Abstract
We identified cell surface markers associated with repression of p16INK4a/cyclin-dependent kinase inhibitor 2A(CDKN2A), a critical determinant in the acquisition of a plastic state. These cell surface markers allowed direct isolation of rare cells from healthy human breast tissue that exhibit extensive lineage plasticity. This subpopulation is poised to transcribe plasticity markers, OCT3/4, SOX2, and NANOG, at levels similar to those measured in human embryonic stem cells and to acquire a plastic state sensitive to environmental programming. In vitro, in vivo, and teratoma assays demonstrated that either a directly sorted (uncultured) or a single-cell (clonogenic) cell population from primary tissue can differentiate into functional derivatives of each germ layer, ectodermal, endodermal, and mesodermal. In contrast to other cells that express OCT3/4, SOX2, and NANOG, these human endogenous plastic somatic cells are mortal, express low telomerase activity, expand for an extensive but finite number of population doublings, and maintain a diploid karyotype before arresting in G1.
Keywords: NT5E/CD73, pluripotency
Current observations are expanding our classic perceptions about phenotypic plasticity, i.e., the ability to be shaped or programmed to a specific cell type. Recent studies indicate that intestine and skin not only contain classic tissue-specific stem cells that maintain tissue homeostasis but also independent cell populations that generate the same range of differentiated cells in response to wound healing (1, 2). We have also learned that stromal cues not only dictate which cell population will respond to specific environmental signals but also which phenotypes will manifest. For example, placement of neural stem cells within the mammary stroma results in mammary cell differentiation (3). Thus, cell lineage plasticity can be directed by chemical and mechanical stromal signals to generate tissues (4).
Extensive studies have demonstrated that repression of p16INK4a/CDKN2A (cyclin-dependent kinase inhibitor 2A) is associated with the acquisition of a plastic state, i.e., the ability of a cell to change phenotypes. In epithelial cells, repression of p16INK4a does not only inactivate cell cycle arrest in response to stress but also allows increased expression of chromatin remodeling proteins that are important for epigenetic plasticity underlying differentiation (5). In Drosophila, the up-regulation of such chromatin remodeling proteins sets the expression pattern of pluripotent cells (6). In undifferentiated murine myoblasts, an increase in chromatin remodeling proteins inhibits differentiation and dictates the decision between progenitor and differentiated states (7). Furthermore, mice engineered for knock-out of B-lymphoma Mo-MLV insertion region 1 homolog (BMI-1), a polycomb protein that inhibits p16INK4a transcription fail to repress p16INK4a activity and fail to generate hematopoietic and neural stem cells (8–10). Given these observations, we reasoned that repression of p16INK4a might also modulate expression of cell surface markers that could be used for the prospective isolation of cells with extensive lineage plasticity.
Results
Selected Cell Surface Markers Regulated by p16INK4a.
Comparative massively parallel RNA sequencing of human mammary epithelial cells with or without naturally repressed p16INK4a allowed us to search for cell surface markers that would offer the opportunity for positive selection, i.e., markers which would exhibit differential expression in cells with p16INK4a activity compared with cells with repressed p16INK4a. We observed that p16INK4a repression is relieved upon differentiation into luminal and myoepithelial cells (Fig. S1A) and used these cell populations for comparison. Most markers, such as IGF1R (insulin-like growth factor 1 receptor), EGFR (epidermal growth factor receptor), cadherins and integrins, failed to demonstrate differential expression in the presence or absence of p16INK4a activity (Fig. S1B). CD133, a gene reported to be expressed in neural and hematopoietic stem/progenitor cells, was significantly down-regulated and thus could not be used for positive selection (11, 12). Strikingly, this analysis identified a dramatic coincident overexpression and down-regulation of CD73 and CD90, respectively (Fig. S1B). To confirm the causal role of p16INK4a in modulating these two ideal candidates, human mammary epithelial cells with low expression of CD73 and high expression of CD90 were transfected with sh p16INK4a (short hairpin to p16INK4a) and assayed for changes in CD73 and CD90 expression (Fig. S1 C and D). Introduction of sh p16INK4a caused a 77% reduction in basal p16INK4a expression (Fig. S1C). The down-regulation of p16INK4a activity was accompanied by a dramatic increase (>100-fold on average) in the number of cells expressing CD73 and repressing CD90 (CD73+CD90− cells; Fig. S1D).
Having identified CD73+CD90− as a signature for cells with p16INK4a repression and a potential for plasticity, we analyzed 10 disease-free human breast tissues (reduction mammoplasties) for the presence of CD73+CD90− cells. All tissues were devoid of visible disease, bacterial, fungal, or viral contamination, and exhibited a normal diploid 46, XX karyotype (SI Methods and Fig. S2A). Freshly isolated single-cell populations were first depleted of the lineage-positive (Lin+) fraction (hematopoietic, endothelial, and fibroblast cells) (Fig. S2B) then fractionated using the gating shown in Fig. S2C. The resultant lineage-negative (Lin−) population was partitioned into four subpopulations, CD73+CD90− (R1) (5.3%), CD73+CD90+ (R2) (1.9%), CD73−CD90− (R3) (84.6%), and CD73−CD90+ (R4) (8.2%) (Fig. S2D and Dataset S1).
Differentiation into Three Germ Lineages.
Analysis of the R1–R4 subpopulations by quantitative RT-PCR (qPCR) array revealed distinctive expression of genes in R1 previously reported to confer multi- and pluripotency (Fig. 1A). In contrast to R2–R4, R1 expressed several (but not all) of these genes at levels comparable to human embryonic stem cells (hESCs). Thus, we examined the capacity of R1–R4 to differentiate into ectodermal, endodermal, and mesodermal lineages.
