Abstract
A comprehensive whole-genome analysis of gene function by transposon mutagenesis and deep sequencing methodology has been implemented successfully in a representative of the Archaea domain. Libraries of transposon mutants were generated for the hydrogenotrophic, methanogenic archaeon Methanococcus maripaludis S2 using a derivative of the Tn5 transposon. About 89,000 unique insertions were mapped to the genome, which allowed for the classification of 526 genes or about 30% of the genome as possibly essential or strongly advantageous for growth in rich medium. Many of these genes were homologous to eukaryotic genes that encode fundamental processes in replication, transcription, and translation, providing direct evidence for their importance in Archaea. Some genes classified as possibly essential were unique to the archaeal or methanococcal lineages, such as that encoding DNA polymerase PolD. In contrast, the archaeal homolog to the gene encoding DNA polymerase B was not essential for growth, a conclusion confirmed by construction of an independent deletion mutation. Thus PolD, and not PolB, likely plays a fundamental role in DNA replication in methanococci. Similarly, 121 hypothetical ORFs were classified as possibly essential and likely play fundamental roles in methanococcal information processing or metabolism that are not established outside this group of prokaryotes.
Keywords: Tn-seq, polyploidy, methanogenesis
Methanogenic archaea are obligate anaerobic prokaryotes and widely distributed in O2-free environments where electron acceptors other than CO2 have been depleted. Methanogenesis is a highly specialized anaerobic respiration with a distinctive biochemistry composed of unusual coenzymes and catalysts whose roles are poorly understood (1). In anaerobic environments, methanogenesis plays a key role by catalyzing the terminal step of carbon mineralization and maintaining an extremely low partial pressure of H2. Methane, the final product of this process, also is a significant greenhouse gas, with about 80% of the atmospheric methane produced by these archaea (2).
Our understanding of the methanogenic archaea is far from complete. For instance, the methanogen Methanococcus maripaludis S2 possesses 1,779 genes, only a few of which have been characterized and an even smaller portion of which has been studied in detail. Close to 800 genes remain annotated as hypothetical proteins awaiting proper identification (3). Much of this uncertainty is shared with other archaea, in which many of the fundamental life processes have not been investigated in the same detail as in the bacteria and eukaryotes. To address these issues, the function of methanococcal genes was evaluated by a saturation mutagenesis technique. Whole-genome libraries of Tn5 transposon mutants were constructed, and the individual mutations were mapped following enrichment of the transposon-chromosomal DNA junctions and Illumina sequencing (4). Because mutations in genes likely to be essential or strongly advantageous for growth are lethal or rapidly lost from the library, they may be identified by their low frequency in the libraries. This methodology has some limitations: gene and pathway redundancy may mask essential processes, polarity may cause genes downstream of an essential gene to falsely appear essential, truncated genes may retain some activity and falsely appear nonessential, and small essential genes may escape detection. Moreover, it is difficult to distinguish genes that are truly essential, i.e., absolutely required for growth under the specified conditions, from those that are advantageous for growth and rapidly diluted out from the libraries by faster-growing mutants. Although definitive assignments of essentiality still require detailed analyses of each gene, the methodology generates hypotheses about the nature of specific genes and a great deal of insight into specific questions regarding methanogens as well as more general questions about the genetics, biochemistry, and physiology of archaea.
Results and Discussion
Mutant Mapping and Criteria for Gene Classification.
M. maripaludis strain S2 was mutated randomly by transformation with a derivative of the Tn5 transposon (Tn5 <KAN-2-pac>) comprising a kanamycin resistance cassette for selection in Escherichia coli and a puromycin resistance cassette for selection in methanogenic archaea. Two high-density mutant collections of ∼60,000 and 30,000 individual mutants were constructed. The locations of the insertions were mapped in the original libraries, which were formed after growth with puromycin selection for about 20 generations (T0), and after growth without puromycin selection for 7 (T1) or 14 generations (T2) in rich or minimal medium (Table S1). Across all libraries, about 92% of the sequence reads were mapped to the genome. Although insertions generally were evenly distributed around the genome, a few locations possessed a high number of reads. Presumably, these reads represented hotspots for the transposon insertion, which is not uncommon for the Tn5 transposon (5), or artifacts of the sequencing process. For a complete list of reads mapped to the genome, refer to Dataset S1.
M. maripaludis is polyploid and contains 30–55 genomes per cell, depending upon the growth phase (6). To prevent accumulation of heterologous genes, gene conversion rapidly homogenizes the genomes following insertions (6). Therefore, in our experimental design, insertions in nonessential genes are expected to rapidly replace the wild-type alleles during growth with puromycin selection (Fig. S1). In fact, mutants homozygous for transposon insertions are isolated readily in the tryptophan operon under permissive conditions (7). Because the full replacement of the wild-type allele is lethal for essential genes, cells are expected to maintain both the wild-type and mutant genes during puromycin selection. When the puromycin selection is removed, the mutant alleles are expected to be lost rapidly, and wild-type alleles are expected to dominate (6). To test this model, the fraction of genomes with insertions was determined by real-time PCR for library 1. The relative copy numbers of the pac cassette served as a marker for the transposon, and the carbon monoxide dehydrogenase large subunit gene (cdhA), which was present in a single copy on the genome, served as a marker for the genome. Following puromycin selection at T0, 0.71 ± 0.20 (± SD) of the genomes possessed the pac cassette by this measure. In complex medium, the ratio dropped to 0.47 ± 0.09 and 0.37 ± 0.03 in the T1 and T2 libraries, respectively, presumably because gene conversion led to the loss of deleterious insertions in cells that were heterozygous and to selection for wild-type cells. In contrast, in minimal medium, the ratio initially dropped to 0.50 ± 0.06 and then increased to 0.78 ± 0.16 in the T1 and T2 libraries, respectively. The increase was attributed to selection for mutants with insertions in a specific gene with faster growth than the wild type under these conditions (see below). Therefore, these results were consistent with the expectations for polyploid cells. Interestingly, polyploidy offers a unique advantage in transposon mutagenesis and deep sequencing (Tn-seq) experiments because it allows direct demonstration of insertions in essential genes during antibiotic selection. In monoploid cells, selection against insertions in essential genes is inferred from the insertion density of nonessential genes and is never directly observed.
