriched for nominally significant (unadjusted P value <0.05) associations in our dataset. In the dataset by Zeller et al. (15), 48 of our 163 proteins were represented by an eQTL. From each of these eQTL, the top SNP was examined for association in our protein data. We found 7.0% of the top SNPs to be associated with a protein, compared with the 5.0% expected by chance (P = 0.18). These results do not indicate a significant enrichment of pQTL overlapping with previously reported eQTL.
In total, by applying a genome-wide significance threshold in KA06 and the requirement of replication in an independent sample (KA09), 11 peptides showed a significant association. However, the distribution of P values in the combined analyses (KA06 and KA09 together) indicate that altogether 60 peptides from 25 proteins of the 163 proteins detected (15%) are influenced by genetic variation acting in cis (FDR q value <0.05). To validate the MS values by an independent method, we used data for APOE levels determined by immunoassay for the same cohorts. The abundance measured by MS of the APOE peptide associated with rs429358 (Table 1) showed a significant correlation (R2 = 0.22, P value = 2.16 × 10−9) with the level of APOE determined by immunoassay, supporting the robustness of the quantitative MS data.
Discussion
Studies of the impact of genetic variability on the protein profile require access to high-throughput methods in both proteomics and genomics. So far, MS methods have been used mainly to quantify a selected number of proteins in a limited number of individuals or for comparison of the protein profile in pools of individuals. In this study, we have used high-resolution and high-throughput MS to quantify more than a thousand peptides in each of over 1,000 individuals from a population with a high degree of relatedness and a known genetic structure. This is not only one of the largest studies of the human plasma proteome, but also a unique study in assessing the association between cis-regulatory SNPs and the abundance of individual peptides. In contrast to most studies where measurements of peptide levels are combined into a single estimate for each protein, we considered the genetic association with abundance level of single peptides, because these may be dramatically influenced by nonsynonymous SNPs in individual peptides. By combining the abundance of peptide values from MS into a single protein value, the effect of amino acid changes on the peptide levels will be obscured and, as a consequence, it may be difficult (or impossible) to determine if an association between protein abundance and a genetic variant is due to a structural variant of the protein or a regulatory effect on transcription or translation. Indeed, for about 50% of the associations on a peptide level, we did identify a possible amino acid change. In addition, most genes give rise to a number of alternatively spliced transcripts, resulting in a set of proteins with different combinations of peptides and, finally, some peptides might map to more than one protein (e.g., proteins with identical or similar regions). Consequently, the analysis undertaken here provides the highest resolution of the plasma proteome.
Peptide amounts showed significant heritability for 35% of the proteins studied, indicating a surprisingly strong impact of genetic variability on the protein profile. The percentage of proteins showing significant heritability was lower than the estimated heritability of transcript levels (6), consistent with a lower correlation between the genetic profile and protein level compared with transcript level (9). Several of the peptides with the highest heritability were also found to show a significant genetic association. Interestingly, some peptides in the quartile with the highest heritability showed no genetic association, possibly due to a more complex genetic contribution. This indicates that additional genetic associations could be identified using a larger cohort size.
