Abstract
The mammalian circadian clock is composed of interlocking feedback loops. Cryptochrome is a central component in the core negative feedback loop, whereas Rev-Erbα, a member of the nuclear receptor family, is an essential component of the interlocking loop. To understand the roles of different clock genes, we conducted a genetic interaction screen by generating single- and double-mutant mice. We found that the deletion of Rev-erbα in F-box/leucine rich-repeat protein (Fbxl3)-deficient mice rescued its long-circadian period phenotype, and our results further revealed that FBXL3 regulates Rev-Erb/retinoic acid receptor-related orphan receptor-binding element (RRE)-mediated transcription by inactivating the Rev-Erbα:histone deacetylase 3 corepressor complex. By analyzing the Fbxl3 and Cryptochrome 1 double-mutant mice, we found that FBXL3 also regulates the amplitudes of E-box–driven gene expression. These two separate roles of FBXL3 in circadian feedback loops provide a mechanism that contributes to the period determination and robustness of the clock.
Circadian rhythms control many physiological processes in almost all eukaryotic organisms (1–6). The current mammalian clock model comprises a core negative feedback loop that includes the Per-Arnt-Sim (PAS) domain-containing helix–loop–helix transcription factors Clock and Bmal1, Period genes (Per1, Per2, and Per3), and Cryptochrome genes (Cry1 and Cry2). The CLOCK:BMAL1 complex activates the transcription of the Period and Cryptochrome genes by binding to E-boxes in their promoters, whereas the PER:CRY complex closes the negative feedback loop by repressing the activity of CLOCK:BMAL1, resulting in endogenous circadian oscillations of Per and Cry mRNA (3, 5, 7). The nuclear receptors Rev-Erbα and RORα are components of another feedback loop that is interlocked with the core negative loop. These receptors function by competitively binding to the Rev-Erb/ROR-binding element (RRE) of Bmal1 to regulate its rhythmic transcription (8–10).
Mutation of FBXL3 (C358S or I364T), a component of a SKP1–CUL1–F-box (SCF) E3 ubiquitin ligase complex, results in ∼26-h-period phenotypes in mice, indicating that FBXL3 plays an important role in circadian period determination (11–13). Previous studies showed that FBXL3 interacts with CRY1 and CRY2, promoting the degradation of both these proteins by the ubiquitin/proteasome system, thus contributing to period length determination (11–13). However, overexpression of CRY1 protein does not lead to period alteration (14), suggesting that the Fbxl3 mutation might affect additional clock components.
To gain further insights into the mammalian clock network, we studied the genetic interactions between different clock genes in the mouse. Our screens revealed an unexpected genetic interaction between Rev-erbα and Fbxl3. Further biochemical analysis showed that FBXL3 regulates the Rev-Erbα–dependent histone deacetylase 3 (HDAC3) repressor complex, suggesting that the action of FBXL3 on Rev-Erbα:HDAC3 is crucial for clock function. Our study further suggested that FBXL3 plays an important role in coordinating E-box and RRE-mediated transcription to modulate the circadian period and robustness of the clock.
Results
Fbxl3 Genetically Interacts with Rev-erbα.
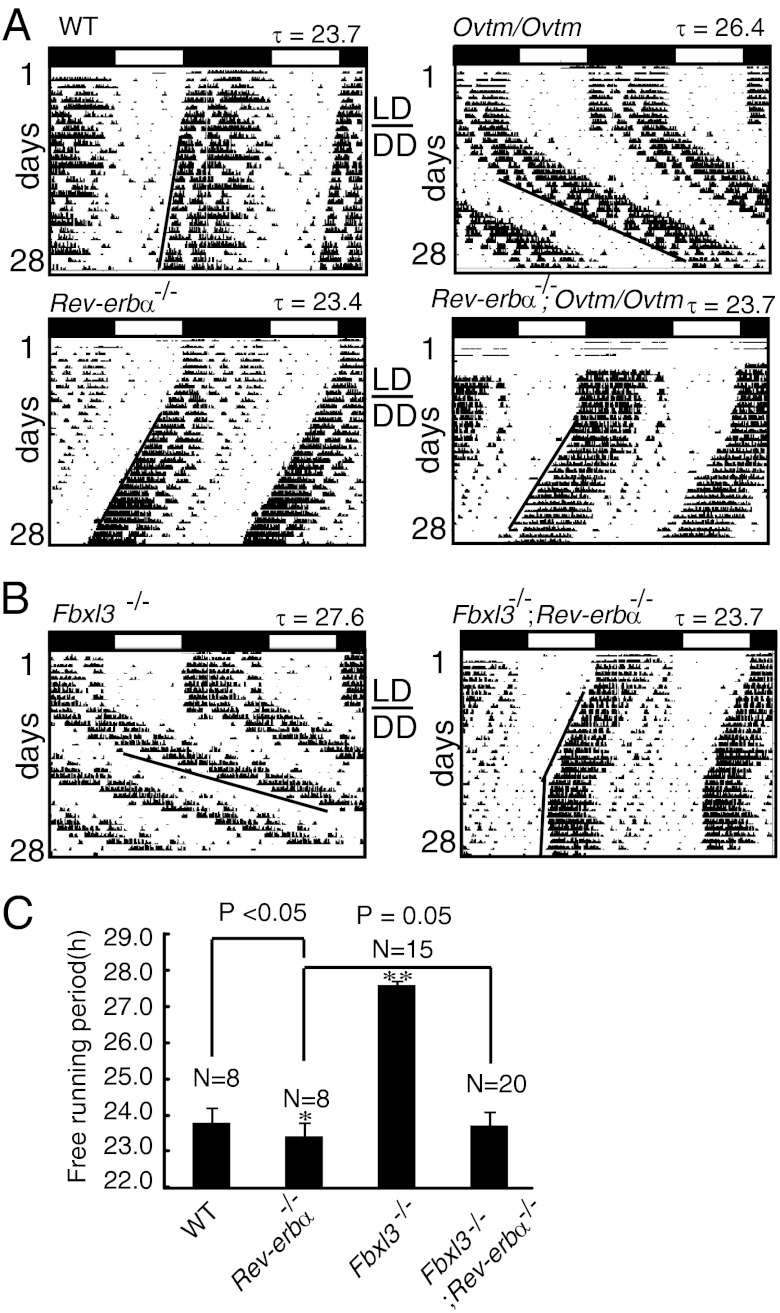
To investigate the genetic interactions between different clock genes, we obtained Clock mutant mice (15), Bmal1-knockout mice (16), PER2S662G and PER2S662D mice (17), Fbxl3 mutant Overtime (Ovtm) mice (11), and Rev-erbα–knockout mice (8), and generated Cry1- and Cry2-knockout mice by homologous recombination as described previously (18, 19). Then, a series of double-mutant mice were generated, and their circadian locomotor activities were examined. We used the formula τpredicted = τx + τy − τwild type (where τx and τy represent the τ of the single-mutant mice) to predict the τ of the double-mutant mice and determine whether there was genetic interplay between the different mutations. Using this criterion, most of these clock genes did not appear to have strong genetic interactions (Fig. S1). The period lengths for Ovtm/Ovtm (11) and Rev-erbα−/− mutant mice (8) were 26.4 ± 0.1 h and 23.4 ± 0.4 h, respectively. Interestingly, we found that the Ovtm/Ovtm;Rev-erbα−/− double-mutant mice exhibited a period close to that of the Rev-erbα single-mutant mice (23.8 ± 0.4 h) (Fig. 1A), suggesting that the long-period phenotype caused by the Ovtm mutation was rescued in the Rev-erbα-null background.
