Abstract
Estrogen receptor β (ERβ) promotes the degradation of hypoxia inducible factor 1α (HIF-1α), which contributes to the ability of this hormone receptor to sustain the differentiation of epithelial and carcinoma cells. Although the loss of ERβ and consequent HIF-1 activation occur in prostate cancer with profound consequences, the mechanism by which ERβ promotes the degradation of HIF-1α is unknown. We report that ERβ regulates the ligand (3β-adiol)-dependent transcription of prolyl hydroxylase 2 (PHD2) also known as Egl nine homolog 1 (EGLN1), a 2-oxoglutarate-dependent dioxygenase that hydroxylates HIF-1α and targets it for recognition by the von Hippel-Lindau tumor suppressor and consequent degradation. ERβ promotes PHD2 transcription by interacting with a unique estrogen response element in the 5′ UTR of the PHD2 gene that functions as an enhancer. PHD2 itself is critical for maintaining epithelial differentiation. Loss of PHD2 expression or inhibition of its function results in dedifferentiation with characteristics of an epithelial–mesenchymal transition, and exogenous PHD2 expression in dedifferentiated cells can restore an epithelial phenotype. Moreover, expression of HIF-1α in cells that express PHD2 does not induce dedifferentiation but expression of HIF-1α containing mutations in the proline residues that are hydroxylated by PHD2 induces dedifferentiation. These data describe a unique mechanism for the regulation of HIF-1α stability that involves ERβ-mediated transcriptional regulation of PHD2 and they highlight an unexpected role for PHD2 in maintaining epithelial differentiation.
The role of estrogen receptors (ERs), which are transcription factors belonging to the steroid/thyroid nuclear receptor superfamily (1–3), in regulating epithelial differentiation is an emerging area of considerable biological interest and pathological relevance. In the prostate, the discovery of ERβ (4, 5) has generated intense interest in the roles played by this ER in several tissues including prostate and breast epithelia (6–11). Increasing evidence supports the hypothesis that ERβ functions to maintain epithelial differentiation in the prostate and breast (9, 10, 12, 13). In the normal prostate, ERβ contributes to epithelial differentiation as evidenced by the observation that ERβ knockout mice exhibit altered differentiation in the ventral prostate, whereas the glands of ERα knockout mice lack these lesions and appear to be normal (12). ERβ in human prostate cancer is of substantial relevance because there is an inverse relationship between the expression of ERβ and highly invasive prostate cancer (9, 14). In pursuit of a functional basis for this relationship, we demonstrated that ERβ sustains an epithelial phenotype and impedes a mesenchymal transition in prostate cancer and we identified a metabolite of dihydrotestosterone, 5α-androstane-3β,17β4-diol (3β-adiol) as the specific ERβ ligand that mediates this function (9). This observation is in agreement with the recent findings that 3β-adiol is a natural ligand for ERβ in prostate (13, 15–17).
A key issue that arises from the above observations is how loss of ERβ promotes a de-differentiated, epithelial mesenchymal transition (EMT) phenotype. We reported that ERβ has a causal role in the genesis of this phenotype because it impedes the expression and activation of hypoxia inducible factor 1 (HIF-1). More specifically, we made the seminal finding that 3β-adiol/ERβ destabilize HIF-1α by promoting its proteasomal degradation (9). Consequently, HIF-1α is stabilized upon loss of ERβ expression or function, enabling HIF-1–mediated transcription. Several HIF-target genes, including VEGF, lysyl oxidase, and TWIST, have the ability to promote epithelial dedifferentiation (9, 18–20). A challenging problem that emerges from these findings is how a nuclear hormone receptor induces the degradation of HIF-1α (21). HIF-1α is degraded in normoxia by a well-established mechanism that involves its hydroxylation on specific prolines by prolyl hydroxylases (PHDs), which target HIF-1α for recognition by the E3 ligase von Hippel-Lindau (VHL) and consequent degradation in the proteosome (22–24). More specifically, HIF-1α is hydroxylated on two conserved proline residues (p402 and p564), which allows for its interaction with VHL E3 ubiquitin ligase for subsequent polyubiquitination and proteasomal degradation (22). The primary PHD that targets HIF-1α under normal conditions is PHD2, also called Egl nine homolog 1 (EGLN1) (22, 25).
In our quest to understand how ERβ destabilizes HIF-α, we pursued the hypothesis that ERβ regulates specific prolyl hydroxylases. The results obtained demonstrate that ERβ regulates the transcription of PHD2 but not other PHD genes and that this regulation provides a mechanism for how a nuclear hormone receptor controls HIF-1α stability. They also reveal an unexpected role for PHD2 in regulating epithelial differentiation.
Results
PHD2 Expression Is Regulated by Ligand-Dependent Activation of ERβ in Epithelial Cells.
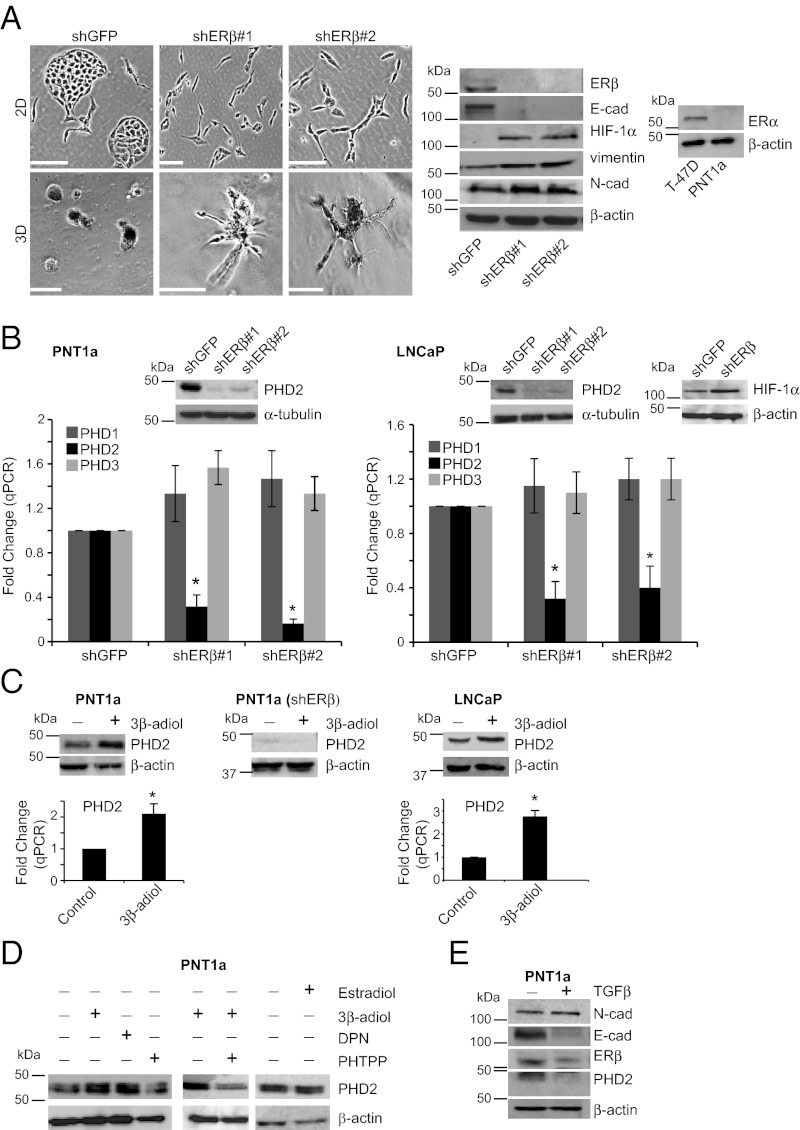
PNT1a cells are immortalized, normal prostate epithelial cells (26) that express ERβ but lack ERα (Fig. 1A). Depletion of ERβ in these cells using shRNAs disrupts their cobblestone, epithelial appearance in 2D culture and promotes a mesenchymal phenotype as evidenced by their morphology, loss of E-cadherin and increased HIF-1α, vimentin, and N-cadherin expression (Fig. 1A). PNT1a cells form compact spheroids in 3D culture that are characteristic of other epithelial cells in 3D (27), but loss of ERβ induces invasive outgrowths (Fig. 1A), similar to that observed for other EMT cells in 3D (28) (29). These data support the hypothesis that ERβ contributes to the maintenance of an epithelial phenotype in normal prostate epithelial cells.