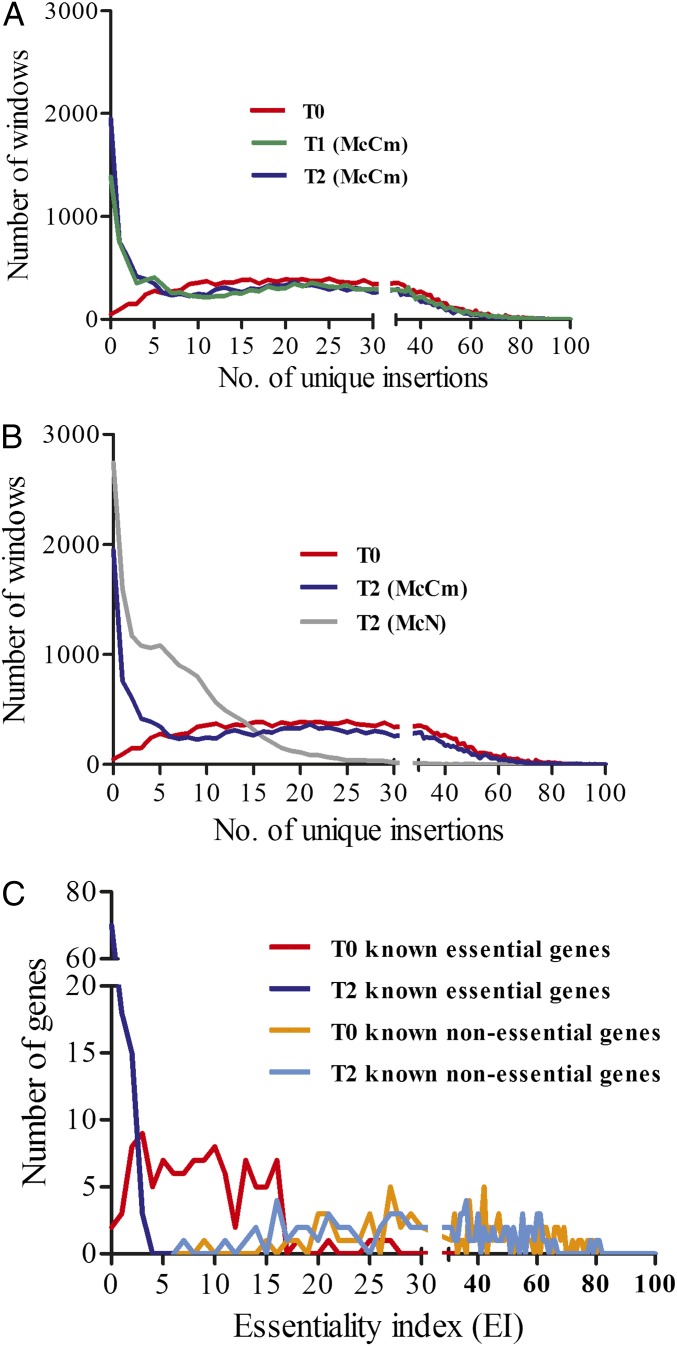
Genes essential to growth were expected to have fewer insertions than nonessential genes. A sliding window method was used to identify the number of insertions in genes (SI Materials and Methods). In this approach, an essentiality index (EI) was calculated based upon the number of insertions within 800-bp windows. The EI then was calibrated empirically without making assumptions about the expected distribution of insertions. In the T0 libraries, insertions were found in nearly all genes, including many expected to be essential for growth, and the number of unique insertions per window resembled a Poisson distribution, with a maximum between 20 and 25 insertions per window (Fig. 1A). This unimodal distribution indicated that there was little selection against insertions in most of the windows. In T1 and T2 cells grown in rich medium without puromycin selection, the number of unique insertions per window acquired a bimodal distribution, with a large increase in the number of windows with no or only few insertions (Fig. 1A). Presumably, these windows encompassed possibly essential or strongly advantageous genes.
Fig. 1.
Distributions of the numbers of unique insertions in 800-bp windows in library 1. (A) Distribution following growth for 7 (T1) and 14 (T2) generations in the absence of antibiotic in McCm. T0 is the initial library. (B) Distribution following growth in rich (McCm) or minimal (McN) medium in the absence of antibiotic. (C) Distribution of the EI for 104 representative essential and 89 representative nonessential genes.