Fig. 1.
Representative actograms of locomotor activity in established mice across the circadian day. Mice were entrained in light/dark 12:12 (LD) for 7–10 d and then released into constant darkness (DD) thereafter. LD-to-DD transition is indicated by a horizontal bar. Black lines represent the phase of activity onset in DD. (A) Wild-type (WT) (n = 8), Ovtm/Ovtm (n = 12), Rev-erbα−/− (n = 8), and Rev-erbα−/−;Ovtm/Ovtm (n = 8). (B) Fbxl3−/− (n = 15) and Fbxl3−/−;Rev-erbα−/− (n = 20). The period was calculated between days 8 and 21 in DD. (C) Period-length quantification of C57BL/6J-backcrossed wild type (WT), Rev-erbα−/−, Fbxl3−/−, and Fbxl3−/−;Rev-erbα−/− mice. Bars indicate the mean ± SD. Statistical significance was set at P < 0.05, as determined by one-way ANOVA followed by Tukey’s post hoc test. *P < 0.05, **P < 0.01.
The Ovtm/Ovtm mice carry mutant alleles of the Fbxl3 gene (11). To fully understand the role of Fbxl3, we generated conditional Fbxl3-knockout (Fbxl3−/−) mice (Fig. S2) and Fbxl3-null mice were developed by breeding with transgenic mice carrying the Cre-recombinase gene under the control of the adenovirus EIIa promoter. The heterozygous Fbxl3+/− mice exhibited a period length of 24.42 h ± 0.1 h, which was 0.7 h longer than that of their wild-type littermates. The homozygous Fbxl3−/− mice exhibited a period of 27.60 ± 0.1 h, which was 1 h longer than that of the Ovtm or Afterhour mice (Fig. 1B) (11, 12). We then compared the levels of FBXL3 proteins in the wild-type and Ovtm/Ovtm liver tissues and found dramatically reduced levels of the mutant FBXL3 proteins (Fig. S3), suggesting that FBXL3 abundance is critical for period length in vivo. As expected, the Fbxl3−/−;Rev-erbα −/− double-mutant mice also exhibited a period near that of Rev-erbα−/− mice (23.7 ± 0.4 h, n = 20 vs. 23.4 ± 0.4 h, n = 8, P = 0.05) (Fig. 1 B and C), even though the Fbxl3-knockout mice had an even longer period than that of the Ovtm/Ovtm mice. These results suggest that the deletion of Rev-erbα masks the defects of Fbxl3 knockout and that Rev-Erbα lies downstream of FBXL3 in a common circadian pathway. Of note, the Fbxl3−/−;Rev-erbα−/− mice showed an unstable onset of activity compared with the onset of activity in the wild-type mice (Fig. 1B and Fig. S4), and 10% (2/20) of the double-mutant mice became arrhythmic after 2–3 wk in constant darkness (DD) (Fig. S4). These observations indicated that these two clock proteins play important roles in regulating the robustness and stability of the clock.
Deletion of Rev-erbα Does Not Rescue Impaired CRY Degradation in Fbxl3−/− Mice.
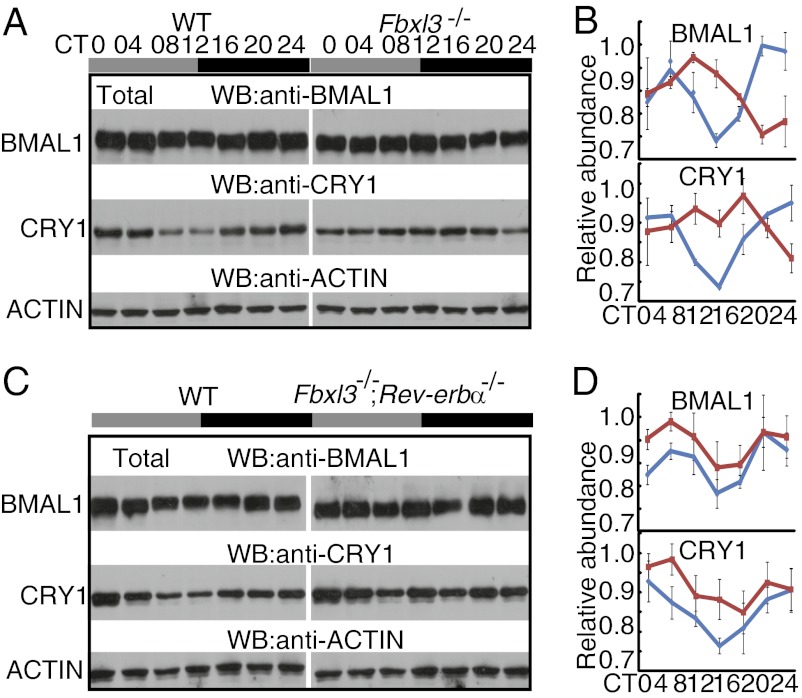
Previous studies have revealed that FBXL3 regulates the stability of CRYs, and the increased accumulation of CRYs in Fbxl3 mutant mice could potentially explain their long-period phenotype (11–13). We therefore asked whether the rescue of the circadian period in the Fbxl3−/−;Rev-erbα−/− mice was due to reduction of CRY levels to near wild-type levels. To examine the levels of different clock proteins, we generated antibodies against the mouse BMAL1, CRY1, and FBXL3 proteins. These antibodies strongly detect the target proteins (Fig. S5). The total level of CRY1 protein was modestly elevated in Fbxl3−/− mice compared with wild-type mice as described previously (11) (Fig. 2 A and B). The levels of both BMAL1 and CRY1 proteins were significantly increased in the Fbxl3−/−;Rev-erbα−/− double-mutant mice (Fig. 2 C and D), indicating that the rescue of the circadian period in Fbxl3−/− mice via the deletion of Rev-erbα was not due to a reduction in CRY1 abundance.
