Abstract
Despite the importance of tryptophan (Trp) radicals in biology, very few radicals have been trapped and characterized in a physiologically meaningful context. Here we demonstrate that the diheme enzyme MauG uses Trp radical chemistry to catalyze formation of a Trp-derived tryptophan tryptophylquinone cofactor on its substrate protein, premethylamine dehydrogenase. The unusual six-electron oxidation that results in tryptophan tryptophylquinone formation occurs in three discrete two-electron catalytic steps. Here the exact order of these oxidation steps in the processive six-electron biosynthetic reaction is determined, and reaction intermediates are structurally characterized. The intermediates observed in crystal structures are also verified in solution using mass spectrometry. Furthermore, an unprecedented Trp-derived diradical species on premethylamine dehydrogenase, which is an intermediate in the first two-electron step, is characterized using high-frequency and -field electron paramagnetic resonance spectroscopy and UV-visible absorbance spectroscopy. This work defines a unique mechanism for radical-mediated catalysis of a protein substrate, and has broad implications in the areas of applied biocatalysis and understanding of oxidative protein modification during oxidative stress.
Keywords: cofactor biosynthesis, tryptophan radical, heme, posttranslational modification, electron transfer
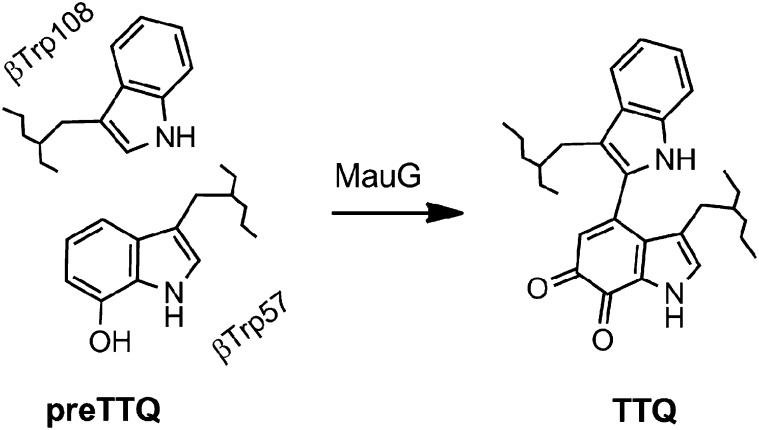
Protein-based radicals, particularly on tyrosine (Tyr) and tryptophan (Trp) residues, have been implicated in a large number of catalytic and electron transfer reactions in biology (1), including the long-range electron transfer reactions required for photosynthesis (2), respiration (3), and DNA synthesis (4) and repair (5). Aberrant formation of protein radicals during oxidative stress is also of special importance. Evidence for the involvement of protein-based and substrate-based radicals in enzyme-catalyzed reactions has increased substantially in recent years, and expanded our knowledge of the scope of chemical reactions accessible to enzymes. The ability of enzymes to catalyze what were previously thought to be unattainable reactions is exemplified by MauG. The biosynthesis of the tryptophan tryptophylquinone (TTQ) cofactor in methylamine dehydrogenase (MADH) requires posttranslational modification of two tryptophan residues. MADH from Paracoccus denitrificans is a 119-kDa α2β2 heterotetramer that contains two active sites and two TTQ cofactors derived from residues βTrp57 and βTrp108 (6). MauG is a c-type diheme enzyme that catalyzes the conversion of a MADH precursor (preMADH) that has one oxygen atom already inserted into the βTrp57 indole ring (βTrp57-OH of preTTQ) (7), to mature TTQ-containing MADH (8) (Fig. 1).
Fig. 1.
Chemical structure of the MADH-based substrate and product of MauG catalysis.
Catalysis by MauG proceeds via a bis-Fe(IV) redox state, which may be generated by reaction of di-Fe(II) MauG with O2 or di-Fe(III) MauG with H2O2 (9). Catalytically active crystals of the MauG–preMADH protein complex show that the site of TTQ formation on β-preMADH is 40.1 Å from the MauG high-spin heme iron that reacts with H2O2 or O2 (Fig. S1A), demonstrating that catalysis involves long-range electron transfer (10). The overall reaction is a six-electron oxidation, and thus three cycles of MauG bis-Fe(IV) formation are required to complete TTQ biosynthesis. The three two-electron steps must equate to (i) formation of the cross-link between βTrp57-OH and βTrp108, (ii) insertion of the second oxygen atom into βTrp57-OH, and (iii) oxidation of the quinol to the quinone. The oxidation of the cross-linked quinol to quinone is believed to be the final step in the reaction (11). Here the order of the first two steps of the reaction is resolved using X-ray crystallography and mass spectrometry, with high-frequency and -field electron paramagnetic resonance (HFEPR) measurements providing details of an unprecedented radical mechanism for the first step. This study fills gaps in the MauG reaction mechanism, and leads to a better understanding of the remarkable chemistry that occurs in this system.
