Abstract
Systemic lupus erythematosus (SLE) is an autoimmune disease that is characterized by the loss of tolerance to nuclear self antigens, the production of pathogenic autoantibodies and damage to multiple organ systems. Over the years, patients with SLE have been managed largely with empiric immunosuppressive therapies, which are associated with substantial toxicities and do not always provide adequate control of the disease. The development of targeted therapies that specifically address disease pathogenesis or progression has lagged, largely because of the complex and heterogeneous nature of the disease, as well as difficulties in designing uniform outcome measures for clinical trials. Recent advances that could improve the treatment of SLE include the identification of genetic variations that influence the risk of developing the disease, an enhanced understanding of innate and adaptive immune activation and regulation of tolerance, dissection of immune cell activation and inflammatory pathways and elucidation of mechanisms and markers of tissue damage. These discoveries, together with improvements in clinical trial design, form a platform from which to launch the development of a new generation of lupus therapies.
SLE is an autoimmune disease predominantly affecting females in which loss of tolerance to nucleic acids and their interacting proteins results in the production of pathogenic autoantibodies that cause inflammation and tissue damage. The disease often begins during childbearing years and is characterized by flares and remissions affecting multiple organs, as well as by the consequences of immuno-suppressive medications used to control the disease and cumulative organ damage. A marked improvement in the survival of patients with lupus over the last 50 years is attributable largely to advances in the medical management of life threatening conditions that can occur during the course of disease, such as infections and renal impairment. In contrast, the development of targeted therapies that specifically address disease pathogenesis and progression has lagged, resulting in a limited therapeutic armamentarium of broad-spectrum immuno-suppressive agents that have substantial toxicities and are not always adequate to control symptoms or prevent disease flares. Recent clinical trials based on rational hypotheses and robust preclinical effects in mouse SLE models have tested the efficacy of new biologic drugs in combination with standard-of-care therapies but have met with only limited success1. The recent approval by the US Food and Drug Administration (FDA) of belimumab, a monoclonal antibody that targets the B cell survival cytokine B cell activating factor (BAFF), for the treatment of moderately active lupus shows that a rational approach may be successful; however, some clinicians have reservations about belimumab because its therapeutic effect is modest and may decrease over time2,3 (Box 1). One reason for the lack of a therapeutic breakthrough in SLE is the difficulty in evaluating therapeutic outcomes, largely because of its complex pathogenesis, the heterogeneity of clinical manifestations and the perceived inadequacies in clinical trial design and in the outcome instruments themselves. In addition, proof-of-concept studies using mouse models have their limitations: the mice are highly homogeneous, therapies are compared to placebo rather than the standard of care, and the timing of treatment (including the use of knockout mice) has mostly been preventive.
BOX 1. Belimumab is the first biologic drug approved by the FDA for the treatment of SLE.
Belimumab is a human monoclonal antibody against the cytokine BAFF (also known as BLyS). BAFF and its homolog APRIL are members of the trimeric TNF family and are expressed by multiple cell types. BAFF binds to three receptors, BAFF-R, TACI and BCMA (B cell maturation antigen), that are expressed by B cells at various developmental stages. BAFF-R is specific for BAFF, whereas TACI and BCMA also bind to APRIL. Strong evidence implicates BAFF, and perhaps APRIL, in the pathogenesis of SLE. In non-autoimmune mice, overexpression of BAFF is sufficient to cause lupus in a T cell–independent manner, and BAFF blockade delays disease onset. Increased serum concentrations of BAFF and APRIL have been detected in patients with SLE, and BAFF concentrations have been reported to correlate with disease activity. BAFF, through its interaction with BAFF-R, regulates selection of the naive B cell repertoire and is required for the survival of mature B cells. BAFF and APRIL, primarily through their interaction with TACI, facilitate class switching of immunoglobulin genes and are involved in the amplification of TLR-mediated signals through the interaction of the TACI signaling pathway with the MyD88 pathway. Either BAFF or APRIL is sufficient to support the survival of plasma cells through their interaction with TACI, BCMA or both, but neither is required for the survival of memory B cells. BAFF receptors are also expressed by T cells, and BAFF may have a T cell co-stimulatory role. BAFF receptors are also expressed by activated monocytes, and BAFF can facilitate the production of proinflammatory cytokines from these cells (reviewed in refs. 143,144).
The clinical efficacy of belimumab was shown at week 52 after the start of treatment in two large phase 3 clinical trials (BLISS-52 and BLISS-76) in serologically positive patients with SLE with moderately active disease but without severe renal or central nervous system involvement2,3. Belimumab significantly depleted the number of transitional and naive B cells but resulted in a transient increase in the number of memory cells145 and had more of an effect on circulating IgM- than IgG-producing plasma cells80. Belimumab resulted in an improvement in serological activity over time145 and, crucially, conferred both steroid-sparing effects and a decrease in severe flares; these disease features are often responsible for cumulative irreversible tissue injury. After these successful trials, belimumab was approved by the FDA, becoming the first new drug to be approved for the treatment of patients with SLE in 50 years. However, despite its clear clinical efficacy, enthusiasm for using belimumab in clinical practice has been tempered by the failure to sustain the primary efficacy outcome at 76 weeks of treatment in patients in the United States, as well as the high cost of the drug. Furthermore, because the mechanism for the efficacy of belimumab is SLE is not entirely clear, it is difficult to define the immunologic parameters of response or predict which patients will respond best145 to treatment. Furthermore, it remains to be determined how patients with more severe manifestations will respond to belimumab.
Despite the disappointing results of the first forays into biologic therapies for SLE, basic scientific discoveries made over the last decade have greatly enhanced our understanding of the pathogenesis of autoimmune diseases; some of these discoveries could evolve into new therapeutic strategies for SLE that complement or even replace the currently used broadly immunosuppressive treatments. These discoveries, encompassing genetics, immune-cell subsets and activation pathways, inflammation, mechanisms of tissue damage and biomarker discovery lay the foundation for improved clinical trials of new systemic and local therapies.
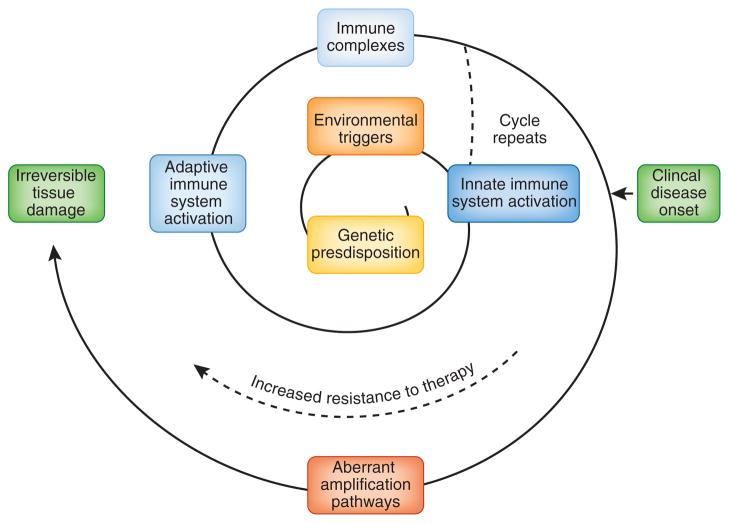
The current view of SLE is that it is a heterogeneous group of diseases in which environmental exposures in a genetically susceptible individual trigger activation of both innate and adaptive immune responses, resulting in loss of tolerance to ubiquitous self antigens. There may be a prolonged preclinical phase that is characterized by the accumulation of an increasing number of autoantibody specificities, followed by inflammation and tissue injury, onset of clinical symptoms, continuing immune amplification and, finally, irreversible tissue damage (Fig. 1). The goal of this review is to summarize recent immunologic discoveries that have a direct bearing on the pathogenesis of SLE and that should form the basis of a new generation of lupus therapies that improve both longevity and quality of life in patients with lupus.
Figure 1.
The spiral of disease progression in SLE. Individuals with identified genetic polymorphisms are at higher risk for SLE compared to the general population. Environmental triggers also probably contribute to the initiation and perpetuation of the disease. Activation of the innate immune system results in enhanced antigen presentation to T cells and the release of proinflammatory cytokines, including type I IFNs. These changes facilitate activation of the adaptive immune system and the development of autoantibodies. Autoantibodies bind to nucleic acids or cellular debris to form complexes that provide further stimulation to innate immune cells through TLRs. Autoreactive B cells act as antigen-presenting cells for the recruitment of more autoreactive T cells. These positive feedback loops that involve the innate and adaptive immune systems amplify clones of autoreactive lymphocytes during the preclinical stage of SLE. The onset of clinical manifestations is associated with systemic inflammation and injury of target organs, resulting in further amplification of immune activation. SLE becomes increasingly resistant to immune-modulating therapies and may eventually progress to irreversible tissue damage. The events that start to occur before or after the clinical onset are shown in green or blue, respectively.
Progress in defining the genetic basis of SLE
Animal studies have shown that genetic alterations in diverse single genes or the simultaneous genetic alteration of multiple genes may lead to the development of lupus-like diseases4,5. In humans, a number of single genetic deficiencies, such as those affecting components of the classical complement activation pathway or intracellular nucleases, are strongly associated with SLE. More recently, genome-wide association studies (GWASs) that use millions of common single nucleotide polymorphisms as genomic markers to type increasingly large data-sets of well-defined patient and control populations have definitively linked a number of genetic variants to disease susceptibility6–8.
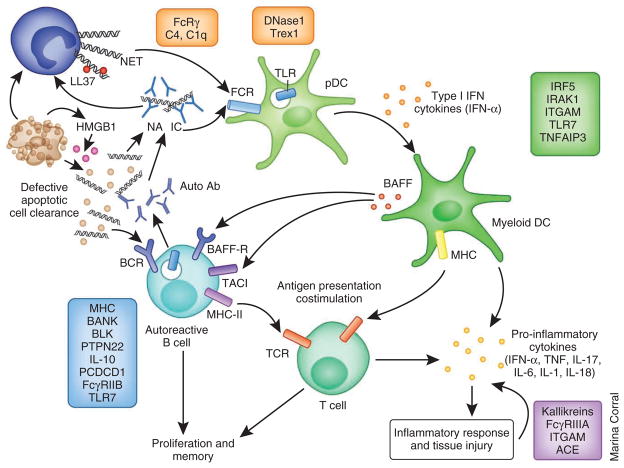
Although this field is still in its early stages, some fundamental concepts have emerged regarding the inheritance of autoimmunity. First, with the exception of the major histocompatibility complex (MHC), the genetic risk loci that have been identified by GWASs involve a conglomeration of relatively common alleles, each of which confers only a modest risk, with odds ratios less than two. Second, some of these polymorphisms are shared between autoimmune disorders. Third, not all of the risk genes cross major racial groups. Fourth, genetic studies have identified functional pathways that are involved in disease pathogenesis7,8. In SLE, three main immune pathways have been identified: aberrant clearance of nucleic-acid–containing debris and immune complexes, excessive innate immune activation involving Toll-like receptors (TLRs) and type I interferons (IFNs) and abnormal T and B lymphocyte activation8. In addition, genetic polymorphisms may be associated with susceptibility to target organ damage9,10 (Fig. 2). These SLE-related pathways overlap with those identified in mouse lupus models and are currently leading to the development of therapeutics that target key components of these pathways.