For 11 peptides, we identified and replicated SNP associations that influence the abundance. In 5 of these 11 peptides, the top SNP is either altering the amino acid sequence of the protein or is in strong LD with a nonsynonymous SNP (Table 1). The strongest association was seen for peptides in the A1AT protein. A recent study demonstrated that A1AT plays an important role in regulating metabolic pathways and that polymorphisms within the SERPINA1 gene (which encodes the A1AT protein) have been associated with atherosclerosis (16). Another set of strong associations were seen for HPT, where several SNPs were associated with abundance of a number of peptides. The strongest association was seen for three SNPs (rs217181, rs217184, and rs77303550) located far apart on chromosome 16 (rs217181 is at position 72114001, 19 kb downstream of the HP gene; rs217184 is at position 72105965, about 11 kb downstream of HP; and rs77303550 is at position 72079657, 9 kb upstream of HP), but in more or less complete LD with each other. These three SNPs identify a large LD block and could themselves, or another variant in high LD with these, be causal. Because these SNPs were associated with a number of peptides in HPT, it is likely that they reflect a genetic polymorphism that regulates the level of the protein, similar to previous studies of eQTL, where a large number of SNPs in the region have been associated with the transcription level of the HP gene (eQTL database: eqtl.uchicago.edu (Generic Genome Browser version 1.68). None of our top SNPs were analyzed in those studies, but the most significant SNP in the eQTL analyses by Zeller et al. (15) is rs6499560, an SNP that is also associated with the peptide levels in our data (P value = 2.92 × 10−13). Haptoglobin is an Alpha-2-glycoprotein that binds free hemoglobin and protects tissues from oxidative damage and has been associated with a large number of disease states, including cardiovascular diseases, diabetes, cancer, and persistence of various infections (17), as well as regulation of serum lipid levels (2, 18). Two different polymorphisms in the HP gene have been suggested to play an important role in protein activity. The first variant is characterized by a 1.7-kb duplication, resulting in a duplication of exon 3 and exon 4 and generating exon 5 and exon 6. The second polymorphism is two neighboring SNPs within the duplicated region (changing two consecutive amino acids). The latter polymorphism is the one measured by two of our most significant peptides in HPT (TEGDGVYTLNDKK and TEGDGVYTLNNEK). Unfortunately, neither our genotyped SNPs nor any of the SNPs in the reference panels used for imputations include the SNPs underlying these protein polymorphisms. The most common variant in Scandinavian populations carries the duplication polymorphism, where exons 3 and 4 encode the TEGDGVYTLNDKK peptide and exons 5 and 6 encode the TEGDGVYTLNNEK peptide (19).
The method we used for peptide quantification is well established and has been described in detail previously (20). What differentiates our quantification study from those previously performed using this methodology is the very large number of samples included. The fact that it is based on such a large series of samples makes it impossible to obtain quantitative estimates using an independent method. To address the robustness of the results using another method, we therefore verified the correlation between one APOE peptide detected by MS and for which we had quantitative protein data available based on an independent method (immunoassay). The results for this limited validation support the validity of the MS method for protein quantification.
Plasma contains a large number of proteins, and their concentrations span at least 10 orders of magnitude (21). This means that it is difficult to measure more than the 100 or so of the most abundant proteins without depletion of the top proteins or enrichment of low-abundant proteins and peptides from low-abundant proteins (22). We used a spectral library for identifying the peptides. Such libraries differ from sequence databases in the sense that they combine spectra identified by different methods, including semitryptic searches or even SNPs. Spectral libraries have recently been proposed (23) as an archiving method for storing and sharing observed peptides and their tandem mass spectra. The libraries used in spectral analyses are still smaller than searching all possible peptides, but do include common variants and peptides resulting from semitryptic cleavage. Consequently, a number of peptides that differ by semitryptic cleavage are included in our dataset. The handling and processing of plasma samples might affect the measurements, such as oxidation of some peptides during handling. However, samples were handled in random order and as consistently as possible using cooled solvent to minimize oxidation and sample-to-sample variability. The critical aspect is that there is no systematic bias due to oxidation or other modifications introduced by the sample handling and genotyping. However, it is important to realize that all these potential uncertainties influence the accuracy of the peptide quantification independently of the underlying cis-regulatory genetic variants and will consequently only reduce the power of identifying biologically significant associations rather than introduce false positive associations.
We compared our results with eQTL, showing that four of five of our associations were overlapping with a known eQTL, whereas previously known eQTL were not enriched for low P values in our dataset. This discrepancy is most likely due to the more cell-specific localization of mRNAs compared with proteins that are often released into the blood stream. However, protein levels are mainly regulated at the level of translation and the correlation between mRNA and protein concentrations is far from perfect (9). In addition, different protein isomers (caused by nsSNPs) might differ in their stability, causing differences in half-life and protein levels, independent of the transcriptional and translational regulation.