To test this hypothesis, the EI of 104 genes assumed to be essential based upon our general knowledge of the physiology and biochemistry of methanococci and 89 genes assumed to be nonessential were examined in rich medium (Fig. 1C and Dataset S2). The EI for the known essential genes in T2 were 0–3, indicating that few insertions remained following growth without puromycin. In contrast, the EI of the nonessential genes was in the range 7–81, with 99% of nonessential genes having an EI > 11. For that reason, an EI ≤ 3 was considered diagnostic of possibly essential genes, and an EI ≥ 11 was considered diagnostic of nonessential genes. Genes with an EI of 4–10 were unassigned and presumably comprised genes advantageous for growth but nonessential as well as small genes whose essentiality cannot be determined reliably by this method. For genes smaller than the window size of 800 bp, insertions at neighboring genes might increase the EI even in the absence of insertions within the gene itself. Similar methods were used to calibrate the EI for library 2, and genes were classified as possibly essential if they satisfied the criteria in at least one library and were unassigned in the second library. Following growth in minimal medium, fewer unique insertions were found, and the EI range was much lower (Fig. 1B). This decrease resulted from a large increase in the number of reads in one specific gene (MMP1511) as discussed below. Calibration of the EI in these libraries yielded cutoffs of the EI for possibly essential and nonessential of ≤ 2 and ≥ 5, respectively.
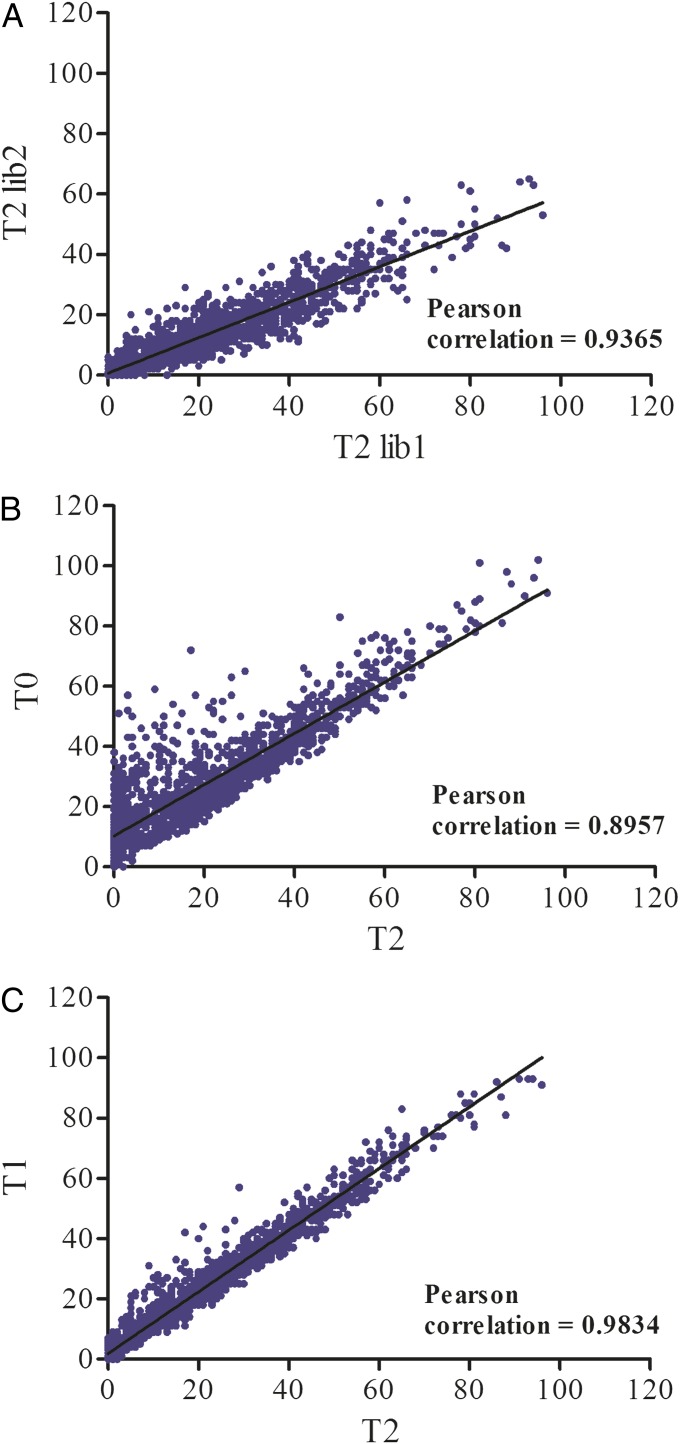
A gene’s index in library 1 was correlated highly with its index in library 2, and so the EI appeared to be reproducible (Fig. 2A). In fact, following growth in rich medium, only genes for seven hypothetical proteins were classified possibly essential in library 2 and nonessential in library 1. In the absence of additional information, these genes were classified as unassigned. For essential and highly advantageous genes, the EI also is expected to depend upon the rate of gene conversion and the fitness of the resulting mutants as well as on stochastic factors affecting the original number of insertions. The mechanism of gene conversion in the euryarchaeotes is not well understood. Although gene conversion is rapid and relatively independent of the fitness of the resulting homozygous mutant, the rate depends in part on the extent of the genetic difference between the genomes (8). Although the Tn5 transposon insertions are all identical, it is possible that the rate of gene conversion also depends upon the sequence surrounding the insertion site. Slowly growing mutants also are being selected against at the same time gene conversion occurs. Therefore, differences in the observed EIs might result from differences in the gene conversion rates as well as the growth rates of the resulting homozygous mutants, and it is not possible to distinguish between truly essential and highly advantageous genes. In contrast, for nonessential genes in which the fitness of the resulting mutants is close to that of the wild type, gene conversion should occur before the formation of the T0 library, and the EI should be constant in the subsequent libraries. Consistent with this prediction, the EI for many genes was correlated highly in the T0, T1, and T2 libraries, especially when the EI was >11 and the genes presumably were nonessential (Fig. 2 B and C).
