Abstract
Glypican-3 (GPC3) has emerged as a candidate therapeutic target in hepatocellular carcinoma (HCC), but the oncogenic role of GPC3 in HCC is poorly understood. Here, we report a human heavy-chain variable domain antibody, HN3, with high affinity (Kd = 0.6 nM) for cell-surface–associated GPC3 molecules. The human antibody recognized a conformational epitope that requires both the amino and carboxy terminal domains of GPC3. HN3 inhibited proliferation of GPC3-positive cells and exhibited significant inhibition of HCC xenograft tumor growth in nude mice. The underlying mechanism of HN3 action may involve cell-cycle arrest at G1 phase through Yes-associated protein signaling. This study suggests a previously unrecognized mechanism for GPC3-targeted cancer therapy.
Keywords: heparan sulfate proteoglycans, liver cancer, monoclonal antibodies, phage display, cell growth
Liver cancer is the fifth most prevalent neoplasm worldwide and is the third most common cause of cancer-related mortality (1, 2). Both the incidence and mortality of liver cancer are rising. More than 80% of liver cancer cases are attributed to hepatocellular carcinoma (HCC), which does not respond to most chemotherapy drugs (3). Currently, surgery is the standard treatment for liver cancer diagnosed at an early stage, which constitutes only 37% of cases. The overall 5-y relative survival rate for patients with liver cancer is only 14% in the United States (www.cancer.org). The development of new drugs targeting different mechanisms of action is an important challenge, one that antibody therapy may address.
Glypican-3 (GPC3, also called “DGSX,” “GTR2-2,” “MXR7,” “OCI-5,” “SDYS,” “SGB,” “SGBS,” and “SGBS1”) is a member of the glypican family of heparan sulfate (HS) proteoglycans that are attached to the cell surface by a glycosylphosphatidylinositol (GPI) anchor (4). The GPC3 gene encodes a 70-kDa core protein, which can be cleaved by furin to produce the N-terminal 40-kDa fragment and the C-terminal 30-kDa fragment (5). Recently reported crystal structures of Drosophila glypican Dally-like (Dlp, a homolog to human GPC1) and human GPC1 show that the N- and C-terminal parts of GPC1 are linked by disulfide bonds (6, 7). Because all the glypicans (including GPC1 and GPC3) have 14 highly conserved cysteine residues, their 3D structures may be similar. It is likely that the N- and C-terminal parts of GPC3 also are linked by disulfide bonds. Two HS chains are attached to the C-terminal portion of GPC3.
A host of studies confirmed that GPC3 is an attractive liver cancer-specific target, because it is highly expressed in HCC but not in normal tissue (8–10). However, the precise biological functions of GPC3 and its role in tumorigenesis still remain elusive. Loss-of-function mutations of GPC3 cause Simpson–Golabi–Behmel syndrome (SGBS), a rare X-linked overgrowth disease (11). GPC3-deficient mice display developmental overgrowth and some of the abnormalities typical of SGBS (12). In transgenic mice, overexpression of GPC3 suppresses hepatocyte proliferation and liver regeneration (13). HCC cells infected with lentivirus expressing soluble GPC3 (sGPC3, a secreted form that lacks the GPI anchoring domain) have a lower cell-proliferation rate (14). This finding suggests that the sGPC3 protein secreted by infected cells may inhibit cell proliferation in an autocrine manner. We produced a recombinant sGPC3 (GPC3ΔGPI, amino acid residues Q25–H559) and found that sGPC3 protein, functioning as a dominant-negative form, can inhibit the growth of HCC in vitro (15). GPC3 knockdown also can inhibit cell proliferation in the HCC cell lines Huh-7 and HepG2 (16).
Recent advances in understanding the signaling pathways that lead to HCC indicate that the Hippo–Yes-associated protein (yap) pathway protects the liver from overgrowth and HCC development. Deregulation of the Hippo pathway is seen frequently in HCC. The oncogene yap, which is the down-stream effector of the Hippo pathway, can be inactivated by phosphorylation; elevated yap protein levels are strongly associated with HCC (17–19). We speculate that yap may be a downstream oncogenic gene involved in GPC3-mediated liver carcinogenesis, but studies showing the possible connection between GPC3 and yap have yet to be reported.
To date, several mouse mAbs against GPC3 have been produced (20–27), and almost all of them target a peptide derived from GPC3. However, none of these antibodies has shown the ability to inhibit cell proliferation or induce apoptosis, possibly because of the difficulty of having a conventional antibody targeting the potentially cryptic functional epitope of GPC3. Because of their small size, domain antibodies are able to target cryptic epitopes on antigens (e.g., in the clefts of enzymes and receptors) (28–30).
In the present study, we were interested in identifying anti-GPC3 mAbs that are able to inhibit cancer cell proliferation and/or survival directly by blocking important and undetermined signaling pathways. We identified a human heavy chain variable (VH) domain antibody (HN3) targeting GPC3 using phage display technology and found that HN3 binds a unique conformational epitope in the core protein of GPC3 with high affinity. Interestingly, the HN3 binding requires both the N and C termini of GPC3. Furthermore, we discovered that HN3 inhibits HCC cell growth in several HCC cell models and that HN3 significantly inhibits the growth of HCC xenograft tumors in nude mice. Our findings show that it is possible to inhibit HCC cell proliferation with an antibody that neutralizes the proliferative function of GPC3.
Results
Knockdown of GPC3 Inhibits HCC Cell Proliferation.
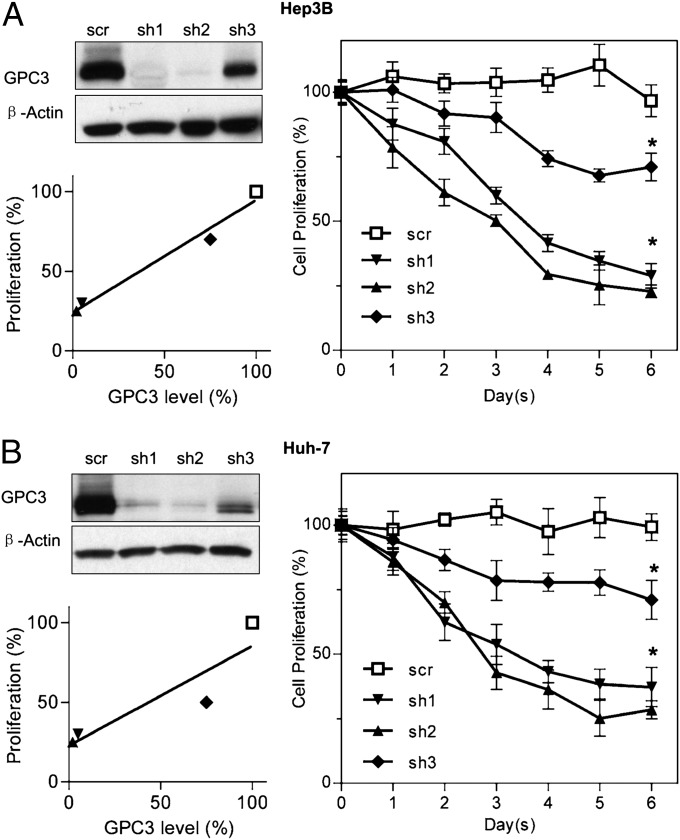
GPC3 is highly and specifically expressed in HCC. In assessing whether HCC cell proliferation could be inhibited by silencing GPC3, a previous study showed that RNAi suppression of GPC3 in HCC led to inhibitory effects on cell growth and cell-cycle progression (16). In this study, we constructed three different shRNAs designated “sh1,” “sh2,” and “sh3.” We found that RNAs sh1 and sh2 reduced GPC3 protein expression by more than 90% in the HCC cell lines Hep3B (Fig. 1A) and Huh-7 (Fig. 1B), inhibiting ∼70% of cell proliferation compared with the scrambled (scr) control. The other shRNA, sh3, reduced GPC3 expression by only 20% in HCC cells and inhibited only 20–30% of cell proliferation. The inhibition of cell proliferation correlated with the efficiency of GPC3 knockdown in both HCC cell models, showing that silencing the GPC3 gene inhibits cell proliferation in liver cancer.
Fig. 1.
