Abstract
Biological clocks regulate various behavioural and physiological traits; slower circadian clocks are expected to slow down the development, suggesting a potential genetic correlation between the developmental period and circadian rhythm. However, a correlation between natural genetic variations in the developmental period and circadian rhythm has only been found in Bactrocera cucurbitae. The number of genetic factors that contribute to this genetic correlation is largely unclear. In this study, to examine whether natural genetic variations in the developmental period and circadian rhythm are correlated in Drosophila melanogaster, we performed an artificial disruptive selection on the developmental periods using wild-type strains and evaluated the circadian rhythms of the selected lines. To investigate whether multiple genetic factors mediate the genetic correlation, we reanalyzed previously published genome-wide deficiency screening data based on DrosDel isogenic deficiency strains and evaluated the effect of 438 genomic deficiencies on the developmental periods. We then randomly selected 32 genomic deficiencies with significant effects on the developmental periods and tested their effects on circadian rhythms. As a result, we found a significant response to selection for longer developmental periods and their correlated effects on circadian rhythms of the selected lines. We also found that 18 genomic regions had significant effects on the developmental periods and circadian rhythms, indicating their potential for mediating the genetic correlation between the developmental period and circadian rhythm. The novel findings of our study might lead to a better understanding of how this correlation is regulated genetically in broader taxonomic groups.
Keywords: circadian rhythm, deficiency screening, disruptive selection, free-running period
Introduction
Biological clocks regulate various behavioural and physiological traits and allow organisms to accommodate to daily and seasonal environmental cycles (Panda et al., 2002; Paranjpe et al., 2004; Mazzoni et al., 2005). The core molecular mechanisms of these clocks are highly conserved across taxa, and the generation of molecular oscillation has been well studied in flies and mammals (Panda et al., 2002; Grima et al., 2004; Chiu et al., 2011; Goda et al., 2011). In general, faster circadian clocks are expected to speed up development and shorten the pre-adult developmental period, whereas slower clocks prolong this period (Paranjpe et al., 2005), suggesting a potential genetic correlation between the developmental period and circadian rhythm.
A genetic correlation between the developmental period and circadian rhythm has been demonstrated in two fly species, Drosophila melanogaster and Bactrocera cucurbitae. In D. melanogaster, period (per) mutants have a wide range of circadian rhythm variations represented by largely different free-running periods (τ) (wild type: τ=24 h, perS: τ=19 h, perL: τ=28 h) that are positively correlated with the developmental periods (perS develops faster than perL regardless of the light conditions; Kyriacou et al., 1990). The positive genetic correlation between the free-running and developmental periods might be mediated by the pleiotropic effects of per mutations. Another example in D. melanogaster is the genetic correlation between the timing of adult emergence and circadian clocks found by Kumar et al., 2007. Flies selected to emerge in the morning showed shorter circadian rhythm than the ones selected to emerge at evening, indicating the regulation of pre-adult period by a circadian clock (Kumar et al., 2007). In B. cucurbitae, Miyatake (1995) performed a disruptive selection on the developmental period and established selected lines with shorter and longer developmental periods. Under constant darkness, Shimizu et al. (1997) then observed that the selected lines with shorter developmental periods had shorter free-running periods, whereas the lines with longer developmental periods had longer free-running periods, indicating a positive genetic correlation between the developmental period and circadian rhythm in this species. In addition, the developmental and free-running periods of B. cucurbitae were also genetically correlated with the timing of mating (Miyatake et al., 2002). This genetic correlation between life-history and behavioural traits might have an important role in ecological diversifications (Miyatake, 2002). However, in a broader range of organisms it is still unknown whether natural genetic variations in the developmental period and circadian rhythm are correlated with each other. In addition, the number of quantitative trait loci other than per that contribute to genetic correlation are largely unclear.
To examine whether the correlation between natural genetic variations in the developmental period and circadian rhythm in B. cucurbitae also exists in D. melanogaster, we performed an artificial disruptive selection on the developmental periods of strains that originated from wild populations. We then evaluated the circadian rhythms represented as the free-running periods of these lines. To map the genomic regions that had effects on the developmental periods, we reanalyzed the genome-wide deficiency mapping data of Takahashi et al. (2011a) and evaluated the effect of 438 isogenic deficient strains covering about 65% of the D. melanogaster genome. We then randomly selected 32 genomic deficiencies with significant effects on the developmental periods, and tested their effects on the free-running periods. As a result, we found a significant response to the selection for longer developmental periods, and their correlated effects to prolong free-running periods in the selected lines. We also found that 253 genomic deficiencies had significant effects on the developmental periods. Of the 32 deficiencies randomly selected from the deficiencies that had effects on the developmental periods, we found 18 deficiencies that had significant effects on the free-running periods. These results clearly show that there was an ample natural genetic variation in developmental period in D. melanogaster, and it had significant correlation with the natural genetic variation in circadian rhythm. The deficiency mapping identified a number of genomic regions that affected the developmental periods and circadian rhythms, suggesting that genetic correlation between them might be mediated by multiple genetic factors.