Fig. 1.
PHD2 expression is regulated by ligand-dependent activation of ERβ in epithelial cells. (A) ERβ ablation induces dedifferentiaton in PNT1a epithelial cells. ERb expression was ablated in PNT1a cells using two independent shRNAs (shERb#1 and shERb#2). These cells and control cells (shGFP) were maintained in either 2D or 3D culture. Control cells exhibit a distinct epithelial phenotype in 2D and a spheroid shape with a distinct polarity in 3D. In contrast, loss of ERβ expression promotes a mesenchymal phenotype in 2D and invasive outgrowths in 3D. (Scale bars, 100 μm.) Morphological changes observed in response to loss of ERβ are accompanied by diminished E-cadherin expression and an increase in mesenchymal markers (N-cadherin, vimentin, and HIF-1α). Immunoblot on the Far Right demonstrates that PNT1a cells lack expression of ERα in comparison with T47D breast carcinoma cells. (B) ERβ ablation suppresses PHD2 expression. ERβ-ablated PNT1a cells were assessed for PHD2 expression by qPCR and immunoblotting. Loss of ERβ expression is accompanied by a marked reduction in PHD2 mRNA and protein compared with the control cells. The reduction of PHD2 expression as a consequence of ERβ ablation is specific because there is no effect on PHD1 and PHD3 mRNA (*P < 0.05). Immunoblot on the Far Right demonstrates that loss of ERβ in LNCaP cells induces HIF-1α. (C) PHD2 expression is induced by an ERβ ligand, 3β-adiol. PNT1a and LNCaP cells were incubated with DMSO (control) or 5 µM 3β-adiol for 2 d. Cells were examined for PHD2 expression by qPCR and immunoblotting. 3β-Adiol enhances PHD2 mRNA and protein expression significantly. In contrast, 3β-adiol had no effect on PHD2 expression in ERβ-ablated PNT1a cells. (D) 3β-Adiol and DPN but not estradiol enhance PHD2 expression. PHTPP attenuates PHD2 expression in either the absence or presence of 3β-adiol. (E) TGF-β–induced EMT suppresses PHD2 expression. PNT1a cells were cultured in the absence or presence of TGF-β (5 ng/mL) for 3 d and harvested for immunoblotting. TGF-β treatment diminishes PHD2 expression significantly with a concomitant decrease in E-cadherin and ERβ.
To examine the putative relationship between ERβ and PHDs, we quantified the expression of PHD1, PHD2, and PHD3 mRNAs in control and ERβ-depleted PNT1a cells. As shown in Fig. 1B, ERβ contributes to the expression of PHD2 but not to either PHD1 or PHD3. This observation is consistent with the loss of PHD2 protein expression observed in ERβ-depleted cells (Fig. 1B). These data were confirmed using LNCaP cells, a differentiated prostate carcinoma cell line derived from a lymph node metastasis (Fig. 1B). LNCaP cells express ERβ but lack ERα (8). Loss of ERβ also promotes the expression of HIF-1α in LNCaP cells (Fig. 1B).
The ligand dependency of ERβ-mediated regulation of PHD2 expression was evaluated by treating both PNT1a and LNCaP cells with 3β-adiol, a natural ligand for ERβ in the prostate (9, 13, 15–17). Indeed, 3β-adiol treatment caused a significant increase in PHD2 mRNA and protein expression in both cell types (Fig. 1C). In contrast, 3β-adiol did not have any effect on PHD2 expression in ERβ-ablated PNT1a cells (Fig. 1C), indicating that the effect of 3β-adiol is ERβ dependent. The ligand specificity of ERβ in regulating PHD2 was substantiated by comparing the effects of the ERβ agonists 3β-adiol and diarylpropionitrile (DPN), the ERβ antagonist 4-[2-Phenyl-5,7-bis(trifluoromethyl)pyrazolo[1,5-a]pyrimidin-3-yl]phenol (PHTPP) and estradiol on PHD2 expression. Both 3β-adiol and DPN enhanced PHD2 expression in PNT1a cells, whereas estradiol had no effect. In contrast, PHTPP attenuated PHD2 expression in either the absence or presence of 3β-adiol providing further evidence that 3β-adiol is the specific ERβ ligand that regulates PHD2 expression (Fig. 1D).
The hypothesis that ERβ contributes to epithelial differentiation by a mechanism that involves PHD2 implies that a physiological stimulus that induces a mesenchymal transition such as TGF-β will impede PHD2 expression or function. Indeed, TGF-β stimulation of PNT1a cells promotes a mescenchymal transition as evidenced by an increase in N-cadherin and a decrease in E-cadherin and ERβ1 (Fig. 1E). Importantly, there is a concomitant decrease in PHD2 expression in TGF-β–treated cells compared with control cells (Fig. 1E).
ERβ Regulates PHD2 Expression via a Unique Estrogen Response Element in the 5′ UTR of the PHD2 Gene.