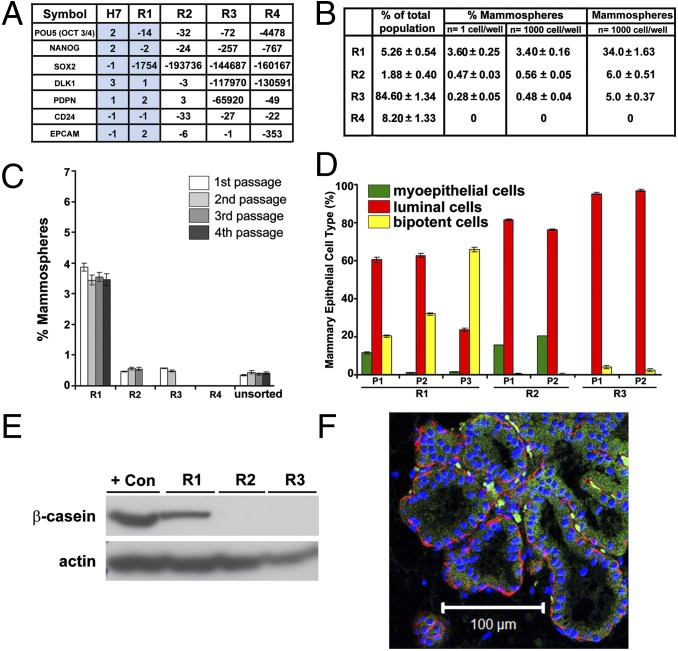
Fig. 1.
Assessment of R1 for ectodermal multilineage potential. (A) Expression of multi- and pluripotency-associated genes in directly sorted R1–R4 subpopulations (n = 4) and hESC H7 (n = 3) assayed by qPCR array. Results are expressed as fold changes compared with hESC H9 (n = 3) and normalized to the housekeeping gene GAPDH. Blue shading highlights the similarities in gene expression levels for H7 and R1. (B) Distribution of R1–R4 subsets within the total epithelial population and ability to form mammospheres from either 1 cell or 1,000 cells. Results are expressed as averaged mean and percentages ±SEM (n = 10). Individual values are shown in Dataset S1. (C) Mammosphere initiating capacity for R1–R4 subpopulations was assessed using 10,000 cells (first passage) and 1,000 cells (subsequent passages) (n = 5). Actual values are shown in the legend of Fig. S3A. (D) Distribution of myoepithelial (α-6-integrin/CD49f+), luminal (MUC-1+), and bipotent (α-6-integrin/CD49f+ and MUC-1+) cells assessed by FACS after dissociation of colonies derived from successive passages of mammospheres. P, passage. Quantitative data are shown in the SI Methods. (E) Human β-casein expression in R1–R3 mammosphere-derived cells by Western blot analysis. Loading control: actin. Positive control: BT-20 cell line. (F) Ducts consisting of a luminal layer expressing CK8/18 (green) and a myoepithelial layer expressing α-SMA (red) stained with human-specific antibodies documenting the human origin of R1-derived structures formed in mouse fat pads. (Scale bar, 100 µm.)
To examine the capacity of directly sorted (uncultured) R1–R4 to differentiate into an ectodermal lineage, R1–R4 were evaluated for breast multipotency using standard techniques of mammosphere formation and multilineage differentiation (Fig. S2E) (13–15). Each subpopulation was cultured under mammosphere-forming conditions (Fig. 1B) and subjected to serial passages (Fig. 1C). Robust and sustained mammosphere generation was observed only for R1 cells and is shown for four passages (Fig. 1 B and C and Fig. S3A). Importantly, only a small fraction (∼3%) of R1 (CD73+CD90−) cells, accounting for only ∼0.15% of total epithelial cells, exhibited this complete and sustained, clonogenic, serial mammosphere-forming capacity in vitro.
First, in vitro lineage differentiation potential was assessed on serial passages of mammosphere-derived cells by flow cytometry and immunocytochemistry analysis of colonies derived from R1–R3 mammospheres from multiple women. Because R4 cells did not generate mammospheres, they could not be assessed for lineage differentiation. Mammospheres were dissociated into single cells and cultured in suspension to test for subsequent mammosphere-forming capacity and multilineage potential by plating on collagen-coated coverslips at colony-producing densities (Fig. S2E). Differentiation was monitored by staining cells for expression of mammary myoepithelial and luminal markers, CD49f (α-6-integrin) and MUC-1 (mucin 1), respectively (Fig. S3B) (16, 17). Flow cytometry identified R1 as the only population with multilineage potential (Fig. 1D). The first mammosphere passage generated colonies representative of all three mammary lineages. With subsequent passages, the bipotent progenitors (CD49f+/MUC-1+) increased over passages 1–3: 20%, 32%, and 66%, respectively, at the expense of the differentiated progeny. Using the complementary morphologic colony-producing assay, R1 mammosphere-derived (single) cells differentiated into three types of colonies: colonies containing exclusively myoepithelial cells (Fig. S3B, R1, Top), exclusively luminal epithelial cells (Fig. S3B, R1, Middle) or bipotent cells (coexpressing both lineage markers) along with cells of both lineages (Fig. S3B, R1, Bottom). This differentiation potential was maintained in subsequent passages (Fig. 1D). In contrast, R2 and R3 mammosphere-derived cells differentiated predominantly into myoepithelial and luminal colonies or only into luminal colonies, respectively (Fig. 1D and Fig. S3B).
In a second assay, acquisition of functional alveolar differentiation was assessed by cellular production of human β-casein with human-specific antibodies (Fig. S2E and Fig. 1E). Cells derived from R1–R3 mammospheres were allowed to differentiate on a collagen substratum, overlaid with reconstituted basement membrane gel devoid of growth factors (Matrigel), supplemented with prolactin. Under these conditions, only R1 mammosphere-derived cells produced β-casein, consistent with alveolar differentiation. R2 and R3 produced luminal cells that failed to differentiate in the presence of prolactin. We speculate that these cells are much like the mature luminal cells reported by Lim et al. that fail to respond to lactogenic cues and to form ducts/lobules in 3D Matrigel culture (18).