Inhibition of HCC cell proliferation by GPC3 knockdown. (A) Hep3B cell-proliferation assay after GPC3 knockdown. sh1, sh2, and sh3 are three different GPC3-knockdown shRNAs. scr is the scrambled control shRNA. Cell proliferation was measured by the WST method and was normalized with unengineered Hep3B cells. GPC3-knockdown efficiency was validated by Western blot and correlated with cell proliferation. Cell proliferation in the three sh groups was significantly lower than in scr control. (B) Huh-7 cell-proliferation assay after GPC3 knockdown. The experimental settings were those used in A. Values represent mean ± SD. *P < 0.001 in A and B.
Isolation of Anti-GPC3 Human VH Domain Antibody HN3.
Based on GPC3-knockdown results, we speculated that an antibody targeting a functional domain of GPC3 may be able to inhibit cell proliferation by neutralizing the GPC3 proliferative function. We hypothesized that the previously developed antibodies did not demonstrate this function because they did not target the cryptic GPC3 functional epitope. Therefore, we explored the potential of a human single-domain antibody by screening a previously developed phage-display VH domain antibody library (31).
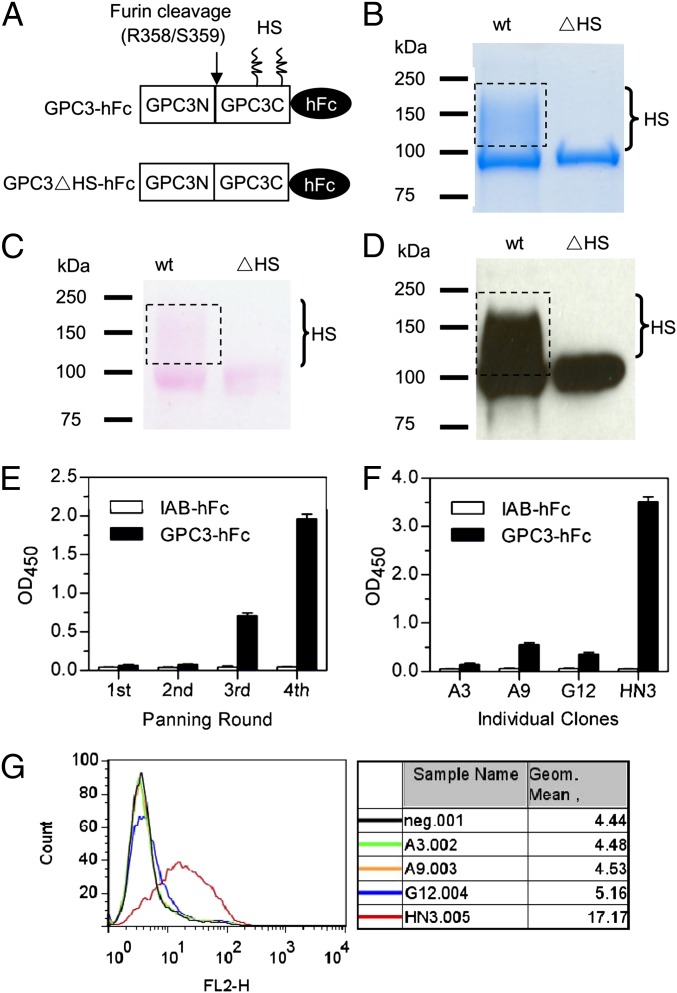
To screen and identify antibodies specific for the core protein or the HS chain of GPC3, we first produced recombinant GPC3-hFc and GPC3∆HS-hFc proteins in human HEK-293F cells (Fig. 2A). We replaced two serine residues with alanine (S495A and S509A, respectively) to abolish the formation of HS chains at the posttranslational glycosylation step. The molecular mass and purity of the proteins were validated by SDS/PAGE separation followed by Coomassie blue staining (Fig. 2B), sugar chain staining (Fig. 2C), and Western blot using a commercial GPC3 antibody that detects a core protein epitope at the C terminus (Fig. 2D). We then screened the phage-display antibody domain library by four rounds of panning on a 96-well ELISA plate coated with recombinant GPC3-hFc protein. GPC3-specific binders were enriched gradually as determined by polyclonal phage ELISA (Fig. 2E). At the end of the fourth round of panning, 96 clones were selected randomly, and 88 of these clones were confirmed to be GPC3 binders by phage ELISA. We selected positive phage clones with GPC3-hFc OD values at least threefold higher than that of IAB-hFc, a control Fc fusion protein specific for MUC16. Subsequent sequencing analysis revealed four representative binders (A3, A9, G12, and HN3). Using the same number of phage, the HN3 clone was determined by both monoclonal phage ELISA (Fig. 2F) and FACS (Fig. 2G) to be the strongest GPC3 binder. Because cell binding is one of the most important criteria for therapeutic antibodies targeting cell-surface proteins, we decided to develop and analyze HN3 further in this study. We successfully identified a human VH domain that binds cell-surface–associated GPC3 molecules.
Fig. 2.
Preparation of recombinant GPC3 proteins and discovery of HN3. (A) Schematic diagram of the primary structure of GPC3-hFc fusions (Q25–S550). The arrow indicates the furin-cleavage site. HS, heparan sulfate chains. GPC3∆HS-hFc is a GPC3 mutant (S495A, S509A) with no HS chain formation. (B) Coomassie-stained SDS/PAGE of wild-type and mutant (∆HS) GPC3. Five micrograms of protein were loaded for each lane. Dotted rectangles indicate the GPC3 fraction with HS modification. (C) SDS/PAGE gel of the wild-type and mutant (∆HS) GPC3 stained with glycoprotein-staining reagent (Pierce). (D) Western blot validation using a commercial anti-GPC3 mAb 1G12. One hundred nanograms of protein were loaded for each lane. (E) Polyclonal phage ELISA from the output phage of each panning round. IAB-hFc, a human Fc control to an irrelevant antigen. (F) Monoclonal phage ELISA of the four candidate GPC3 binders. Values in E and F represent mean ± SD. (G) Phage FACS analysis of the four clones. Neg. indicates background staining with secondary antibody only (anti-M13 phycoerythrin conjugate).
Binding Properties of HN3 Domain Antibody.
To construct an HN3 domain antibody, the VH coding sequence was recovered from the phage and cloned into an expression vector as a VH-hFc molecule (i.e., VH fused with human IgG1 Fc; Fig. 3A), a widely used therapeutic format. For large-scale production of HN3 protein, a CHO-S stable cell line was generated with around 110 mg/L productivity in volumetric titer. SDS/PAGE analysis showed that the purified HN3 formed a dimer with a molecular mass of ∼100 kDa (Fig. 3A), slightly larger than expected (theoretical size, 82 kDa). The difference in size likely results from the glycosylation of the hFc part (32). Under reducing conditions, the molecular mass was ∼40 kDa.
Fig. 3.