Results
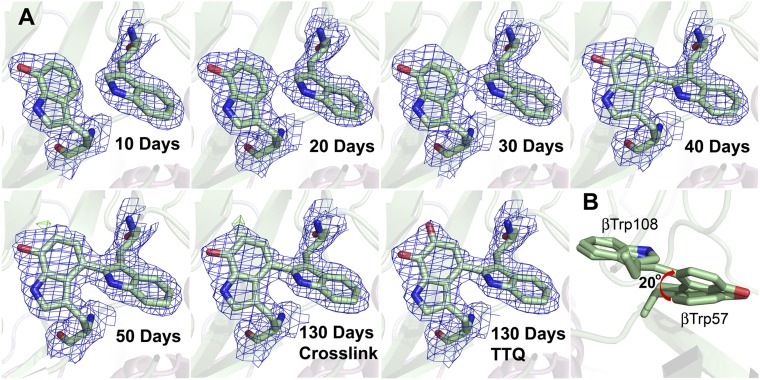
It was previously shown that addition of excess H2O2 to crystals of the MauG–preMADH complex resulted in formation of TTQ in crystallo (10). X-ray crystallography cannot resolve quinol to quinone oxidation, but can distinguish cross-link formation from oxygen insertion. To determine the order of these events in MADH maturation, the residual peroxide contaminants within commercial PEG were exploited. PEG solutions slowly accumulate organic peroxides through autoxidation of the polyoxyethylene bonds, and can release hydrogen peroxide as part of this process (12). The MauG–preMADH crystals were grown using 22–26% (wt/vol) PEG 8000 as precipitant, and thus as the crystals aged within the crystallization trays they were exposed to the slow release of H2O2. As a result, catalytic turnover in MauG–preMADH crystals was observed over time. X-ray diffraction data from crystals harvested after 10, 20, 30, 40, 50, and 130 d clearly show a progression of changes at the preTTQ site that is consistent with the cross-link forming first, followed by addition of the second oxygen to βTrp57-OH (Fig. 2A; Table S1 and S2). Electron density changes indicate cross-link formation between βTrp57-OH and βTrp108 is complete after ∼40 d, and requires βTrp57-OH to undergo a rotation of ∼20° (Fig. 2B). Significant positive-difference electron density appears for the second O atom of TTQ only after cross-linking has occurred.
Fig. 2.
(A) Electron density for preTTQ site in MauG–preMADH crystals of different ages. Electron density maps for the crystals aged for 10, 40, 50 and 130 d were generated from the respective refined structures. The 20- and 30-d electron density maps were calculated using the refined 10-d model (preTTQ) to enable visualization of the development of cross-link density. The two final refined models for the 20- and 30-d data contain a mix of preTTQ and cross-linked preTTQ (Tables S1 and S2). The 130-d cross-link electron density image was calculated using the refined 50-d model (cross-linked preTTQ) to enable visualization of the positive-difference electron density for the second oxygen. The final refined model at 130 d contains 100% TTQ. 2Fo–Fc, blue, contoured at 1.0σ; Fo–Fc, green, contoured at +4σ. (B) The observed rotation in βTrp57-OH during cross-link formation. The figure was produced using PyMOL (www.pymol.org).
To ensure these events were indeed MauG-dependent reactions and not simply due to direct reaction with H2O2, a crystal of the complex between inactive W199F MauG and preMADH was aged. The W199F MauG mutant can form the bis-Fe(IV) state on reaction with H2O2, but cannot catalyze TTQ biosynthesis, as hole hopping through Trp199 is required for preTTQ oxidation (Fig. S1A) (13, 14). In contrast to the native MauG–preMADH crystals, the W199F MauG–preMADH crystals showed no changes at the preTTQ site after 60 d (Fig. S1B; Table S3). Therefore, the sequential reactions of preMADH observed in the crystals are MauG dependent.