The third in vitro differentiation assay (Fig. S2E) compared the ability of R1–R3 mammosphere-derived cells to organize into two types of multicellular structures observed in vivo: small acinar-like structures of luminal origin and solid spherical colonies of myoepithelial origin (19, 20). Single cells from dissociated mammospheres (R1–R3) were cultured at colony-producing densities in 3D Matrigel. Only R1 mammosphere-derived cells generated both structures: 26% branched, ductal–acinar structures and 74% acinar-like structures (Fig. S3C). Thus, R1 exhibited true multilineage differentiation potential in vitro, whereas R2 and R3 were significantly restricted in potential.
Finally, we evaluated the in vivo ability of R1–R4 to enrich for mammary gland reconstituting potential as previously described (14). R1–R4 were sorted and directly transplanted into cleared, humanized mammary fat pads of female nonobese diabetic (NOD)/SCID mice. Only R1 had outgrowth potential, as shown by duct formation upon implantation of as few as 5,000 cells. R2–R4 failed to repopulate the mammary gland (Fig. S4A). As observed in the human mammary tree, these acinar and ductal-lobular outgrowths were composed of an inner luminal cell layer and an outer myoepithelial cell layer (Fig. 1F and Fig. S4B). The human origin of these epithelial outgrowths was validated with human-specific antibodies for cytokeratin 8/18 (CK8/18) (luminal cells) and α-smooth muscle actin (α-SMA) (myoepithelial cells) (Fig. 1F). To test whether these human ductal structures had undergone complete functional differentiation, mice were mated and mammary glands were harvested at day 18 of pregnancy. Human β-casein was expressed within human-origin luminal cells lining the acini and secreted into the lumina of human ductal structures only in R1 outgrowths (Fig. S4 C and D). These results established the mammary gland multipotent capacity of R1 both in vitro and in vivo.
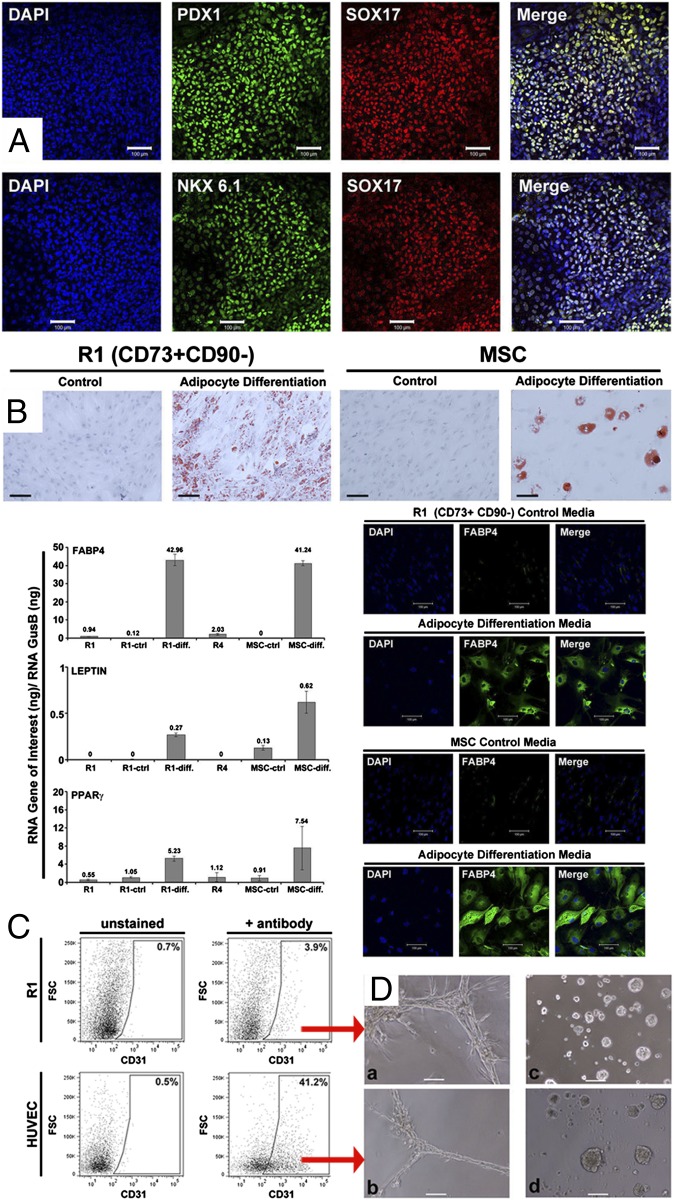
To examine the capacity of R1–R4 subpopulations to differentiate into endodermal lineages, directly sorted (uncultured) R1–R4 were placed under conditions that allow hESCs to differentiate into definitive endoderm (21) and immunostained for expression of transcription factors SOX17 (sex determining region Y-box 17) and FOXA2 (forkhead box A2). R3 and R4 failed to survive under these conditions. R2 survived but failed to proliferate and showed very weak cytoplasmic expression of SOX17 and no expression of FOXA2. Only R1 displayed definitive endoderm phenotypes, with 40% of cells exhibiting SOX17 nuclear expression. Under proper conditions, R1 cells expressing SOX17 could further differentiate toward the pancreatic lineage as demonstrated by the expression of the pancreatic differentiation markers PDX1 (pancreatic and duodenal homeobox 1) and NKX6.1 (NK6 homeobox 1) in cell clusters (Fig. 2A).
Fig. 2.