Fig. 2.
Impaired CRY1 and BMAL1 accumulation in Fbxl3-/− and Fbxl3−/−; Rev-erbα−/− liver tissues. (A and C) Protein oscillation profiles of BMAL1 and CRY1 in total extracts from wild-type and Fbxl3−/− (A) and wild-type and Fbxl3−/−;Rev-erbα−/− liver tissues (C). Samples and blots were processed in parallel in paired experiments and exposed on the same film. Each figure shows a representative example from three independent experiments. All tissues were collected at 4-h intervals during the first day in constant darkness (thereafter). Gray bars represent subjective day, and black bars represent subjective night. Actin served as the loading control (B and D). Quantification of BMAL1 and CRY1 in the total extracts from the wild-type (blue line) and Fbxl3−/− (red line) (B) and wild-type (blue line) and Fbxl3−/−;Rev-erbα−/− (red line) liver tissues (D) using National Institutes of Health ImageJ. Data were taken from three independent experiments. Bars indicate the mean ± SD. Two-way ANOVA showed significant statistical differences between WT and Fbxl3−/− mice for CRY1 (P < 0.01) and between WT and Fbxl3−/−;Rev-erbα−/− mice for BMAL1 (P < 0.01) and CRY1 (P < 0.05) in liver tissues.
Then, we examined the binding of CRY1 protein to the E-box regions of Per2 and Cry1 over a circadian cycle in liver tissues using chromatin immunoprecipitation (ChIP) assays. In wild-type tissues, the binding of CRY1 proteins to both the E-box regions exhibited circadian rhythmicity. However, CRY1 binding to the E-box region from circadian time (CT)12 to CT20 was considerably higher in Fbxl3−/− mice compared with their wild-type littermates (Fig. 3A). Furthermore, the immunoprecipitation assays showed that there was little interaction between CRY1 and FBXL3 from CT12 to CT20 in wild-type tissues (Fig. 3B), likely due to FBXL3-mediated CRY1 degradation. Again, the level of CRY1 binding to the E-box regions in the Fbxl3−/−;Rev-erbα−/− double-mutant mice was similar to that in Fbxl3−/− mice (Fig. 3A), showing that the rescued period length in Fbxl3−/−;Rev-erbα−/− mice was not due to reductions in CRY protein levels or its reduced binding to the E-box regions. To determine the activity of CRY1 on the E-box, we examined the binding of RNA polymerase II (RNAPII) to the Per1 and Per2 promoters to assay for dynamic changes of transcriptional activity (20). Consistent with CRY1 accumulation, the RNAPII binding signals were substantially lower in Fbxl3−/−;Rev-erbα−/− and Fbxl3−/− liver tissues (Fig. S6).
Fig. 3.
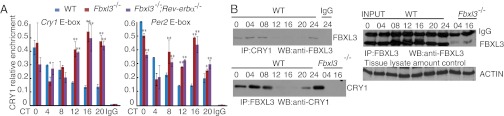
Altered CRY1 binding to the E-box regions in Fbxl3-/−and Fbxl3−/−;Rev-erbα−/− liver tissues. (A) CRY1 binding to the E-box regions of the Cry1 and Per2 promoter/enhancer loci across a circadian cycle (mean ± SD, n = 3). IgG served as an internal control. RRE site in the first intron of Cry1 from wild-type samples served as the negative control locus (data not shown). Chromatin samples from the livers of wild-type (blue), Fbxl3−/− (red) and Fbxl3−/−;Rev-erbα−/− (purple) mice were analyzed by ChIP (each time point represents an average of three to four mice for each genotype). Two-way ANOVA demonstrated significant statistical differences between the WT and Fbxl3−/− and WT and Fbxl3−/−;Rev-erbα−/− liver tissues for CRY1 binding. Differences between the Fbxl3−/− and Fbxl3−/−;Rev-erbα−/− were subtle relative to the other groups. *P < 0.05, **P < 0.01. (B) Interaction between FBXL3 and CRY1 exhibits circadian oscillation. Liver extracts were IPed with anti-CRY1 or anti-FBXL3, and IgG or Fbxl3 knockout samples were used as negative controls. Immune complexes were Western blotted using anti-FBXL3 or anti-CRY1 antibodies. CRY1 profiles in wild-type tissues (Fig. 2A, Middle) and FBXL3 profile (Fig. 3B) served as input. Actin served as the tissue lysate control.
Dampened E-box–Driven Gene Expression in the Fbxl3−/−;Rev-erbα−/− Mice Are CRY1 Dependent.