To demonstrate that the in crystallo results reflect the order of the reaction in solution, whole-protein mass spectrometry of MauG, preMADH, and H2O2 in a 2:1:2 stoichiometry was compared with a control sample without H2O2; this equates to one MauG:one preTTQ:one H2O2, because preMADH contains two active sites. Reverse-phase chromatography on a C4 column allows for the separation of the β-subunit of MADH from both MauG and α-MADH before mass spectrometry (Fig. S2). Comparison of the chromatogram of the MauG–preMADH sample to the H2O2 treated sample shows the appearance of a new peak eluting earlier than β-preMADH (Fig. S2A). Mass spectrometry demonstrated the presence of two distinct species within this peak (Fig. S2B). The majority species is consistent with the mass of TTQ-containing β-MADH (15,009.73 ± 0.20 Da), and the assignment is confirmed by this chromatographic fraction having an absorbance maximum at 440 nm (Fig. S2D) (15). The other species present is 2-Da lighter (14,995.60 ± 0.05 Da) than β-preMADH in the control (14,997.54 ± 0.02 Da) and H2O2-treated samples (14,997.32 ± 0.26 Da; Fig. S2B), which is consistent with a β-MADH species containing the cross-link between βTrp57-OH and βTrp108 in the absence of the second oxygen atom. None of the chromatographic peaks from the H2O2-treated sample contained a species with a mass consistent with either quinol or quinone, but no cross-link (Table S4); this confirms that cross-link formation occurs first, followed by insertion of a second hydroxyl group into βTrp57-OH, and finally oxidation to the quinone.
Previous work demonstrated that when bis-Fe(IV) MauG was treated with stoichiometric preMADH, a protein-based radical was observed by electron paramagnetic resonance spectroscopy (9). To characterize this radical species in the context of TTQ formation, X-band EPR and HFEPR were used. After mixing MauG, preMADH and H2O2 in a 1:1:1 ratio, regardless of the order of addition, the diferric EPR signal of MauG returns, and a radical intermediate is detectable with a spin quantitation of 1.4–1.6 equivalents by X-band EPR, suggesting that two electrons were transferred to the bis-Fe(IV) site resulting in formation of two protein-based radicals.
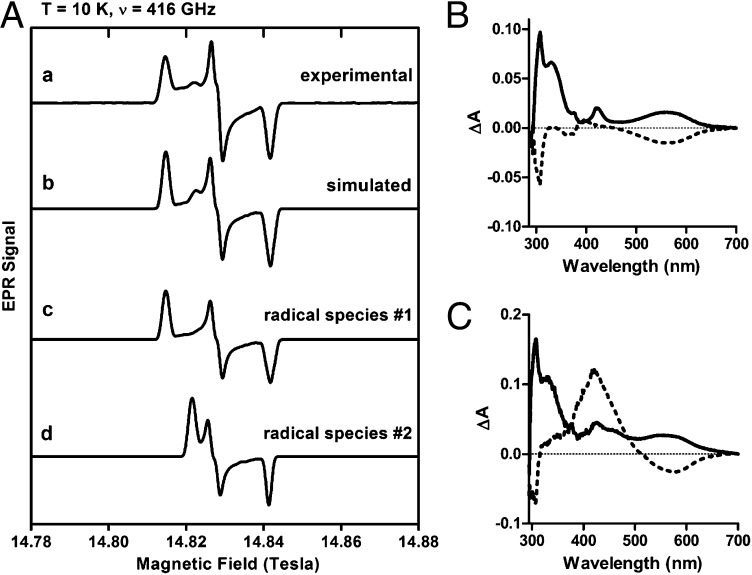
Trp and monohydroxylated Trp radicals, as well as other aromatic radicals such as Tyr, are indistinguishable by conventional X-band EPR spectroscopy (frequency ∼9 GHz). Therefore, HFEPR at 108–416 GHz and correspondingly high fields (up to 15 T) was used to determine the nature of the radical intermediate preceding cross-link formation in the reaction of preMADH with bis-Fe(IV) MauG (Fig. S3). Trp and Tyr radicals are differentiable by HFEPR because oxygen substitution on the phenyl ring of tyrosine significantly increases the g-tensor anisotropy (Δg = |gx − gz|) (16). Specifically, Tyr radicals exhibit a much larger g-anisotropy (Δg > 5 ×10−3 for isolated radicals) than Trp radicals (Δg < 1.5 × 10−3; Table S5) (16–21). A similar g-tensor anisotropy effect would be expected for a hydroxylated Trp radical because the radical spin density is typically delocalized over the entire indole ring (22–27). Therefore, βTrp57-OH of preMADH would be expected to have a Δg value greater than that of Trp radicals and closer to that of Tyr radicals. Fig. 3A describes a diradical intermediate composed of two distinct radicals at 416 GHz (T = 10 K). A single radical species cannot account for all of the resonances of the HFEPR data (Fig. S4). One radical species (species 1) exhibits principal g values of gx = 2.00216, gy = 2.00398, gz = 2.00581, and Δg = 0.00365; the other (species 