Assessment of R1 for endodermal and mesodermal lineage potential. (A) Immunostaining of R1 cells for PDX1, SOX17, and NKX6.1 after 12 d of pancreatic differentiation: 40% cells were SOX17+, almost all SOX17+ cells also expressed PDX1 and NKX6.1. (B) Oil Red O staining of R1 cells and MSCs (experimental control) after 9 d of adipogenic differentiation. Transcript levels of fatty acid binding protein 4 (FABP4), LEPTIN, and PPARγ (normalized to glucuronidase B) (GUSB) in directly sorted R1, R1 in expansion medium (R1-ctrl), R1 in adipogenic differentiation medium (R1-diff), MSC in expansion medium (MSC-ctrl), MSC in adipogenic differentiation medium (MSC-diff), and directly sorted R4. R4 cells fail to grow under R1 expansion conditions. Expression differences were validated at the protein level for FABP4 by immunostaining. (C) Analysis of expression of the endothelial marker CD31 in R1 cells after 7 d of endothelial differentiation by FACS. Positive control: human umbilical vein endothelial cells (HUVEC) cells. (D) Cord formation capacity evaluated after 24 h in endothelial Matrigel differentiation assay by phase-contrast microscopy for (a) CD31+ R1 cells from C, (b) CD31+ HUVEC from C, (c) primary human mammary epithelial cells, or (d) primary mammary epithelial cells after 24 h of culture in mammary epithelial cell growth medium (MEGM) as a negative control. (Scale bars, 100 μm.) These results were obtained in five of five analyses.
To determine mesodermal potential, directly sorted (uncultured), R1–R4 cells were exposed to differentiation media previously reported to induce adult human mesenchymal stem cells (MSCs) and hESCs toward adipocyte (22) or endothelial (23) cell lineages. Under adipogenic conditions, only R1 cells attached and grew, produced lipid-filled adipocytes, and uniformly coexpressed the adipocyte-specific markers FABP4 (fatty acid binding protein 4), LEPTIN, and PPARγ (peroxisome proliferator-activated receptor gamma), as observed with positive control MSCs (Fig. 2B). Expression differences were validated at the protein level for FABP4 (Fig. 2B). Under endotheliogenic conditions, R1 survived, whereas R2–R4 died. A fraction of R1 cells expressed the endothelial marker CD31 and subsequently generated tubule-forming endothelial cells (Fig. 2 C and D).
To further document the lineage plasticity of R1 toward mesodermal derivatives, R1 cells were cultured on human placental fibroblast feeders and grown under conditions that promote embryoid body formation and hESC differentiation into cardiomyocytes (24). In these conditions, spontaneous beating of cardiomyocytes was observed in focal patches within the culture (∼5%) (Movie S1).
Clonal Evidence for Extensive Lineage Plasticity of Rare Somatic Cells.
The power of assessing differentiation potential in directly sorted, uncultured R1 cells is that the results cannot be attributed to cell expansion in culture. However, with this approach, one cannot ascertain if the R1 subfraction consists of single cells, each of which has the ability to generate all three germ lineages or if, alternatively, the population represents a collection of cells, each with restricted potency for a single-germ lineage. To distinguish between these two possibilities, we isolated single cells from the R1 population and assessed their potential for generating multiple-tissue lineages.
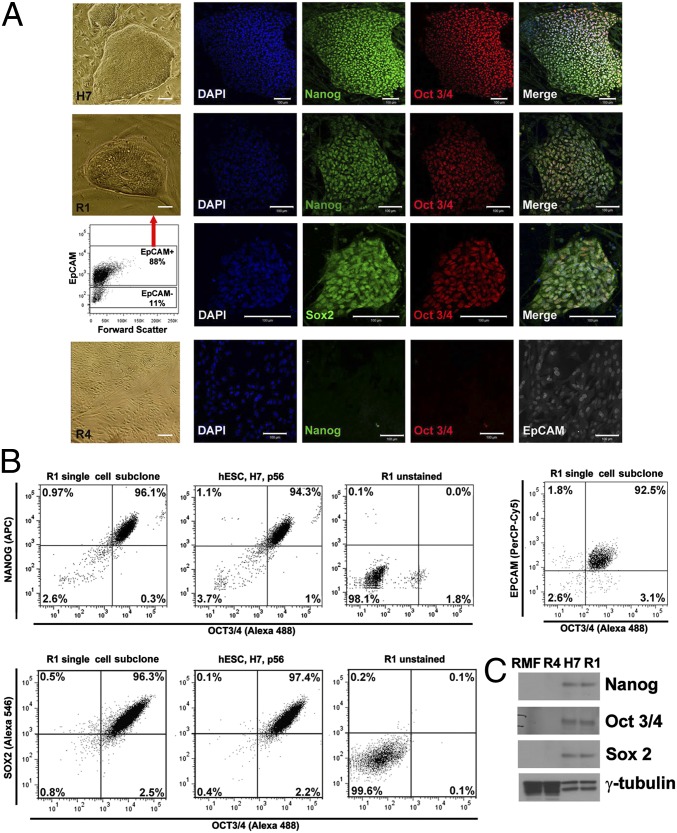
R1 cells, isolated from primary tissue, were separated into EpCAM+ (epithelial cell adhesion molecule-positive) and EpCAM− (epithelial cell adhesion molecule-negative) fractions and placed under conditions known to allow expansion of hESCs (25). Under these conditions, ∼3% of R1 cells from the EpCAM+ fraction formed single-cell–derived colonies by day 14 (Fig. 3A) with robust induction of the canonical pluripotency genes, NANOG, OCT3/4, and SOX2. Immunostaining provided expression levels and confirmed nuclear localization (Fig. 3A). FACS analysis demonstrated that >95% of cells within each colony coexpressed the three pluripotency proteins and the epithelial cell surface marker EpCAM (Fig. 3B). Marker expression was also confirmed at the protein and transcript level using Western blot analysis (Fig. 3C) and qPCR (Fig. 5A and Dataset S2), respectively. Expression of these pluripotency markers was not observed within R2 and R3 (Dataset S2) nor within R4 (Fig. 3 A and C and Dataset S2). Importantly, EpCAM− R1 cells also failed to exhibit any of these phenotypes (Fig. 3A).
Fig. 3.