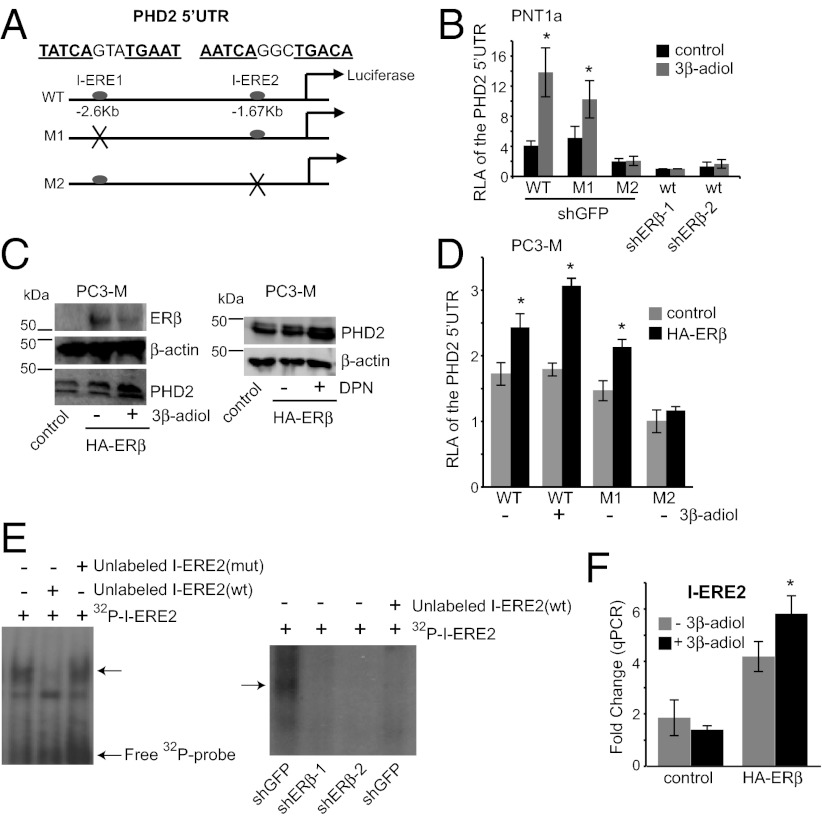
We next sought to define the mechanism by which ERβ regulates PHD2 expression. Given that ERβ is a transcription factor, we searched for estrogen response elements (EREs) in the 5′ UTR of the PHD2 gene that could mediate ERβ binding and transcriptional activation. Although we found no consensus ERE (GGTCAnnnTGACC) within the 5′ UTR up to −4.0 kb from the transcription start site, we discovered two imperfect EREs (I-EREs) located at −2597 bp (TATCAnnnTGAAT) and −1670 bp (AATCAnnnTGACA). To assess the functional importance of I-ERE1 and I-ERE2, we cloned the 927-base-pair (bp) sequence containing these two elements into a luciferase reporter construct driven by the SV40 promoter (Fig. 2A). These two elements were also mutated by site-directed mutagenesis to generate M1 mutant (TAAAAnnnAAAAT) and M2 mutant (AAAAAnnnAAACA) constructs (Fig. 2A). These three luciferase constructs were transfected into PNT1a and luciferase reporter assays were performed after the cells were incubated in the absence or presence of 3β-adiol for 20 h. As shown in Fig. 2B, the M2 mutant exhibited a significant reduction in luciferase activity compared with the M1 mutant and wild-type constructs either in the absence or presence of 3β-adiol. Loss of ERβ expression resulted in a significant reduction in the activity of the wild-type construct that was comparable to that of the M2 mutant. More importantly, ligand-induced transcriptional activation was detected only with the wild-type and M1 constructs. These data indicate that I-ERE2 but not I-ERE1 functions as an enhancer for ERβ regulation of PHD2 transcription.
Fig. 2.
ERβ regulates PHD2 transcription via a unique ERE, located in the 5′ UTR of the PHD2 gene. (A) Schematic of the wild-type and mutant luciferase reporter constructs used. A 927-bp fragment (−2597 bp to −1670 bp), containing two variant EREs (I-ERE1: TATCAgtaTGAAT and I-ERE2: AATCAggcTGACA), was cloned into a pGL3-promoter construct (WT). M1 represents the mutated version of I-ERE1 (TAAAAGTAAAAAT), whereas M2 represents the mutated version of I-ERE2 (AAAAAGGCAAACA). These constructs were used for subsequent experiments. (B) PNT1a cells were transfected with WT, M1, or M2 luciferase constructs. ERβ-ablated cells (shERβ-1 and shERβ-2) were transfected with the WT construct together with Renilla for normalization. Cells were subsequently incubated for 20 h in the absence or presence of 3β-adiol. Note that the relative luciferase activity (RLA) of the M2 construct is significantly less than that of the WT and M1 constructs and comparable to that of the wild-type construct in ERβ-ablated cells in the absence or presence of 3β-adiol (*P < 0.05). However, only the wild-type and M1 constructs exhibit ligand responsiveness (two- to threefold increase in luciferase activity). (C) Ligand-activated ERβ promotes PHD2 expression in PC3-M cells. PC3-M cells, which have an undetectable level of ERβ, were infected with an empty vector or an HA-ERβ expression vector. ERβ expressing cells (lanes 2 and 3) were incubated with DMSO (control), 3β-adiol (5 µM), or DPN (30 nM) for 24 h and harvested for immunoblotting. ERβ expression alone increases PHD2 expression and this effect is enhanced by either 3β-adiol or DPN. (D) Analyses of ERE-luciferase activity in PC3-M cells. PC3-M cells that express an HA-ERβ or an empty vector (control) were transfected with either WT (±3β-adiol), M1, or M2 constructs together with Renilla. Expression of HA-ERβ increases luciferase activity, which is enhanced by 3β-adiol (*P < 0.05). This enhancement is not apparent with M2. (E) EMSA assays to detect ERβ–DNA complex formation in vitro. Nuclear extracts containing ERβ from 3β-adiol–treated PNT1a cells or ERβ-ablated cells were incubated with 32P-labeled I-ERE2 (WT) in the absence or presence of unlabeled competitors. A distinct protein–DNA complex was detected (arrow) in PNT1a cells but not in ERβ-ablated cells (shERβ-1 and shERβ-2) and this complex was displaced by unlabeled I-ERE2 (WT) but not I-ERE2 (mut) confirming its specificity. (F) ChIP analysis of HA-ERβ–expressing PC3-M cells. PC3-M (+HA-ERβ) or PC3-M cells (empty vector) were incubated in the absence or presence of 3β-adiol for 48 h. Either an HA antibody or IgG (control) were used to precipitate the chromatin. Enrichment on the ERE2 locus by HA was normalized to IgG by qPCR. I-ERE2 mRNA levels exhibit a fourfold induction in HA-ERβ–expressing cells and a sixfold induction upon treatment with 3β-adiol. In contrast, no enrichment in I-ERE2 mRNA is apparent in control PC3-M cells (*P < 0.05) under the same conditions.