Our results demonstrate the potential of combining high-throughput methods in proteomics and genomics to understand the effect of genetic variability on the protein profile. This opens up the possibility for systematic studies of the functional importance of specific genetic variants on the protein. An evaluation of the contribution of genetic variability on the protein profile may also be important for specific proteins selected as biomarkers for disease prediction, because genetic factors may affect the risk estimate in an individual-specific manner. The ability to detect single amino acid changes should be of great interest in biomarker research. In our data, we detected variants that have previously been associated with disease in man. For instance, rs1047286 showing an association with peptide levels in the C3 gene, has previously been associated with increased risk of age-related macular degeneration (24). The peptide with an amino acid change caused by this polymorphism was detected in our data (Table 1). Similarly, the SNP rs429358 identified in our data is located in the APOE gene and tags the ɛ4 variant, a variant associated with increased risk of Alzheimer's disease (25). Further developments of the methodology for MS proteome analyses to include a larger number of peptides and peptides with lower abundance levels, would make it possible to cover more of the human plasma proteome and more precise quantitation. Also, the SNP array data should be complemented by sequence data to also include rare variants and structural variation. Nevertheless, the results of our study demonstrate the presence of genetic variants with a strong impact on the protein profile and the ability to annotate their functional relevance in affecting the level of gene products.
Methods
Clinical Materials.
The Northern Sweden Population Health Study (NSPHS) was initiated in 2006 to provide a health survey of the population in the parish of Karesuando, county of Norrbotten, Sweden, and to study the medical consequences of lifestyle and genetics. This parish has about 1,500 inhabitants who meet the eligibility criteria in terms of age (≥15 y), of which 719 individuals participated in the study (KA06 cohort). As a second phase of the NSPHS, another 350 individuals from a neighboring village (Soppero) were recruited in 2009 (KA09 cohort). For each participant in the NSPHS, blood samples were taken (serum and plasma) and stored at −70 °C. DNA has been extracted for genetic analyses. APOE levels for verification purposes have been measured previously using multiplex immunoassay.
Ethical Considerations.
The NSPHS study was approved by the local ethics committee at the University of Uppsala (Regionala Etikprövningsnämnden, Uppsala, 2005:325) in compliance with the Declaration of Helsinki (26). All participants gave their written informed consent to the study including the examination of environmental and genetic causes of disease. In cases where the participant was not of age, a legal guardian signed additionally. The procedure that was used to obtain informed consent and the respective informed consent form have recently been discussed in light of present ethical guidelines (27).
Plasma Protein Digestion.
Aliquots of 5 μL of plasma from 1,060 individuals, diluted to 20 μL in 50 mM ammonium bicarbonate (NH4HCO3) were centrifuged 16,000 × g for 10 min in 5 °C. From the supernatant, 16 μL was digested following the previously published protocol (28). Briefly the plasma samples were transferred into a 96-well plate. A volume of 10 μL of 45 mM aqueous dithiothreitol was added to all samples and the mixtures were incubated at 50 °C for 15 min to reduce the disulfide bridges. The samples were cooled down to room temperature and 10 μL of 100 mM aqueous iodoacetamide was added and the mixtures were incubated for an additional 15 min at room temperature in darkness to carabamidomethylate the cysteines. Finally, a volume of 10 µL of 50 mM NH4HCO3 was added together with trypsin to yield a final trypsin/protein concentration of 0.8% (wt/wt). The tryptic digestion was performed at 37 °C overnight in darkness for 12 h. The digestion reaction was quenched by addition of 5 µL of 10% (vol/vol) trifluoroacetic acid. The samples were then centrifuged for 20 min to spin down undigested material and stored at −80 °C before capillary liquid chromatographic MS/MS analysis.
MS.
The plasma protein digests were analyzed using an Fourier transform ion cyclotron resonance (FTICR)-ion trap cluster (20). First, 2 µL of a reference plasma tryptic digest was loaded and desalted on a PepMap C18 trap column (5 mm, 300 mm i.d.; Dionex), separated by a 150-min reversed-phase chromatographic gradient from 4% to 33% (vol/vol) acetonitrile in 0.05% formic acid and a constant flow rate of 4 mL/min using a ChromXP C18 column (15 cm, 300 µm i.d.; Eksigent) connected to a splitless NanoLC-Ultra 2D Plus system (Eksigent) and analyzed by tandem MS (collision-induced dissociation) in an amaZon speed ion trap (Bruker Daltonics) to identify peptides and measure their elution times. Each individual sample was then separately measured on a 12 T solariX FTICR mass spectrometer (Bruker) and a shorter (30 min) but otherwise identical gradient and chromatographic system to quantify the peptides identified in the ion trap. Quantitative information from FTICR-MS was extracted for all peptides identified. The data were searched against the 2011 National Institute of Standards and Technology (NIST) human ion trap spectral library (http://peptide.nist.gov) using SpectraST (23) with default settings, allowing a mass measurement error of 2.5 Da and assuming all cysteines were carbamidomethylated. All peptide values were normalized as described in SI Methods.