Expression of pluripotency markers NANOG, OCT3/4, and SOX2 in single-cell–derived R1 colonies. (A) Phase contrast images of hESCs H7 (Top row), EpCAM+-R1–derived colonies (two Middle rows) and R4 cells (Bottom row) cultured on feeders and immunofluorescence analysis documenting expression of NANOG, OCT3/4, and SOX2. Scatterplot shows EpCAM distribution in R1 sorted cells (third row). R4 cells lack expression of NANOG and OCT3/4 but express the epithelial marker EpCAM. (B) Coincident expression of NANOG, OCT3/4, and SOX2 in individual cells assessed by FACS after 14 d of culture on feeder layer. Left: R1 single cell subclones; Center: hESCs H7; Right: R1 unstained (isotype control). Upper three panels: coanalysis of NANOG and OCT3/4. Lower three panels: coanalysis of SOX2 and OCT3/4. Far Right Upper panel: 93% of cells from R1 single cell subclones coexpressed EpCAM and OCT3/4. (C) Western blot analysis of NANOG, OCT3/4, and SOX2 in single-cell–derived R1 colonies on feeders after 14 d. H7, positive control; reduction mammary fibroblasts (RMFs) and R4 cells, negative controls. Loading control: γ-tubulin. Results were obtained in five of five analyses.
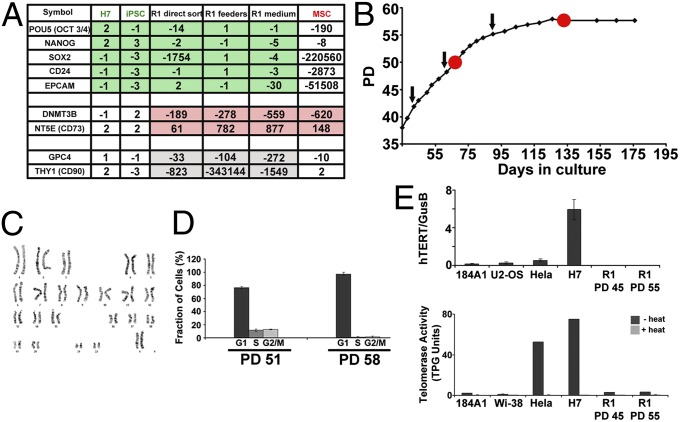
Fig. 5.
R1-derived clones are mortal and distinct from hESCs, iPSCs, and MSCs. (A) Expression of selected genes was compared between breast-derived R1 cells [either uncultured (n = 4) or expanded on feeder layers (n = 3) or in media (n = 3)], hESCs (n = 2), human iPSCs (n = 3), and human MSCs (n = 4) by qPCR-array analysis. Gene expression values in R1 cells, under the three conditions described above, are similar to each other and share several key expression characteristics with pluripotent hESCs and human iPSCs (OCT3/4, NANOG, SOX2, CD24 and EpCAM) (green shaded values) easily distinguishable from MSCs. Likewise, these cells share other expression characteristics with MSCs (DNMT3B and NT5E/CD73) (red shaded values) easily distinguishable from hESC and human iPSCs. Several genes (GPC4 and THY1/CD90) (gray shaded values) exhibit unique expression levels in the three breast-derived R1 cells compared with hESC, iPSCs, and MSCs verifying their unique identity. Transcript expression levels, normalized to GAPDH expression, are relative to H9 human ES cells. Full details of analysis are found in Dataset S2. (B) Growth curve of a single R1 cell expanded in culture demonstrating that cells eventually enter senescence. Black arrows indicate passage doublings (PDs) 43, 50, and 56 at which karyotypes were confirmed to be diploid 46, XX (normal). (C) Representative karyotype at PD 50. (D) Red dots (on curve in B) indicate PDs [mid (PD 51) and late (PD 58)] at which cell cycle was analyzed. FACS analysis using propidium iodide (PI) staining demonstrated a G1 arrest at late passage. (E) Expression of hTERT normalized to GUSB and telomerase activity evaluated using the TRAPeze XL Telomerase Detection kit were measured in the indicated cell lines. TPG, total products generated. Cells from five of five individual patient samples arrested at late passage and maintained diploid 46, XX karyotypes.
To examine the extent of plasticity of R1 cells at a clonal level, progeny of R1 single-cell–derived subclones were manually divided into three parts, placed in each of the in vitro differentiation assays described above, and assessed for potency. These single-cell–derived R1 subclones generated all three previously described lineage derivatives: ectodermal mammary cell multilineage derivatives, endodermal pancreatic derivatives, and beating cardiomyocytes (Fig. S5 and Movie S2). Thus, directly sorted R1 cells and single-cell–derived R1 subclones are equally potent in generating all three germ-line derivatives in vitro.
To confirm the origin and individual identity of R1 cells, we used short tandem repeat (STR) forensic analysis to compare markers in flow cytometry-isolated cells and a matched mesodermally differentiated R1 derivative (beating cardiomyocytes) derived from two independent breast tissues. Each pair of parental and differentiated samples exhibited identical genetic markers for a given donor, each being distinct from markers for hESC and K562 control cell lines (Table S1). Collectively, these data demonstrate that a single endogenous plastic somatic (ePS) cell can express OCT3/4, SOX2, and NANOG proteins and generate all three germ lineages when exposed to proper conditions.
R1 Cells Can Form Teratomas.
Given the extensive lineage plasticity of R1 cells in vitro, we assessed their plasticity in vivo. Because human blastocyst rescue is not possible for ethical reasons, we instead used a teratoma assay. R1–R4 cells were grafted under the renal capsules of immunocompromised mice and assessed for the generation of multiple human tissue derivatives using human-specific antibodies (Fig. S6). We found that the R1 fraction of cells, either sorted directly (uncultured) from breast tissue (Fig. 4A) or expanded from a single-cell subclone from breast tissue (Fig. 4B and Fig. S7A), generated teratomas with representation of all three germ layers. The differentiated structures of ecto-, endo-, and mesodermal lineages were equally diverse and extensive in teratomas whether derived from clonogenic or directly sorted (uncultured) R1 cells and were indistinguishable from the structures in teratomas generated by the positive control, hESC H7 (Fig. S7 A and B).