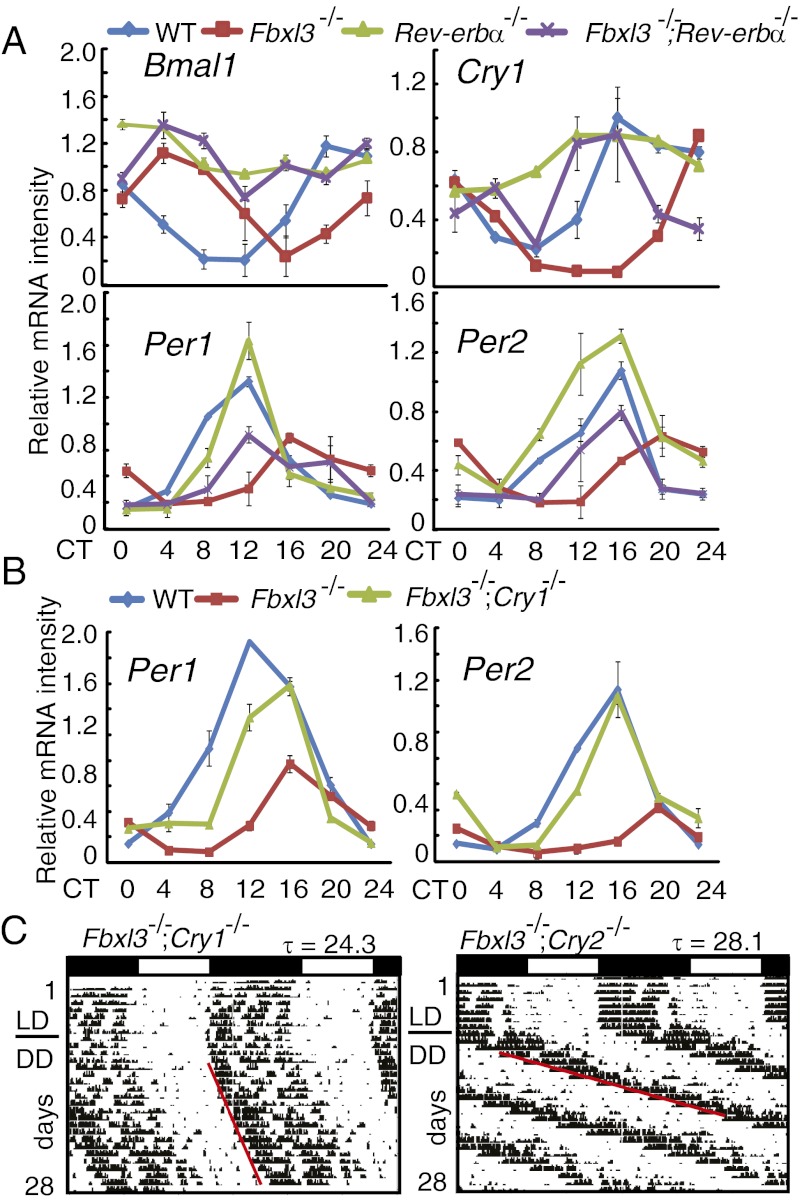
It was previously reported that the rhythms of circadian gene transcripts are severely dampened in the Fbxl3 mutant mice (11). We examined the mRNA levels of Bmal1, Cry1, Per1, and Per2 over a circadian cycle in these mutant liver tissues (Fig. 4A) and found near-constant and high Bmal1 expression levels in Fbxl3−/−;Rev-erbα−/− mice and similar levels in Rev-erbα−/− liver tissues. In addition, the phase of the Cry1 mRNA rhythm was also dramatically advanced in Fbxl3−/−;Rev-erbα−/− mice compared with the Fbxl3−/− mice. Furthermore, the amplitudes of the Per1 and Per2 mRNAs were attenuated to a similar extent in Fbxl3−/−;Rev-erbα−/− and Fbxl3−/− liver (Fig. 4A). These results were consistent with the similar enhancement of CRY1 binding to the E-box regions in both mutant mice (Fig. 3A), suggesting that the enhanced suppression of E-box–driven transcription in the Fbxl3−/− mice was not rescued in the Fbxl3−/−;Rev-erbα−/− double-mutant mice.
Fig. 4.
Deletions of Rev-erbα and Cry1 have different effects on circadian gene expression and locomotor in the Fbxl3-deficient background. (A) Expression of the Bmal1, Cry1, Per2, and Per1 clock genes in liver tissues of wild-type (blue), Fbxl3−/− (red), Rev-erbα−/− (green), and Fbxl3−/−;Rev-erbα−/− (purple) mice were measured by quantitative RT-PCR and normalized to Gapdh. Expression levels were plotted as arbitrary units. Each value represents the mean ± SD (n = 3). Two-way ANOVA demonstrated significant statistical differences among the WT, Fbxl3−/−, and Fbxl3−/−;Rev-erbα−/− liver tissues for the expression of Bmal1, Cry1, Per1, and Per2 (P < 0.01). (B) mRNA levels of Per1 and Per2 in the livers of wild-type (blue), Fbxl3−/− (red), and Fbxl3−/−;Cry1−/− (green) mice were measured by quantitative RT-PCR. Two-way ANOVA demonstrated significant statistical differences among the WT, Fbxl3−/−, and Fbxl3−/−;Cry1−/− liver tissues for the expression of Per1 and Per2 (P < 0.01). (C) Representative actograms from Fbxl3−/−;Cry1−/− (n = 8) and Fbxl3−/−;Cry2−/− (n = 6) mice.
To determine whether the effects of FBXL3 on the E-box–mediated transcriptional inhibition were dependent on the CRY proteins, we generated Fbxl3−/−;Cry1−/− double-mutant mice and profiled the mRNA levels of Per1 and Per2 in the livers. Unlike the Fbxl3−/− single-mutant mice and the Fbxl3−/−;Rev-erbα−/− mice, the circadian amplitudes of the E-box–driven transcripts in the Fbxl3−/−;Cry1−/− liver tissues were all restored to levels comparable with those in wild-type tissues (Fig. 4B). These results suggest that the role of FBXL3 in regulating the amplitude of circadian clock is mainly through its action on CRY1.
To further evaluate the contribution of CRYs to the long-period phenotype of the Fbxl3−/− mice, we determined the locomotor activities of the Fbxl3−/−;Cry1−/− and Fbxl3−/−;Cry2−/− double-mutant mice. Consistent with a previous report (18, 19), Cry1−/− mice displayed a period length of 22.5 ± 0.3 h, whereas the Cry2−/− mice had a period of 24.5 ± 0.3 h. The period lengths were predicated to be ∼26.4 h and ∼28.4 h for the Fbxl3−/−;Cry1−/− and Fbxl3−/−;Cry2−/− double-mutant mice, respectively, if there are no genetic interactions between FBXL3 and the CRYs. The observed period for the Fbxl3−/−;Cry2−/− double-mutant mice (28.2 ± 0.1 h) was very close to the predicted period, suggesting a lack of genetic interaction for the period determination between these two genes (Fig. 4C). However, the Fbxl3−/−;Cry1−/− double-mutant mice had a 24.3 ± 0.4 h period (Fig. 4C), suggesting that the long-period phenotype of the Fbxl3 mutant was only partially dependent on Cry1 (Cry1−/− mice: 22.5 ± 0.3 h).