Figure 2.
The role of SLE risk alleles in the pathogenesis of SLE. A panoply of genetic variations has been linked to SLE susceptibility. Polymorphisms in genes involved in the immune clearance of apoptotic particles and nucleic-acid–containing immune complexes (clearance functions, with examples shown in orange) may induce the enhanced activation of pDCs and autoreactive B cells, leading to the production of type I IFN and the expansion of autoreactive effector cells, respectively. Polymorphisms in genes involved in innate immunity (with examples shown in green) regulate the induction of, as well as the response to, type I IFN. Abnormal function of innate immune cells in turn activates the adaptive immune system. Both the innate and adaptive immune systems contribute to the inflammatory response and tissue damage. A third major group of polymorphic genes is involved in ligand recognition, receptor signaling and other immunological functions of adaptive immune cells (with examples shown in blue). Dysregulation of the adaptive immune system results in loss of tolerance and the production of autoantibodies, which in turn bind to nuclear antigens and activate innate immune cells, completing a positive feedback loop that amplifies the pathogenic processes in SLE. Polymorphic alleles may also influence the severity of organ damage (with examples shown in purple). Ab, antibody; IRAK1, interleukin-1 receptor-associated kinase 1; ITGAM, integrin α M; TNFAIP3, tumor necrosis factor, α-induced protein 3; BANK, B cell scaffold protein with ankyrin repeats; BLK, B lymphoid tyrosine kinase; PCDCD1, programmed cell death 1; NA, nucleic acid; IC, immune complex; ACE, angiotensin I converting enzyme (peptidyl-dipeptidase A) 1.
Applying this new information to individual patients is now a major challenge. As many of the identified single nucleotide polymorphisms are not within coding regions, attribution of disease risk to a single gene requires extensive resequencing of large regions of DNA from many patients, which is a daunting and costly task. Furthermore, the risk variants identified so far account for only a small proportion of the overall heritability of disease7, highlighting the limitations of the GWAS approach11, which identifies only common polymorphisms with frequencies within the population of greater than 1–5%. The finding that 3′ repair exonuclease 1 (TREX1) is mutated in 0.5–2% of patients with SLE but was not identified in GWAS exemplifies the limitations of the GWAS approach for detecting variants at the lower end of this range. New methodology to identify rare variants, copy number variations and epistatic changes are being developed, but these alterations will only be found in a small fraction of affected individuals. Moreover, it is not easy to establish a link between genetic variations, gene function and disease pathogenesis. Many of the risk alleles identified by GWASs are common variants with subtle effects that are compatible with normal immunological function; some variants may even have been evolutionarily selected because they confer protection against certain pathogens. Even for genes with coding-region mutations, such as TREX1 and PTPN22, functional studies have either not yet elicited definitive mechanisms by which the disease-associated polymorphism leads to increased risk for SLE or have yielded conflicting results12,13. More studies will be required to understand how disease-associated variants affect immune responses and interact with each other to contribute to the risk of SLE. Nevertheless, as the function of the variants is clarified and potentially pathogenic pathways are identified, personalized interventions may become possible.
Although the practical applicability of individual genetic profiling seems distant, pilot studies are currently underway to explore the prediction of risk in family members and the prediction of disease phenotype and severity in individual patients10,14. Testing is already being used in the clinic for gene variants that affect the metabolism of the immunosuppressive drug azathioprine to help predict the risk for toxicity associated with the use of this drug in patients15. Undoubtedly, other applications of the new genetic knowledge will be forthcoming.
The reasons for gender bias in SLE
SLE is nine times more common in women than in men. Studies in mice have shown multiple effects of both male and female sex steroids on the immune system16; studies in knockout mice have clearly attributed the immune-activating effects of estrogen to signaling by the estrogen receptor α. However, a clear correlation of female sex hormone concentrations and disease activity has not been established in human patients, reflecting the complexity of the contributions from genetic and environmental factors. The X chromosome harbors many genes of immunologic interest. In gonad-matched mice, the presence of XX confers a greater susceptibility to pristane-induced lupus than does XY17. Similarly, in humans, the XXY phenotype is associated with an increased prevalence of SLE. Although there are decreased concentrations of protective T helper type 2 (TH2) cytokines in XX mice than in XY mice, the genetic components of the XX chromosome complement associated with lupus risk are not clear. As candidate genes on the X chromosome are identified and validated in gonad-matched mice, new therapies for female patients with SLE may emerge.
The innate immune system is activated by nucleic acids
Safe clearance of ubiquitous cell debris is a normal immune function that is crucial for the maintenance of self tolerance; multiple pathways exist for clearing the billions of cell corpses generated daily. Apoptotic cells are engulfed by phagocytes through receptors that recognize altered cell membrane components or self antigens opsonized by ‘natural’ IgM autoantibodies that are present in the sera of healthy individuals, early complement components or both, inducing a suppressive program that facilitates the disposal of debris with little consequent inflammation18. In addition, nucleases break down circulating nucleic acids, preventing their recognition by innate receptors19. Defective clearance mechanisms result in secondary necrosis and an overload of self antigens that, instead of being safely consumed by phagocytes, access proinflammatory receptors, such as activating Fc receptors (FcRs) or TLRs on or inside innate immune cells.
The best studied innate cell in SLE is the plasmacytoid dendritic cell (pDC), which produces large amounts of type I IFNs in response to nucleic-acid–containing immune complexes (NA-IC) and can migrate to injured sites20,21. The pathogenic role of type I IFNs in SLE is supported by a signature of IFN-induced genes in the peripheral blood of patients22, an association with risk alleles involved in TLR and IFN pathways23, disease acceleration by exogenous IFN-α in several lupus models24,25 and disease amelioration in some lupus-prone mouse strains that have been rendered deficient for the type I IFN receptor26,27. Type I IFNs have multiple proinflammatory functions in both innate and adaptive immune cells28 and can also activate and damage endothelial cells29. Identification of a neutrophil signature in the blood of patients with lupus led to the recent discovery that neutrophils exposed to NA-IC are prone to die by NETosis: these dying cells extrude DNA webs (neutrophil extracellular traps, NETs) that are associated with small alarmin peptides, such as the calthelicidin LL37, that protect the DNA from nucleases. LL37-complexed DNA in turn activates pDCs, enhancing the release of type I IFNs30,31. It is now crucial to determine whether this amplification mechanism prevails in vivo and whether NET formation is required for disease to develop. Even basophils activated by IgE autoantibodies have been reported to have a proinflammatory role in lupus32. The relative contribution of each of these cell types to lupus pathogenesis remains to be determined.
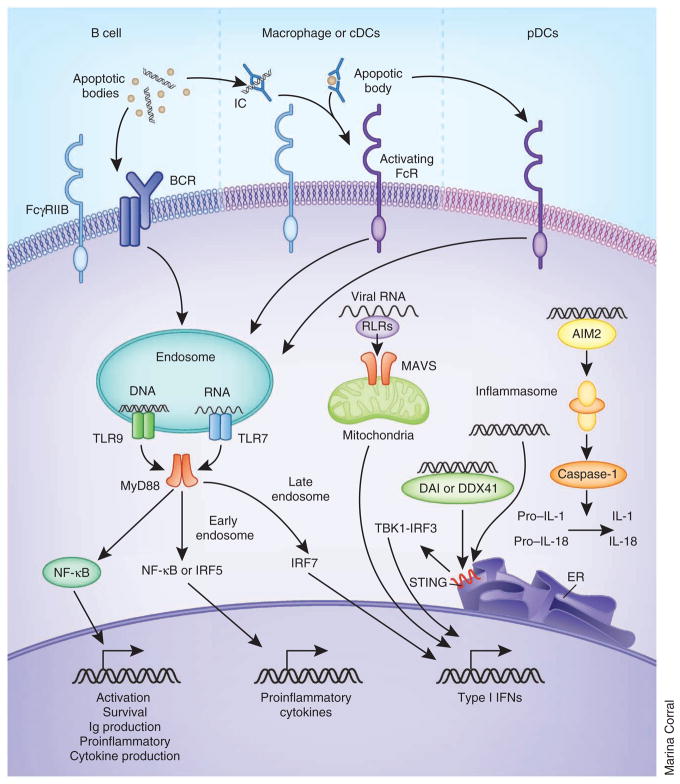
Some TLRs are on the cell surface and interact with the circulating products of tissue injury and inflammation; in contrast, nucleic-acid–specific TLRs (TLR3, TLR7, TLR8 and TLR9) are segregated in an unprocessed form within intracellular vesicles, where they are subsequently cleaved before interacting with internalized ligands33. Great progress has been made in understanding the intracellular trafficking of nucleic acid antigens, TLR signaling mechanisms and downstream consequences of TLR signaling (Fig. 3). NA-IC and opsonized cellular debris are internalized by binding to activating FcRs on innate cells34; the uptake and intracellular trafficking of self antigens may be facilitated by LL37 (ref. 35) or high mobility group box 1 (HMGB1)36, an intracellular protein that is released by dying cells or activated monocytes and dendritic cells. Vesicles containing the internalized antigens then fuse with TLR-containing endosomes to form large autophagosomes in which processed TLRs, antigens and signaling molecules can interact37. On interaction with their ligands and a series of adaptor molecules, TLRs initiate transcription of either type I IFNs or inflammatory cytokines, depending on where the ligand binding occurs. In pDCs, TLR binding in early endosomes preferentially induces phosphorylation of interferon regulatory factor 7 (IRF7), which initiates the transcription of type I IFNs. In contrast, TLR ligation in late endosomes favors the activation of nuclear factor of κ light polypeptide gene enhancer in B cells (NF-κB) or mitogen-activated protein kinase (MAPK) signaling pathways, resulting in the abundant production of proinflammatory cytokines37, such as interleukin-6 (IL-6), IL-12, tumor necrosis factor α (TNF-α) or BAFF, that serve to bridge innate and adaptive immunity, contributing to the overall inflammatory process of SLE38. Nucleic-acid–containing debris can also be taken up by the B cell receptor (BCR) of auto-reactive B cells, resulting in cell activation and differentiation and the expression of transmembrane activator and calcium modulator ligand interactor (TACI), the receptor for the B cell survival molecules BAFF and APRIL, a proliferation ligand39.