2) has g tensors of gx = 2.00216, gy = 2.00402, gz = 2.00486, and Δg = 0.00270. Radical species 2 is consistent with a Trp radical, whereas species 1 is consistent with the anticipated βTrp57-OH radical. The presence of two distinct radical species is further verified by HFEPR data measured at a higher temperature (Fig. S5). In the HFEPR spectrum of the frozen solution, the two radicals are not in a 1:1 ratio. The solution kinetics associated with the two-electron oxidation of preMADH (klim = 0.8 s−1) shows that full diradical formation will occur within the dead time of mixing the HFEPR sample (28). Therefore, any inequality must arise from differing decay rates or stabilities of the two radicals; this is most likely due to the two radicals having different stabilities during the freezing process. The pH of common biological buffers is temperature dependent (29–31). Even a modest change in pH could differentially affect the stability of one of the radicals. A similar mismatched ratio has been previously observed in the bis-Fe(IV) intermediate of MauG, likely for the same reason (9). It is also possible that the radicals may have different power saturation properties, which cannot be tested due to technical limitations of the HFEPR technique.
Fig. 3.
(A) 10 K HFEPR spectrum at 416 GHz and correspondingly high field (14–15 Tesla) of the preMADH diradical intermediate (signal a). Spectral simulation (signal b) shows two free radical species, one with g tensors of 2.00216, 2.00398, 2.00581, and Δg = 0.00365 (signal c) and the other 2.00216, 2.00402, 2.00486, and Δg = 0.00270 (signal d). The final simulated spectrum presents a decomposition of 85% radical species 1, and 15% radical species 2. Absorbance difference spectra of the diradical intermediate of preMADH obtained under single turnover (B) and pre-steady state (C) conditions. Solid line is the difference of spectra taken after addition of H2O2—before addition. Broken line is the difference of spectra taken after addition of hydroxyurea—after H2O2 addition.
Evidence for the Trp-based diradical species was also obtained by monitoring solution single-turnover and steady-state kinetic reactions of MauG and preMADH by UV-visible absorption spectroscopy. When MauG plus preMADH were mixed with stoichiometric H2O2 for 30 s, the formation of absorption features at 560 and 310 nm characteristic of Trp-based cation radicals in solution (32) and in protein (33) were observed (Fig. 3B). Subsequent addition of hydroxyurea, a radical scavenger, quenched these features, confirming that they represent radical species. The shoulder at 330 nm, which was not sensitive to hydroxyurea, is likely a small amount of quinol MADH (15). The reaction was also studied with excess H2O2 present to generate steady-state conditions (Fig. 3C). Conditions were such that after 36 s the reaction was still in the pre–steady-state phase. At this time, the 560- and 310-nm absorption features were again observed, and subsequent addition of hydroxyurea quenched these features. Appearance of an absorption feature at 440 nm following hydroxyurea treatment is likely a small amount of quinone MADH that formed during the ∼10-s interval between recording the initial difference spectrum and addition of the radical scavenger. These data illustrate formation of the di-Trp radical in solution on a time scale that parallels the mixing/freezing time to prepare the EPR samples. The pre–steady-state Trp-based cation radical spectral features disappear with the appearance of the 440-nm absorbance that indicates TTQ formation, and confirms the relevance of the intermediate to the catalytic cycle.