Fbxl3 Deficiency Is Associated with Retention of HDAC3 Complexes at RRE Sites.
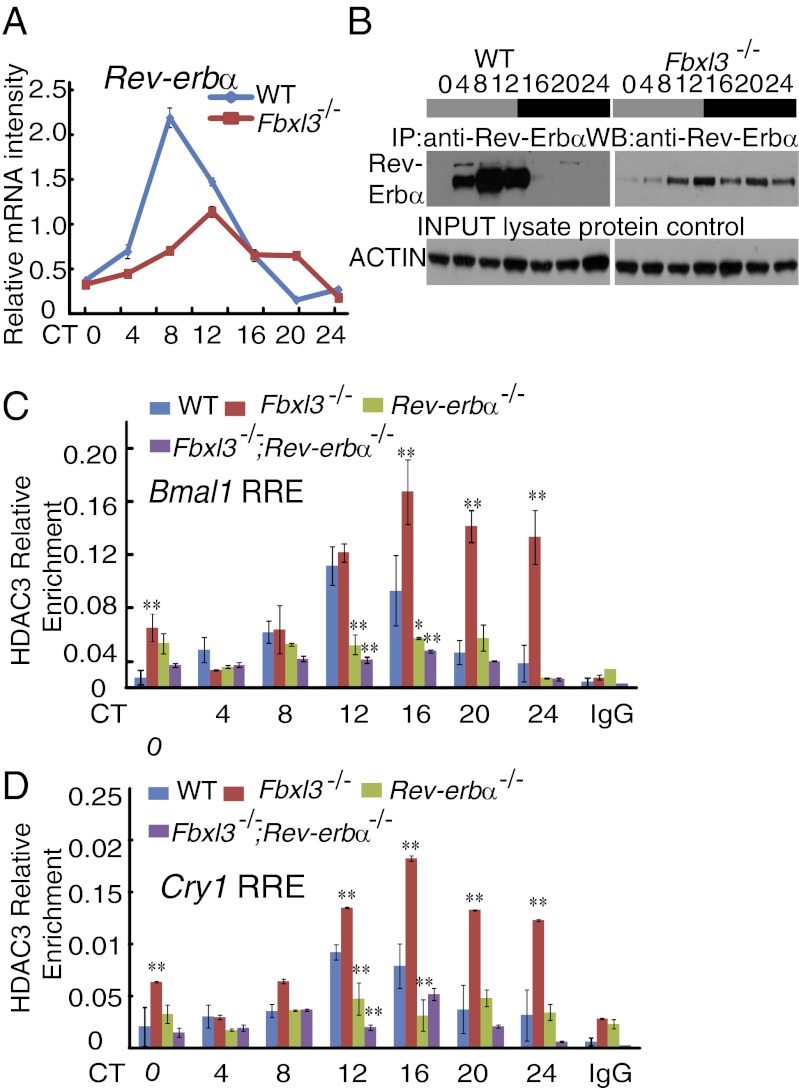
The rescue of the Fbxl3−/− circadian period length by the deletion of Rev-Erbα suggested that Rev-Erbα may act downstream of FBXL3 (21). We thus examined the Rev-erbα mRNA and the Rev-Erbα protein profiles in the Fbxl3−/− liver tissues. In agreement with previous observations (8, 22), the levels of Rev-erbα mRNA and Rev-Erbα protein exhibited robust circadian rhythms in wild-type tissues (Fig. 5 A and B). We observed reduced levels of Rev-erbα mRNA in Fbxl3−/− mice. Correspondingly, the peak abundance of Rev-Erbα protein were also reduced; however, the protein accumulation pattern was remarkable altered as Rev-Erbα protein could be detected at the points when it was not present under normal circumstances (Fig. 5B), indicating that the loss of Fbxl3 may impair the clearance of the Rev-Erbα protein.
Fig. 5.
Impaired HDAC3 clearances from the RRE sites in Fbxl3−/− mice. (A) Quantitative analysis of the Rev-Erbα mRNA levels in the livers of wild-type and Fbxl3−/− mice. Relative levels of RNA were estimated by quantitative RT-PCR and normalized to Gapdh. Data represent the mean ± SD (n = 3). (B) Representative circadian profiles of Rev-Erbα protein in the liver of wild-type and Fbxl3−/− mice. Rev-Erbα protein was detected with an anti–Rev-Erbα antibody after IP with anti–Rev-Erbα serum. No signal after prolonged exposure could be detected from CT16 to CT24 in wild-type liver tissues. Actin served as control. (C and D) Altered HDAC3 binding to the RRE regions in the Bmal1 (C) and Cry1 promoter/enhancer locus (D) (mean ± SD, n = 3). Each figure shows a representative example from three independent experiments. Two-way ANOVA demonstrated significant statistical differences between the WT and Fbxl3−/−, WT and Fbxl3−/−;Rev-erbα−/−, and Fbxl3−/− and Fbxl3−/−;Rev-erbα−/− liver tissues for HDAC3 binding. Notably, the differences in HDAC3 binding between Rev-erbα−/− and Fbxl3−/−;Rev-erbα−/− samples were abolished (P > 0.05). *P < 0.05, **P < 0.01.
Rev-Erbα was shown to recruit HDAC3-containing complexes to RREs to repress transcription (23, 24). The impaired clearance of Rev-Erbα in the Fbxl3−/− mice prompted us to investigate the recruitment of HDAC3 to the RREs by ChIP. As shown in Fig. 5C, the binding of HDAC3 to the Bmal1 RRE showed a robust rhythm in wild-type liver samples, with peak levels occurring from CT8 to CT16. In the Fbxl3−/− mice, however, HDAC3 binding persisted at high levels in this region from CT12 to CT24 (Fig. 5C). Similar results were observed for the RRE of Cry1 (Fig. 5D), suggesting that the RREs of both Cry1 and Bmal1 are regulated by a similar mechanism. Moreover, HDAC3 binding was virtually absent from the RRE regions in either the Rev-erbα−/− or Fbxl3−/−; Rev-erbα−/− mice (Fig. 5 C and D), consistent with the observation that Rev-Erbα was required for recruitment of the repressor complex at these RRE loci (24). These data suggested that loss of Fbxl3 leads to impaired clearance of Rev-Erbα and retention of HDAC3 at the RREs.