Figure 3.
Recognition of nucleic acids by innate immune cells triggers cytokine production. Nucleic acids or apoptotic particles can be taken up by B cells through the BCR, and immune complexes containing these antigens are taken up by monocytes, myeloid dendritic cells and pDCs through FcR-mediated recognition and internalization. Within endosomes, DNA and RNA then interact with TLR9 and TLR7, respectively. Viral RNA can also be delivered to endosomes by autophagosome formation in pDCs. The ligation of TLRs recruits the adaptor protein MyD88, which activates the NF-κB signaling cascade in B cells and leads to B cell activation and survival, as well as various effector functions. In pDCs, the recruitment of MyD88 preferentially triggers an IRF7-mediated signaling pathway, which initiates type I IFN production. In other cell types, MyD88 recruitment in late endosomes leads to inflammatory cytokine production. Cytosolic DNA and RNA can be recognized by sensors that, through adaptors, lead to type I IFN production. Cytosolic DNA can also be recognized by AIM2, which activates caspase-1, leading to proteolytic cleavage of pro–IL-1 and pro–IL-18 into active forms. Immune complexes may signal directly through both activating and inhibitory FcRs whose relative expression on the cell surface varies with cell activation status. How these positive and negative signals are integrated with each other and with TLR-mediated signals has not yet been fully elucidated. cDC, conventional dendritic cell; RLR, RIG-I like receptors; DDX41, a member of the DEXDc family of helicases; TBK1, TANK-binding kinase 1; ER, endoplasmic reticulum; Ig, immunoglobulin.
It is now clear that IgG antibodies to double-stranded DNA are produced only when TLR9 is accessed40, whereas antibodies to RNA require TLR7 (ref. 40). Nevertheless, TLR9 deficiency exacerbates SLE in mouse models, perhaps because it competes with TLR7 for transport to intracellular compartments; absence of this competition skews the autoantibody specificity to a more pathogenic anti-RNA profile41. TLR7 deficiency has modest protective42 effects, but full protection from disease is only achieved when both TLR7 and TLR9, or their adaptor protein myeloid differentiation primary response gene 88 (MyD88), are absent41. Notably, susceptibility to SLE in mice overexpressing TLR7 is conferred predominantly by excess TLR7 expression in B cells, illustrating the overlap of innate receptor function with adaptive lymphocyte populations43. Other nucleic acid sensors detect cytosolic DNA and RNA44 (Fig. 3). Cytosolic RNA derived from viruses, a possible trigger for disease onset in some cases45, is recognized by retinoic acid inducible receptor (RIG-I)-like family receptors, which signal through a mitochondrial adaptor protein, the mitochondrial antiviral signaling protein (MAVS), to initiate type I IFN transcription. Cytosolic DNA sensors are less well defined and may include DNA-dependent activator of interferon regulatory factor (DAI), absent in melanoma 2 (AIM2) and stimulator of interferon genes (STING). Although DAI was the first cytosolic DNA sensor identified46, its absence does not prevent cytosolic-DNA–induced signaling, leading to considerable skepticism about its role in intracellular nucleic acid surveillance47. Recent studies have provided convincing results that support the role of STING as either an adaptor protein or a direct component of the cytosolic-DNA–sensing machinery that is negatively regulated by the exonuclease Trex1 (refs. 48,49). AIM2 is different from other sensors in that its interaction with DNA activates the inflammasome, leading to the production of active IL-1β and IL-18.
Although more work needs to be done to delineate how the various innate immune activation pathways interact and are regulated, and whether innate immune activation is required for both disease initiation and perpetuation, this new understanding offers opportunities for cell-specific intervention or for intervention that is targeted to shared pathways, such as TLR signaling or type I IFN release.
The adaptive immune system becomes activated in SLE
Adaptive immunity involves the clonal expansion of lymphocytes and the generation of long-lived effector T and B cells. Self-reactive lymphocytes are continuously generated by somatic gene processes that establish the large repertoire of T and B cell receptors required for protection against pathogens. Although relatively low affinity natural IgM autoantibodies, made by specific innate B cell subsets, help to prevent immune responses to the products of cell death, potentially pathogenic self-reactive lymphocytes are removed at specific developmental checkpoints (deletion), become unresponsive to external stimulation (anergy), replace their receptors (receptor editing) or are suppressed by regulatory molecules or cells50. In SLE, these regulatory mechanisms may be genetically defective, environmentally altered or both. Substantial evidence points to clonal expansion of autoreactive B lymphocytes in the preclinical phase of SLE51, and most mouse studies have shown a requirement for T cells in disease initiation.
T lymphocytes are activated on recognition of a peptide–MHC complex on activated antigen-presenting cells. Polymorphisms in MHC molecules control the size and diversity of the peptide repertoire that is presented to T cells and thus have a major impact on T cell activation and the subsequent immune responses. The strongest genetic risk alleles for SLE have been mapped across the MHC region in humans6, implying that loss of T cell tolerance in SLE might be triggered by aberrant MHC presentation of particular peptides. In addition, peripheral T cells from patients with SLE have altered signaling and a faster T cell calcium flux than those of healthy individuals as a result of replacement of the principle signaling molecule of the TCR complex, cluster of differentiation 3 ζ (CD3-ζ), by the FcR ζ chain52, resulting in the use of the adaptor molecule spleen tyrosine kinase (SYK) rather than the usual ζ chain (TCR) associated protein kinase (ZAP70) and activation of the downstream kinase calcium/calmodulin-dependent protein kinase type IV (CAMK4) that, through the transcription factor cAMP response element modulator α (CREM-α), enhances production of IL-17 and blocks production of IL-2. Other abnormalities of T cell function in SLE that contribute to excessive activation or failure of regulation include T cell mitochondrial dysfunction leading to oxidative stress53, a decrease in the cytotoxic activity of CD8 cells54 and increased expression of the co-stimulatory molecule CD40 ligand (CD40L) and of the adhesion molecule CD44 on CD4+ T cells. CD44 ligation results in the activation of ρ-kinase coiled-coil–containing domain protein kinase (ROCK), which promotes the production of IL-17 and IL-21. The identification of key signaling molecules in SLE T cells, such as SYK, CAMK4 and ROCK, has resulted in successful proof-of-principle use of inhibitors in SLE models55–57 that may now be translated to human use.
The role of newly discovered T cell subsets in SLE pathogenesis is also being investigated. Dysregulation of follicular T helper (T FH) cells that promote B cell differentiation in germinal centers is associated with the development of SLE in mouse models58, and expansion of a circulating TFH cell population has been reported in patients with active SLE59. In SLE models, abundant TFH-like cells are also located outside the germinal centers, where they support extrafollicular B cell differentiation60. Other reported abnormalities include a decrease in the number of regulatory T cells during active SLE and an expansion of the CD3+CD4−CD8− cell population61 that produces the proinflammatory cytokine IL-17 and may contribute to local inflammation in the kidneys. Given the extensive heterogeneity among patients with SLE, it is plausible that genetically diverse individuals exposed to varying environments will differ in the mechanism for disease initiation and/or the phenotype or function of dominant effector T cell subsets and will therefore require different therapeutic strategies. For example, in some individuals, excessive activation may predominate, whereas in others there may be a failure of regulation.
Defective B cell tolerance is another hallmark of SLE. Central and peripheral checkpoints that remove self-reactive immature B cells are defective in patients with SLE50,62. Enhanced BCR signaling can lead to autoreactivity, probably as a result of excessive B cell activation after the late transitional stage, when BCR signals start to activate the cells rather than tolerize them. Signals from several molecules, including CD19, intracellular TLRs and the BAFF receptor (BAFF-R) interact with BCR signals to enhance B cell activation and survival at this stage, and an excess of any of these molecules may predispose to SLE63. In addition, an SLE-susceptibility–related polymorphism in PTPN22 is associated with early B cell tolerance defects through a mechanism that has not yet been determined64,65.
B cell tolerance can also be broken during antigen activation. An area of intense interest is the regulation of the germinal center, where clonal expansion of B cells occurs, along with somatic mutation, class switching and differentiation to long-lived effector cells. Germinal center selection is clearly defective in SLE, allowing autoreactive B cells to differentiate into pathogenic memory and plasma cells66. Recent live microscopy studies of secondary lymphoid organs have revealed mechanisms for the efficient sampling of antigens and the intricate orchestration of sequential reciprocal interactions between B cells, T cells and follicular dendritic cells before and during the germinal center reaction67. Despite the enormous gains in knowledge that have been made about the anatomy of germinal centers, a convincing explanation of how the autoreactive B cells that are inevitably generated by somatic mutation are regulated at or after the germinal center stage has yet to emerge. A recapitulation of some of the negative selection events found in early B cell development is probable. The loss of germinal center tolerance in SLE could be the result of an excess of T cell help, defects in B cell signaling or death, an excess of self antigens resulting from inadequate clearance of apoptotic cells68 or defects in the function of various regulatory cells, including B cells69 or T cells70,71. The association of SLE with an acquired decrease in the expression of the inhibitory receptor FcγRIIB has been experimentally attributed to an intrinsic failure of regulation of activated B cells in the post–germinal-center phase72.
Autoreactive B cells can also be expanded outside germinal centers. Excess inflammatory signals, such as IL-12 (ref. 73), TLR ligation74 or CD40 ligation75, preferentially promote B cell expansion in extra-follicular foci in which short-lived plasma cells with a limited degree of somatic mutation arise. Little is currently known about the regulation of autoreactive B cells during the extrafollicular response or the relative contribution of this response to the pool of pathogenic effector cells in SLE.
Despite a decrease in total circulating B cells in many patients with SLE, long-lived autoreactive effector memory and plasma cells are often increased in number and, because of their quiescent state, may be resistant to immunosuppression. Molecules such as BAFF or APRIL, IL-6, transforming growth factor β (TGF-β), chemokine (C-X-C motif) ligand 12 (CXCL12) and vascular cell adhesion molecule (VCAM) contribute to bone marrow ‘survival niches’ that support long-lived plasma cells76,77. Inflamed organs express many of these molecules and can become sites for disorganized lymphoid follicles or even germinal centers in which normal regulation might be disturbed, as well as a new niche for plasma cells76. It is worth noting that plasma cells are resistant to B cell depletion therapy with rituximab78, and neither class-switched memory B cells nor long-lived plasma cells are eliminated by BAFF blockade79,80.