Fig. 2.
Reproducibility and stability of the EI. (A) Correlation of a gene’s index in library 1 with its index in library 2. (B) Correlation of the EI in the T0 and T2 of library 1. (C) Correlation of the EI in the T1 and T2 of library 1.
To evaluate the effect of polarity on the functional categorization of genes, the EI of genes in the 209 operons previously identified in M. maripaludis S2 were examined (9). If the insertion of the Tn5 transposon interrupts transcript, genes upstream of a possibly essential gene will always appear possibly essential. However, this was not the case. Twelve of the operons possessed possibly essential genes with an EI ≤3 downstream of nonessential genes. The apparent lack of polarity is consistent with earlier observations of transcriptional read-through from the pac cassette (10).
To validate the EI further, this classification scheme was compared with the experimental evidence for the essentiality of 63 genes from published studies (Dataset S3). For 62 of these genes, the EI correctly predicted whether the gene was required for growth under the conditions used here. For example, independent genetic evidence demonstrated that three genes of the Energy conserving hydrogenase a (ehaHIJ), which encode the cation translocator of this enzyme complex, were essential (11). Their low EIs in the current studies were consistent with this conclusion. However, for the diaminopimelate aminotransferase gene dapL, which encodes a large bifunctional enzyme required for lysine biosynthesis, the EI remained in the range of 4–5, even following growth in minimal medium without lysine. These insertions were concentrated in regions outside the catalytic and the pyridoxal 5′-phosphate binding sites of the gene, suggesting that a truncated protein with some functionality may have been formed. Although these results confirm the overall validity of this approach, they demonstrate the need for cautious interpretation of mutations within largely uncharacterized genes.
Possibly Essential Genes for Growth in Rich Medium.
In rich medium at T2, 526 genes were classified as possibly essential for growth. Many of the essential genes were required for fundamental biological processes, such as methanogenesis, replication, transcription, and translation (Fig. S2). Of the remaining genes, 834 genes were classified as nonessential and 419 could not be assigned confidently. A complete list of the EIs for the M. maripaludis genes under all the conditions examined is given in Dataset S4.
By these criteria, genes from many different functional categories were possibly essential for growth in rich medium (Table 1). In addition, over 45% of the protein-encoding genes are annotated as hypothetical proteins, and 121 genes were possibly essential (Fig. S2). In contrast, most of the genes assigned to cellular processes, signal transduction, or transport were nonessential. Some specific categories are analyzed in more detail in SI Materials and Methods.
Table 1.
Known protein-encoding genes related to fundamental biological processes in M. maripaludis S2
| Biological process | Subprocess* | Possibly essential† | Unassigned† | Nonessential† |
| Energy metabolism | Methanogenesis (32) | mer, mch, fwdHFGDAC, mcrBDCGA, mtrEDCBAGH, or900, ftr, atwA | fwdB | hmd, fmdE1, mtd, fmdE2ACB1B2 |
| ATP generation (9) | atpHIKECFABD | |||
| Membrane-bound hydrogenases (34) | ehaABCDEFGHIJKLMNOT | ehbQ, ehbJ, 1626, ehbF | ehaPQRS, ehbCD, ehbN, ehbA, ehbOMLK, ehbG, ehbE | |
| Cytoplasmic hydrogenases (17) | frcD, vhuUAGD, | vhuB | frcBG, frcA, vhcDGAB, fruADGB | |
| Replication | DNA polymerases (3) | DP1, DP2 | polB | |
| Replication factors (23) | priL, mcm1, priS, rfcB, rfcA, rpa2, topA, lig, top6B, rpa1, pcnA, top6A, pcnA gins15 | rnhA | mcm2, mcm3, mcm4, rpa3, dnaG, fen1, rnhB, smc1 | |
| Transcription | RNA polymerase (12) | rpoP, rpoL, rpoE’, rpoD,rpoN, rpoK, rpoHB2B1A1A2 | rpoF | |
| Transcription factors (8) | tfe, tfb, tbp, nusA, nusG | 1015, spt4 | tfs | |
| Translation | tRNA synthetase and related proteins (26) | 0212,0255, tyrS, metS, alaS, thrS, valS, 0688, proS, leuS, serS, gltX, argS, pheT, lysS, ileS, pheS, trpS, hisS, aspS | 0002, truA, 0377, 0693, 0816, cysS | |
| Ribosomal proteins (60) | rplX, rpl31e, rpl40e, rpl37ae, rpl12p, rpl1P, rps17E, rpl34e, rpl24e, rps28e, rpl7ae, rps2P, rps3ae rps6e, rpl10e, rps13p, rps4p, rps11p, rpl18e, rpl13p, rps9p, rps12p, rps7p, rps10p, rpl22p, rps3p, rpmC, rps17p, rpl14p, rpl24p, rps4e, rpl5p, rps14p, rps8p, rpl6p, rpl32e, rpl19e, rpl18p, rps5p, rpl30p, rpl15p, rpl3p, rpl4p, rplW, rpl2p, rps19p, rps15p, rps8e, rpl44e | rpl21e, rps19e, rplP0, rpl15, rps24e, rps27ae, rpl37e, rpl30e, rpl11p, rps27e | rpl39e | |
| Translation factors (14) | aIF6, infB, aIF2β, aIF1A, aIF5A, 1131, aIF2γ, aEF2,selB, aEF1α, aEF1β | aIF2α | 0738, aIF2BI |
*The number in parentheses corresponds to the total number of known genes for that specific subprocess.