Discussion
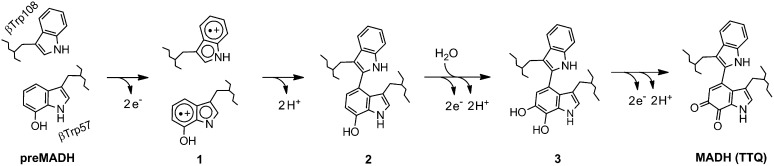
The radical intermediate associated with the first two-electron oxidation of preMADH was characterized by HFEPR, which identified two distinct radical species; one of these is consistent with a preMADH βTrp57-OH radical, whereas the other appears to be a Trp radical. The characterization of this radical intermediate, and the order of the reaction in crystallo, allows a working chemical reaction mechanism to be proposed for TTQ biosynthesis (Fig. 4). In this mechanism, the first two oxidizing equivalents from the bis-Fe(IV) intermediate oxidize preMADH to produce an unprecedented Trp-based diradical (1), which subsequently loses two protons and combines to form a cross-link (2). The second cycle of oxidation by a regenerated bis-Fe(IV) MauG incurs an attack by a water molecule, leading to oxygen insertion, as defined by X-ray crystallography, to form the quinol MADH (3). The final bis-Fe(IV) MauG catalyzes oxidation of reduced quinol TTQ to the oxidized and catalytically active TTQ cofactor. Although a βTrp108 radical makes sense from a mechanistic standpoint, the possibility that the second Trp radical may be localized on another Trp residue cannot be excluded.
Fig. 4.
Proposed chemical reaction mechanism for the six-electron oxidation of preMADH leading to the formation of TTQ and mature MADH.
The ability to resolve the g-values for the two radicals by HFEPR signifies that there is little or no magnetic coupling between them; this may have various origins, such as geometric factors, the specific nature of coupling (through space or via bonds), and the fact that both radicals are indicated by absorbance data to be cationic. We know through theoretical work that the spin distribution on a cation radical (of 3-methylindole) is different from that on a neutral radical (34), which may also affect the magnetic coupling between them.
Generally, for the cross-linking reaction between the cation radicals to proceed, they need to become deprotonated and couple. These processes are likely interconnected and essentially concurrent. Although the precise mechanism of deprotonation remains to be elucidated, it is known to be a spontaneous process for Trp-based protein radicals (20). It should be noted that the Fe ions in MauG would not affect the coupling within the diradical, analogous to the situation described for the R2 protein of Escherichia coli ribonucleotide reductase (33). The nearest heme Fe in MauG is 19 Å away from βTrp108 and βTrp57-OH, which is much longer than the Trp radical and di-iron center separation in R2. A hydrogen bond connection postulated to play a role in R2 is also absent in the MauG–preMADH complex. Rather, the electron transfer from preMADH to MauG has been shown to occur through hole-hopping (13, 14).
The present study begins to unravel the remarkable chemistry that occurs during this biosynthetic reaction. The occurrence and stability of the di-Trp radical intermediate is unprecedented. The longevity may be a consequence of the required rotation of Trp57-OH to react with Trp108 (Fig. 2B), or a rate-limiting proton loss that precedes radical coupling. The nature of the proton acceptors is as yet unknown. The role of the protein environment surrounding the two Trp residues in preMADH clearly plays an important role in facilitating the reaction; this may be considered an unusual form of substrate-assisted catalysis coupled with long-range electron transfer to the MauG high-valent hemes.
Methods
Detailed methods can be found in SI Methods.
Protein Expression and Purification.
Recombinant MauG (35) was purified from P. denitrificans as described previously. PreMADH (7) was expressed in Rhodobacter sphaeroides and purified as described previously (36).
UV-Visible Absorbance Spectroscopy.
The single-turnover reactions contained 10 μM preMADH, 1.0 μM MauG, and 10 μM H2O2. Hydroxyurea was then added at a concentration of 2.0 mM. The pre–steady-state reaction contained 30 μM preMADH, 3.0 μM MauG, and 1.0 mM H2O2. Hydroxyurea was added to 3.0 mM.
EPR Spectroscopy.
The HFEPR sample was prepared in 10 mM potassium phosphate buffer (pH 7.5) containing 5–10% (vol/vol) glycerol by first adding MauG (135 µL, 1.43 mM) to preMADH (170 µL, 1.17 mM). Then, H2O2 (final concentration = 1.22 mM) was added and the mixture frozen. HFEPR spectra were recorded at the EMR Facility at the National High Magnetic Field Laboratory in Tallahassee, FL (37). HFEPR simulations were performed using DOUBLET. The samples for continuous-wave X-band EPR quantitative analyses had final protein concentrations of 150 µM for both MauG and preMADH with addition of 1 equivalent of H2O2. The X-band EPR spectra were recorded on a Bruker ER200D spectrometer at 100-kHz modulation frequency using a 4116DM resonator.