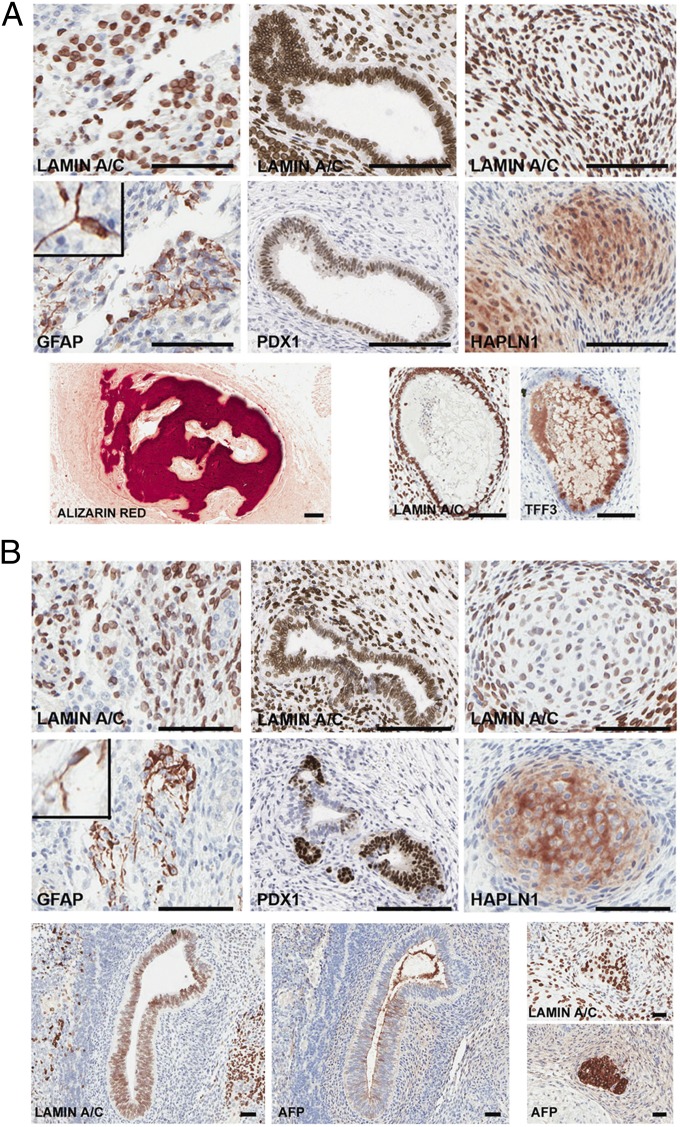
Fig. 4.
R1 cells form teratomas in immunocompromised mice. R1 cells directly isolated from reduction mammoplasty (A) or a single-cell–derived R1 clone expanded in culture (B) were grafted under the renal capsule of 7- to 8-wk-old female SCID/beige mice. Teratomas, harvested 16 wk after injection, were paraffin embedded, serial sectioned, and stained for the panhuman-specific marker lamin A/C to document the human origin of these structures and for lineage-specific markers. Neuroepithelial (ectodermal) marker, GFAP; pancreatic (endodermal) marker, PDX1; or cartilage (mesodermal) marker, HAPLN1 validated human-specific tissue formation in teratomas. Insets: magnified GFAP+ cells. Representative views show bone stained with Alizarin Red and a gastrointestinal structure with goblet cells expressing TFF3 (A, Lower). Additional views of structures expressing the endodermal marker AFP and corresponding staining for human lamin A/C are shown (B, Lower). (Scale bars, 150 µm.) Directly sorted R1 cells generated teratomas in two of five analyses, failed to generate any mass or structures in two of five analyses and gave rise to a mesoderm derivative (cartilage) in one of five analyses. In one of one attempt, a single-cell–derived R1 subclone formed a teratoma and gave rise to representative three germ layers.
Staining with a panhuman lamin A/C antibody confirmed the human origin of the differentiated structures within the teratomas, whereas staining with human-specific, tissue-specific antibodies confirmed their lineage and functional status. GFAP+ (glial fibrillary acidic protein-positive) neuroepithelial cells as well as PDX1+ structures, reminiscent of those seen in normal tissues (Fig. S6), were documented (Fig. 4 A and B and Fig. S7B). The generation of cartilage structures was validated by the expression of HAPLN1 (hyaluronan and proteoglycan link protein 1) marker (Fig. 4 A and B and Figs. S6 and S7B), bone using Alizarin Red stain, intestinal goblet cells by expression of the gastrointestinal transcription factor TFF3 (trefoil factor 3, intestinal) (Fig. 4A and Fig. S6), and gut derivatives by AFP(alpha-fetoprotein)-positive staining (Fig. 4B and Fig. S6). None of these markers are routinely expressed in the human breast (Fig. S6). Together, these data demonstrate that ePS cells, either directly sorted from tissue (uncultured) or expanded in a clonogenic fashion, exhibit lineage plasticity and can generate derivatives of all three developmental lineages.
R2–R4 populations, as well as premalignant mammary cells (184A1), failed to form any cell mass. Metastatic mammary cells (MDA-MB-231) used in the same assay formed a malignant tumor with no differentiated structures (Fig. S7C) nor expression of tissue-specific markers other than those typical for breast tissue. Importantly, these data demonstrate that ePS cells can generate derivatives of all three developmental lineages and that these cells are not malignant.
ePS Cells Are Distinct from hESCs, Induced Pluripotent Stem Cells, and MSCs.