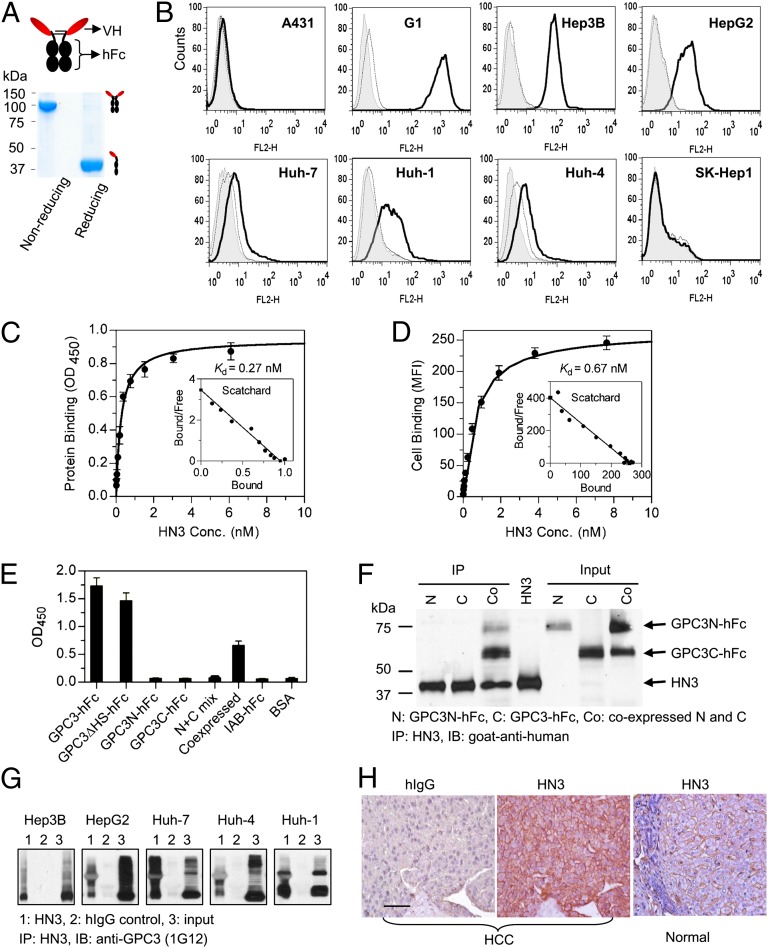
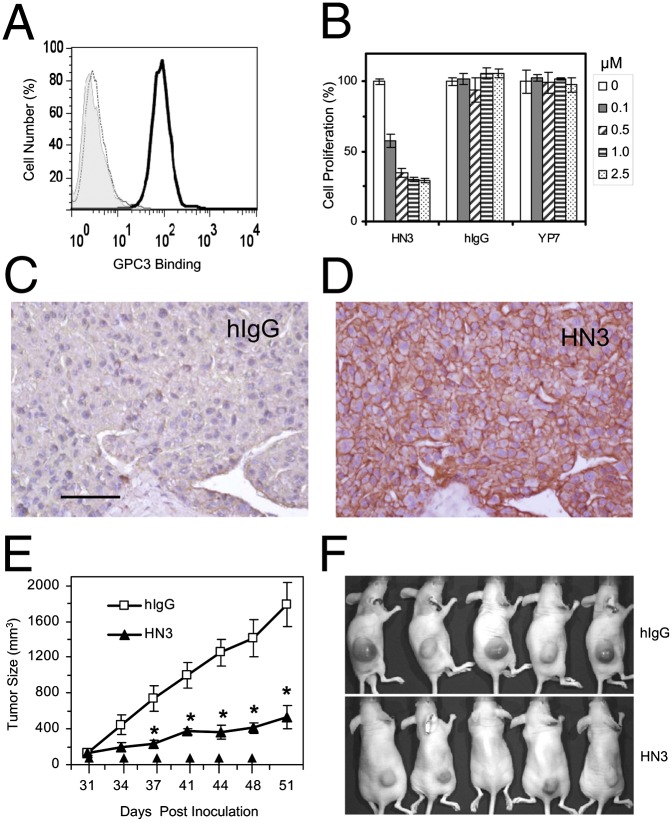
Binding properties of HN3 antibody. (A) SDS/PAGE analysis of purified HN3 antibody. Five micrograms of purified protein were loaded in each lane. (B) FACS analysis of cell binding on A431 and SK-Hep1 (GPC3−), G1 (transfected A431 that highly expresses GPC3), and a panel of HCC (GPC3+) cells. Shaded area is background staining with secondary antibody only (goat anti-human phycoerythrin conjugate). The thin dotted curve shows staining with irrelevant hIgG control, and the heavy solid line shows HN3 staining. (C) Protein-binding affinity was measured by ELISA using purified GPC3-hFc. (D) Measurement of cell-binding affinity by FACS using the G1 cell line. (E) Epitope-mapping ELISA showing that HN3 binds to the full-length core protein of GPC3. GPC3-hFc is full-length wild-type GPC3 (amino acids 25–550), GPC3∆HS-hFc is a mutant GPC3 (S495A, S509A) that does not contain HS chains, and GPC3N-hFc and GPC3C-hFc are N-terminal (amino acids 25–358) and C-terminal (amino acids 359–550) fragments of GPC3, respectively. N+C mix is the simple mixture of N- and C-terminal fragments. “Coexpressed” designates the preparation of N- and C-terminal fragments that was made by cotransfecting 293F cells with two separate plasmids, each encoding the N- or C-terminal fragment. Values in C–E represent mean ± SD. (F) Immunoprecipitation assay using coexpressed GPC3 N- and C-terminal fragments. (G) Immunoprecipitation assay. Native GPC3 proteins were immunoprecipitated from the HCC cell lysate incubated with HN3 but not hIgG control. (H) HCC tissue immunohistochemistry staining with HN3. (Scale bar: 100 µm.)
We then tested the binding of HN3 antibody to five native HCC cell lines (Hep3B, HepG2, Huh-7, Huh-4, and Huh-1), two GPC3-negative cell lines (A431 and SK-Hep1), and an A431-derived cell line (G1) developed in our laboratory. The G1 cell line stably and highly expresses GPC3 on the cell surface. HN3 showed specific binding to HCC and G1 cells but no binding to GPC3-negative A431 and SK-Hep1 cells (Fig. 3B). We measured the binding affinities of HN3 to GPC3 protein and GPC3-positive cells by ELISA and flow cytometry. The calculated Kd values were 0.27 nM for GPC3 protein (Fig. 3C) and 0.67 nM for GPC3-positive cells (Fig. 3D).
To analyze the binding site recognized by HN3, we made the N-terminal (residues 25–358) and C-terminal (residues 359–550) fragments of GPC3 fused with hFc. Epitope mapping ELISA results showed that HN3 bound to wild type GPC3 comparably as it did to mutant GPC3ΔHS and that HN3 did not bind to either the N-terminal or C-terminal fragment of GPC3 alone (Fig. 3E), indicating that the HN3 epitope is formed by regions from both N- and C-terminal domains of GPC3. Interestingly, HN3 also binds the coexpressed N- and C-terminal fragments but not to mixtures of N- and C-terminal fragments made by simply mixing the fragments together. The coexpression was performed by cotransfecting two separate plasmids encoding N- or C-terminal fragments in human HEK-293 cells. In addition to ELISA, we performed immunoprecipitation experiments. We found HN3 could immunoprecipitate the complex consisting of both N- and C-terminal fragments from the culture supernatant, but it could not immunoprecipitate either the N- or C-terminal fragment alone (Fig. 3F). This result indicates that HN3 may recognize the heterodimers consisting of both N- and C-terminal domains of GPC3 instead of the N- or C-terminal domain homodimers.
We also evaluated the binding of HN3 to native GPC3 proteins of HCC cells. HN3 did not recognize denatured GPC3 on Western blot, but native GPC3 proteins could be immunoprecipitated by incubating the cell lysates of five native HCC cell lines with HN3 (Fig. 3G). Immunohistochemistry showed that HN3 specifically stained tissue from HCC patients but did not stain healthy tissue (Fig. 3H). Taken together, these results show that HN3 is a high-affinity domain antibody specific for cell-surface GPC3 and recognizes a conformational epitope in the GPC3 core protein that requires both N- and C-terminal domains.
HN3 Inhibited HCC Cell Proliferation.