Additional insight into the mechanism by which ERβ regulates PHD2 transcription was obtained using PC3-M prostate carcinoma cells, a metastatic variant of PC3 cells (30). These cells have a mesenchymal phenotype and express levels of ERβ below the detectable limit (Fig. 2C). We expressed HA-tagged ERβ (HA-ERβ) in PC3-M cells and observed an increase in PHD2 expression that was amplified by either 3β-adiol or DPN treatment (Fig. 2C). The activity of the wild-type, M1, and M2 luciferase constructs described above was compared in control and HA-ERβ PC3-M cells. Expression of HA-ERβ significantly enhanced the transcriptional activity of the wild-type construct. In contrast, mutation of ERE2 (M2), but not ERE1 (M1) abolished this enhancement, indicating a functional link between the I-ERE2 site and ERβ. Treatment of HA-ERβ PC3-M with 3β-adiol further potentiated the transcriptional activity of the wild-type construct but not the M2 construct, whereas the control cells did not respond to this hormone (Fig. 2D).
To establish that I-ERE2 is a bona fide binding site for ERβ in vitro and in vivo, we performed EMSA and ChIP analyses. A distinct protein–DNA complex formation was detected in extracts of 3β-adiol–treated PNT1a cells but not ERβ-ablated cells in EMSA assays (Fig. 2E). This complex formation could be displaced by unlabeled wild-type I-ERE2 oligonucleotide, but not by a mutated version confirming its specificity. ChIP analysis was performed using control and HA-ERβ PC3-M cells, and the results were quantified by qPCR. We detected a fourfold HA/IgG enrichment on the I-ERE2 locus in HA-ERβ PC3-M cells with no significant enrichment observed in the control cells (Fig. 2F). Treatment of these cells with 3β-adiol resulted in a sixfold induction in binding, indicating that the binding is enhanced by ligand activation. These data strongly support the conclusion that ERβ regulates PHD2 transcription by binding to a unique ERE (I-ERE2) that serves as an enhancer element located 1.67 kb away from the transcription start site.
PHD2 Sustains Epithelial Differentiation.
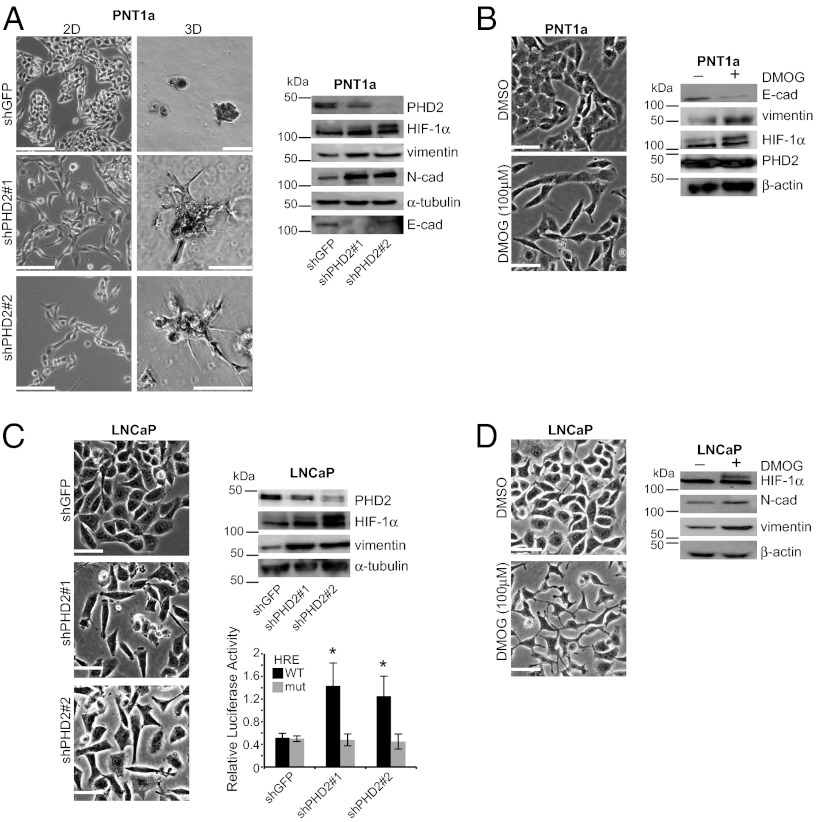
Our findings raised the unexpected possibility that PHD2 itself has a causal role in epithelial differentiation. To address this possibility, we depleted PHD2 expression in PNT1a and LNCaP cells. As shown in Fig. 3A, loss of PHD2 expression in PNT1a cells resulted in the acquisition of a mesenchymal morphology in 2D culture and invasive outgrowths in 3D culture. These morphological changes were accompanied by diminished E-cadherin expression and increased expression of HIF-1α, vimentin, and N-cadherin. The ability of PHD2 to maintain epithelial differentiation was substantiated using dimethyloxoglutarate glycine (DMOG), an inhibitor of PHD2 enzymatic activity (31, 32). DMOG treatment of PNT1a cells for 2 d resulted in a mesenchymal phenotype with the same changes in the expression of epithelial and mesenchymal markers observed with loss of PHD2 (Fig. 3B). Similar results were observed in LNCaP cells for both PHD2 depletion and DMOG treatment (Fig. 3 C and D). These data indicate that loss of PHD2 expression or activity promotes epithelial dedifferentiation.
Fig. 3.
PHD2 sustains epithelial differentiation. (A) PHD2 ablation promotes dedifferentiation. PHD2-ablated cells (shPHD2#1 and shPHD2#2) and control cells (shGFP) were maintained in either 2D or 3D cultures. Note that the morphology of PHD2-ablated cells is similar to that of ERβ-ablated cells in Fig. 1. (Scale bars, 100 μm.) Immunoblot analysis of these cells indicates that loss of PHD2 induces a decrease in E-cadherin expression and an increase in mesenchymal markers. (B) Inhibition of PHD2 activity induces epithelial dedifferentiation. PNT1a cells were incubated for 2 d with DMSO (control) or 100 µM DMOG, a PHD2 inhibitor. DMOG-treated cells exhibit a mesenchymal phenotype with concomitant increase in vimentin and HIF-1α expression compared with controls. (C) PHD2 ablation induces epithelial dedifferentiation and promotes HIF-1α activity in LNCaP cells. PHD2-ablated LNCaP cells (shPHD2#1 and shPHD2#2) exhibit a mesenchymal phenotype compared with control cells. Loss of PHD2 expression also induces HIF-1α and vimentin expression. These cells were transfected with a reporter gene carrying either a wild-type HRE or a mutated HRE. A significant induction in luciferase activity is seen in PHD2-ablated cells compared with control cells with the wild type HRE-reporter gene but not with the mutant HRE reporter (*P < 0.05). (D) Inhibition of PHD2 activity induces dedifferentiation in LNCaP cells. LNCaP cells were incubated with either DMSO (−) or 100 µM DMOG (+) for 2 d. DMOG-treated cells exhibit a mesenchymal morphology and an increase in vimentin and HIF-1α expression compared with control cells.