FBXL3 Regulates the Rev-Erbα:HDAC3 Mediated Suppression.
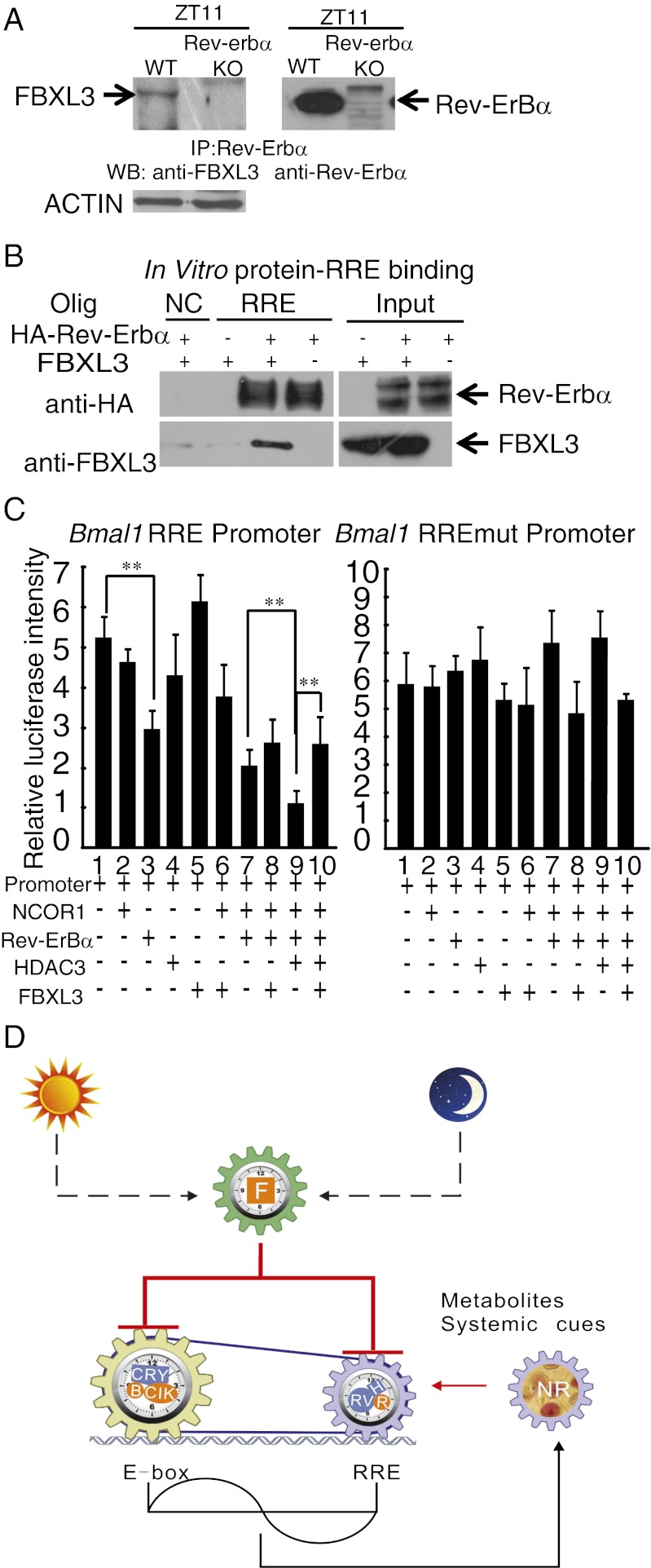
The above results suggested that the presence of FBXL3 may regulate Rev-Erbα:HDAC3 activity on the RRE. To determine whether FBXL3 is a component of the Rev-Erbα:HDAC3 complex, we examined the interaction of endogenous FBXL3 and Rev-Erbα by co-IP experiments. We found that FBXL3 can interact with Rev-Erbα under physiological conditions (Fig. 6A). To determine whether FBXL3 could be recruited directly to the RRE region together with Rev-Erbα, we performed an in vitro protein–DNA binding assay. Streptavidin magnetic beads with tethered concatemers containing the RRE sequence were incubated with cell extracts from human embryonic kidney 293T cells expressing Myc-tagged FBXL3 with or without HA-tagged Rev-Erbα. The purified RRE-binding proteins were then analyzed by Western blotting (WB). As shown in Fig. 6B, FBXL3 associated with the RRE oligonucleotide-coupled beads only in the presence of Rev-Erbα, and FBXL3 did not affect the ability of Rev-Erbα to bind DNA, suggesting that Rev-Erbα mediates the recruitment of FBXL3 to RREs. Finally, to analyze whether FBXL3 regulates the transcriptional suppression function of the HDAC3:Rev-Erbα complex, we examined the ability of FBXL3 to regulate a Bmal1 promoter-driven luciferase reporter. As shown in Fig. 6C, the expression of HDAC3 significantly augmented the Rev-Erbα–mediated inhibition of Bmal1 promoter activity (lane 3 vs. 9). The coexpression of FBXL3 blocked this effect (Fig. 6C, lane 9 vs. 10), indicating that FBXL3 inhibits the repressor activity of the Rev-Erbα:HDAC3 complex. The effect of FBXL3 was abolished when the RRE sites on the Bmal1 promoter were mutated (25) (Fig. 6C), indicating that the RRE sites are necessary for FBXL3-mediated transcriptional regulation and that FBXL3 acts by targeting the RRE-associated Rev-Erbα:HDAC3 complex.
Fig. 6.