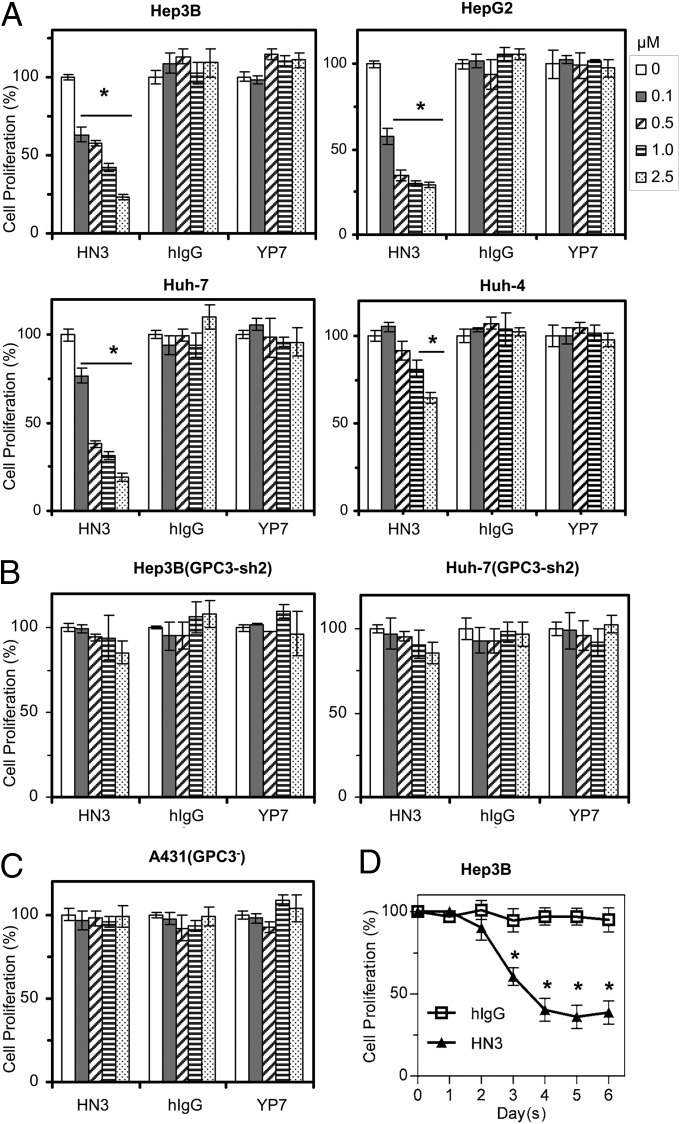
To determine whether HN3 could inhibit HCC growth directly, we performed cell growth-inhibition assays on four GPC3-positive HCC cell lines and the GPC3-negative cell line A431. We found that HN3 potently inhibited the growth of Hep3B, HepG2, and Huh-7 cells and partially inhibited the growth of Huh-4 cells (Fig. 4A). The maximal inhibition was about 70% of untreated cells in the Hep3B, HepG2, and Huh-7 cell lines. HN3 did not affect the growth of the GPC3-knockdown Hep3B and Huh-7 cells (Fig. 4B) or of the GPC3-negative A431 cell line (Fig. 4C). A representative time course of Hep3B cell proliferation is shown in Fig. 4D. In comparison with HN3, YP7, another high-affinity GPC3 antibody specific for the C terminus of GPC3 developed in our laboratory (24), did not inhibit cell proliferation (Fig. 4A), indicating that the ability of a GPC3 antibody to inhibit cell growth is strictly epitope dependent.
Fig. 4.
Inhibition of HCC cell proliferation by treatment with HN3 antibody. (A) Cell-proliferation assay with HN3 treatment on GPC3-positive cells. Cells were incubated for 5 d with 0–2.5 µM HN3 or hIgG or YP7 (GPC3 mouse antibody that recognizes a C-terminal epitope). Cell proliferation was measured by the WST method and was normalized to untreated cells. (B) HN3 treatment on GPC3-knockdown Hep3B and Huh-7 cells. (C) Cell-proliferation assay with HN3 treatment on GPC3-negative A431 cells. Values represent mean ± SD in all panels. *P < 0.001 compared with no antibody treatment (0 µM) in A, B, and C. (D) A representative time course of Hep3B response upon HN3 treatment (1 µM). Error bars indicate SD. *P < 0.001 compared with hIgG control.
HN3 Induced Cell-Cycle Arrest.
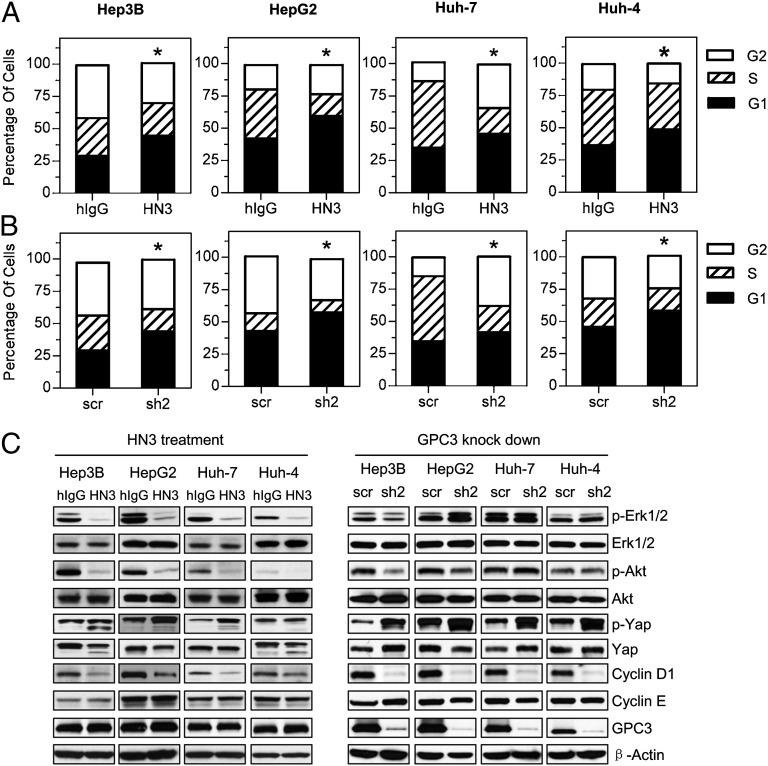
To understand the underlying mechanism of HN3 activity, we investigated cell-cycle progression after HN3 treatment. In the four HCC cell lines tested (Hep3B, HepG2, Huh-7, and Huh-4), HN3 treatment significantly increased the G1 population (Fig. 5A), indicating that HN3 can induce cell-cycle arrest in the G1 phase. GPC3 knockdown by shRNA in these four cell lines also resulted in G1 arrest (Fig. 5B).
Fig. 5.
HN3 induced cell-cycle arrest at G1 phase. (A) Cell-cycle analysis after HN3 treatment. HCC cells were incubated with 1 µM of HN3 or human IgG isotype control (hIgG). *P < 0.05, HN3 vs. hIgG in G1 phase. (B) Cell-cycle analysis after GPC3 knockdown. *P < 0.05, scrambled control (scr) vs. GPC3 knockdown in G1 phase. (C) Western blot analysis of HCC treated with HN3- or GPC3-knockdown cells.
The Hippo pathway recently was found to be a potent tumor-suppressing pathway in HCC (17–19). To analyze the mechanism of HN3 activity at molecular level, we investigated the expression of several major cell proliferation/cell cycle-related markers in the Erk, Akt, and Hippo signaling pathways in HN3-treated HCC cells (Fig. 5C). We found that phosphorylated Erk (p-Erk) and Akt (p-Akt) were decreased significantly in the four HN3-treated HCC cell lines (Hep3B, HepG2, Huh-7, and Huh-4), whereas phosphorylated yap (p-yap), the inactive form of yap, was increased in treated Hep3B, HepG2, and Huh-7 cells. The total level of yap was decreased in treated Hep3B and Huh-4 cells. Expression of cyclin D1, a target gene of yap, was decreased in the four treated HCC cell lines, as is consistent with G1 phase arrest induced by HN3. Because the Erk/Akt pathways also regulate the Hippo pathway by phosphorylating yap or the upstream components (33–37), the molecular changes in Erk, Akt, and yap collectively indicate that yap may play an important role in HN3 function. Similar molecular changes were observed in GPC3-knockdown Hep3B, HepG2, Huh-7, and Huh-4 cells, except that p-Erk and p-Akt levels were decreased only in Hep3B-knockdown cells (Fig. 5C).
Regulating Yap Bioavailability Affected HN3 Activity Dramatically.
To analyze further the role of yap in HN3 activity, we knocked down yap and overexpressed a constitutively active yap mutant, yap-S127A (38), in Hep3B cells. When yap was knocked down by yap-specific shRNA, cell proliferation was decreased by about 50% compared with scrambled control (Fig. 6A). Interestingly, HN3 treatment of yap-knockdown cells did not inhibit cell proliferation any further, even at high concentrations (2.5 and 5.0 μM) (Fig. 6B), indicating that yap stable knockdown may induce acquired resistance to the HN3 treatment. Overexpression of yap-S127A was found to increase cell proliferation by about 50% (Fig. 6C) and completely abolished HN3 antagonist activity (Fig. 6D), i.e., HN3 did not inhibit the proliferation of cells overexpressing yap-S127A. Western blot analysis showed that yap knockdown decreased the cyclin D1 level (Fig. 6E), whereas yap overexpression increased cyclin D1 levels. Taken together, these results indicate that yap is involved in the proliferative effect of GPC3; therefore, yap is a downstream target of the HN3 domain antibody.