We sought to determine molecular commonalities and distinctions between this newly characterized ePS cell population and well-characterized hESCs and human induced pluripotent stem cells (iPSCs). Additionally, because CD73 is a cell surface marker in MSCs, we also included MSCs in our comparison. To this end, we measured transcript levels of 43 selected pluripotency, stress, and reprogramming genes in R1 populations directly sorted from reduction mammoplasties (n = 4), single cell (clonogenic) expanded R1 populations grown on feeder layers (n = 3), or in media (n = 3) by qRT-PCR. Expression levels were compared with those from independent samples of hESCs (n = 2), iPSCs (n = 3), and MSCs (n = 4) (Dataset S2). R1, although sharing some commonalities with hESCs and iPSCs, exhibited distinct other commonalities with MSCs (Fig. 5A, green and red shaded values, respectively). Two critical shared characteristics among ePS cells, hESCs, and iPSCs, that distinguished them from differentiated cells or MSCs, was high expression of pluripotency genes OCT3/4, SOX2, and NANOG and high expression of the epithelial marker, EpCAM (Fig. 5A and Dataset S2). Importantly, only the CD73+CD90− subpopulation expressing EpCAM contained ePS cells verifying their epithelial nature and the requirement for EpCAM as an enrichment marker for ePS (Fig. 3A). However, unlike hESCs or iPSCs, and similar to MSCs, ePS cells expressed much reduced levels of the epigenetic plasticity marker DNMT3B [DNA (cytosine-5-)-methyltransferase 3 beta] (Fig. 5A and Dataset S2). Furthermore, R1 cells clearly displayed reduced expression of CD90, this latter phenotype distinguishing them from hESCs, iPSCs, and MSCs (Fig. 5A and Dataset S2). Similar results were obtained using either single-cell–derived clonal populations grown on feeder layers or in media or directly sorted uncultured populations (Fig. 5A and Dataset S2). Thus, ePS cells exhibited a unique expression profile that supported their extensive plasticity and clearly distinguished them from hESCs, iPSCs, and MSCs. Finally, also in distinction to immortal hESCs and iPSCs, ePS cells were mortal, grew for up to 58 population doublings (Fig. 5B), and maintained a diploid karyotype before arresting in G1 (Fig. 5 C and D). The ePS population exhibited very low levels of human telomerase reverse transcriptase (hTERT) and telomerase activity, comparable to those observed in differentiated cells but much lower than those observed in hESCs or malignant cells (Fig. 5E).
Discussion
We have identified a unique population of somatic cells isolated from disease-free human breast tissue that exhibits remarkable lineage plasticity. Our studies demonstrate (i) lineage plasticity without cell culture, (ii) clonal evidence of lineage plasticity, (iii) cell-type–specific gene expression, and (iv) functionality of all three lineage derivatives (ectodermal, secretion of human milk in transplanted mice; mesodermal, lipid-accumulating adipocytes, tubule-forming endothelial cells, and beating cardiomyocytes; and endodermal, intestinal goblet cells). Finally, exclusion of cell–cell fusion or contamination events (through STR analysis and karyotyping of multiple cell populations before and after differentiation) confirms the origin of the cells. Taken together, these studies provide morphological, molecular, and functional evidence of pluripotency of these cells.
Notably, we demonstrate here the mortal and nonmalignant state of the ePS cells isolated from breast tissue obtained from multiple disease-free women, both parous and nonparous, and of various ages and ethnicities. The ePS cells, either directly isolated from human tissue or expanded in culture, exhibit normal diploid 46, XX karyotypes, low telomerase expression and activity, and ultimately enter replicative senescence, distinguishing them from immortal, genomically unstable tumor cells and from hESCs and iPSCs. Of particular note, a single-cell–derived clone can generate all three lineage derivatives both in vitro and in vivo. Whereas ePS cells exhibit enough proliferation potential to generate the fully expanded and differentiated tissues of teratomas, the mortality of these cells suggests a short-term contribution to tissue function under yet-to-be-determined conditions such as tissue replacement during wound healing (1, 2). Similar to hESCs and other stem cells (3, 26), ePS cells are directed to express different cell fates by an instructive microenvironment (Figs. 1F and 4). Thus, this work not only addresses the plastic state of the ePS cells and their proliferative potential but also highlights the importance of the stromal environment (niche) in programming their cell fate.
Importantly, the ePS cell population described here differs from previously characterized tissue-specific mammary gland stem cell populations (18) and plastic populations (27–29) in their range of cell fate choices, growth requirements, expression of selected genes, and cell surface markers (Figs. 1 and 5). These studies do not challenge the long-accepted concept of lineage commitment of tissue-specific adult stem cells and do not suggest that transdifferentiation or dedifferentiation is occurring. Rather, the cells we describe here exist in the body devoid of commitment. In time, studies will determine if the expression of pluripotency proteins can be found in vivo (as seen in vitro) and if their function underlies this plasticity phenotype. Encouragingly, rare events have been described by pathologists (choristomas, heterotopias, metaplasias, or ectopic tissues) that consist of normal tissues in abnormal locations, such as bone in the colon and eye (30, 31), liver in gallbladder (32) and pancreas in brain (33). Thus, cells with extensive lineage plasticity exist in healthy tissues and evidence for their plasticity is sometimes observed.
The observation that repressed p16INK4a, a key tumor suppressor gene, links epigenetic (5) and phenotypic plasticity, suggests functional links between tumor suppressor genes and plastic states that are yet to be determined and may some day be exploited for disease intervention. We anticipate that ePS cells may be used in the future as a non-embryonic resource to study acquisition of pluripotency, self-repair, replacement and complications in regenerative medicine and cancer.
Methods
Breast tissues obtained from disease-free women exhibited a normal diploid karyotype (Fig.5 and Fig. S2). Tissues were dissociated and epithelial cell clusters (organoids) were further digested to generate single-cell suspensions. All procedures are described in SI Methods.