FBXL3 regulates Rev-Erbα:HDAC3 complex-mediated suppression. (A) Coimmunoprecipitation of FBXL3 and Rev-Erbα in liver tissues at zeitgeber time (ZT)11. Immunoprecipitates of Rev-Erbα from the wild-type and Rev-erbα knockout (KO) liver tissues were probed with the FBXL3 (Left) and Rev-Erbα (Right) antibodies. Actin served as a control for protein amount. (B) DNA (RRE)-conjugated magnetic beads were incubated with cell lysates containing FBXL3 and/or HA–Rev-Erbα. FBXL3 and HA–Rev-Erbα were detected by WB of the eluants. NC, negative control (nonrelated oligo-DNA); RRE, oligo-DNA containing RRE sequences. (C) Effects of FBXL3 on the Rev-Erbα:HDAC3 complex-mediated suppression of Bmal1 promoter-driven luciferase. Bar graphs depict the relative luciferase activities (mean ± SD) from three independent experiments. Statistical analysis was performed using one-way ANOVA followed by Tukey’s test. **P < 0.01. A total of 251 ng DNA containing 50 ng of each vector together with 1 ng of renilla luciferase expressing vector was transfected into U2OS (a human osteosarcoma cell line) cells. Luciferase activity was measured using the dual-luciferase report assay system. (Left) Bmal1 promoter with wild-type RRE. (Right) Bmal1 promoter containing the mutant RRE as previously described (25). (D) Model illustrating FBXL3 on E-box and RRE codependency for effective circadian period determination. FBXL3 releases E-box repressor complexes through degradation of chromatin on CRY1. It also removes repressors at the RRE sites. Inhibition duration by E-box and RRE-regulated complexes on these elements may regulate circadian period. BMAL1 (B), CLOCK (Clk), FBXL3 (F), HDAC3 (H), Rev-Erbα (RV), Rorα (R), and nuclear receptors (NR).
Discussion
Forward and reverse genetic studies have successfully discovered many key molecular components of the mammalian circadian clock (8, 11, 12, 15, 16, 26, 27). However, the genetic deletion of individual core clock components, including Per1, Per2, Cry1, Cry2, Clock, and Rev-erbα, resulted in only modest (1–2 h) period changes (8, 18, 19, 26, 28, 29), suggesting that the mammalian clock functions as a network to maintain its periodicity and robustness. In the past, efforts have been made to analyze circadian phenotypes by disrupting paralogous clock genes such as Cry1/2, Per1/2, and Rev-erbα/β in mice (22, 26, 30, 31). Cry/Per double-mutant mice have also been analyzed (32). The inactivation of two or more clock components provides unique insights into the genetic mechanisms of circadian rhythms. In this report, we tested genetic interactions between different clock mutant mice without preconceived ideas about the roles of these genes in the circadian network. Although most of the double mutants we generated were functionally independent, we found a strong genetic interaction between Fbxl3 and Rev-erbα, which led us to uncover an important role of Fbxl3 in regulating the Rev-Erbα:HDAC3 complex.
Previous studies showed that FBXL3 mediated the degradation of CRY (11, 13) and our findings further suggest that FBXL3 plays an important role in controlling CRY1 binding to chromatin in a rhythmic manner. In the Fbxl3−/− and Fbxl3−/−;Rev-erbα−/− mice, the increased binding of the CRY1 protein to the E-box regions of the Per2 and Cry1 genes was observed from CT12 to CT20, at which the FBXL3 and CRY1 interaction could barely be detected in wild-type mice (Fig. 3B). Furthermore, the E-box–driven transcription of Per1 and Per2 exhibited similar low-amplitude rhythms in Fbxl3−/− and Fbxl3−/−;Rev-erbα−/− liver tissues. In addition, we assayed Per1 and Per2 mRNA profiles in the cerebellum. As described previously (11), the expression levels were reduced to a larger extent in Fbxl3−/− and Fbxl3−/−; Rev-erbα−/− mice (Fig. S7). We do not exclude the possibility that tissue-specific regulators also play an important role in attaining the robustness of the clock. However, these results are not in conflict with the notion that FBXL3 facilitates depletion of CRY1 from E-box, and deletion of Rev-erbα has less effort on the amplitudes of Per1 and Per2 expression. Importantly, the deletion of Cry1 in an Fbxl3-null background appeared to restore the levels of Per1 and Per2 transcription and partially rescued its period length. Taken together, these observations indicate that the impairment of CRY degradation on chromatin can account for the dampened E-box–driven transcription and part of the long-period phenotype of the Fbxl3−/− mice.
We found that the deletion of Rev-erbα rescued the very-long-period phenotype caused by Fbxl3 deficiency, indicating a genetic interaction between these two genes. We examined the relationship between Fbxl3 and Rev-erbα in these mutant mice and found that the expression of Rev-erbα was dramatically reduced in the Fbxl3−/− mice (Fig. 5A), whereas the clearance of Rev-Erbα protein is impaired. The deletion of Fbxl3 results in increased HDAC3 binding at the RRE regions and deletion of Rev-erbα abolishes the retention of HDAC3 (Fig. 5 C and D). In addition, FBXL3 was also found to alleviate HDAC3 repression for a Bmal1 RRE reporter (Fig. 6C). These results suggested that FBXL3 directly regulates the RRE-mediated transcription by regulating the Rev-Erbα:HDAC3 complex, providing a molecular explanation for the genetic interaction between FBXL3 and Rev-Erbα. Therefore, FBXL3 serves as a critical regulatory link between the negative and positive circadian feedback loops. Previous synthetic promoter studies showed that RREs, E/E′ boxes, and D boxes work in a coordinated fashion to generate substantial phase delay during circadian oscillation that is important for establishing a normal circadian period (33, 34). Our results provided genetic evidence that: (i) deletion of Fbxl3 impairs the amplitudes of E-box–driven gene transcription, (ii) the abnormal retention of HDAC3 further contributes to a phase delay in the RRE-mediated transcriptional expression. We thus proposed that suppression at both E-boxes and RREs in the Fbxl3−/− mice may contribute to its extremely long period length (Fig. 6D). This work highlights the important role of FBXL3 in connecting two circadian feedback loops and suggests that the proper control and balance of transcriptional activity at the E-boxes of Pers/Crys and RREs of Bma1/Crys contribute significantly to the period determination of the clock in mammals.
Materials and Methods
Generation of Single- and Double-Mutant Mice.