Fig. 6.
The effect of yap deregulation on HN3 function. (A) Inhibition of cell proliferation by yap knockdown. Yap-sh, yap shRNA; scr, scrambled control. Proliferation was normalized with unengineered cells. *P < 0.001 between yap-sh and scr control. (B) HN3 treatment of yap-knockdown cells. Proliferation was normalized with unengineered cells without antibody treatment. (C) Promotion of cell proliferation by cells overexpressing yap-S127A. *P < 0.001, yap-S127A vs. mock control. (D) HN3 treatment of cells overexpressing yap-S127A. Values in A–D represent mean ± SD. (E) Western blot analysis of molecular changes after HN3 treatment on yap-knockdown cells or cells overexpressing yap-S127A.
HN3 Inhibited Tumor Growth in Vivo.
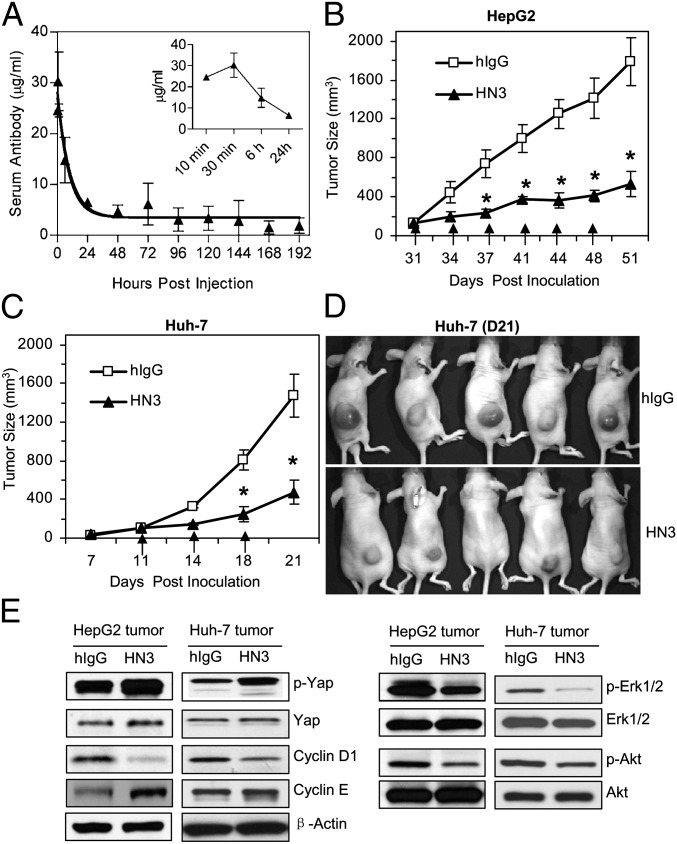
The ability of HN3 to reduce HCC proliferation in vitro prompted us to investigate its in vivo efficacy. We measured the half-life of HN3 antibody by ELISA using mouse sera. After a single i.v. injection of 3 mg/kg HN3, HN3 reached its peak concentration (28.70 ± 2.2 µg/mL) 30 min after antibody injection and then gradually decreased to a steady level (4.68 ± 1.27 µg/mL) at 48 h (Fig. 7A). The terminal half-life was 92.08 ± 23.77 h, and the clearance rate was 0.043 ± 0.005 mL⋅h−1⋅kg−1 as determined by Phoenix WinNonlin software (version 5.2.1; Pharsight Corp).
Fig. 7.
Inhibition of HCC xenograft tumor growth by HN3 antibody. (A) Pharmacokinetics of HN3 antibody. Serum concentrations of the antibody were measured by ELISA following a single i.v. injection of 3 mg/kg. The serum antibody concentration data were fitted with a two-compartment model using Phoenix WinNonlin software. The observed mean serum antibody concentration (in micrograms per milliliter) ± SD of four animals for each time point was plotted as a function of time. (B) Inhibition of HepG2 cells inoculated into nude mice. After the tumor size reached 100 mm3, HN3 antibody or hIgG control was delivered i.v. at 60 mg/kg body weight, twice a week. Arrows indicate antibody injections. Error bars indicate SE. *P < 0.05, HN3 vs. hIgG in B and C. (C) Inhibition of Huh-7 cells. Experimental settings are as in B. (D) Photographs of a representative group of mice treated with HN3 antibody. hIgG is pooled human IgG used as control. (E) Western blot analysis of molecular changes in HN3-treated and control tumors.
We then inoculated nude mice s.c. with HepG2 and Huh-7 cells. After the tumor size reached about 100 mm3, the mice were treated twice weekly by i.v. injection of 60 mg/kg HN3 antibody. The tumor size was measured and compared with the control group. HN3 treatment significantly inhibited the tumor growth in mice injected with both HepG2 (Fig. 7B) and Huh-7 cells (Fig. 7C). At the end of the study, the average tumor size was 400–500 mm3 in the treated group and 1,500–1,700 mm3 in the control group. (Photographs of a representative group of mice treated with HN3 antibody are shown in Fig. 7D). Molecular changes in HN3-treated tumors also were observed (Fig. 7E). Although total yap expression did not change, p-yap levels were increased in treated tumors. We also found the decreases in p-Erk and cyclin D1 expression. The in vivo tumor data support our in vitro mechanistic results.
In our animal testing, we did not observe antibody-related toxicity or behavioral abnormalities in the treated mice. Treated mice consumed food and socialized similarly as control group mice. Both body weights and organ weights (heart, liver, kidney, lung, and spleen) were unchanged. Taken together, our animal testing results indicate that the HN3 single-domain antibody is active in vivo and should be evaluated further as a candidate therapeutic for the treatment of liver cancer.
Discussion
Currently, several GPC3 antibodies have been developed. Almost all the existing antibodies were raised against a peptide derived from GPC3, and one such antibody is being evaluated in clinical trials, although none of the GPC3 antibodies have demonstrated direct inhibition of HCC cell proliferation. We have isolated HN3, a VH single-domain antibody targeting a conformation epitope in GPC3, by phage display. HN3 binds cell-surface–associated GPC3 molecules with subnanomolar affinity but does not bind denatured full-length GPC3. Interestingly, the binding of HN3 requires both the N- and C-terminal domains of GPC3, although this binding is independent of the HS chains on GPC3. Our results strongly suggest that HN3 recognizes a unique conformational structure present in the native form of GPC3 core protein on cancer cells. The present study shows that antibodies targeting GPC3 can inhibit cell proliferation.
Antibody-based therapy targeting GPC3 has been explored recently. The mouse mAb GC33, which recognizes a C-terminal peptide, has been produced, and humanized GC33 is being evaluated currently in clinical trials for liver cancer therapy (http://clinicaltrials.gov/ct2/show/NCT00746317). HN3 has three unique properties that GC33 lacks: it can inhibit HCC cell proliferation directly; it is a single-domain antibody; and it is a fully human protein. Although Fvs from murine sources can be used to make chimeric or humanized mAbs for clinical trials, the problems of humans developing anti-murine antibodies and other side effects must be resolved. Immunotherapy for GPC3-expressing cancers cannot be exploited fully until a human antibody with high affinity against GPC3-expressing cancer cells, such as the one described here, is developed. Because of these characteristics and its unique mechanism, HN3 is an attractive addition to the existing GPC3-targeted liver cancer therapies. Further studies comparing the efficacy of HN3 with that of GC33 in both preclinical and clinical settings are needed.
The HN3 domain antibody binds a conformation epitope that requires both the N- and C-terminal domains of the GPC3 core protein. This feature distinguishes HN3 from all the known mAbs that recognize either the N or C terminus of GPC3. The conformation of the HN3-binding site may affect a newly discovered GPC3 function, because HN3 shows direct inhibition of cell proliferation. Future GPC3 structural studies will be needed to reveal the precise structure that HN3 recognizes.