Mass Spectrometry.
Reaction mixtures contained 25–55 μL of 20 μM preMADH, 40 μM MauG, and 0–120 μM H2O2. Samples were incubated for 1 h before injection onto a C4 column (Phenomenex) connected to a HPLC system (Waters). Fractions were collected, dried, and reconstituted in 50% acetonitrile, 0.1% formic acid for mass spectrometry. Data were acquired on a QSTAR XL (AB Sciex) quadrupole TOF mass spectrometer with the IonSpray electrospray source.
Crystallization, X-Ray Data Collection, and Structure Determination.
The WT MauG and W199F MauG complexes with preMADH were crystallized as previously reported (10, 14). WT MauG–preMADH crystals were harvested every 10 d for 50 d, and once at 130 d following crystallization tray setup. The W199F MauG–preMADH crystal was harvested after 60 d. X-ray diffraction data were collected at the Advanced Photon Source (APS), Argonne National Laboratory, Argonne, IL. The data were processed with HKL2000 (38) (Tables S1–S3). Refinement was carried out using REFMAC (39) in the CCP4 program suite (40) starting from the model of WT MauG–preMADH (PDB ID code 3L4M). Model-building was carried out in COOT (41, 42), and calculated electron density maps were generated using PHENIX (43) (Tables S1–S3).
Supplementary Material
Acknowledgments
We thank Ed Hoeffner for KSBL support; LeeAnn Higgins and Bruce Witthun for CMSP support; and the staff at APS sector 23 for their support, especially Michael Becker, Steve Corcoran, Venugopalan Nagarajan, and Derek Yoder; and Dr. Andrew Ozarowski of the National High Magnetic Field Laboratory for providing the EPR simulation program used in this study. HFEPR studies were supported by the National High Magnetic Field Laboratory, which is funded by the National Science Foundation (NSF) through Cooperative Agreement DMR 1157490, the State of Florida, and the Department of Energy. This work was supported by National Institutes of Health Grants GM-66569 (to C.M.W.), GM-41574 (to V.L.D.), GM-97779 (to E.T.Y.), NSF Grant MCB-0843537 (to A.L.), the Georgia Cancer Coalition Distinguished Scholar Program (A.L.), and Minnesota Partnership for Biotechnology and Medical Genomics Grant SPAP-05-0013-P-FY06 (to C.M.W.). Mass spectrometry was conducted at the Center for Mass Spectrometry and Proteomics at the University of Minnesota. Computer resources were provided by the Basic Sciences Computing Laboratory of the University of Minnesota Supercomputing Institute. X-ray data were collected at the Kahlert Structural Biology Laboratory at the University of Minnesota and the National Institute of General Medical Sciences and the National Institute of Cancer Institute Structural Biology Facility-Collaborative Access Team (GM/CA-CAT) at the APS, Argonne National Laboratory. GM/CA-CAT has been funded in whole or in part by National Cancer Institute Grant Y1-CO-1020 and National Institute of General Medical Science Grant Y1-GM-1104. Use of the APS was supported by the US Department of Energy, Basic Energy Sciences, Office of Science Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4FA1, 4FA4, 4FA5, 4FA9, 4FAN, and 4FAV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215011110/-/DCSupplemental.