An important consideration is whether loss of PHD2 expression results in the expression of functional HIF-1α in normoxia. For this reason, we examined HIF-1α transcriptional activity in PHD2-depleted LNCaP cells using a reporter construct containing the hypoxia response element (HRE) fused to the luciferase gene. As shown in Fig, 3C, PHD2-ablated cells exhibited higher luciferase activity compared with the control cells (shGFP) and this enhanced activity was not observed using the mutated reporter construct. These data indicate that the HIF-1α induced as a consequence of PHD2 ablation is functional in normoxia.
PHD2 Expression Can Rescue an Epithelial Phenotype in Dedifferentiated Cells.
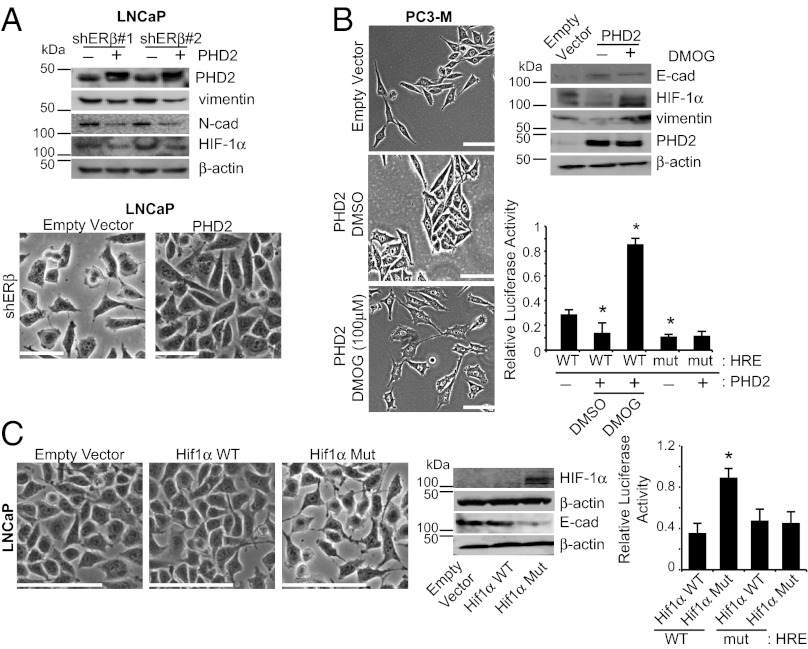
If PHD2 has a causal role in epithelial dedifferentiation, its exogenous expression in dedifferentiated cells should promote a more differentiated phenotype. Indeed, expression of PHD2 in ERβ-ablated LNCaP cells resulted in a more cobblestone, epithelial morphology and a reduction in the expression of mesenchymal markers (N-cadherin, vimentin, and HIF-1α) (Fig. 4A). This conclusion was verified using PC3-M cells, which express very low levels of ERβ and PHD2. Expression of PHD2 in these cells induced a dramatic shift to an epithelial phenotype. Importantly, this transition was reversed by treatment with the PHD2 inhibitor DMOG, providing additional evidence for the specificity of this phenomenon (Fig. 4B). Moreover, the expression of PHD2 correlates well with the level of functional HIF-1α expression as demonstrated by immunoblotting and luciferase assays (Fig. 4B).
Fig. 4.
PHD2 expression can rescue an epithelial phenotype in dedifferentiated cells. (A) Expression of PHD2 in ERβ-ablated cells induces a mesenchymal to epithelial transition. Two clones of ERβ-ablated LNCaP cells (shERβ#1 and shERβ#2) were infected with a PHD2 expression vector or empty vector (control). Expression of PHD2 promotes a more epithelial morphology with and a decrease in mesenchymal markers (vimentin, N-cadherin, and HIF-1α). (B) Expression of PHD2 in dedifferentiated cells induces epithelial differentiation that is dependent on PHD2 activity. PC3-M cells were infected with a PHD2 expression vector or empty vector (control). Note that expression of PHD2 promotes an epithelial phenotype with an increase in E-cadherin and a reduction of HIF-1α and vimentin. Subsequent treatment of the cells with DMOG (100 µM) for 3 d induces dedifferentiation and an increase in HIF-1α and vimentin. The changes in HIF-1α expression in response to PHD2 and DMOG inhibition are consistent with the activity of an HRE reporter (bar graph). (C) Expression of a HIF-1α mutant that is resistant to hydroxylation by PHD2 promotes dedifferentiation in cells with an epithelial phenotype. LNCaP cells were infected with expression vectors containing either wild-type HIF-1α, a mutant HIF-1α, or an empty vector (control). Expression of the mutant HIF-1α promotes dedifferentiation in contrast to wild-type HIF-1α. These morphologic changes are consistent with the changes in HIF-1α and E-cadherin expression (immunoblot) and the activity of an HRE reporter (bar graph: *P < 0.05). (Scale bars, 100 μm.)
Our data infer that PHD2 sustains epithelial differentiation because it targets HIF-1α for degradation and prevents HIF-mediated transcription. To test this hypothesis, a HIF-1α construct containing mutations in the two proline residues (p402 and p564) that are required for PHD2-mediated hydroxylation was expressed in LNCaP cells. Expression of a wild-type HIF-1α construct, which was used as a control, had no effect on their epithelial morphology (Fig. 4C). In contrast, expression of the mutant HIF-1α construct resulted in a mesenchymal transition with concomitant loss of E-cadherin expression (Fig. 4C). These morphological observations are supported by the fact that wild-type HIF-1α but not the proline mutant is degraded and that the proline mutant retains HIF-1 functional activity (Fig. 4C).
Discussion
The data we present implicate ERβ regulation of PHD2 transcription as a mechanism by which this nuclear hormone receptor destabilizes HIF-1α and inhibits its function, given the central role of this prolyl hydroxylase in targeting HIF-1α for degradation (22). The transcriptional regulation of a specific prolyl hydroxylase by ERβ is relevant to our understanding of how this receptor contributes to epithelial biology and cancer. Specifically, ERβ has been implicated in the differentiation of epithelial cells as indicated by the data presented here and previous studies (9, 10, 12). We argue that the ERβ regulation of PHD2-mediated HIF-1α degradation is a major factor in the ability of ERβ to prevent dedifferentiation and an EMT. This conclusion is particularly apt for prostate cancer because we reported that high-grade prostate cancer, which exhibits an EMT phenotype, is characterized by elevated HIF-1α expression and decreased expression of ERβ (9). Moreover, high-grade prostate cancers exhibit markedly elevated levels of the HIF-target gene VEGF in tumor cells (9), consistent with reports from our laboratory and others that autocrine VEGF signaling in tumor cells promotes a dedifferentiated, EMT phenotype (9, 33–36). Clearly, other HIF-target genes may contribute to this dedifferentiated state characteristic of high-grade prostate tumors, but we suggest that ERβ regulation of PHD2 has a key role in their regulation.