SNP Genotyping and Imputation of Genome-Wide Data.
DNA samples were genotyped according to the manufacturer's instructions on Illumina Infinium HumanHap300v2 (n = 700) of Illumina Omni Express (n = 350) SNP bead microarrays. Analyses of genotype raw data and quality control (QC) were performed using the GenABEL package (29) and are described in SI Methods and Fig. S4. A total of 1,032 individuals passed the QC of which 1,029 were also included in the protein quantification. Genotype data were imputed with a prephasing approach using IMPUTE (version 2.2.2) (30) in KA06 and KA09 separately, using the 1000 Genomes Phase I integrated variant set (National Center for Biotechnology Information, build b37, March 2012) (31) as reference panel as described in SI Methods). After QC of imputed data 7.83 M and 8.78 M SNPs remained in KA06 and KA09, respectively. For comparing our results to previously reported eQTL, we downloaded lymphocyte eQTL by Zeller et al. (15) from eQTL database (http://eqtl.uchicago.edu/Home.html). The coordinates for this dataset were lifted from HG18 to HG19 using the University of California Santa Cruz liftOver tool (32) and matched to our imputed data by position.
Statistical Analyses.
Correlation between peptides and covariates were performed using a linear regression model implemented in the function glm in the stats library in R. FDRs were estimated using the fdrtool function implemented in the fdrtool R library (33). Because the NSPHS is a population-based study including related individuals, special care was taken to avoid bias due to relatedness. All association analyses were performed using the R package GenABEL or ProbABEL (29), which has been developed to enable statistical analyses of genetic data of related individuals. It includes functions for measuring correlation coefficients between variables among related individuals (using a linear mixed-effects model), estimating the heritability (using a polygenic model) and performing genetic association analyses (34) by adjusting for pedigree structure using the kinship matrix. Before association analyses, the values for each peptide were adjusted for age and BMI and the residuals were transformed using the rank-based inverse normal transformation. Only peptides that were detected in more than 400 individuals were analyzed. To identify cis-pQTL, a region of 100 kb upstream and 100 kb downstream of the gene coding for the protein was studied, resulting in an average of 819 SNPs (ranging from 291 to 7,671) analyzed for each protein. To be able to replicate our findings, we used the KA06 cohort as a discovery cohort and KA09 for replication. For each peptide, the significance for each SNP was adjusted for using Bonferroni correction for multiple testing with respect to the number of SNPs tested per peptide (but not the total number of peptides tested). The top SNP from each peptide with Bonferroni adjusted significant P value was then evaluated in KA09. The significance threshold in the replication phase was corrected for multiple testing using the Bonferroni correction both with respect to the number of SNPs and the number of peptides tested.
Supplementary Material
Acknowledgments
We thank district nurse Svea Hennix for data collection, Inger Jonasson for logistics and coordination of the health survey, Bo Ek for sample preparation, and all the participants from the community for their interest and willingness to contribute to this study. The NSPHS was funded by the Swedish Medical Research Council (Project K2007-66X-20270-01-3), and the Foundation for Strategic Research. NSPHS, as part of European Special Populations Research Network, was also supported by European Commission Sixth Framework Programme Specific Targeted Research Projects Grant 01947 (LSHG-CT-2006-01947). This work has also been supported by the Swedish Research Council (Grant 621-2011-4423), the Science for Life Laboratory, Uppsala, and the Swedish Society for Medical Research (SSMF). Part of the genotyping was performed by the SNP&SEQ Technology Platform in Uppsala, which is supported by Uppsala University, Uppsala University Hospital, Science for Life Laboratory, Uppsala, and the Swedish Research Council (Contracts 80576801 and 70374401). The computations were performed on resources provided by Swedish National Infrastructure for Computing (SNIC) through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project p2011202.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217238110/-/DCSupplemental.