Clock mutant mice (15) and Overtime mice (11) were obtained from the J.S. Takahashi laboratory at Northwestern University, Evanston, IL. Rev-Erbα knockout mice (8) were obtained from the Ueli Schibler laboratory at the University of Geneva, Geneva. For the conditional Fbxl3 knockout mice, the targeting vector was constructed (Fig. S2). The Cry1−/− and Cry2−/− mice were generated as previously reported (18, 19). Briefly, a LacZ-neo cassette was used to replace exons 6–10 of Cry1 and exons 6–11 of Cry2 in the targeting vectors. Other single-mutant mice used in these studies have been described previously (27, 35).
Animal Care and Behavioral Analysis.
All of the animals were backcrossed at least five generations before the first pilot study to assure a largely C57BL/6J background. Measurements of the free running period were performed as previously described (27). We used the formula τpredicted = τx + τy − τwild type (x and y represent different single-mutant mice, respectively) to predict τ of the double-mutant mice. This formula was proposed based on the assumption that there is no genetic interplay between the different mutations. Therefore, if the experimental period is close to the predicted value, the two tested mutations are very likely to function independently of each other. All animal studies were carried out in an Association for Assessment and Accreditation of Laboratory Animal Care International accredited SPF animal facility, and all animal protocols were approved by the Animal Care and Use Committee of the Model Animal Research Center, Nanjing University.
Antibodies, Western Blotting, Coimmunoprecipitation, and Chromatin Immunoprecipitation.
Anti-BMAL1, anti-CRY1, and anti-FBXL3 antibodies were generated using synthetic peptides as immunogens (Signalway Antibody). Peptide sequences for each antibody were as follows (underline represents amino acid sequences for each antibody): BMAL1, CSSSILGENPHIGIDMIDNDQGSSSPSNDEA; CRY1, CSQGSGILHYAHGDSQQTHSLKQGRSSAGTG; and FBXL3, CMKRGGRDSDQDSAEEGTAEKPKRPRTTQRE. Anti–Rev-Erbα serum was also generated by immunizing rabbits with purified protein corresponding to the N-terminal 126 AA of mouse Rev-Erbα. Antibodies against the following proteins were obtained from the indicated suppliers: HDAC3 (Santa Cruz; sc11417) and Rev-Erbα (Cell Signaling; 2124). The coimmunoprecipitation experiments were performed as previously described (36) using mouse liver extracts.
For the detection of Rev-Erbα, the liver extracts were immunoprecipitated with the antiserum and then immunoblotted with antibodies purchased from Cell Signaling as described above. For the detection of FBXL3, the liver extracts were immunoprecipitated and immunoblotted with the same antibody.
ChIP assays were performed as described previously (36), except that hypotonic buffer (Active Motif; 100505) was used to obtain the nuclear extracts. Rabbit IgGs from nonimmunized rabbits were used as the negative control. For ChIP on E-box sites (at the Cry1 and Per2 promoter), the first intron of Cry1 was used as the control locus: for ChIP on RRE sites (at the Bmal1 and Cry1 promoter/enhancer), the Cry1 promoter was used as the control site. All of the primers were validated for SYBR-based real-time PCR (Table S1).
Tissue Collection, RNA Isolation, RT-PCR, and mRNA Expression Analyses.
Tissue collection, RNA isolation and RT-PCR were carried out as previously described (27). Details are available in SI Text.
In Vitro DNA-Mediated Pull Down.
Details are available in SI Text.
Statistical Analyses.
One-way or two-way analysis of variance (ANOVA) followed by the Tukey test was performed for multiple group comparisons using Graphpad Prism software to determine the significant differences between different genotypes. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Ueli Schibler for the Rev-erbα mice and J. S. Takahashi for the Clock and Overtime mice. This work was supported by Ministry of Science and Technology Grant 2010CB945100 and 2013CB945203 (to Y.X.), National Science Foundation of China Grants 31171343 and 31230049 (to Y.X.), and National Institutes of Health Grants GM068496 and GM062591 (to Y.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302560110/-/DCSupplemental.
References
- 1.Hastings MH, Reddy AB, Maywood ES. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4(8):649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 2.Allada R, Emery P, Takahashi JS, Rosbash M. Stopping time: The genetics of fly and mouse circadian clocks. Annu Rev Neurosci. 2001;24:1091–1119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- 3.Lowrey PL, Takahashi JS. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 5.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 6.Sharma VK. Adaptive significance of circadian clocks. Chronobiol Int. 2003;20(6):901–919. doi: 10.1081/cbi-120026099. [DOI] [PubMed] [Google Scholar]
- 7.Schibler U, Naef F. Cellular oscillators: Rhythmic gene expression and metabolism. Curr Opin Cell Biol. 2005;17(2):223–229. doi: 10.1016/j.ceb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 9.Sato TK, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43(4):527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 10.Liu AC, et al. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4(2):e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siepka SM, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129(5):1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godinho SI, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316(5826):897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 13.Busino L, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316(5826):900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 14.Chen R, et al. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell. 2009;36(3):417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King DP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89(4):641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bunger MK, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103(7):1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, et al. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128(1):59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thresher RJ, et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282(5393):1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 19.Vitaterna MH, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96(21):12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421(6919):177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 21.Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8(6):437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 22.Bugge A, et al. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26(7):657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alenghat T, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456(7224):997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19(6):1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 25.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447(7143):477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 26.Bae K, et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30(2):525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, et al. Interaction of MAGED1 with nuclear receptors affects circadian clock function. EMBO J. 2010;29(8):1389–1400. doi: 10.1038/emboj.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Debruyne JP, et al. A clock shock: Mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50(3):465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 29.Zheng B, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105(5):683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 30.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485(7396):123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98(2):193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 32.Oster H, Yasui A, van der Horst GT, Albrecht U. Disruption of mCry2 restores circadian rhythmicity in mPer2 mutant mice. Genes Dev. 2002;16(20):2633–2638. doi: 10.1101/gad.233702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueda HR, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37(2):187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 34.Ukai-Tadenuma M, et al. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144(2):268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 35.Gu X, et al. The circadian mutation PER2(S662G) is linked to cell cycle progression and tumorigenesis. Cell Death Differ. 2012;19(3):397–405. doi: 10.1038/cdd.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107(7):855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.