Our findings provide insight into the relationship between ERβ and hypoxia, especially in the context of prostate cancer. O2 is the primary regulator of PHD2 in tissues, and hypoxic conditions inhibit the activity of this enzyme (22). Our data, however, reveal that the function of this enzyme is inhibited in normoxia by loss of ERβ expression with a concomitant activation of HIF-1α. This finding is highly relevant to HIF-1 regulation in prostate cancer because there is evidence that clinically relevant hypoxia is not evident in these tumors and it has been suggested that activation of HIF-1α may be independent of tissue hypoxia (37). We propose that loss of PHD2 as a consequence of ERβ depletion contributes to HIF-1α activation, a proposition strengthened by the inverse correlation observed between ERβ and HIF-1α expression that we observed in prostate cancer (9).
We conclude from our data that ERβ regulates PHD2 transcription by interacting with a specific sequence located 1.67 kb upstream from the transcription start site that functions as a transcriptional enhancer. This enhancer is a unique variant of the consensus ERE sequence consistent with the reports that some variant EREs or half sites of the consensus ERE can function as cis-acting elements or enhancers for estrogen receptors (38, 39). Our finding also agrees with ChIP-seq data, indicating that a small percentage of ER binding sites are located within the promoter of target genes and that most are located in intergenic sites or within intragenic regions (>1 kb from either end of transcripts) (38, 39). Based on this information, it is possible that ERβ regulates PHD2 transcription by long-range chromatin interactions that involve interaction with other transcription factors such as Activator Protein1 (AP1) factors, Specific Protein1 (Sp1), Forkhead Box Protein A (FOXA), and cAMP response element binding protein 1 (CREB1) that exhibit ChIP enrichment within the PHD2 promoter region based on analysis of the ENCODE database (http://genome.ucsc.edu/). AP1 is particularly relevant because genome-wide mapping of ERβ-binding regions revealed that AP1 is recruited to a significant fraction of ERβ-binding regions (40). We also demonstrated that 3β-adiol is a specific ERβ ligand that stimulates PHD2 transcription. In contrast, estrogen was unable to stimulate PHD2 expression. These data support previous studies indicating that 3β-adiol, but not estrogen, promotes an epithelial phenotype in prostate cells (9, 16, 17). It is worth noting, however, that the PHD1 (EGLN2) gene is estrogen inducible in breast carcinoma cells (41).
Finally, our work bears on the potential role of PHD2 as a therapeutic target for cancer. PHD2 is particularly amenable to therapeutic targeting because it is an enzyme (2-oxoglutarate-dependent dioxygenase) that can be inhibited by small molecules such as DMOG (25, 31). The rationale for its targeting in cancer is based on the interesting observation that genetic inactivation of PHD2 in endothelial cells improves response to chemotherapy by normalizing blood vessels and enhancing perfusion (32, 42). This function of PHD2 in cancer, however, should be tempered by our findings because PHD2 inhibition has the potential to promote a more dedifferentiated, EMT-like phenotype associated with aggressive behavior. Indeed, it has been suggested that the EMT can increase the frequency of cancer stem cells, which could increase the propensity for tumor recurrence (43). In a more positive direction, our data strengthen the rationale for drug discovery efforts aimed at sustaining ERβ expression and diminishing HIF-1 activation, given that HIF-1 has been implicated in the pathogenesis of highly aggressive cancers (9, 23, 44). This strategy should be tumor specific, however, because HIF-1α can function as a tumor suppressor gene in some cancers (45).
Methods
Cell Culture and Reagents.
Immortalized, normal prostate epithelial cells, PNT1a were obtained from M. Littmann (Baylor College of Medicine, Houston). The human prostate cancer cell line, LNCaP was obtained from American Type Culture Collection (ATCC). PC3-M cells were obtained from R. C. Bergan (Northwestern University, Chicago). LNCaP cells were cultured in DMEM with 10% FBS, and 1% (wt/vol) streptomycin and penicillin at 37 °C in an incubator supplied with 5% CO2, whereas PNT1a and PC3-M cells were cultured in RPMI medium with the same supplements. TGF-β, 3β-adiol, DPN, PHTPP, and DMOG experiments were performed by incubating cells with recombinant human TGF-β (5 ng/mL; Peprotech), 3β-adiol (5 µM; Sigma), or the following compounds from Tocris Bioscience: DPN (30 nM), PHTPP (10 μM), and DMOG (100 µM) for 2–3 d.
Lentiviruses (pLKO.1) containing ERβ (TRCN0000003325, TRCN0000003326, TRCN0000003327, and TRCN0000003328) and PHD2 shRNA oligonucleotides (TRCN0000001044 and TRCN0000021793) or pLKO-shGFP control were purchased from Open Biosystems, titrated according to the manufacturer's instructions, and used to infect PNT1a and LNCaP cells following standard protocols. Stable cell transfectants were generated by puromycin selection (0.5 μg/mL for PNT1a and 2 μg/mL for LNCaP cells). The resultant ERβ or PHD2-depleted cells were used for subsequent experiments. To generate HA-tagged ERβ, the ERβ coding sequence was fused with HA (hemagglutin) at the N terminus with a linker sequence GGG (Gly). The resultant PCR product was confirmed by sequencing and cloned into the p-MSCV-IRES-GFP (pMIG) retroviral vector (Addgene; plasmid 9044). PC3-M cells were infected with HA-tagged ERβ or HA-tagged retroviruses. The expression efficiency was determined by quantifying the percentage of GFP-positive cells. Retroviral vectors expressing wild-type HIF-1α (Addgene; plasmid 19365: HA-HIF1α-WT-pBabe-puro) or mutant HIF-1α (Addgene; plasmid 19005: HA-HIF1α P402A/P564A-pBabe-puro), were used to generate LNCaP cells expressing wild-type or mutant HIF-α. To generate PHD2 expressing ERβ-ablated cells, an HA-EglN1-pcDNA3 construct (Addgene; plasmid 18963) was used to transfect LNCaP cells and cells were selected by G418 (5 μg/mL). The reporter gene, p11w, which contains the wild-type HRE and the mutated version, p11m fused to luciferase, were obtained from ATCC. The Renilla luciferase plasmid was purchased from Promega.