References
- 1.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464(7289):768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12(10):683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 5.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39(10):1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 6.Göring HH, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39(10):1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 7.Illig T, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42(2):137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suhre K, et al. A genome-wide association study of metabolic traits in human urine. Nat Genet. 2011;43(6):565–569. doi: 10.1038/ng.837. [DOI] [PubMed] [Google Scholar]
- 9.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 10.Melzer D, et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4(5):e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garge N, et al. Identification of quantitative trait loci underlying proteome variation in human lymphoblastoid cells. Mol Cell Proteomics. 2010;9(7):1383–1399. doi: 10.1074/mcp.M900378-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lourdusamy A, et al. AddNeuroMed Consortium Alzheimer’s Disease Neuroimaging Initiative Identification of cis-regulatory variation influencing protein abundance levels in human plasma. Hum Mol Genet. 2012;21(16):3719–3726. doi: 10.1093/hmg/dds186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kersey PJ, et al. The International Protein Index: An integrated database for proteomics experiments. Proteomics. 2004;4(7):1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 14. The UniProt Consortium (2012) Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 40(Database issue):D71–5. [DOI] [PMC free article] [PubMed]
- 15.Zeller T, et al. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS ONE. 2010;5(5):e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inouye M, et al. Novel loci for metabolic networks and multi-tissue expression studies reveal genes for atherosclerosis. PLoS Genet. 2012;8(8):e1002907. doi: 10.1371/journal.pgen.1002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter K, Worwood M. Haptoglobin: A review of the major allele frequencies worldwide and their association with diseases. Int J Lab Hematol. 2007;29(2):92–110. doi: 10.1111/j.1751-553X.2007.00898.x. [DOI] [PubMed] [Google Scholar]
- 18.Igl W, et al. EUROSPAN Consortium Modeling of environmental effects in genome-wide association studies identifies SLC2A2 and HP as novel loci influencing serum cholesterol levels. PLoS Genet. 2010;6(1):e1000798. doi: 10.1371/journal.pgen.1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teige B, Olaisen B, Teisberg P. Haptoglobin subtypes in Norway and a review of HP subtypes in various populations. Hum Hered. 1992;42(2):93–106. doi: 10.1159/000154048. [DOI] [PubMed] [Google Scholar]
- 20.Palmblad M, van der Burgt YE, Mostovenko E, Dalebout H, Deelder AM. A novel mass spectrometry cluster for high-throughput quantitative proteomics. J Am Soc Mass Spectrom. 2010;21(6):1002–1011. doi: 10.1016/j.jasms.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Anderson NL, Anderson NG. The human plasma proteome: History, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 22.Anderson NL, et al. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3(2):235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 23.Lam H. 2011. Building and searching tandem mass spectral libraries for peptide identification. Mol Cell Proteomics: 10(12):R111. 008565.
- 24.Thakkinstian A, et al. Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: A HuGE review and meta-analysis. Am J Epidemiol. 2011;173(12):1365–1379. doi: 10.1093/aje/kwr025. [DOI] [PubMed] [Google Scholar]
- 25.Zuo L, et al. Variation at APOE and STH loci and Alzheimer’s disease. Behav Brain Funct. 2006;2:13. doi: 10.1186/1744-9081-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anonymous World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA. 2000;284(23):3043–3045. [PubMed] [Google Scholar]
- 27.Mascalzoni D, et al. EUROSPAN consortium Comparison of participant information and informed consent forms of five European studies in genetic isolated populations. Eur J Hum Genet. 2010;18(3):296–302. doi: 10.1038/ejhg.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergquist J, Palmblad M, Wetterhall M, Håkansson P, Markides KE. Peptide mapping of proteins in human body fluids using electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Mass Spectrom Rev. 2002;21(1):2–15. doi: 10.1002/mas.10016. [DOI] [PubMed] [Google Scholar]
- 29.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: An R library for genome-wide association analysis. Bioinformatics. 2007;23(10):1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 30.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abecasis GR, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hinrichs AS, et al. The UCSC Genome Browser Database: Update 2006. Nucleic Acids Res. 2006;34(Database issue):D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strimmer K. fdrtool: A versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24(12):1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- 34.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81(5):913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.