Given the multiple roles of B cells in disease pathogenesis and the absolute requirement for B cells in disease initiation, a large amount of effort has been devoted to B cell–directed therapies in SLE, with phase 3 studies having been completed for the BAFF antagonist belimumab, the B cell–depleting agent rituximab (anti-CD20) and the T cell co-stimulatory antagonist abatacept (CTLA4Ig), which blocks the development of TFH cells (see Boxes 1 and 2 and Table 1 and refs. 81–83 for comprehensive reviews). Of these, only belimumab has thus far been successful in placebo-controlled clinical trials, reflecting the complexity of treating a chronic disease in which adaptive immune system activation and clonal expansion have already occurred.
BOX 2. Reasons why clinical trials in SLE are challenging to perform and interpret.
Challenges in trial design
-
Outcome measures not uniform:
No consensus on the best way to measure responses to therapy.
Few validated biomarkers available.
Disease may remit spontaneously.
Therapies are compared with the standard of care, resulting in high response rates in the control group.
Heterogeneity in disease stage
Pleiomorphic role for cytokines during different disease stages.
Initiation: there may be a long preclinical phase during which the therapeutic window for some interventions may be closed.
Progression: recruitment of long-lived effector cells, redundant inflammatory molecules, abnormal signaling pathways and epigenetic changes.
End organ damage: irreversible tissue damage and fibrosis may progress despite immune quiescence.
Heterogeneity in disease mechanisms
Multigenic origin with extensive intrapatient heterogeneity.
Different pathogenic mechanisms and effector cell types among patients with lupus.
Different injury mechanisms in various target organs, even within the same patient.
Homeostatic responses to therapy
B cell depletion enhances B cell release from the bone marrow and impairs negative selection against autoreactivity.
Plasma cell depletion may result in a prolonged immunoglobulin half-life.
Depletion of naive and activated lymphocytes may lead to an increase in the number of memory cells.
Table 1.
Therapeutic strategies for SLE
| Lessons from animal models of SLE128,142 | |
|
| |
| Knockout mice can be used to study disease initiation. | Molecules involved in the clonal expansion of lymphocytes and/or germinal center functions are necessary for the initiation of disease in most models. Some molecules, such as type I IFNs and FcR, are involved in disease initiation or tissue damage only in certain strains, showing the effect of genetic heterogeneity on disease mechanisms. Some molecules, such as TLR9, TNF-α and IL-10, have pleiomorphic functions and therefore may be either protective or pathogenic, depending on the disease stage. |
| Specific targeting of immune pathways has proved less effective in attenuating disease than is observed in the knockouts. | Nearly all drugs are more effective at preventing disease onset than treating established disease. As disease progresses, higher drug doses and combination therapies are required to achieve remission. Different strains have different stringencies for therapeutic responses, and not all strains respond to each therapy. |
|
| |
| Recent approaches to SLE therapies81–83 | |
|
| |
| Innate immunity | Approaches include targeting TLRs and their downstream signaling molecules and effector cytokines, improving clearance mechanisms using exogenous IgM and delivering exogenous DNase to compensate for intrinsic defects of this enzyme in SLE. Some aspects of TLR signaling are protective against autoimmunity in animal models. Circulating DNA in SLE may be resistant to digestion. These approaches are mostly experimental or are in development for human use. Phase 2 studies of IFN-α inhibition are pending. |
| Adaptive immunity—B cells | Approaches include B cell depletion, modulation of co-receptor function and alteration of B cell selection. These approaches rarely target all subsets of B cells and may lead to counterproductive homeostatic expansion of other subsets. Approaches that alter selection may have different effects on naive and antigen-activated immunoglobulin repertoires and may need to be given continuously to achieve clinical efficacy. Clinical trials have so far only shown efficacy for belimumab; new trials of individual treatments and drug combinations are underway. |
| Adaptive immunity—T cells | Approaches include co-stimulation blockade, cytokine inhibitors, kinase inhibitors and induction of regulatory subsets. Clinical trials of co-stimulatory inhibitors have so far either failed to show efficacy or were terminated because of toxicity; new trials of additional agents or drug combinations are underway. Effects of broad inhibitors of T cell activation may result in immune suppression. |
| Tissue injury and inflammation | Approaches include inhibitors of the complement cascade, cytokine blockade and nonimmunological approaches to preserve renal function. Nonimmunological approaches may prevent end-stage organ damage but have no effects on systemic immune dysfunction. Because of the pleiomorphic effects of cytokines, cytokine blockade may improve some aspects of disease but worsen others. Early clinical studies of cytokine blockade have produced promising results but have also shown adverse effects in several cases. Many new approaches are in development. |
New therapies for SLE primarily target one of four major pathogenic pathways: innate immunity, B cells, T cells or tissue injury and inflammation. Although animal studies provide valuable insights into the therapeutic potential of experimental drugs for SLE, additional considerations are needed when predicting the probable outcomes of treatments in human patients.
Target organ damage is mediated by diverse mechanisms
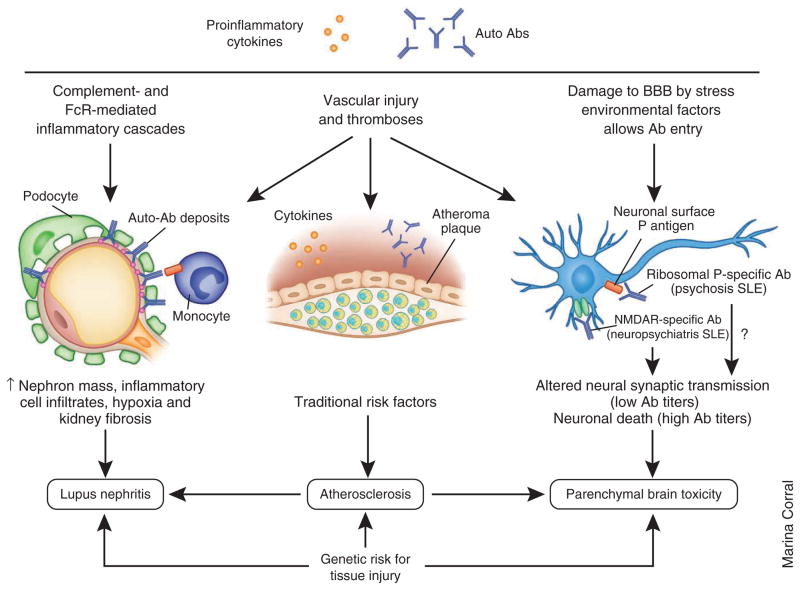
Although autoantibodies are a hallmark of SLE, the mechanisms of tissue injury by autoantibodies are variable, and the pathogenic specificities that direct autoantibodies to particular target organs can be difficult to identify. Depositing autoantibodies may directly injure or activate cells, initiate the complement cascade or activate innate receptors, leading to local inflammation. Furthermore, isotype differences in FcR and complement-binding activities influence antibody pathogenicity, and the cellular inflammatory response can vary in different organs or in patients with different genotypes, resulting in heterogeneous types of injury. This section is focused on recent advances in understanding the mechanisms that mediate injury in the brain, kidney and blood vessels (Fig. 4). Involvement of these vital organs contributes to substantial morbidity and mortality in SLE.
Figure 4.
Mechanisms for organ damage. Organ damage is caused by immune activation and inflammation but is also influenced by genetic and nonimmunologic environmental factors. Autoantibodies and circulating inflammatory mediators trigger tissue injury in target organs by a variety of mechanisms. In the kidneys, immune complex deposition induces complement- and FcR-mediated inflammatory cascades that lead to the activation or injury of renal resident cells, which in turn release inflammatory mediators, leading to the recruitment of inflammatory cells. Long term renal damage is caused by ongoing inflammation, vascular injury by systemic and local mediators, hypoxia and fibrosis. Nephritis occurs in approximately 50% of adult and 80% of pediatric patients with SLE, and the rate of end-stage renal disease in the United States caused by SLE seems to be increasing, especially in minority patients. In the cardiovascular system, autoantibodies and soluble inflammatory mediators cause vascular endothelial injury by inducing endothelial apoptosis or activation. Recruitment of monocytes to the injured site is also crucial for plaque formation. Other proatherogenic factors in addition to traditional risk factors include oxidized low-density lipoproteins, antibodies to oxidized lipids and proinflammatory high-density lipoproteins. The 10-year risk for a coronary event or stroke is 7.5- to 17-fold increased in patients with SLE compared with healthy individuals. In the central nervous system, autoantibodies access the brain when the BBB is attenuated by inflammatory mediators or environmental factors, such as cigarette smoke or neurotransmitters released by stress. Once deposited in the brain, autoantibodies may induce neuron apoptosis or alter neuronal synaptic transmission. Neurologic injury can also result from secondary causes, such as thrombosis or infections. All these pathogenic changes lead to a wide range of neuropsychiatric manifestations of SLE, including progressive cognitive dysfunction. Neuropsychiatric manifestations may occur relatively early in the disease process, and they affect up to 40% of patients.
SLE is characterized by wide range of neuropsychiatric syndromes. Although many of these manifestations are the result of secondary causes, autoantibodies may directly cause vascular or parenchymal injury; the association of phospholipid-specific antibodies with intracranial clotting events is well known84. Recent studies have begun to unravel the pathogenic events involved in parenchymal injury. Antibodies directed to the N-methyl-D-aspartate receptor (NMDAR) and to ribosomal P protein can be detected in the cerebrospinal fluid of patients with neuropsychiatric SLE; serum P-specific antibodies are predictive of future SLE psychosis85. In experimental mice, a low concentration of NMDAR-specific antibody alters neural synaptic transmission, whereas a high concentration of this antibody induces neuronal death, providing some insight into the observations that cognitive dysfunction is transient in some patients and is permanent in others86. Neuronal damage mediated by NMDAR-specific antibody occurs without inflammation and is independent of complement activation or FcR ligation87. The mechanisms for tissue damage mediated by ribosomal P-specific antibodies are not as well characterized, but studies in mice have shown direct deposition of these autoantibodies in various regions of the brain and their cross reactivity with neuronal surface P antigen88, suggesting a toxicity mechanism similar to that postulated for NMDAR-specific antibodies. Furthermore, the integrity of the brain-blood barrier (BBB) needs to be compromised for these autoantibodies to enter the brain and exert their pathogenic effects89. Depending on the mechanism by which the BBB is breached, autoantibodies may enter different anatomic sites, thereby causing different neuropsychiatric manifestations. These findings suggest that therapies directed at protection of the BBB and brain parenchyma may be effective in neuropsychiatric SLE.