†Genome annotation code number (MMP#) is used for genes that do not possess a gene symbol. Gene names are defined in Dataset S4.
Replication.
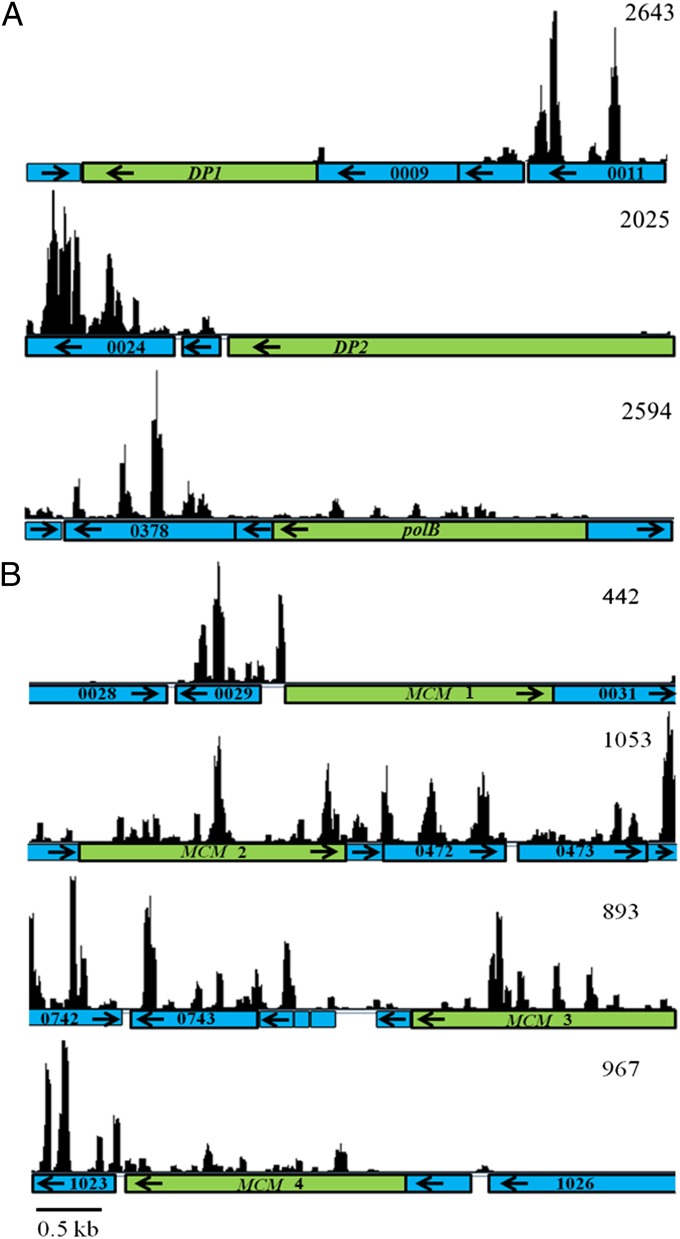
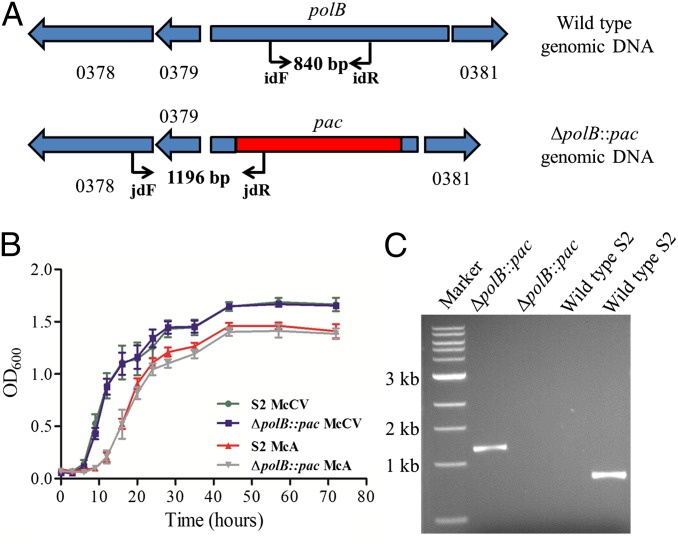
Because most mutations in the replicative system are lethal, replication in the Archaea has been studied largely in vitro, and the Tn-seq approach is especially informative. Of the 26 genes assigned to replication in methanococci, 16 were possibly essential. Like other euryarchaeotes, methanococcal genomes encode two replicative DNA polymerases (12). DNA polymerase B (polB) is proposed to synthesize the leading strand, and DNA polymerase D (polD) is proposed to synthesize the lagging strand during replication (13). In addition, mutations in genes for both polymerases in Halobacterium were lethal (14). However, only the genes for PolD, MMP0008 and 0026, were possibly essential in methanococci (Fig. 3A). To confirm this observation, a deletion mutant was constructed in which most of the polB gene—encompassing most of the DNA polymerase domain, including the two essential aspartyl residues and all of the exonuclease domain—was replaced with the pac cassette (Fig. 4 and Figs. S3 and S4). Growth of this mutant was indistinguishable from the wild type in rich and minimal medium (Fig. 4B). These results suggested that PolD is the major replicative DNA polymerase in methanococci and PolB plays a secondary role or is redundant with another protein. In support of this conclusion, in Thermococcus kodakarensis PolD can be coisolated with proliferating cell nuclear antigen (PCNA) and other proteins of the archaeal replication fork (15). Although PolB coisolated with the replication proteins minichromosome maintenance 2 (MCM2) and replication protein A-3 (RPA3), most of its associations were with proteins whose roles have yet to be defined. Methanococci also possess MMP1230, which possesses a nucleotidyltransferase domain common in DNA polymerase homologs. This gene is found in all methanogens and in Archaeoglobus but is missing in other related euryarchaeotes. Although essential, its distribution is more consistent with a role in tetrahydromethanopterin (H4MPT) metabolism, which is shared by these archaea, than in replication. In eukaryotes and archaea, MCM proteins form homo- or heteromeric complexes that play fundamental roles in the initiation and progression of the replication fork. Four probable MCM homologs are found in M. maripaludis, but only three of them are expressed (16). Only one of these expressed MCM genes, MMP0030, was possibly essential (Fig. 3B). Similar results have been found in T. kodakarensis, in which three MCM homologs are present but only one is essential (17). In contrast, in eukaryotes, all six of the MCM homologs are essential for the initiation of DNA synthesis (18). These results suggest that M. maripaludis forms a homomeric MCM complex or a heteromer, in which MMP0030 predominates. Similar MCM complexes have been observed in other methanogenic archaea, such as Methanothermobacter thermautotrophicus (19).
Fig. 3.