In addition to GPC3-targeted immunotherapy, EGF receptor (EGFR)-targeted antibody therapy by the FDA-approved agent cetuximab is being evaluated in clinical trials for liver cancer (ClinicalTrials.gov identifier NCT00980239). Cetuximab inhibits HCC cell proliferation in HepG2 and Huh-7 cells at doses of 10–1,000 µg/mL (39, 40). The maximal inhibition was 57% in HepG2 cells and 20% in Huh-7 cells at doses of 1,000 µg/mL (40). Results in HepG2 and Huh-7 cells are believed to be particularly indicative of clinical HCC activity because they have preserved differentiated liver characteristics, as evidenced by the production of many human plasma proteins (41, 42). HepG2 is a well-differentiated wild-type p53 HCC cell line (41), and Huh-7 is a highly differentiated human HCC cell line with mutated p53 (42). Cetuximab was much less potent in p53-mutated Huh-7 cells (39, 40). Unlike cetuximab, HN3 inhibits both HepG2 and Huh-7 cell proliferation, indicating that HN3 function is independent of p53. The maximal inhibition by HN3 is about 70% in Hep3B, HepG2, and Huh-7 cells, and the corresponding concentration for maximal inhibition is in the range of 0.5–2.5 µM (50–250 µg/mL). Taken together, these results indicate that HN3 may be more potent than cetuximab in HCC and that the antitumor activity of HN3 is not related to the mutation of p53.
Based on our experimental data, we hypothesize that the molecular mechanism underlying HN3-related inhibition of cell proliferation in HCC involves cell-cycle arrest through inactivating yap. We measured the changes of yap expression after HN3 treatment by Western blot and found that yap was inactivated after HN3 treatment. When yap was knocked down in Hep3B cells, cell proliferation was decreased significantly. Overexpression of yap-S127A promoted cell proliferation and completely abolished HN3 activity.
The present report describes the generation and characterization of a high-affinity single-domain antibody against cell-surface–associated GPC3. Because HN3 has a human origin and displays high affinity (Kd = 0.67 nM), it is expected to be less immunogenic than murine mAb and to be efficient in targeting GPC3-expressing tumors. Furthermore, HN3 is a unique mAb against GPC3 that shows direct inhibition of HCC growth, and it should be further evaluated as a therapeutic candidate for the treatment of liver cancer.
Materials and Methods
Cell Lines.
Five HCC cell lines (Hep3B, HepG2, Huh-7, Huh-4, and Huh-1) and the GPC3-negative cell line SK-Hep1 were maintained as adherent monolayer cultures in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS (HyClone), 1% L-glutamine, and 1% penicillin-streptomycin (Invitrogen) and were incubated in 5% CO2 with a balance of air at 37 °C. Cells were harvested and the medium was changed twice a week. GPC3-negative A431 cells (human epithelial carcinoma cell line) were engineered to express high levels of GPC3 by transfection with a plasmid encoding GPC3. Both A431 and the stably transfected cells (G1) were maintained in DMEM.
Protein Reagents.
YP7, a GPC3 mouse antibody developed in our laboratory, recognizes a C-terminal epitope (amino acids 510–560) (24). YP7 was used for comparison with HN3 in cell-proliferation assays. IAB-hFc, a 64-aa fragment of mesothelin (amino acids 296–359) fused with human Fc at the C terminus (43), was used as an irrelevant hFc control for GPC3-hFc in HN3 binding-property assays. Pooled human IgG (hIgG; Sigma-Aldrich) was used as HN3 control in cell-proliferation assays and in vivo animal studies.
Antibodies and Western Blot.
Antibody for GPC3 (1G12; sc-65443) was obtained from Santa Cruz Biotechnology, Inc. Antibodies for detecting Erk1 (1171–1), Erk2 (1586–1), Akt1 (1085–1), and Akt2 (T1933), were obtained from Epitomics. Antibodies for p-Erk1/2 (9101), p-Akt (4060), yap (4912), and p-yap (4911) were obtained from Cell Signaling. Antibody for cyclin D1 was obtained from Invitrogen (33–3500). Unless specifically stated, cells were cultured in T-25 or T-75 flasks for 5 d at a final confluence of 70% with or without HN3 treatment. Cells were collected and lysed with lysis buffer [50 mM Tris⋅HCl (pH7.5), 50 mM NaCl, 5 mM EDTA, 1% Triton X-100] mixed with mixture proteinase inhibitors from Roche Applied Science. Protein concentration of the cell lysate was measured by the Coomassie Plus (Bradford) Protein Assay (23236; Thermo Scientific). Thirty micrograms of total protein were run on reducing SDS/PAGE for Western blot analysis.
DNA Oligos and Plasmids.
For GPC3 knockdown, three pairs of DNA oligomers were chemically synthesized, in vitro annealed, and subsequently cloned into a knockdown vector pGreenPuro from System Biosciences according to the manufacturer’s instructions. The insert DNA sequences coding GPC3 shRNA are indicated below. The GPC3 sequence of the sense strand is underlined, and the complementary sequence is underlined and in boldface.
GPC3sh1: 5′-GATCCGGAGCTCAAGTTCTTAATTATCTTCCTGTCAGAATAATTAAGAACTTGAGCTCCTTTTTG-3′
GPC3sh2: 5′GATCCACTGCAAGTCACTAGGATCTTCTTCCTGTCAGAAAGATCCTAGTGACTTGCAGTTTTTTG-3′
GPC3sh3: 5′-GATCCATTCTCAACAACGCCAATATACTTCCTGTCAGATATATTGGCGTTGTTGAGAATTTTTTG-3′
The universal control for knockdown is a scrambled sequence that does not target any mammalian expressed sequence. Oligo for the sequence of scrambled shRNA is
5′-GATCCGCGTAATAACGATGTCTCTACTTCCTGTCAGATAGAGACATCGT
TA-3′
The insert DNA sequence coding yap shRNA is
5′-GATCCGCTCAGCATCTTCGACAGTCTCTTCCTGTCAGAAGACTGTCGAA
GATGCTGAGC TTTTTG-3′
Yap Overexpression.
A constitutively active yap mutant, yap-S127A (38), was used to transform Hep3B cells. The yap-S127A coding sequence (GenBank accession number: NM_001130145.2) was cloned into lentiviral vector pLenti6_3_v5 (Invitrogen). The resultant expression vector was packaged into lentivirus that was used to infect Hep3B cells.
Preparation of Recombinant GPC3.
The coding sequence for GPC3 is based on GenBank accession number NP_004475.1. To make full-length mature GPC3-hFc, the predicted N-terminal secretion signal and C-terminal GPI attachment peptide were removed, a sequence coding for amino acids 25–550 was fused with hFc (hindge-CH2-CH3) at the C terminus, and an IL-2 secretion signal (amino acids 1–20) was added at the N terminus. The expression cassette was cloned into expression vector pVRC8400 and then was transfected into 293F cells (Invitrogen). Protein purification was accomplished with a protein A affinity column from GE Healthcare. Different GPC3 mutant or truncation forms were generated: the GPC3∆HS-hFc (S495A and S509A), GPC3N-hFc (Q25–R358), and GPC3C-hFc (S359–S550).
Phage Display and Panning Method.
A combinatorial human VH domain library, with an estimated size of 2.5 × 1010, was previously constructed (31). The antigen used for panning the library was full-length GPC3-hFc, which was expressed and purified from 293F suspension cells. Library bacterial stock was inoculated into 2.5 L of 2YT medium containing 2% (wt/vol) glucose and 100 µg/mL ampicillin and was cultured at 37 °C with shaking at 250 rpm. When the cells reached midlog phase (OD600 between 0.4–0.8), superinfection was performed by adding helper phage M13KO7 at 5 × 109 pfu/mL After 1 h of continued growth, the cells were resuspended in 2.5 L of 2YT medium containing 100 µg/mL ampicillin and 50 µg/mL kanamycin and were incubated at 25 °C overnight. After the cells were centrifuged and filtered with a 0.22-µm membrane, the supernatant was stored at 4 °C for panning.