Lupus nephritis is initiated by the deposition of immune complexes that may activate complement90, engage activating FcRs on mononuclear cells91 and/or activate resident renal cells, such as mesangial cells and endothelial cells, through FcRs or TLRs92,93. Mesangial cells produce inflammatory mediators, whereas endothelial cells either undergo apoptosis or facilitate the influx of inflammatory cells into the interstitium94; these infiltrating cells may then undergo local expansion and differentiation. Renal inflammation is associated with overexpression of diverse cytokines and chemokines that are upregulated at various stages of lupus nephritis and at different renal sites95. TH1 and TH17 cells have been detected in human proliferative lupus kidneys96, and both TNF-α and IL-18 are local effector cytokines in at least some patients97. A relatively new development has been the identification of subsets of infiltrating renal macrophages and dendritic cells that mediate aberrant tissue remodeling and whose presence is associated with poor outcome98,99. As glomerular cells become injured and die, the decline of nephron mass and function is followed by the extension of the inflammatory process into the tubulo-interstitium, with consequent damage to the renal tubules100. If the inciting injury remains active and the sites of tissue repair continue to be hypoxic, reparative processes become chronic and lead to the amplification of inflammation and fibrosis101. Eventually, deterioration of kidney function progresses even if the initiating inflammatory process has been adequately contained. Genetic polymorphisms may influence disease severity and the rate of progression; for example, kallikrein gene polymorphisms have recently been identified as susceptibility alleles for lupus nephritis9, and an FcγRIIIA allele associated with excess activation confers risk for progression to end-stage disease102. These findings suggest that the progression of renal impairment in SLE shares many common mechanisms with other non-immune–mediated renal diseases and that strategies currently being applied in other diseases to prevent tissue hypoxia and remodeling may also be useful in SLE.
Atherosclerotic cardiovascular disease is increasingly being recognized as a major cause of death in patients with SLE; its high prevalence in SLE cannot be fully explained by traditional risk factors103,104. Persistent inflammation, circulating immune complexes, autoantibodies and corticosteroid use have all been proposed to contribute to the pathogenesis of atherosclerosis in SLE105. Vascular endothelial injury is the primary event in atherosclerosis and can be exacerbated in SLE by autoantibody- or cytokine-mediated endothelial apoptosis or by endothelial cell activation that induces the local production of pro-inflammatory cytokines and the upregulation of adhesive molecules that facilitate the local recruitment and activation of monocytes that help form plaque105. Another key observation in SLE is an imbalance of proinflammatory and anti-inflammatory lipids. Proinflammatory oxidized low-density lipoproteins, as well as antibodies to oxidized cardiolipin and oxidized low-density lipoproteins, have been detected in patients with SLE, with at least some subsets of these antibodies being proatherogenic106–108. High-density lipoproteins, instead of being protective, become proinflammatory in some patients with SLE109. These pathological events facilitate the development of foam cells and are linked to a high risk of plaque formation in SLE105. Disappointingly, initial studies of statins have failed so far to show any substantial protection from atherosclerosis progression in patients with SLE110,111. It is therefore imperative to determine which patients are most at risk for atherosclerosis progression and how best to protect the endothelium in these patients from inflammatory triggers.
Emerging areas in SLE
Although the last decade has brought the discovery of many new areas of knowledge relevant to the treatment of SLE, there is still a large amount of unexplored territory. Technologic advances that allow the sequencing of individual genomes should uncover rarer variants that are associated with SLE and improve our understanding of the scope of the immune abnormalities associated with disease susceptibility, patterns and outcomes. Similarly, the delineation of the mechanisms of gene regulation is another key to understanding disease progression. Epigenetic alterations, such as DNA hypomethylation and histone modification, affect a wide range of immunological events, including cell death, cell activation and inflammation, and have been implicated in the pathogenesis of lupus112,113. In addition, the aberrant expression of microRNAs that regulate gene expression within normal and inflammatory environments has been associated with lupus114,115. More studies are needed to understand the full complement and function of microRNAs to exploit them as therapeutic targets. Systems biology approaches and high-throughput screening technologies should make all these areas more amenable to investigation using small amounts of human cells or tissue from well-phenotyped patients. Another technical advance has been the development of live-tissue imaging, which has opened up a dynamic understanding of biology within secondary lymphoid tissue and inflamed organs. Although this technology has mostly been applied to normal immune function, its application to diseased states will likely increase in the future.
The cellular regulation of immune responses is a new area of interest. The contribution of commensal gut flora to lupus is currently unknown, but the ability of particular commensal organisms to regulate proinflammatory TH17 cell responses suggests that these organisms could influence target organ inflammation116. In addition, there is an ever growing repertoire of regulatory cell types whose function might be exploited in vivo. Finally, early studies are using both hematopoietic117 and mesenchymal stem cells118,119 to help reconstitute a predisease immune repertoire.
With the increasing recognition that genetic and phenotypic heterogeneity affects the clinical manifestations of SLE and patients’ responses to therapies, there has been much interest in developing biomarkers for individualized disease risk prediction and prognosis, detecting impending disease flares, defining what constitutes a therapeutic response and predicting whether an individual patient is likely to mount a therapeutic response to a specific agent. This is the subject of a recent comprehensive review120. These types of biomarkers include measures of soluble molecules or sets of molecules in serum or urine, gene expression profiles from peripheral blood cells or tissues and functional assays of immune cell activation. Major challenges in biomarker development are the collection of longitudinal reliable and reproducible clinical data, as well as optimal specimen collection and storage. Interpretation of urine data needs to take into account the degree of proteinuria and the stability of the biomarker protein over time under conditions of variable pH, as well as the biology of the biomarker (size, tubular filtration, reabsorption and secretion characteristics, the presence of a cleaved form, and so on). Nevertheless, major methodologic advances in both generating data-sets and data analysis have been made, as exemplified by the ability to group biomarkers into modules with predictive power121,122. In addition, several biomarkers of renal flare with somewhat improved sensitivity or specificity over the currently used serologic and urine measures have been identified in a limited number of longitudinal studies123–125.
The promise and challenges of developing therapies for SLE
Systemic inflammation, NA-IC and tissue injury amplify immune activation in SLE by profoundly affecting both innate and adaptive immune cells. Furthermore, chronic inflammation in SLE activates aberrant signaling cascades that may impair normal regulatory control mechanisms or confer resistance to immunosuppressive drugs126,127. Some of these aberrant phenotypes may subsequently become fixed by epigenetic changes. With so many inflammatory pathways in play in active SLE, intense therapeutic intervention is often needed, and it can be difficult to balance the risks of prolonged or excessive immune suppression against those of smoldering inflammation. Mouse studies have shown that preventing clonal expansion of autoreactive B and T cells is effective in early disease, but this approach may fail once escalation of the immune response occurs; more aggressive drug combinations are then required to induce remission128. These findings suggest that earlier intervention and continuous maintenance therapy may reduce morbidity from the incremental tissue damage and drug toxicity that accompanies disease flares; the current therapeutic paradigm is to reserve immunosuppressive interventions for disease flares and withdraw the use of most medications during quiescence. As safer drugs are developed, this paradigm will probably change.
Another challenge is the variability in design and primary outcome measures in lupus clinical trials, making it impossible to compare the results of new experimental agents with each other or even to ascertain the therapeutic benefit of a single agent. One striking example is the different efficacy results obtained when alternative outcome measures were used to analyze the results of a trial of the T cell co-stimulatory inhibitor abatacept (CTLA4Ig) for SLE nephritis129. In addition, most clinical trials are of short duration, so the long-term benefits of even a modest decrease in flares or corticosteroid dose cannot be measured.
Apart from the contentious issue of trial design, heterogeneity in disease mechanisms among different individuals, disease stages and organ systems may greatly influence responses to therapeutic interventions. Given the multiple cell subsets and cytokines involved, as well as the emergence of abnormal and redundant cell activation pathways that make it difficult to achieve a therapeutic response by targeting single pathways and the capacity for ongoing organ damage even after immune quiescence, it is not surprising that the effects of new therapies in broad SLE populations compared with the current standard-of-care therapies, which have measurable efficacy, have been rather modest (Box 2). This highlights the need for determining whether a particular therapeutic approach will be appropriate for particular genotypic or phenotypic patient subsets; this is currently not possible, but it is an area that is ripe for exploration.
Despite these major mechanistic and logistic challenges, new immunologic discoveries have yielded a treasure trove of potential therapeutic avenues (Table 1). Targeting the innate immune system by enhancing clearance of DNA, RNA and cellular debris, inhibiting the activation of pattern recognition receptors or blocking downstream signaling events and cytokines may prevent cell activation by self antigens130. Components of the adaptive immune system are also attractive targets, which has led to the development of drugs that deplete or functionally inhibit particular B or T cell subsets or strategies that alter B cell tolerance or differentiation54,81. Cell-surface molecules can be targeted by monoclonal antibodies that either facilitate cell depletion or transmit activating or inhibitory signals into selected cells. Monoclonal antibodies can also target soluble molecules, particularly cytokines, thus altering the function of particular cell subsets or inhibiting molecules that directly cause tissue damage38. Small molecules are being designed that enter cells and inhibit components of cell-signaling cascades131. Moreover, an enhanced understanding of immune regulation suggests the possibility of harnessing normal immune protective mechanisms to improve tolerance and protect target organs without the disadvantages that accompany excessive immunosuppression. As clinical trial design improves, the place of each intervention will need to be carefully determined.
In mouse models, late-stage disease that is resistant to single immune interventions can effectively be controlled with combination therapies132,133. Uncontrolled case series have suggested that combination therapies are also effective in humans despite concerns about toxicities, for example, the combination of rituximab, cyclophosphamide and prednisone is reportedly effective in some individuals with severe disease that are unresponsive to traditional immunosuppression134, and a controlled trial of a combination treatment using abatacept and cyclophosphamide for nephritis is ongoing. However, it must be remembered that in humans, most clinical trials are currently designed to test new therapies on a background of standard maintenance therapies.
In addition to immune modulation, an improved understanding of the pathogenic processes in chronic organ injury is bringing new strategies to light. In a model of lupus in the central nervous system, a peptide mimic of the NMDAR receptor is able to block antibody-mediated damage to the brain parenchyma135. In lupus nephritis, the realization that immune complex deposition can be dissociated from renal inflammation132,136 has prompted the exploration of mechanisms for the local recruitment of inflammatory cells and their soluble products, as well as mechanisms of tissue repair. Preserving renal function by inhibiting angiotensin activity, targeting local effector cytokines, inhibiting late complement components, addressing tissue hypoxia and using natural inhibitors of fibrosis are all avenues currently being explored to prevent chronic renal diseases94,101,137–140. Although optimal control of inflammation and aggressive management of traditional atherosclerosis risk factors seem to be prudent interventions in all patients, new strategies for the early detection of atherosclerotic lesions may direct potentially toxic therapies only to those patients most at risk for events141. Strict blood pressure control, appropriate nutrition and avoidance of environmental insults all provide long-term benefits to patients, and mechanisms need to be put in place to address the disparities that prevent delivery of this care. Furthermore, the identification of useful biomarkers may improve the ability to diagnose and treat disease flares, categorize patients for clinical trials and evaluate therapeutic responses.