Supplementary Material
Acknowledgments
We thank Drs. Arnold Kriegstein and Robert Blelloch and members of the T.D.T. laboratory for discussions; Dr. Susan Fisher [Eli and Edythe Broad Center of Regeneration Medicine and Stem Cell Research, University of California, San Francisco (UCSF)] for providing human placental fibroblasts; Drs. Judy Tjoe and James Marx (Comprehensive Breast Health Center Aurora Sinai Medical Center, Milwaukee); Drs. David Baer, Karen M. Yokoo, Benjamin Hornik, and Jane H. Kim (Kaiser Foundation Research Institute, Oakland, CA) and Ms. Kerry Wiles (Cooperative Human Tissue Network, Nashville, TN) for providing breast tissue samples; Matthew Donne and Don Home (UCSF) for assistance with hESC culture conditions and teratoma assays; Jo Dee Fish and Caroline Miller (Gladstone Histology Core, UCSF) for assistance with immunohistochemistry; and Dr. Charles Streuli for providing the human-specific β-casein antibody. We thank the UCSF National Institutes of Health Reference Epigenomics Mapping Consortium (UCSF-NIH-REMC; ref. 34), Dr. Joe Costello, principal investigator, for the use of the public database. This work was supported by National Cancer Institute Grants CA097214, CA107584, ES017154, and CA135626; Avon Foundation Research Grant 07-2007-074; Cancer League, Inc.; and California Institute for Regenerative Medicine Grant RS1-00444-1 (to T.D.T.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218682110/-/DCSupplemental.
References
- 1.Ito M, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11(12):1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 2.Roth S, et al. Paneth cells in intestinal homeostasis and tissue injury. PLoS ONE. 2012;7(6):e38965. doi: 10.1371/journal.pone.0038965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bussard KM, Smith GH. The mammary gland microenvironment directs progenitor cell fate in vivo. Int J Cell Biol. 2011;2011:451676. doi: 10.1155/2011/451676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 5.Reynolds PA, et al. Tumor suppressor p16INK4A regulates polycomb-mediated DNA hypermethylation in human mammary epithelial cells. J Biol Chem. 2006;281(34):24790–24802. doi: 10.1074/jbc.M604175200. [DOI] [PubMed] [Google Scholar]
- 6.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14(2):155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18(21):2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fasano CA, et al. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1(1):87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397(6715):164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 10.Park IK, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 11.Uchida N, et al. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci USA. 2000;97(26):14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin AH, et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90(12):5002–5012. [PubMed] [Google Scholar]
- 13.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17(10):1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuperwasser C, et al. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci USA. 2004;101(14):4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Proia DA, Kuperwasser C. Reconstruction of human mammary tissues in a mouse model. Nat Protoc. 2006;1(1):206–214. doi: 10.1038/nprot.2006.31. [DOI] [PubMed] [Google Scholar]
- 16.Rudland PS, Barraclough R, Fernig DG, Smith JA. 1997. Mammary stem cells in normal development and cancer. Stem Cells, ed Potten CS (Academic, San Diego), pp 147–232.
- 17.Stingl J, Eaves CJ, Zandieh I, Emerman JT. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res Treat. 2001;67(2):93–109. doi: 10.1023/a:1010615124301. [DOI] [PubMed] [Google Scholar]
- 18.Lim E, et al. kConFab Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 19.Petersen OW, Rønnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci USA. 1992;89(19):9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simian M, et al. The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development. 2001;128(16):3117–3131. doi: 10.1242/dev.128.16.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroon E, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26(4):443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 22.Ng F, et al. PDGF, TGF-beta, and FGF signaling is important for differentiation and growth of mesenchymal stem cells (MSCs): Transcriptional profiling can identify markers and signaling pathways important in differentiation of MSCs into adipogenic, chondrogenic, and osteogenic lineages. Blood. 2008;112(2):295–307. doi: 10.1182/blood-2007-07-103697. [DOI] [PubMed] [Google Scholar]
- 23.Levenberg S, Golub JS, Amit M, Itskovitz-Eldor J, Langer R. Endothelial cells derived from human embryonic stem cells. Proc Natl Acad Sci USA. 2002;99(7):4391–4396. doi: 10.1073/pnas.032074999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu C, Police S, Rao N, Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res. 2002;91(6):501–508. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- 25.Genbacev O, et al. Serum-free derivation of human embryonic stem cell lines on human placental fibroblast feeders. Fertil Steril. 2005;83(5):1517–1529. doi: 10.1016/j.fertnstert.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 26.Eirew P, et al. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med. 2008;14(12):1384–1389. doi: 10.1038/nm.1791. [DOI] [PubMed] [Google Scholar]
- 27.De Coppi P, et al. Isolation of amniotic stem cell lines with potential for therapy. Nat Biotechnol. 2007;25(1):100–106. doi: 10.1038/nbt1274. [DOI] [PubMed] [Google Scholar]
- 28.Krause DS, et al. Multi-organ, multi-lineage engraftment by a single bone marrow-derived stem cell. Cell. 2001;105(3):369–377. doi: 10.1016/s0092-8674(01)00328-2. [DOI] [PubMed] [Google Scholar]
- 29.Thomas S, et al. Human neural crest cells display molecular and phenotypic hallmarks of stem cells. Hum Mol Genet. 2008;17(21):3411–3425. doi: 10.1093/hmg/ddn235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Daraji WI, Abdellaoui A, Salman WD. Osseous metaplasia in a tubular adenoma of the colon. J Clin Pathol. 2005;58(2):220–221. doi: 10.1136/jcp.2004.020685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buch AC, Chopra YV, Jadhav PS, Pradhan PM. Intraocular osseous metaplasia. Med J DY Patil Univ. 2012;5:73–75. [Google Scholar]
- 32.Catani M, et al. Ectopic liver nodules: A rare finding during cholecystectomy. G Chir. 2011;32(5):255–258. [PubMed] [Google Scholar]
- 33.Heller RS, Tsugu H, Nabeshima K, Madsen OD. Intracranial ectopic pancreatic tissue. Islets. 2010;2(2):65–71. doi: 10.4161/isl.2.2.10580. [DOI] [PubMed] [Google Scholar]
- 34.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28(10):1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.