Distribution of reads across selected genes of the M. maripaludis S2 genome. The y axis is the number of reads within a window size of three. The numbers in the top right corners represent the maximum number of reads in each segment. (A) The DP1 and DP2 genes (green) encode the small and large subunits of DNA polymerase type II (PolD), respectively. polB (green) encodes DNA polymerase B (PolB). (B) The four homologs for the MCM genes (green), which encode the minichromosome maintenance proteins. Surrounding genes are represented in blue. Numbers indicate the MMP identification, and arrows indicate the direction of transcription. Initial plots were generated using Artemis.
Fig. 4.
Characterization of the ΔpolB::pac mutant strain. (A) Genetic maps of the gene polB in the wild-type strain and the replacement of almost the complete pol B gene by the pac cassette in the ΔpolB::pac mutant strain S123. Numbers indicate the MMP identification, and the black arrows indicate primers used for PCR amplification. (B) Growth curves of the wild-type and ΔpolB::pac mutant strain in complex medium (McCV) and minimal medium plus acetate (McA). The error bars indicate the SD of four independent replicates. (C) Genotypic characterization of the ΔpolB::pac mutant strain by PCR amplification. Lanes: 1, standard 1-kb ladder (New England Biolab); 2 and 3, PCR amplifications of a junction fragment between genomic DNA and the pac cassette using primers jdF and jdR and an internal portion of the polB gene using primers idF and idR, respectively, for genomic DNA of ΔpolB::pac mutant strain S123; 3 and 4, same PCR amplifications of genomic DNA from the wild-type strain M. maripaludis S2.
The genes that encode the large and small subunits of a homolog of the eukaryotic primase (MMP0009 and MMP0071) both were possibly essential. The Pyrococcus furiosus homolog to this primase was characterized previously (20). In contrast, a homolog of the bacterial DnaG-type primase, MMP1286, was nonessential. A homolog of this protein in T. kodakarensis (TK1410) copurified with the exosome complex (15), and the presence of both primase activities has been reported in Sulfolobus solfataricus (21). Other possibly essential genes related to replication were the processivity factors PCNA (MMP1126 and 1711), DNA topoisomerases (MMP0956, 0989, and 1437), and DNA ligase (MMP0970). Three homologs for RPA are present in M. maripaludis S2 (16), but only two of them were possibly essential (MMP0616 and 1032). Lastly, the flap endonuclease gene (MMP1313, fen1) was nonessential.
Two other genes of M. maripaludis S2 that encode hypothetical proteins share homology with proteins that copurified with the replication fork complex of T. kodakarensis (15). Although their functions are unknown in both archaea, MMP0668 (homolog to TK1313) and MMP1392 (homolog to TK0358) were both possibly essential and may play important roles in archaeal replication.
Hypothetical proteins.
A total of 121 hypothetical proteins were possibly essential (Dataset S5). Of these, 56 are conserved throughout the order Methanococcales, 19 are conserved by all methanogenic archaea, 4 are conserved by the Euryarchaeota phylum, and only 1 is conserved throughout Archaea (MMP0694). This gene possesses several domains involved in RNA metabolism, suggesting an involvement in posttranscriptional RNA modifications. The remaining 41 possibly essential hypothetical proteins are conserved at the family, genus, or species level, possibly playing major roles in the adaptations defining the lifestyle of M. maripaludis.
Possibly Essential Genes for Growth in Minimal Medium.