For panning, a 96-well ELISA plate (Maxisorb; Nunc/Thermo Fisher Scientific) was used to capture various amounts of purified GPC3-hFc in PBS buffer at 4 °C overnight. After the coating buffer was decanted, the plate was treated with blocking buffer [2% (wt/vol) BSA in PBS] at room temperature for 1 h. Then 30 µL of preblocked phage supernatant (typically containing 1010–1011 pfu) in 30 µL blocking buffer was added per well for 1 h at room temperature to allow binding. After four washes with PBS containing 0.05% Tween-20, bound phages were eluted with 100 mM triethylamine. We used 5 µg immobilized GPC3 for the first round of panning and 0.5 µg for the second, third, and fourth rounds of panning. After four rounds of panning, single colonies were picked and identified as GPC3 binders by phage ELISA and phage FACS.
HN3 Expression and Purification.
The coding sequence of HN3 was cloned into the lentiviral expression vector pLenti6_3_v5 (Invitrogen). The resultant vector was packaged into lentivirus that was used to infect CHO-S cells. After antibody productivity screening of 960 clones by ELISA and subsequent characterization, a stable cell line (9B2) was generated. The volumetric titer of the secreted HN3 in batch culturing was about 110 mg/L. Purification was carried out with a protein A column (GE Healthcare) according to the manufacturer’s instructions.
ELISA.
Purified GPC3 was used to coat a 96-well plate at 1 μg/mL in PBS buffer, 50 μL per well, at 4 °C overnight. After the plate was blocked with 2% BSA in PBS buffer, biotinylated HN3 antibody solution was added, and the plate was incubated at room temperature for 1 h to allow binding to occur. After the plate was washed twice with PBS buffer containing 0.05% Tween 20, the binding was detected by a streptavidin-HRP conjugate (Invitrogen).
To measure the HN3 affinity, various amounts of HN3 were incubated. The A450 values were associated with corresponding HN3 concentration, and the Kd value was determined by Prism 5.0 software (GraphPad Software) using the two binding sites (hyperbola) method.
To measure the concentration of mouse serum HN3 antibody, the plate was coated with AffiniPure F(ab′)2 fragment goat anti-human IgG (Jackson ImmunoResearch Inc.), dilutions of mouse serum were added to the plate, and detection was performed by a secondary goat anti-human antibody conjugated with HRP (Jackson ImmunoResearch Inc.). Both the capturing and detecting antibodies were validated to have no cross-reactivity with mouse serum.
For phage ELISA, a 96-well ELISA plate (Maxisorb; Nunc/Thermo Fisher Scientific) was coated with 50 µL of GPC3-hFc (5 µg/mL) overnight at 4 °C. After blocking, 30 µL of preblocked phage supernatant (typically 1010–1011 pfu) was added to the plate. Binding was detected by HRP-conjugated mouse anti-M13 antibody (GE Healthcare).
Flow Cytometry Method.
Cells were harvested in cell dissociation solution (Invitrogen), washed, and resuspended in ice-cold PBS containing 5% (wt/vol) BSA. One million cells per milliliter were incubated with 10 μg/mL of HN3 antibody and hIgG isotype control (Sigma-Aldrich). Binding was detected with goat anti-human IgG conjugated with phycoerythrin (Sigma-Aldrich). The fluorescence associated with the live cells was measured using a FACS Calibur (BD Biosciences).
To measure cellular binding affinity, various amounts of HN3 antibody were incubated with G1 cells. The geometric mean values were associated with the corresponding HN3 concentration, and the Kd value was determined using Prism 5.0 software (GraphPad Software) using the two binding sites (hyperbola) method.
For the phage FACS, 30 µL of phage supernatant (∼1010 phages) was preblocked with FACS buffer (5% BSA in PBS) for 1 h on ice and then was mixed with 106 G1 cell suspension and incubated for 1 h on ice. The binding was detected by a mouse anti-M13 primary antibody and phycoerythrin-conjugated goat anti-mouse secondary antibody (Sigma-Aldrich).
Cell-Cycle Analysis.
Cells were treated with 1 µM HN3 for 5 d and then were collected and fixed with 70% (vol/vol) ethanol overnight. Fixed cells were stained with propidium iodide and analyzed by FACS. The cell-cycle distribution of different phases was analyzed with FlowJo v 9.0 (Tree Star, Inc.).
Cell-Proliferation Assay.
Cell growth was assessed by WST assay using the Cell Counting Kit-8 (Dojindo). Five hundred microliters of cells were seeded on a 24-well plate (1 × 104 cells per well), and HN3, YP7, or hIgG was added at the indicated concentrations. The cells were incubated at 37 °C for 5 d; then the cell-viability assay was performed according to the manufacturer's instructions.
HN3 Pharmacokinetics in Mice.
Twelve mice were assigned to three groups with four mice in each group. Each animal received a single i.v. dose of 3 mg/kg of HN3 antibody via the tail vein. Blood samples were collected from four mice at 10 min, 30 min, 6 h, and at 1, 2, 3, 4, 5, 6, 7, 8 d after dose. Serum antibody concentrations were detected using ELISA. Group mean serum antibody concentration–time profiles were used to estimate pharmacokinetic parameters using Phoenix WinNonlin software (version 5.2.1; Pharsight Corp.). A two-compartment elimination model with i.v. bolus input and first-order elimination, and with microconstants, was used to describe the observed data. Concentrations were weighted using iterative reweighting (1/y) and the Gauss–Newton (Levenberg and Hartley) algorithm. All procedures used in mouse pharmacokinetic and drug-efficacy studies followed the protocol (LMB-059) approved by the Institutional Animal Care and Use Committee at the National Institutes of Health.
In Vivo Efficacy Study.
Ten million Huh-7 and HepG2 cells were injected s.c. into nude mice. After a tumor formed and reached a size of 100 mm3, the treatment was started by i.v. delivery of HN3 antibody, twice a week, 60 mg/kg body weight. Tumor dimensions were determined twice a week with a caliper. Tumor volume in cubic millimeters was calculated by the formula: (a) × (b2) × 0.5, where a is tumor length and b is tumor width in millimeters.
Statistical Analysis.
All statistical analyses were conducted using GraphPad Prism5 software (GraphPad Software, Inc). Differences between groups were analyzed using the two-tailed Student t test of means. HN3-binding curves were plotted using nonlinear least square fit. Kd values were calculated by using the two binding sites (hyperbola) method.
Acknowledgments
We thank Shawn Spencer for calculating the HN3 half-life in vivo; our colleagues Yen Phung (National Cancer Institute; NCI) for generating the YP7 mAb and Heungnam Kim (NCI) for establishing the A431/G1 cell line used in the present project; and the National Institutes of Health (NIH) Fellows Editorial Board for editorial assistance. This work was supported by Grants Z01 BC 010891 and ZIA BC 010891 from the Intramural Research Program, Center for Cancer Research, NCI, NIH (to M.H.).
Footnotes
Conflict of interest statement: M.F., W.G., D.S.D., and M.H. are the coinventors on a patent application filed by the National Institutes of Health/National Cancer Institute on subject matter disclosed in this manuscript.
See Author Summary on page 4447 (volume 110, number 12).
This article is a PNAS Direct Submission.