Conclusions
Although SLE is a heterogeneous group of disease states linked together by the formation of autoantibodies directed to ubiquitous cellular debris, a comprehensive model of disease pathogenesis is beginning to evolve based on genetic studies and new immunologic discoveries that paint autoreactivity both as a necessity and a bane of immune responses. The disease is multigenic and involves a loss of tolerance in both innate and adaptive immune pathways that fails to be controlled by sequential regulatory mechanisms. Multiple triggers are probably involved in disease initiation and perpetuation. Tissue injury depends on antigen specificity and antigen access, as well as the degree of systemic inflammation and the strength of local inflammatory responses. Continuous exposure to excess nucleic-acid– containing material amplifies the disease process. Epigenetic changes may establish persistently aberrant activation pathways and maintain the inflammatory phenotype of long-lived effector cells. This broad range of immunologic abnormalities in SLE provides many opportunities for therapeutic intervention but is also responsible for the extensive heterogeneity and redundancy of inflammatory mechanisms that will probably require tailored or combined approaches. Improvements in clinical trial design together with the integration of genetic and biomarker information are new challenges that should be addressed as cooperative groups are formed to prioritize therapeutic approaches in the relatively limited number patients that meet the criteria for clinical trials. These strategies, together with discovery-based approaches using well-characterized human samples and the appropriate animal models, should translate into a decrease in morbidity and mortality in patients with SLE in the coming decades.
Acknowledgments
This work was supported by US National Institutes of Health grants R01 DK085241-01, R01 AI083901 and R21 AR057930. The authors thank T. Rothstein and A. Boneparth for critical reading of the manuscript.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Lateef A, Petri M. Biologics in the treatment of systemic lupus erythematosus. Curr Opin Rheumatol. 2010;22:504–509. doi: 10.1097/BOR.0b013e32833b475e. [DOI] [PubMed] [Google Scholar]
- 2.Navarra SV, et al. Efficacy and safety of belimumab in patients with active systemic lupus erythematosus: a randomised, placebo-controlled, phase 3 trial. Lancet. 2011;377:721–731. doi: 10.1016/S0140-6736(10)61354-2. [DOI] [PubMed] [Google Scholar]
- 3.Furie R, et al. A phase III, randomized, placebo-controlled study of belimumab, a monoclonal antibody that inhibits B lymphocyte stimulator, in patients with systemic lupus erythematosus. Arthritis Rheum. 2011;63:3918–3930. doi: 10.1002/art.30613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morel L, et al. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proc Natl Acad Sci USA. 2000;97:6670–6675. doi: 10.1073/pnas.97.12.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lauwerys BR, Wakeland EK. Genetics of lupus nephritis. Lupus. 2005;14:2–12. doi: 10.1191/0961203305lu2052oa. [DOI] [PubMed] [Google Scholar]
- 6.Harley JB, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6:683–692. doi: 10.1038/nrrheum.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flesher DL, Sun X, Behrens TW, Graham RR, Criswell LA. Recent advances in the genetics of systemic lupus erythematosus. Expert Rev Clin Immunol. 2010;6:461–479. doi: 10.1586/eci.10.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu K, et al. Kallikrein genes are associated with lupus and glomerular basement membrane-specific antibody-induced nephritis in mice and humans. J Clin Invest. 2009;119:911–923. doi: 10.1172/JCI36728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez E, et al. Phenotypic associations of genetic susceptibility loci in systemic lupus erythematosus. Ann Rheum Dis. 2011;70:1752–1757. doi: 10.1136/ard.2011.154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: a review of statistical methods and recommendations for their application. Am J Hum Genet. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, et al. The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness. Nat Genet. 2011;43:902–907. doi: 10.1038/ng.904. [DOI] [PubMed] [Google Scholar]
- 13.Rieck M, et al. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol. 2007;179:4704–4710. doi: 10.4049/jimmunol.179.7.4704. [DOI] [PubMed] [Google Scholar]
- 14.Taylor KE, et al. Risk alleles for systemic lupus erythematosus in a large case-control collection and associations with clinical subphenotypes. PLoS Genet. 2011;7:e1001311. doi: 10.1371/journal.pgen.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Askanase AD, et al. Use of pharmacogenetics, enzymatic phenotyping, and metabolite monitoring to guide treatment with azathioprine in patients with systemic lupus erythematosus. J Rheumatol. 2009;36:89–95. doi: 10.3899/jrheum.070968. [DOI] [PubMed] [Google Scholar]
- 16.Rubtsov AV, Rubtsova K, Kappler JW, Marrack P. Genetic and hormonal factors in female-biased autoimmunity. Autoimmun Rev. 2010;9:494–498. doi: 10.1016/j.autrev.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith-Bouvier DL, et al. A role for sex chromosome complement in the female bias in autoimmune disease. J Exp Med. 2008;205:1099–1108. doi: 10.1084/jem.20070850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravichandran KS, Lorenz U. Engulfment of apoptotic cells: signals for a good meal. Nat Rev Immunol. 2007;7:964–974. doi: 10.1038/nri2214. [DOI] [PubMed] [Google Scholar]
- 19.Marínez Valle F, Balada E, Ordi-Ros J, Vilardell-Tarres M. DNase 1 and systemic lupus erythematosus. Autoimmun Rev. 2008;7:359–363. doi: 10.1016/j.autrev.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Rönnblom L, Alm GV. The natural interferon-α producing cells in systemic lupus erythematosus. Hum Immunol. 2002;63:1181–1193. doi: 10.1016/s0198-8859(02)00757-7. [DOI] [PubMed] [Google Scholar]
- 21.Blanco P, Palucka AK, Gill M, Pascual V, Banchereau J. Induction of dendritic cell differentiation by IFN-α in systemic lupus erythematosus. Science. 2001;294:1540–1543. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 22.Bennett L, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauer JW, et al. Elevated serum levels of interferon-regulated chemokines are biomarkers for active human systemic lupus erythematosus. PLoS Med. 2006;3:e491. doi: 10.1371/journal.pmed.0030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathian A, Weinberg A, Gallegos M, Banchereau J, Koutouzov S. IFN-α induces early lethal lupus in preautoimmune (New Zealand Black × New Zealand White) F1 but not in BALB/c mice. J Immunol. 2005;174:2499–2506. doi: 10.4049/jimmunol.174.5.2499. [DOI] [PubMed] [Google Scholar]
- 25.Ramanujam M, et al. Interferon-α treatment of female (NZW × BXSB)F(1) mice mimics some but not all features associated with the Yaa mutation. Arthritis Rheum. 2009;60:1096–1101. doi: 10.1002/art.24414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nacionales DC, et al. Deficiency of the type I interferon receptor protects mice from experimental lupus. Arthritis Rheum. 2007;56:3770–3783. doi: 10.1002/art.23023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agrawal H, et al. Deficiency of type I IFN receptor in lupus-prone New Zealand mixed 2328 mice decreases dendritic cell numbers and activation and protects from disease. J Immunol. 2009;183:6021–6029. doi: 10.4049/jimmunol.0803872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Thacker SG, et al. The detrimental effects of IFN-α on vasculogenesis in lupus are mediated by repression of IL-1 pathways: potential role in atherogenesis and renal vascular rarefaction. J Immunol. 2010;185:4457–4469. doi: 10.4049/jimmunol.1001782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Romo GS, et al. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra20. doi: 10.1126/scitranslmed.3001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lande R, et al. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra19. doi: 10.1126/scitranslmed.3001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charles N, Hardwick D, Daugas E, Illei GG, Rivera J. Basophils and the T helper 2 environment can promote the development of lupus nephritis. Nat Med. 2010;16:701–707. doi: 10.1038/nm.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 34.Boulé MW, et al. Toll-like receptor 9–dependent and –independent dendritic cell activation by chromatin-immunoglobulin G complexes. J Exp Med. 2004;199:1631–1640. doi: 10.1084/jem.20031942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lande R, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 36.Tian J, et al. Toll-like receptor 9–dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 37.Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- 38.Rönnblom L, Elkon KB. Cytokines as therapeutic targets in SLE. Nat Rev Rheumatol. 2010;6:339–347. doi: 10.1038/nrrheum.2010.64. [DOI] [PubMed] [Google Scholar]
- 39.Leadbetter EA, et al. Chromatin–IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 40.Berland R, et al. Toll-like receptor 7–dependent loss of B cell tolerance in pathogenic autoantibody knockin mice. Immunity. 2006;25:429–440. doi: 10.1016/j.immuni.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 41.Christensen SR, Shlomchik MJ. Regulation of lupus-related autoantibody production and clinical disease by Toll-like receptors. Semin Immunol. 2007;19:11–23. doi: 10.1016/j.smim.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christensen SR, et al. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 43.Fossati L, et al. The Yaa gene-mediated acceleration of murine lupus: Yaa− T cells from non-autoimmune mice collaborate with Yaa+ B cells to produce lupus autoantibodies in vivo. Eur J Immunol. 1995;25:3412–3417. doi: 10.1002/eji.1830251231. [DOI] [PubMed] [Google Scholar]
- 44.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 45.Harley JB, Harley IT, Guthridge JM, James JA. The curiously suspicious: a role for Epstein-Barr virus in lupus. Lupus. 2006;15:768–777. doi: 10.1177/0961203306070009. [DOI] [PubMed] [Google Scholar]
- 46.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 47.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Z, et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gall A, et al. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Control systems and decision making for antibody production. Nat Immunol. 2010;11:681–688. doi: 10.1038/ni.1900. [DOI] [PubMed] [Google Scholar]