A total of 664 possibly essential genes were found in minimal medium at T2. The classification of most genes was the same as in complex medium. Many of the genes that became possibly essential in minimal medium did not have an obvious role in biosynthesis, including genes for two ferredoxins, MMP0389 and 1140, and 87 hypothetical proteins. Many genes that encode proteins involved in stress response also became possibly essential, including MMP0684, which encodes the heat shock protein Hsp20; MMP0264, which encodes a mechanosensitive ion channel; and MMP0585, which encodes the universal stress protein (Usp). Finally, two homologs of the S-adenosyl methionine or SAM proteins, MMP0560 and 1221, became possibly essential in minimal medium.
In contrast to the other insertions, the abundance of reads in the gene MMP1511, which encodes an alanine/sodium symporter, increased from 0.6% of the total reads in the T0 libraries to 85% in the T2 libraries following growth in minimal medium. This gene is the last gene transcribed in an operon that includes alanine dehydrogenase and alanine racemase (22). However, the number of reads in these other genes did not increase. This result indicated that the inactivation of the symporter stimulated the growth rate in minimal medium. Indeed, the relative fitness of the mutant, calculated by the Malthusian parameter (23), was 1.5 times that of mutants of nonessential genes. Because this effect was not observed in rich medium, it appears to depend upon the absence of amino acids.
Conclusions
The Tn-seq technology was successfully implemented in M. maripaludis, generating a comprehensive database of possibly essential and nonessential genes for an archaeon. Although these results provide fresh insights into numerous metabolic and molecular pathways of these unique prokaryotes, it is important to note that the classifications of essentiality are hypotheses about the nature of specific genes. Definitive assignments of essentiality require detailed analysis of each gene, which is not possible in a global survey of the genome. Many of the genes for the fundamental processes in archaeal replication, transcription, and translation, which have been identified largely based upon their similarity to eukaryotic homologs, proved possibly essential for growth. These results provide direct evidence for these roles and for a close relationship between archaea and eukaryotes. In contrast, the gene for the archaeal-specific DNA polymerase PolD was possibly essential, suggesting that it performs a fundamental role in replication. Thus, archaeal replication also possesses unique features. Interestingly, PolD is absent from the genomes of the crenarchaeotes, suggesting that PolB is the replicative DNA polymerase in this phylum. If true, this observation implies an unanticipated variability in archaeal replication. Similarly, many genes encoding hypothetical proteins proved to be possibly essential. Because many of these genes were found only in specific phylogenetic or physiological groups, they may enable important but unidentified functions unique to these archaea. For instance, many of these genes may be involved in coenzyme biosynthesis or other currently poorly described processes in methanogenesis. The further functional analysis of these genes will help unveil many of the unsolved mysteries of the third domain of life.
Finally, it is remarkable that the number of possibly essential genes in this methanogenic archaeon is very similar to that found in bacteria. In M. maripaludis, 526 genes were classified as possibly essential for growth on rich medium, which correspond to 30% of the total genes in the genome of M. maripaludis. Many of these genes encoded monomer biosynthesis and reflected the inability of these lithotrophs to use organic nutrients. For the heterotrophic bacteria tested, the number of essential genes ranges from 271 to 642 genes (24). These results imply that the total number of genes required for growth in laboratory media is fairly small. If only limited functionality is needed for growth under these conditions, only a few genes will appear possibly essential. Certainly, the number of genes required to encode the core informational processes is only a small fraction of the total. Alternatively, essentiality of function often is masked by the inherent redundancy of biological systems so that many mutations are compensated by alternative pathways.
Materials and Methods
Additional details of the methods are described in SI Materials and Methods.
In Vivo Transposon Mutagenesis.
Production of stable transposomes was achieved by incubation of 100 ng of the Tn5 <KAN-2-pac> transposon with 2 μL of EZ-Tn5 Transposase (1 U/μL; Epicentre) and transformed into M. maripaludis S2. After transformation, cells were spread onto rich medium + coenzyme M or McCm agar plates supplemented with puromycin and incubated for 6 d in the presence of 100 kPa of H2/CO2 [80:20, (vol/vol)] at 37 °C. Puromycin-resistant colonies were washed off the plates and stored at −80 °C.
Mutant Library Passages.
An aliquot of 300 μL from the mutant library frozen stock was diluted in 5 mL of minimal or McN medium to an absorbance (600 nm) of 0.5–0.6. This suspension was defined as T0. From this dilution, 4 × 107 viable cells were inoculated into 20 mL of either McCm or McN medium. In both cases, ampicillin was added. In the first passage (T1), cells were grown to an absorbance (600 nm) of 0.5–0.6 at 37 °C. For the second passage (T2), cells were transferred to fresh medium and grown to an absorbance of 0.5–0.6. Thus, each passage comprised about seven generations and a 100-fold amplification of the cell number. Genomic DNA was extracted from 5 mL of each passage using the ZR Fungal/Bacterial DNA MiniPrep (Zymo Research) and resuspended in TLE buffer [10 mM Tris⋅HCl buffer (pH 8) and 0.1 mM EDTA].
High-Throughput Insertion Tracking by Deep Sequencing (HITS or Tn-seq).
Five micrograms of genomic DNA was sheared to an average fragment size of 500 bp, and Illumina DNA libraries were prepared by ligating specific indexed linkers to the DNA fragments. Transposon–chromosome junctions were enriched using a biotinylated probe and PCR. Sequencing of the enriched DNA fragment library was performed at the Genome Services Laboratory at the Hudson Alpha Institute for Biotechnology, Huntsville, Alabama. Custom primer single-end sequencing (50 bp) was carried out on a HiSeq Flow Cell v1.5 using a HiSeq 2000 sequencer.