References
- 1.Stubbe J, van Der Donk WA. Protein radicals in enzyme catalysis. Chem Rev. 1998;98(2):705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 2.Nelson N, Yocum CF. Structure and function of photosystems I and II. Annu Rev Plant Biol. 2006;57:521–565. doi: 10.1146/annurev.arplant.57.032905.105350. [DOI] [PubMed] [Google Scholar]
- 3.Svistunenko DA, Wilson MT, Cooper CE. Tryptophan or tyrosine? On the nature of the amino acid radical formed following hydrogen peroxide treatment of cytochrome c oxidase. Biochim Biophys Acta. 2004;1655(1-3):372–380. doi: 10.1016/j.bbabio.2003.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Stubbe J, Nocera DG, Yee CS, Chang MC. Radical initiation in the class I ribonucleotide reductase: Long-range proton-coupled electron transfer? Chem Rev. 2003;103(6):2167–2201. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 5.Aubert C, Vos MH, Mathis P, Eker APM, Brettel K. Intraprotein radical transfer during photoactivation of DNA photolyase. Nature. 2000;405(6786):586–590. doi: 10.1038/35014644. [DOI] [PubMed] [Google Scholar]
- 6.Davidson VL. Generation of protein-derived redox cofactors by posttranslational modification. Mol Biosyst. 2011;7(1):29–37. doi: 10.1039/c005311b. [DOI] [PubMed] [Google Scholar]
- 7.Pearson AR, et al. Further insights into quinone cofactor biogenesis: Probing the role of MauG in methylamine dehydrogenase tryptophan tryptophylquinone formation. Biochemistry. 2004;43(18):5494–5502. doi: 10.1021/bi049863l. [DOI] [PubMed] [Google Scholar]
- 8.Wang YT, et al. MauG-dependent in vitro biosynthesis of tryptophan tryptophylquinone in methylamine dehydrogenase. J Am Chem Soc. 2005;127(23):8258–8259. doi: 10.1021/ja051734k. [DOI] [PubMed] [Google Scholar]
- 9.Li XH, et al. A catalytic di-heme bis-Fe(IV) intermediate, alternative to an Fe(IV)=O porphyrin radical. Proc Natl Acad Sci USA. 2008;105(25):8597–8600. doi: 10.1073/pnas.0801643105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen LMR, Sanishvili R, Davidson VL, Wilmot CM. In crystallo posttranslational modification within a MauG/pre-methylamine dehydrogenase complex. Science. 2010;327(5971):1392–1394. doi: 10.1126/science.1182492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li XH, Jones LH, Pearson AR, Wilmot CM, Davidson VL. Mechanistic possibilities in MauG-dependent tryptophan tryptophylquinone biosynthesis. Biochemistry. 2006;45(44):13276–13283. doi: 10.1021/bi061497d. [DOI] [PubMed] [Google Scholar]
- 12.Hamburger R, Azaz E, Donbrow M. Autoxidation of polyoxyethylenic non-ionic surfactants and of polyethylene glycols. Pharm Acta Helv. 1975;50(1-2):10–17. [PubMed] [Google Scholar]
- 13.Choi M, Shin S, Davidson VL. Characterization of electron tunneling and hole hopping reactions between different forms of MauG and methylamine dehydrogenase within a natural protein complex. Biochemistry. 2012;51(35):6942–6949. doi: 10.1021/bi300817d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarboush NA, et al. Mutagenesis of tryptophan199 suggests that hopping is required for MauG-dependent tryptophan tryptophylquinone biosynthesis. Proc Natl Acad Sci USA. 2011;108(41):16956–16961. doi: 10.1073/pnas.1109423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husain M, Davidson VL, Gray KA, Knaff DB. Redox properties of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans. Biochemistry. 1987;26(13):4139–4143. doi: 10.1021/bi00387a059. [DOI] [PubMed] [Google Scholar]
- 16.Bleifuss G, et al. Tryptophan and tyrosine radicals in ribonucleotide reductase: A comparative high-field EPR study at 94 GHz. Biochemistry. 2001;40(50):15362–15368. doi: 10.1021/bi010707d. [DOI] [PubMed] [Google Scholar]
- 17.Dorlet P, et al. High-field EPR study of tyrosyl radicals in prostaglandin H(2) synthase-1. Biochemistry. 2002;41(19):6107–6114. doi: 10.1021/bi015871f. [DOI] [PubMed] [Google Scholar]
- 18.Liu AM, Barra AL, Rubin H, Lu GZ, Gräslund A. Heterogeneity of the local electrostatic environment of the tyrosyl radical in Mycobacterium tuberculosis ribonucleotide reductase observed by high-field electron paramagnetic resonance. J Am Chem Soc. 2000;122(9):1974–1978. [Google Scholar]