- 51.Arbuckle MR, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349:1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 52.Moulton VR, Tsokos GC. Abnormalities of T cell signaling in systemic lupus erythematosus. Arthritis Res Ther. 2011;13:207. doi: 10.1186/ar3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perl A, et al. T-cell and B-cell signaling biomarkers and treatment targets in lupus. Curr Opin Rheumatol. 2009;21:454–464. doi: 10.1097/BOR.0b013e32832e977c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crispín JC, Kyttaris VC, Terhorst C, Tsokos GC. T cells as therapeutic targets in SLE. Nat Rev Rheumatol. 2010;6:317–325. doi: 10.1038/nrrheum.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng GM, Liu L, Bahjat FR, Pine PR, Tsokos GC. Suppression of skin and kidney disease by inhibition of spleen tyrosine kinase in lupus-prone mice. Arthritis Rheum. 2010;62:2086–2092. doi: 10.1002/art.27452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ichinose K, Juang YT, Crispin JC, Kis-Toth K, Tsokos GC. Suppression of autoimmunity and organ pathology in lupus-prone mice upon inhibition of calcium/calmodulin-dependent protein kinase type IV. Arthritis Rheum. 2011;63:523–529. doi: 10.1002/art.30085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bahjat FR, et al. An orally bioavailable spleen tyrosine kinase inhibitor delays disease progression and prolongs survival in murine lupus. Arthritis Rheum. 2008;58:1433–1444. doi: 10.1002/art.23428. [DOI] [PubMed] [Google Scholar]
- 58.Linterman MA, et al. Follicular helper T cells are required for systemic autoimmunity. J Exp Med. 2009;206:561–576. doi: 10.1084/jem.20081886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simpson N, et al. Expansion of circulating T cells resembling follicular helper T cells is a fixed phenotype that identifies a subset of severe systemic lupus erythematosus. Arthritis Rheum. 2010;62:234–244. doi: 10.1002/art.25032. [DOI] [PubMed] [Google Scholar]
- 60.Odegard JM, et al. ICOS-dependent extrafollicular helper T cells elicit IgG production via IL-21 in systemic autoimmunity. J Exp Med. 2008;205:2873–2886. doi: 10.1084/jem.20080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crispín JC, et al. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J Immunol. 2008;181:8761–8766. doi: 10.4049/jimmunol.181.12.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wardemann H, Nussenzweig MC. B-cell self-tolerance in humans. Adv Immunol. 2007;95:83–110. doi: 10.1016/S0065-2776(07)95003-8. [DOI] [PubMed] [Google Scholar]
- 63.Liu Z, Davidson A. BAFF and selection of autoreactive B cells. Trends Immunol. 2011;32:388–394. doi: 10.1016/j.it.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arechiga AF, et al. Cutting edge: the PTPN22 allelic variant associated with autoimmunity impairs B cell signaling. J Immunol. 2009;182:3343–3347. doi: 10.4049/jimmunol.0713370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Menard L, et al. The PTPN22 allele encoding an R620W variant interferes with the removal of developing autoreactive B cells in humans. J Clin Invest. 2011;121:3635–3644. doi: 10.1172/JCI45790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cappione A, III , et al. Germinal center exclusion of autoreactive B cells is defective in human systemic lupus erythematosus. J Clin Invest. 2005;115:3205–3216. doi: 10.1172/JCI24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gonzalez SF, et al. Trafficking of B cell antigen in lymph nodes. Annu Rev Immunol. 2011;29:215–233. doi: 10.1146/annurev-immunol-031210-101255. [DOI] [PubMed] [Google Scholar]
- 68.Kranich J, et al. Follicular dendritic cells control engulfment of apoptotic bodies by secreting Mfge8. J Exp Med. 2008;205:1293–1302. doi: 10.1084/jem.20071019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blair PA, et al. CD19+CD24hiCD38hi B cells exhibit regulatory capacity in healthy individuals but are functionally impaired in systemic lupus erythematosus patients. Immunity. 2010;32:129–140. doi: 10.1016/j.immuni.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 70.Campbell DJ, Koch MA. Treg cells: patrolling a dangerous neighborhood. Nat Med. 2011;17:929–930. doi: 10.1038/nm.2433. [DOI] [PubMed] [Google Scholar]
- 71.Kim HJ, et al. CD8+ T regulatory cells express the Ly49 Class I MHC receptor and are defective in autoimmune prone B6-Yaa mice. Proc Natl Acad Sci USA. 2011;108:2010–2015. doi: 10.1073/pnas.1018974108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brownlie RJ, et al. Distinct cell-specific control of autoimmunity and infection by FcgammaRIIb. J Exp Med. 2008;205:883–895. doi: 10.1084/jem.20072565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim SJ, et al. Increased IL-12 inhibits B cell differentiation to germinal center plasma cells and promotes differentiation to short-lived plasmablasts. J Exp Med. 2008;205:2437–2448. doi: 10.1084/jem.20070731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Herlands RA, William J, Hershberg U, Shlomchik MJ. Anti-chromatin antibodies drive in vivo antigen-specific activation and somatic hypermutation of rheumatoid factor B cells at extrafollicular sites. Eur J Immunol. 2007;37:3339–3351. doi: 10.1002/eji.200737752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Erickson LD, et al. Short-circuiting long-lived humoral immunity by the heightened engagement of CD40. J Clin Invest. 2002;109:613–620. doi: 10.1172/JCI14110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cassese G, et al. Inflamed kidneys of NZB/W mice are a major site for the homeostasis of plasma cells. Eur J Immunol. 2001;31:2726–2732. doi: 10.1002/1521-4141(200109)31:9<2726::aid-immu2726>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 77.Tokoyoda K, Hauser AE, Nakayama T, Radbruch A. Organization of immunological memory by bone marrow stroma. Nat Rev Immunol. 2010;10:193–200. doi: 10.1038/nri2727. [DOI] [PubMed] [Google Scholar]
- 78.Merrill JT, et al. Efficacy and safety of rituximab in moderately-to-severely active systemic lupus erythematosus: the randomized, double-blind, phase II/III systemic lupus erythematosus evaluation of rituximab trial. Arthritis Rheum. 2010;62:222–233. doi: 10.1002/art.27233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Benson MJ, et al. Cutting edge: the dependence of plasma cells and independence of memory B cells on BAFF and APRIL. J Immunol. 2008;180:3655–3659. doi: 10.4049/jimmunol.180.6.3655. [DOI] [PubMed] [Google Scholar]
- 80.Jacobi AM, et al. Effect of long-term belimumab treatment on B cells in systemic lupus erythematosus: extension of a phase II, double-blind, placebo-controlled, dose-ranging study. Arthritis Rheum. 2010;62:201–210. doi: 10.1002/art.27189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Looney RJ, Anolik J, Sanz I. A perspective on B-cell–targeting therapy for SLE. Mod Rheumatol. 2010;20:1–10. doi: 10.1007/s10165-009-0213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aringer M, et al. Current state of evidence on “off label” therapeutic options for systemic lupus erythematosus, including biological immunosuppressive agents, in Germany, Austria, and Switzerland—a consensus report. Lupus. 2012;21:386–401. doi: 10.1177/0961203311426569. [DOI] [PubMed] [Google Scholar]
- 83.Hahn BH. Targeted therapies in systemic lupus erythematosus: successes, failures and future. Ann Rheum Dis. 2011;70 (suppl 1):i64–i66. doi: 10.1136/ard.2010.142208. [DOI] [PubMed] [Google Scholar]
- 84.de Laat B, Mertens K, de Groot PG. Mechanisms of disease: antiphospholipid antibodies—from clinical association to pathologic mechanism. Nat Clin Pract Rheumatol. 2008;4:192–199. doi: 10.1038/ncprheum0740. [DOI] [PubMed] [Google Scholar]
- 85.Lauvsnes MB, Omdal R. Systemic lupus erythematosus, the brain, and anti-NR2 antibodies. J Neurol. 2012;259:622–629. doi: 10.1007/s00415-011-6232-5. [DOI] [PubMed] [Google Scholar]
- 86.Faust TW, et al. Neurotoxic lupus autoantibodies alter brain function through two distinct mechanisms. Proc Natl Acad Sci USA. 2010;107:18569–18574. doi: 10.1073/pnas.1006980107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.DeGiorgio LA, et al. A subset of lupus anti-DNA antibodies cross-reacts with the NR2 glutamate receptor in systemic lupus erythematosus. Nat Med. 2001;7:1189–1193. doi: 10.1038/nm1101-1189. [DOI] [PubMed] [Google Scholar]
- 88.Matus S, et al. Antiribosomal-P autoantibodies from psychiatric lupus target a novel neuronal surface protein causing calcium influx and apoptosis. J Exp Med. 2007;204:3221–3234. doi: 10.1084/jem.20071285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kowal C, et al. Cognition and immunity; antibody impairs memory. Immunity. 2004;21:179–188. doi: 10.1016/j.immuni.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 90.Turnberg D, Cook HT. Complement and glomerulonephritis: new insights. Curr Opin Nephrol Hypertens. 2005;14:223–228. doi: 10.1097/01.mnh.0000165887.75501.24. [DOI] [PubMed] [Google Scholar]
- 91.Bergtold A, Gavhane A, D’Agati V, Madaio M, Clynes R. FcR-bearing myeloid cells are responsible for triggering murine lupus nephritis. J Immunol. 2006;177:7287–7295. doi: 10.4049/jimmunol.177.10.7287. [DOI] [PubMed] [Google Scholar]
- 92.Anders HJ, Schlondorff D. Toll-like receptors: emerging concepts in kidney disease. Curr Opin Nephrol Hypertens. 2007;16:177–183. doi: 10.1097/MNH.0b013e32803fb767. [DOI] [PubMed] [Google Scholar]
- 93.Manderson AP, Botto M, Walport MJ. The role of complement in the development of systemic lupus erythematosus. Annu Rev Immunol. 2004;22:431–456. doi: 10.1146/annurev.immunol.22.012703.104549. [DOI] [PubMed] [Google Scholar]
- 94.Woodruff TM, Nandakumar KS, Tedesco F. Inhibiting the C5-C5a receptor axis. Mol Immunol. 2011;48:1631–1642. doi: 10.1016/j.molimm.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 95.Vielhauer V, Anders HJ, Schlondorff D. Chemokines and chemokine receptors as therapeutic targets in lupus nephritis. Semin Nephrol. 2007;27:81–97. doi: 10.1016/j.semnephrol.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 96.Kitching AR, Holdsworth SR. The emergence of TH17 cells as effectors of renal injury. J Am Soc Nephrol. 2011;22:235–238. doi: 10.1681/ASN.2010050536. [DOI] [PubMed] [Google Scholar]
- 97.Ernandez T, Mayadas TN. Immunoregulatory role of TNFα in inflammatory kidney diseases. Kidney Int. 2009;76:262–276. doi: 10.1038/ki.2009.142. [DOI] [PubMed] [Google Scholar]