For 3D Matrigel cultures, a base layer of Matrigel (BD Bioscience; CB-40230; 200 μL per well) was overlaid in duplicate wells of a 24-well dish containing 104 cells suspended in 300 μL of a 2:1 mixture of PBS and Matrigel. The Matrigel was overlaid with complete serum-containing medium (0.5 mL per well), which was changed every 3 d.
Biochemical Analyses.
For immunoblotting, total cellular proteins were extracted in RIPA buffer (Boston Bioproducts) containing protease inhibitors (Complete Mini; Roche), cleared by centrifugation, and quantified using the Bradford assay (BioRad). Blots were incubated at 4 °C overnight with primary Abs against ERα or ERβ (GeneTex), E-cadherin (Invitrogen), N-cadherin (Invitrogen), vimentin (Dako), HIF-1α (Novus), PHD2 (Abcam), and β-actin (Sigma) and immune complexes were detected using enhanced chemiluminescence (ECL; Pierce). For quantitative real-time RT-PCR (qPCR), total RNA was extracted from cells using the TRI reagent (Sigma) and was reverse transcribed using reverse transcription reagents (Applied Biosystems) and analyzed by SYBR Green Master (Rox) (Roche) using a real-time PCR system (ABI; Prism 7900HT Sequence Detection system) according to the manufacturer's protocol. The expression of target genes was normalized to GAPDH and analyzed by the comparative cycle threshold method (ΔΔCt).
Analysis of PHD2 Transcription.
The 5′ UTR region containing the two variant EREs (−2597 bp to −1670 bp, relative to the transcriptional start site) was PCR amplified from human genomic DNA using forward primer: 5′-AAGATATTGCTTTACGTATGTGC and reverse primer 5′-TGCCTCATGATGTGATGTAA. The PCR amplified fragment was confirmed by sequencing and cloned at the Xho1–HindIII site into the pGL3-promoter (Promega). Site-directed mutagenesis was used to mutate the imperfect ERE2 sequence from (AATCAGACTGACT) to (AAAAAGACCAACT) using QuikChange XL Site-Directed Mutagenesis kit (Agilent Technologies).
For luciferase assays, PNT1a, LNCaP, or PC3-M cells were transfected with the desired plasmids and the Renilla luciferase construct to normalize for transfection efficiency. After 6–7 h of incubation, transfection medium was removed and replaced with fresh medium. After 18–20 h of incubation, luciferase assays were performed using Dual Glo luciferase assay system (Promega) according to the manufacturer’s protocol. Relative light units were calculated as the ratio of Firefly luciferase to Renilla luciferase activity (normalized luciferase activity).
ChIP was performed using the ChIP-IT Express kit (53008; Active Motif) with minor modifications (46). Briefly, the attached cells were first cross-linked by 2 mM disuccinimidyl glutarate (DSG) for 45 min on a rotating platform at RT. The cells were then washed and subject to a second cross-link using 1% formaldehyde for 15 min at RT with rotation. Subsequent steps for ChIP analysis were performed according to the manufacturer’s protocol. For chromatin precipitation, 2 µg of HA antibody (Roche; Anti-HA clone 3F10) or rat isotype IgG (eBioscience; 16-4301-81) was used. End-point real-time PCR was performed using the following primer pairs: I-ERE2 (forward) 5′ TAATGTAGGAATCAGGCTGACACC; I-ERE2 (reverse) 5′ AGTTGCCGTGTATTTCCTATTACAT. Note that we used a 58.5 οC annealing temperature to increase PCR sensitivity and specificity.
EMSA.
Nuclear extracts were prepared from PNT1a cells or ERβ-ablated PNT1a cells treated with 3β-adiol (5 µM) for 16–18 h. An oligonucleotide containing the I-ERE2 sequence (GGAATCAGGCTGACACCACCTCAA) was end labeled with 32P gamma-ATP. Nuclear extracts were incubated with 32P probe in the absence or presence of unlabeled competitor oligonucleotides. The mutated version of the probe (GGAAAAAGGCAAACACCACCTCAA) was used as a control. Protein–DNA complex formation was detected by 6% PAGE.
Statistical Analysis.
Data are presented as the mean from three separate experiments ± SD. The Student t test was used to determine the significance of independent experiments. The criterion P < 0.05 was used to determine statistical significance.
Acknowledgments
National Institutes of Health Grant CA159856 (to A.M.M.) supported this work.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Tsai MJ, O’Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 2.Morani A, Warner M, Gustafsson JA. Biological functions and clinical implications of oestrogen receptors alfa and beta in epithelial tissues. J Intern Med. 2008;264(2):128–142. doi: 10.1111/j.1365-2796.2008.01976.x. [DOI] [PubMed] [Google Scholar]
- 3.Kumar MV, Tindall DJ. Transcriptional regulation of the steroid receptor genes. Prog Nucleic Acid Res Mol Biol. 1998;59:289–306. doi: 10.1016/s0079-6603(08)61035-1. [DOI] [PubMed] [Google Scholar]
- 4.Kuiper GG, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson JA. Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci USA. 1996;93(12):5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuiper GG, Gustafsson JA. The novel estrogen receptor-beta subtype: Potential role in the cell- and promoter-specific actions of estrogens and anti-estrogens. FEBS Lett. 1997;410(1):87–90. doi: 10.1016/s0014-5793(97)00413-4. [DOI] [PubMed] [Google Scholar]
- 6.Prins GS, et al. Estrogen receptor-beta messenger ribonucleic acid ontogeny in the prostate of normal and neonatally estrogenized rats. Endocrinology. 1998;139(3):874–883. doi: 10.1210/endo.139.3.5827. [DOI] [PubMed] [Google Scholar]
- 7.Herynk MH, Fuqua SA. Estrogen receptors in resistance to hormone therapy. Adv Exp Med Biol. 2007;608:130–143. doi: 10.1007/978-0-387-74039-3_10. [DOI] [PubMed] [Google Scholar]
- 8.Lau KM, LaSpina M, Long J, Ho SM. Expression of estrogen receptor (ER)-alpha and ER-beta in normal and malignant prostatic epithelial cells: Regulation by methylation and involvement in growth regulation. Cancer Res. 2000;60(12):3175–3182. [PubMed] [Google Scholar]
- 9.Mak P, et al. ERbeta impedes prostate cancer EMT by destabilizing HIF-1alpha and inhibiting VEGF-mediated snail nuclear localization: Implications for Gleason grading. Cancer Cell. 2010;17(4):319–332. doi: 10.1016/j.ccr.2010.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas C, et al. ERβ1 represses basal-like breast cancer epithelial to mesenchymal transition by destabilizing EGFR. Breast Cancer Res. 2012;14(6):R148. doi: 10.1186/bcr3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas C, Gustafsson JA. Targeting PES1 for restoring the ERα/ERβ ratio in breast cancer. J Clin Invest. 2012;122(8):2771–2773. doi: 10.1172/JCI65133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imamov O, et al. Estrogen receptor beta regulates epithelial cellular differentiation in the mouse ventral prostate. Proc Natl Acad Sci USA. 2004;101(25):9375–9380. doi: 10.1073/pnas.0403041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dondi D, et al. Estrogen receptor beta and the progression of prostate cancer: Role of 5alpha-androstane-3beta,17beta-diol. Endocr Relat Cancer. 2010;17(3):731–742. doi: 10.1677/ERC-10-0032. [DOI] [PubMed] [Google Scholar]