References
- 1.Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: Worldwide incidence and trends. Gastroenterology. 2004;127(5) Suppl 1:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 3.Cao H, Phan H, Yang LX. Improved chemotherapy for hepatocellular carcinoma. Anticancer Res. 2012;32(4):1379–1386. [PubMed] [Google Scholar]
- 4.Filmus J, Selleck SB. Glypicans: Proteoglycans with a surprise. J Clin Invest. 2001;108(4):497–501. doi: 10.1172/JCI13712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho M, Kim H. Glypican-3: A new target for cancer immunotherapy. Eur J Cancer. 2011;47(3):333–338. doi: 10.1016/j.ejca.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim MS, Saunders AM, Hamaoka BY, Beachy PA, Leahy DJ. Structure of the protein core of the glypican Dally-like and localization of a region important for hedgehog signaling. Proc Natl Acad Sci USA. 2011;108(32):13112–13117. doi: 10.1073/pnas.1109877108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svensson G, Awad W, Håkansson M, Mani K, Logan DT. Crystal structure of N-glycosylated human glypican-1 core protein: Structure of two loops evolutionarily conserved in vertebrate glypican-1. J Biol Chem. 2012;287(17):14040–14051. doi: 10.1074/jbc.M111.322487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumhoer D, et al. Glypican 3 expression in human nonneoplastic, preneoplastic, and neoplastic tissues: A tissue microarray analysis of 4,387 tissue samples. Am J Clin Pathol. 2008;129(6):899–906. doi: 10.1309/HCQWPWD50XHD2DW6. [DOI] [PubMed] [Google Scholar]
- 9.Llovet JM, et al. A molecular signature to discriminate dysplastic nodules from early hepatocellular carcinoma in HCV cirrhosis. Gastroenterology. 2006;131(6):1758–1767. doi: 10.1053/j.gastro.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 10.Zhu ZW, et al. Enhanced glypican-3 expression differentiates the majority of hepatocellular carcinomas from benign hepatic disorders. Gut. 2001;48(4):558–564. doi: 10.1136/gut.48.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pilia G, et al. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat Genet. 1996;12(3):241–247. doi: 10.1038/ng0396-241. [DOI] [PubMed] [Google Scholar]
- 12.Cano-Gauci DF, et al. Glypican-3-deficient mice exhibit developmental overgrowth and some of the abnormalities typical of Simpson-Golabi-Behmel syndrome. J Cell Biol. 1999;146(1):255–264. doi: 10.1083/jcb.146.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu B, et al. Suppression of liver regeneration and hepatocyte proliferation in hepatocyte-targeted glypican 3 transgenic mice. Hepatology. 2010;52(3):1060–1067. doi: 10.1002/hep.23794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zittermann SI, Capurro MI, Shi W, Filmus J. Soluble glypican 3 inhibits the growth of hepatocellular carcinoma in vitro and in vivo. Int J Cancer. 2010;126(6):1291–1301. doi: 10.1002/ijc.24941. [DOI] [PubMed] [Google Scholar]
- 15.Feng M, Kim H, Phung Y, Ho M. Recombinant soluble glypican 3 protein inhibits the growth of hepatocellular carcinoma in vitro. Int J Cancer. 2011;128(9):2246–2247. doi: 10.1002/ijc.25549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun CK, Chua MS, He J, So SK. Suppression of glypican 3 inhibits growth of hepatocellular carcinoma cells through up-regulation of TGF-β2. Neoplasia. 2011;13(8):735–747. doi: 10.1593/neo.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee KP, et al. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107(18):8248–8253. doi: 10.1073/pnas.0912203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu L, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA. 2010;107(4):1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou D, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16(5):425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Midorikawa Y, et al. Glypican-3, overexpressed in hepatocellular carcinoma, modulates FGF2 and BMP-7 signaling. Int J Cancer. 2003;103(4):455–465. doi: 10.1002/ijc.10856. [DOI] [PubMed] [Google Scholar]
- 21.Capurro M, et al. Glypican-3: A novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125(1):89–97. doi: 10.1016/s0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- 22.Hippo Y, et al. Identification of soluble NH2-terminal fragment of glypican-3 as a serological marker for early-stage hepatocellular carcinoma. Cancer Res. 2004;64(7):2418–2423. doi: 10.1158/0008-5472.can-03-2191. [DOI] [PubMed] [Google Scholar]
- 23.Yamauchi N, et al. The glypican 3 oncofetal protein is a promising diagnostic marker for hepatocellular carcinoma. Mod Pathol. 2005;18(12):1591–1598. doi: 10.1038/modpathol.3800436. [DOI] [PubMed] [Google Scholar]
- 24.Phung Y, Gao W, Man YG, Nagata S, Ho M. High-affinity monoclonal antibodies to cell surface tumor antigen glypican-3 generated through a combination of peptide immunization and flow cytometry screening. MAbs. 2012;4(5):592–599. doi: 10.4161/mabs.20933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishiguro T, et al. Anti-glypican 3 antibody as a potential antitumor agent for human liver cancer. Cancer Res. 2008;68(23):9832–9838. doi: 10.1158/0008-5472.CAN-08-1973. [DOI] [PubMed] [Google Scholar]
- 26.Nakano K, et al. Generation of a humanized anti-glypican 3 antibody by CDR grafting and stability optimization. Anticancer Drugs. 2010;21(10):907–916. doi: 10.1097/CAD.0b013e32833f5d68. [DOI] [PubMed] [Google Scholar]
- 27.Nakano K, et al. Anti-glypican 3 antibodies cause ADCC against human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2009;378(2):279–284. doi: 10.1016/j.bbrc.2008.11.033. [DOI] [PubMed] [Google Scholar]
- 28.Henderson KA, et al. Structure of an IgNAR-AMA1 complex: Targeting a conserved hydrophobic cleft broadens malarial strain recognition. Structure. 2007;15(11):1452–1466. doi: 10.1016/j.str.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23(9):1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 30.Stijlemans B, et al. Efficient targeting of conserved cryptic epitopes of infectious agents by single domain antibodies. African trypanosomes as paradigm. J Biol Chem. 2004;279(2):1256–1261. doi: 10.1074/jbc.M307341200. [DOI] [PubMed] [Google Scholar]
- 31.Chen W, Zhu Z, Feng Y, Xiao X, Dimitrov DS. Construction of a large phage-displayed human antibody domain library with a scaffold based on a newly identified highly soluble, stable heavy chain variable domain. J Mol Biol. 2008;382(3):779–789. doi: 10.1016/j.jmb.2008.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stork R, et al. N-glycosylation as novel strategy to improve pharmacokinetic properties of bispecific single-chain diabodies. J Biol Chem. 2008;283(12):7804–7812. doi: 10.1074/jbc.M709179200. [DOI] [PubMed] [Google Scholar]
- 33.Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11(1):11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 34.Kilili GK, Kyriakis JM. Mammalian Ste20-like kinase (Mst2) indirectly supports Raf-1/ERK pathway activity via maintenance of protein phosphatase-2A catalytic subunit levels and consequent suppression of inhibitory Raf-1 phosphorylation. J Biol Chem. 2010;285(20):15076–15087. doi: 10.1074/jbc.M109.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D, et al. Regulation of proapoptotic mammalian ste20-like kinase MST2 by the IGF1-Akt pathway. PLoS ONE. 2010;5(3):e9616. doi: 10.1371/journal.pone.0009616. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.O’Neill E, Kolch W. Taming the Hippo: Raf-1 controls apoptosis by suppressing MST2/Hippo. Cell Cycle. 2005;4(3):365–367. doi: 10.4161/cc.4.3.1531. [DOI] [PubMed] [Google Scholar]
- 37.Romano D, et al. Proapoptotic kinase MST2 coordinates signaling crosstalk between RASSF1A, Raf-1, and Akt. Cancer Res. 2010;70(3):1195–1203. doi: 10.1158/0008-5472.CAN-09-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizuno T, et al. YAP induces malignant mesothelioma cell proliferation by upregulating transcription of cell cycle-promoting genes. Oncogene. 2012;31(1):1–6. doi: 10.1038/onc.2012.5. [DOI] [PubMed] [Google Scholar]
- 39.Chen W, et al. NSC 74859-mediated inhibition of STAT3 enhances the anti-proliferative activity of cetuximab in hepatocellular carcinoma. Liver Int. 2012;32(1):70–77. doi: 10.1111/j.1478-3231.2011.02631.x. [DOI] [PubMed] [Google Scholar]
- 40.Huether A, Höpfner M, Baradari V, Schuppan D, Scherübl H. EGFR blockade by cetuximab alone or as combination therapy for growth control of hepatocellular cancer. Biochem Pharmacol. 2005;70(11):1568–1578. doi: 10.1016/j.bcp.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 41.Aden DP, Fogel A, Plotkin S, Damjanov I, Knowles BB. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature. 1979;282(5739):615–616. doi: 10.1038/282615a0. [DOI] [PubMed] [Google Scholar]
- 42.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42(9):3858–3863. [PubMed] [Google Scholar]
- 43.Kaneko O, et al. A binding domain on mesothelin for CA125/MUC16. J Biol Chem. 2009;284(6):3739–3749. doi: 10.1074/jbc.M806776200. [DOI] [PMC free article] [PubMed] [Google Scholar]