- 98.Bethunaickan R, et al. A unique hybrid renal mononuclear phagocyte activation phenotype in murine systemic lupus erythematosus nephritis. J Immunol. 2011;186:4994–5003. doi: 10.4049/jimmunol.1003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hill GS, et al. Predictive power of the second renal biopsy in lupus nephritis: significance of macrophages. Kidney Int. 2001;59:304–316. doi: 10.1046/j.1523-1755.2001.00492.x. [DOI] [PubMed] [Google Scholar]
- 100.Schlondorff DO. Overview of factors contributing to the pathophysiology of progressive renal disease. Kidney Int. 2008;74:860–866. doi: 10.1038/ki.2008.351. [DOI] [PubMed] [Google Scholar]
- 101.Deelman L, Sharma K. Mechanisms of kidney fibrosis and the role of antifibrotic therapies. Curr Opin Nephrol Hypertens. 2009;18:85–90. doi: 10.1097/MNH.0b013e32831c50a1. [DOI] [PubMed] [Google Scholar]
- 102.Alarcón GS, et al. Time to renal disease and end-stage renal disease in PROFILE: a multiethnic lupus cohort. PLoS Med. 2006;3:e396. doi: 10.1371/journal.pmed.0030396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Esdaile JM, et al. Traditional Framingham risk factors fail to fully account for accelerated atherosclerosis in systemic lupus erythematosus. Arthritis Rheum. 2001;44:2331–2337. doi: 10.1002/1529-0131(200110)44:10<2331::aid-art395>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 104.Symmons DP, Gabriel SE. Epidemiology of CVD in rheumatic disease, with a focus on RA and SLE. Nat Rev Rheumatol. 2011;7:399–408. doi: 10.1038/nrrheum.2011.75. [DOI] [PubMed] [Google Scholar]
- 105.Narshi CB, Giles IP, Rahman A. The endothelium: an interface between autoimmunity and atherosclerosis in systemic lupus erythematosus? Lupus. 2011;20:5–13. doi: 10.1177/0961203310382429. [DOI] [PubMed] [Google Scholar]
- 106.Lopez LR, et al. Oxidized low-density lipoprotein and β2-glycoprotein I in patients with systemic lupus erythematosus and increased carotid intima-media thickness: implications in autoimmune-mediated atherosclerosis. Lupus. 2006;15:80–86. doi: 10.1191/0961203306lu2267oa. [DOI] [PubMed] [Google Scholar]
- 107.Matsuura E, Kobayashi K, Hurley BL, Lopez LR. Atherogenic oxidized low-density lipoprotein/β2-glycoprotein I (oxLDL/β2GPI) complexes in patients with systemic lupus erythematosus and antiphospholipid syndrome. Lupus. 2006;15:478–483. doi: 10.1191/0961203306lu2337oa. [DOI] [PubMed] [Google Scholar]
- 108.Skaggs BJ, Hahn BH, Sahakian L, Grossman J, McMahon M. Dysfunctional, pro-inflammatory HDL directly upregulates monocyte PDGFRβ, chemotaxis and TNFα production. Clin Immunol. 2010;137:147–156. doi: 10.1016/j.clim.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McMahon M, et al. Dysfunctional proinflammatory high-density lipoproteins confer increased risk of atherosclerosis in women with systemic lupus erythematosus. Arthritis Rheum. 2009;60:2428–2437. doi: 10.1002/art.24677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schanberg LE, et al. Use of atorvastatin in systemic lupus erythematosus in children and adolescents. Arthritis Rheum. 2012;64:285–296. doi: 10.1002/art.30645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Petri MA, Kiani AN, Post W, Christopher-Stine L, Magder LS. Lupus Atherosclerosis Prevention Study (LAPS) Ann Rheum Dis. 2011;70:760–765. doi: 10.1136/ard.2010.136762. [DOI] [PubMed] [Google Scholar]
- 112.Ceribelli A, Yao B, Dominguez-Gutierrez PR, Chan EK. Lupus T cells switched on by DNA hypomethylation via microRNA? Arthritis Rheum. 2011;63:1177–1181. doi: 10.1002/art.30192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pan Y, Sawalha AH. Epigenetic regulation and the pathogenesis of systemic lupus erythematosus. Transl Res. 2009;153:4–10. doi: 10.1016/j.trsl.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 114.Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011;157:163–179. doi: 10.1016/j.trsl.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ceribelli A, et al. MicroRNAs in systemic rheumatic diseases. Arthritis Res Ther. 2011;13:229. doi: 10.1186/ar3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 117.Illei GG, et al. Current state and future directions of autologous hematopoietic stem cell transplantation in systemic lupus erythematosus. Ann Rheum Dis. 2011;70:2071–2074. doi: 10.1136/ard.2010.148049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Choi EW, et al. Reversal of serological, immunological and histological dysfunction in systemic lupus erythematosus mice by long-term serial adipose tissue-derived mesenchymal stem cell transplantation. Arthritis Rheum. 2012;64:243–253. doi: 10.1002/art.33313. [DOI] [PubMed] [Google Scholar]
- 119.Liang J, et al. Allogenic mesenchymal stem cells transplantation in refractory systemic lupus erythematosus: a pilot clinical study. Ann Rheum Dis. 2010;69:1423–1429. doi: 10.1136/ard.2009.123463. [DOI] [PubMed] [Google Scholar]
- 120.Mok CC. Biomarkers for lupus nephritis: a critical appraisal. J Biomed Biotechnol. 2010;2010:638413. doi: 10.1155/2010/638413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bauer JW, et al. Interferon-regulated chemokines as biomarkers of systemic lupus erythematosus disease activity: a validation study. Arthritis Rheum. 2009;60:3098–3107. doi: 10.1002/art.24803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chaussabel D, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hinze CH, et al. Neutrophil gelatinase-associated lipocalin is a predictor of the course of global and renal childhood-onset systemic lupus erythematosus disease activity. Arthritis Rheum. 2009;60:2772–2781. doi: 10.1002/art.24751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rovin BH, et al. Urine chemokines as biomarkers of human systemic lupus erythematosus activity. J Am Soc Nephrol. 2005;16:467–473. doi: 10.1681/ASN.2004080658. [DOI] [PubMed] [Google Scholar]
- 125.Rubinstein T, et al. Urinary neutrophil gelatinase-associated lipocalin as a novel biomarker for disease activity in lupus nephritis. Rheumatology (Oxford) 2010;49:960–971. doi: 10.1093/rheumatology/kep468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guiducci C, et al. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature. 2010;465:937–941. doi: 10.1038/nature09102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yuan W, DiMartino SJ, Redecha PB, Ivashkiv LB, Salmon JE. Systemic lupus erythematosus monocytes are less responsive to interleukin-10 in the presence of immune complexes. Arthritis Rheum. 2011;63:212–218. doi: 10.1002/art.30083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ramanujam M, Davidson A. Targeting of the immune system in systemic lupus erythematosus. Expert Rev Mol Med. 2008;10:e2. doi: 10.1017/S1462399408000562. [DOI] [PubMed] [Google Scholar]
- 129.Wofsy DS, Shropshire SM, Hillson JL, Diamond B. Abatacept for lupus nephritis: alternative outcome measures support opposing interpretations of data From a multicenter, randomized, double-blind, placebo-controlled phase II/III study. ACR presentation. 2011 Nov 8;2474 [Google Scholar]
- 130.Lenert PS. Classification, mechanisms of action, and therapeutic applications of inhibitory oligonucleotides for Toll-like receptors (TLR) 7 and 9. Mediators Inflamm. 2010;2010:986596. doi: 10.1155/2010/986596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kyttaris VC, Tsokos GC. Targeting lymphocyte signaling pathways as a therapeutic approach to systemic lupus erythematosus. Curr Opin Rheumatol. 2011;23:449–453. doi: 10.1097/BOR.0b013e328349a242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schiffer L, et al. Short term administration of costimulatory blockade and cyclophosphamide induces remission of systemic lupus erythematosus nephritis in NZB/W F1 mice by a mechanism downstream of renal immune complex deposition. J Immunol. 2003;171:489–497. doi: 10.4049/jimmunol.171.1.489. [DOI] [PubMed] [Google Scholar]
- 133.Daikh DI, Wofsy D. Cutting edge: reversal of murine lupus nephritis with CTLA4Ig and cyclophosphamide. J Immunol. 2001;166:2913–2916. doi: 10.4049/jimmunol.166.5.2913. [DOI] [PubMed] [Google Scholar]
- 134.Ng KP, et al. B cell depletion therapy in systemic lupus erythematosus: long-term follow-up and predictors of response. Ann Rheum Dis. 2007;66:1259–1262. doi: 10.1136/ard.2006.067124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bloom O, et al. Generation of a unique small molecule peptidomimetic that neutralizes lupus autoantibody activity. Proc Natl Acad Sci USA. 2011;108:10255–10259. doi: 10.1073/pnas.1103555108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Clynes R, Dumitru C, Ravetch JV. Uncoupling of immune complex formation and kidney damage in autoimmune glomerulonephritis. Science. 1998;279:1052–1054. doi: 10.1126/science.279.5353.1052. [DOI] [PubMed] [Google Scholar]
- 137.Gambaro G, Kong NC. Glycosaminoglycan treatment in glomerulonephritis? An interesting option to investigate. J Nephrol. 2010;23:244–252. [PubMed] [Google Scholar]
- 138.Nguyen TQ, Goldschmeding R. Bone morphogenetic protein-7 and connective tissue growth factor: novel targets for treatment of renal fibrosis? Pharm Res. 2008;25:2416–2426. doi: 10.1007/s11095-008-9548-9. [DOI] [PubMed] [Google Scholar]
- 139.Renner B, et al. Binding of factor H to tubular epithelial cells limits interstitial complement activation in ischemic injury. Kidney Int. 2011;80:165–173. doi: 10.1038/ki.2011.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tse KC, et al. Angiotensin inhibition or blockade for the treatment of patients with quiescent lupus nephritis and persistent proteinuria. Lupus. 2005;14:947–952. doi: 10.1191/0961203305lu2249oa. [DOI] [PubMed] [Google Scholar]
- 141.McMahon M, Hahn BH, Skaggs BJ. Systemic lupus erythematosus and cardiovascular disease: prediction and potential for therapeutic intervention. Expert Rev Clin Immunol. 2011;7:227–241. doi: 10.1586/eci.10.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stohl W, et al. Belimumab reduces autoantibodies, normalizes low complement, and reduces select B-cell populations in patients with systemic lupus erythematosus. Arthritis Rheum. 2012 Jan 24; doi: 10.1002/art.34400. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu Z, Davidson A. BAFF inhibition: a new class of drugs for the treatment of autoimmunity. Exp Cell Res. 2011;317:1270–1277. doi: 10.1016/j.yexcr.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mackay F, Schneider P. Cracking the BAFF code. Nat Rev Immunol. 2009;9:491–502. doi: 10.1038/nri2572. [DOI] [PubMed] [Google Scholar]
- 145.Stohl W, et al. Belimumab reduces autoantibodies, normalizes low complement, and reduces select B-cell populations in patients with systemic lupus erythematosus. Arthritis Rheum. 2012 Jan 24; doi: 10.1002/art.34400. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]