- 14.Leav I, et al. Comparative studies of the estrogen receptors beta and alpha and the androgen receptor in normal human prostate glands, dysplasia, and in primary and metastatic carcinoma. Am J Pathol. 2001;159(1):79–92. doi: 10.1016/s0002-9440(10)61676-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weihua Z, Lathe R, Warner M, Gustafsson JA. An endocrine pathway in the prostate, ERbeta, AR, 5alpha-androstane-3beta,17beta-diol, and CYP7B1, regulates prostate growth. Proc Natl Acad Sci USA. 2002;99(21):13589–13594. doi: 10.1073/pnas.162477299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerini V, et al. The androgen derivative 5alpha-androstane-3beta,17beta-diol inhibits prostate cancer cell migration through activation of the estrogen receptor beta subtype. Cancer Res. 2005;65(12):5445–5453. doi: 10.1158/0008-5472.CAN-04-1941. [DOI] [PubMed] [Google Scholar]
- 17.Muthusamy S, et al. Estrogen receptor β and 17β-hydroxysteroid dehydrogenase type 6, a growth regulatory pathway that is lost in prostate cancer. Proc Natl Acad Sci USA. 2011;108(50):20090–20094. doi: 10.1073/pnas.1117772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins DF, et al. Hypoxia promotes fibrogenesis in vivo via HIF-1 stimulation of epithelial-to-mesenchymal transition. J Clin Invest. 2007;117(12):3810–3820. doi: 10.1172/JCI30487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci USA. 2008;105(17):6392–6397. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang MH, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10(3):295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 21.Loda M, Kaelin WG., Jr Prostate cancer: Beta control your hormones. Cancer Cell. 2010;17(4):311–312. doi: 10.1016/j.ccr.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40(2):294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaelin WG., Jr New cancer targets emerging from studies of the Von Hippel-Lindau tumor suppressor protein. Ann N Y Acad Sci. 2010;1210:1–7. doi: 10.1111/j.1749-6632.2010.05781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berthon P, Cussenot O, Hopwood L, Leduc A, Maitland N. Functional expression of sv40 in normal human prostatic epithelial and fibroblastic cells: Differentiation pattern of nontumorigenic cell-lines. Int J Oncol. 1995;6(2):333–343. doi: 10.3892/ijo.6.2.333. [DOI] [PubMed] [Google Scholar]
- 27.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5(9):675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 28.Gibbons DL, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23(18):2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ocaña OH, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22(6):709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Kozlowski JM, et al. Metastatic behavior of human tumor cell lines grown in the nude mouse. Cancer Res. 1984;44(8):3522–3529. [PubMed] [Google Scholar]
- 31.Elvidge GP, et al. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: The role of HIF-1alpha, HIF-2alpha, and other pathways. J Biol Chem. 2006;281(22):15215–15226. doi: 10.1074/jbc.M511408200. [DOI] [PubMed] [Google Scholar]
- 32.Leite de Oliveira R, et al. Gene-targeting of Phd2 improves tumor response to chemotherapy and prevents side-toxicity. Cancer Cell. 2012;22(2):263–277. doi: 10.1016/j.ccr.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 33.Bates RC, et al. Flt-1-dependent survival characterizes the epithelial-mesenchymal transition of colonic organoids. Curr Biol. 2003;13(19):1721–1727. doi: 10.1016/j.cub.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Goel HL, et al. VEGF/neuropilin-2 regulation of Bmi-1 and consequent repression of IGF-IR define a novel mechanism of aggressive prostate cancer. Cancer Discov. 2012;2(10):906–921. doi: 10.1158/2159-8290.CD-12-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang AD, et al. Vascular endothelial growth factor receptor-1 activation mediates epithelial to mesenchymal transition in human pancreatic carcinoma cells. Cancer Res. 2006;66(1):46–51. doi: 10.1158/0008-5472.CAN-05-3086. [DOI] [PubMed] [Google Scholar]
- 36.Lichtenberger BM, et al. Autocrine VEGF signaling synergizes with EGFR in tumor cells to promote epithelial cancer development. Cell. 2010;140(2):268–279. doi: 10.1016/j.cell.2009.12.046. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Parra R, et al. Investigation on tumor hypoxia in resectable primary prostate cancer as demonstrated by 18F-FAZA PET/CT utilizing multimodality fusion techniques. Eur J Nucl Med Mol Imaging. 2011;38(10):1816–1823. doi: 10.1007/s00259-011-1876-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vivar OI, et al. Estrogen receptor beta binds to and regulates three distinct classes of target genes. J Biol Chem. 2010;285(29):22059–22066. doi: 10.1074/jbc.M110.114116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grober OM, et al. Global analysis of estrogen receptor beta binding to breast cancer cell genome reveals an extensive interplay with estrogen receptor alpha for target gene regulation. BMC Genomics. 2011;12:36. doi: 10.1186/1471-2164-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao C, et al. Genome-wide mapping of estrogen receptor-beta-binding regions reveals extensive cross-talk with transcription factor activator protein-1. Cancer Res. 2010;70(12):5174–5183. doi: 10.1158/0008-5472.CAN-09-4407. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Q, et al. Control of cyclin D1 and breast tumorigenesis by the EglN2 prolyl hydroxylase. Cancer Cell. 2009;16(5):413–424. doi: 10.1016/j.ccr.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazzone M, et al. Heterozygous deficiency of PHD2 restores tumor oxygenation and inhibits metastasis via endothelial normalization. Cell. 2009;136(5):839–851. doi: 10.1016/j.cell.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bos R, et al. Levels of hypoxia-inducible factor-1 alpha during breast carcinogenesis. J Natl Cancer Inst. 2001;93(4):309–314. doi: 10.1093/jnci/93.4.309. [DOI] [PubMed] [Google Scholar]
- 45.Shen C, et al. Genetic and functional studies implicate HIF1α as a 14q kidney cancer suppressor gene. Cancer Discov. 2011;1(3):222–235. doi: 10.1158/2159-8290.CD-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian B, Yang J, Brasier AR. Two-step cross-linking for analysis of protein–chromatin interactions. Methods Mol Biol. 2012;809:105–120. doi: 10.1007/978-1-61779-376-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]