Supplementary Material
Acknowledgments
We thank Travis Glenn and Roger Nilsen for valuable technical assistance. This project has been funded by contracts from the National Institutes of Health and the U.S. Department of Energy. J.M. was supported by National Science Foundation Grant BDI-0950266.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220225110/-/DCSupplemental.
References
- 1.Hedderich R, Whitman WB. Physiology and biochemistry of the methane-producing Archaea. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E, editors. The Prokaryotes. New York: Springer; 2006. 3rd Ed, Vol 2, pp. 1050–1079. [Google Scholar]
- 2.Reeburgh WS. Global methane biogeochemistry. In: Keeling RF, editor. Treatise on Geochemistry. Vol 4. Oxford, UK: Elsevier; 2003. pp. 65–89. [Google Scholar]
- 3.Hendrickson EL, et al. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J Bacteriol. 2004;186(20):6956–6969. doi: 10.1128/JB.186.20.6956-6969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci USA. 2009;106(38):16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goryshin IY, Miller JA, Kil YV, Lanzov VA, Reznikoff WS. Tn5/IS50 target recognition. Proc Natl Acad Sci USA. 1998;95(18):10716–10721. doi: 10.1073/pnas.95.18.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hildenbrand C, Stock T, Lange C, Rother M, Soppa J. Genome copy numbers and gene conversion in methanogenic archaea. J Bacteriol. 2011;193(3):734–743. doi: 10.1128/JB.01016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porat I, Whitman WB. Tryptophan auxotrophs were obtained by random transposon insertions in the Methanococcus maripaludis tryptophan operon. FEMS Microbiol Lett. 2009;297(2):250–254. doi: 10.1111/j.1574-6968.2009.01689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lange C, Zerulla K, Breuert S, Soppa J. Gene conversion results in the equalization of genome copies in the polyploid haloarchaeon Haloferax volcanii. Mol Microbiol. 2011;80(3):666–677. doi: 10.1111/j.1365-2958.2011.07600.x. [DOI] [PubMed] [Google Scholar]
- 9.Yoon SH, et al. Parallel evolution of transcriptome architecture during genome reorganization. Genome Res. 2011;21(11):1892–1904. doi: 10.1101/gr.122218.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porat I, et al. Disruption of the operon encoding Ehb hydrogenase limits anabolic CO2 assimilation in the archaeon Methanococcus maripaludis. J Bacteriol. 2006;188(4):1373–1380. doi: 10.1128/JB.188.4.1373-1380.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lie TJ, et al. Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis. Proc Natl Acad Sci USA. 2012;109(38):15473–15478. doi: 10.1073/pnas.1208779109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cann IK, Ishino Y. Archaeal DNA replication: Identifying the pieces to solve a puzzle. Genetics. 1999;152(4):1249–1267. doi: 10.1093/genetics/152.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henneke G, Flament D, Hübscher U, Querellou J, Raffin JP. The hyperthermophilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J Mol Biol. 2005;350(1):53–64. doi: 10.1016/j.jmb.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 14.Berquist BR, DasSarma P, DasSarma S. Essential and non-essential DNA replication genes in the model halophilic Archaeon, Halobacterium sp. NRC-1. BMC Genet. 2007;8:31. doi: 10.1186/1471-2156-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Santangelo TJ, Cuboňová L, Reeve JN, Kelman Z. Affinity purification of an archaeal DNA replication protein network. MBio. 2010;1(5):e00221-10. doi: 10.1128/mBio.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walters AD, Chong JP. Methanococcus maripaludis: an archaeon with multiple functional MCM proteins? Biochem Soc Trans. 2009;37(Pt 1):1–6. doi: 10.1042/BST0370001. [DOI] [PubMed] [Google Scholar]
- 17.Pan M, Santangelo TJ, Li Z, Reeve JN, Kelman Z. Thermococcus kodakarensis encodes three MCM homologs but only one is essential. Nucleic Acids Res. 2011;39(22):9671–9680. doi: 10.1093/nar/gkr624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kearsey SE, Labib K. MCM proteins: Evolution, properties, and role in DNA replication. Biochim Biophys Acta. 1998;1398(2):113–136. doi: 10.1016/s0167-4781(98)00033-5. [DOI] [PubMed] [Google Scholar]
- 19.Fletcher RJ, et al. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat Struct Biol. 2003;10(3):160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- 20.Liu L, et al. The archaeal DNA primase: Biochemical characterization of the p41-p46 complex from Pyrococcus furiosus. J Biol Chem. 2001;276(48):45484–45490. doi: 10.1074/jbc.M106391200. [DOI] [PubMed] [Google Scholar]
- 21.Zuo Z, Rodgers CJ, Mikheikin AL, Trakselis MA. Characterization of a functional DnaG-type primase in archaea: Implications for a dual-primase system. J Mol Biol. 2010;397(3):664–676. doi: 10.1016/j.jmb.2010.01.057. [DOI] [PubMed] [Google Scholar]
- 22.Moore BC, Leigh JA. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J Bacteriol. 2005;187(3):972–979. doi: 10.1128/JB.187.3.972-979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woods RJ, et al. Second-order selection for evolvability in a large Escherichia coli population. Science. 2011;331(6023):1433–1436. doi: 10.1126/science.1198914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juhas M, Eberl L, Glass JI. Essence of life: Essential genes of minimal genomes. Trends Cell Biol. 2011;21(10):562–568. doi: 10.1016/j.tcb.2011.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.