- 19.Smith AT, Doyle WA, Dorlet P, Ivancich A. Spectroscopic evidence for an engineered, catalytically active Trp radical that creates the unique reactivity of lignin peroxidase. Proc Natl Acad Sci USA. 2009;106(38):16084–16089. doi: 10.1073/pnas.0904535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoll S, et al. Hydrogen bonding of tryptophan radicals revealed by EPR at 700 GHz. J Am Chem Soc. 2011;133(45):18098–18101. doi: 10.1021/ja208462t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokoyama K, Smith AA, Corzilius B, Griffin RG, Stubbe J. Equilibration of tyrosyl radicals (Y(356)⋅, Y(731)⋅, Y(730)⋅) in the radical propagation pathway of the Escherichia coli class la ribonucleotide reductase. J Am Chem Soc. 2011;133(45):18420–18432. doi: 10.1021/ja207455k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walden SE, Wheeler RA. First evidence of anchimeric spin delocalization in tryptophan radical cation. J Am Chem Soc. 1997;119(13):3175–3176. [Google Scholar]
- 23.Connor HD, Sturgeon BE, Mottley C, Sipe HJ, Jr, Mason RP. L-tryptophan radical cation electron spin resonance studies: Connecting solution-derived hyperfine coupling constants with protein spectral interpretations. J Am Chem Soc. 2008;130(20):6381–6387. doi: 10.1021/ja0780277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernini C, et al. EPR parameters of amino acid radicals in P. eryngii versatile peroxidase and its W164Y variant computed at the QM/MM level. Phys Chem Chem Phys. 2011;13(11):5078–5098. doi: 10.1039/c0cp02151b. [DOI] [PubMed] [Google Scholar]
- 25.Lendzian F, et al. Electronic structure of neutral tryptophan radicals in ribonucleotide reductase studied by EPR and ENDOR spectroscopy. J Am Chem Soc. 1996;118(34):8111–8120. [Google Scholar]
- 26.Pogni R, et al. A tryptophan neutral radical in the oxidized state of versatile peroxidase from Pleurotus eryngii: A combined multifrequency EPR and density functional theory study. J Biol Chem. 2006;281(14):9517–9526. doi: 10.1074/jbc.M510424200. [DOI] [PubMed] [Google Scholar]
- 27.Un S. The g-values and hyperfine coupling of amino acid radicals in proteins: comparison of experimental measurements with ab initio calculations. Magn Reson Chem. 2005;43(Spec no):S229–S236. doi: 10.1002/mrc.1660. [DOI] [PubMed] [Google Scholar]
- 28.Lee SY, Shin S, Li XH, Davidson VL. Kinetic mechanism for the initial steps in MauG-dependent tryptophan tryptophylquinone biosynthesis. Biochemistry. 2009;48(11):2442–2447. doi: 10.1021/bi802166c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams-Smith DL, Bray RC, Barber MJ, Tsopanakis AD, Vincent SP. Changes in apparent pH on freezing aqueous buffer solutions and their relevance to biochemical electron-paramagnetic-resonance spectroscopy. Biochem J. 1977;167(3):593–600. doi: 10.1042/bj1670593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Douzou P. Cryobiochemistry: An Introduction. London: Academic; 1977. [Google Scholar]
- 31.Mazur P. Cryobiology: The freezing of biological systems. Science. 1970;168(3934):939–949. doi: 10.1126/science.168.3934.939. [DOI] [PubMed] [Google Scholar]
- 32.Solar S, Getoff N, Surdhar PS, Armstrong DA, Singh A. Oxidation of tryptophan and N-methylindole by N3⋅, Br2−, and (Scn)2− radicals in light-water and heavy-water solutions—a pulse-radiolysis study. J Phys Chem. 1991;95(9):3639–3643. [Google Scholar]
- 33.Baldwin J, et al. Mechanism of rapid electron transfer during oxygen activation in the R2 subunit of Escherichia coli ribonucleotide reductase. 1. Evidence for a transient tryptophan radical. J Am Chem Soc. 2000;122(49):12195–12206. [Google Scholar]
- 34.Jensen GM, Goodin DB, Bunte SW. Density functional and MP2 calculations of spin densities of oxidized 3-methylindole: Models for tryptophan radicals. J Phys Chem. 1996;100(3):954–959. [Google Scholar]
- 35.Wang YT, et al. MauG, a novel diheme protein required for tryptophan tryptophylquinone biogenesis. Biochemistry. 2003;42(24):7318–7325. doi: 10.1021/bi034243q. [DOI] [PubMed] [Google Scholar]
- 36.Graichen ME, et al. Heterologous expression of correctly assembled methylamine dehydrogenase in Rhodobacter sphaeroides. J Bacteriol. 1999;181(14):4216–4222. doi: 10.1128/jb.181.14.4216-4222.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hassan AK, et al. Ultrawide band multifrequency high-field EMR technique: A methodology for increasing spectroscopic information. J Magn Reson. 2000;142(2):300–312. doi: 10.1006/jmre.1999.1952. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 40.Bailey S. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. COOT: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12, Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.