Materials and methods
Selection experiments
Flies
We obtained 20 wild strains of D. melanogaster that had been collected from across the Japanese islands and maintained in EHIME-Fly, the laboratory for Drosophila resources at Ehime University. We used the same strains that were described in Tsujino and Takahashi (2012), and complete details of the strains can be found in that publication. We mixed four individuals (two females and two males) from each strain to produce a base population of 80 individuals. In this manner, we produced three independent base populations originated from the same set of flies that were reared for three generations at 23 °C under constant light in incubators (MIR-254 or MIR-154; SANYO, Osaka, Japan) in 250-ml plastic bottles containing 50 ml of fly medium containing dried yeast, soy flour, cornmeal, agar, malt extract and dextrose.
Artificial selection on the developmental periods
The developmental period in our study was characterized by days from oviposition of the eggs to their eclosion. We established three ‘short' lines that were selected for shorter developmental periods and three ‘long' lines that were selected for a longer developmental periods by mixing 30 females and 30 males from each base population. During each selection round, we collected all the emerged flies and calculated their developmental periods. Collections were made every 12 h to ensure the virginity of females. We ranked all the emerged females and males on the basis of their developmental periods, and established the next generation using the top 30 females and 30 males for each short line, and the bottom 30 females and 30 males for each long line. The average number of emerged adults was 283.44 throughout the selection, indicating that our current selection procedure selected on an average 21% of individuals from the top or the bottom of the trait score distribution in each generation. We mixed the selected females and males, and maintained them together for a few days to allow them to mate freely. We then transferred the flies to experimental 250-ml plastic bottles and allowed the flies to oviposit for 12 h to maintain the larval density in the plastic bottles at a sufficiently low level to avoid intense intra-specific competition. We incubated the bottles until the flies of the next generation emerged. We reared the flies in the incubators at 23 °C under constant light conditions. Three control lines were also established from the three base populations and were maintained in the same way as the selection lines except for the selection process. We measured the developmental periods of the control lines every five generations.
Locomotor activity rhythm assay of the artificially selected lines
To examine whether artificial selection on the developmental periods had an effect on the circadian rhythms, we measured the locomotor activity of the short, long and control strains at the 25th generation by evaluating the free-running periods. Flies aged 3–7 days after eclosion were entrained for 4 days in cycles of 12-h light and 12-h darkness at 25 °C in incubators. The locomotor activity of these flies was monitored using a DAM2 system (TriKinetics, Waltham, MA, USA) for 10 days in constant darkness. To characterize the rhythmicity of the locomotor activity of these flies, we performed a χ2 periodogram analysis using Clocklab software (Actimetrics, Wilmette, IL, USA) that identified rhythmic flies and determined their free-running periods (τ).
Statistical analysis
To evaluate the divergence in the developmental periods of the short and long lines, we performed a one-way analysis of variance (ANOVA) repeatedly for every generation using the developmental periods as a dependent variable, and the selection treatments (short or long) as an independent variable. We used the mean developmental period of each line in this analysis and regarded three lines of each treatment as biological replicates.
We also tested the effect of artificial selection on the free-running periods at the 25th generation using a one-way ANOVA. In this analysis, we compared the control lines with the long and short lines in a pairwise manner. We used τ scores as the dependent variables and the treatments (control/long or control/short) as independent variables.
To confirm the normality and equality of variance of the data sets used for the above analyses, we performed the Kolmogorov–Smirnov test and F test. When the data sets did not fulfil the requirements of ANOVA, we did not apply ANOVA.
Reanalysis of deficiency screening data to identify genomic regions with effects on the developmental periods
To map genomic regions with effects on the developmental periods, we reanalyzed the deficiency screening data of Takahashi et al. (2011a) in which they solely focused on temporal variation in the developmental periods and not on the mean developmental period. Takahashi et al. (2011a) used DrosDel isogenic deficiency strains and evaluated the developmental period defined as days from oviposition of the eggs to their eclosion. The breakpoints of the deletions were determined at a single base-pair resolution, allowing high-resolution mapping of the candidate genomic regions. The control strain (DSK001: w1118iso; 2iso; 3iso) was isogenized for the X, second and third chromosomes, and all the deficiency strains shared the same genetic background as the control strain (Ryder et al., 2004, 2007). In our study, we reanalyzed the developmental period data of 438 DrosDel deficiency strains that covered about 65% of the whole genome region (Appendix 1). Additional details of the deletion strains are available on the DrosDel web page (http://www.drosdel.org.uk/).
Deficiency effects on the locomotor activity rhythms
We randomly chose 32 deficiencies whose effects on the developmental periods were detected by deficiency screening and evaluated their effect on the locomotor activity rhythms. Because of the homozygous lethality of most deficiencies, we tested deficiency-control heterozygotes (Df/+) for the locomotor activity rhythms, as in Takahashi et al., 2011a. We introduced 100 eggs from each of the crosses between the control strain and the deletion strains into a glass vial along with a standard cornmeal agar medium (details are described in Takahashi et al., 2011b). We crossed females of the control strain with males of each deficiency strain to control the maternal effect. The eggs were reared at 23 °C under constant light in incubators. We genotyped emerging adults (target genotype, Df/+ nontarget genotype, balancer/+) and collected flies for locomotor activity measurements. To obtain control individuals (+/+), we collected 100 eggs from strain DSK001 and reared them as described above. We then monitored the locomotor activity of these control flies in the same way as we did for the selection experiment to determine their free-running periods (τ).
Statistical analysis
To evaluate the effects of deletions on the mean developmental periods and free-running periods, we performed pairwise comparisons between +/+ and each Df/+ using one-way ANOVA. We used average vial-level scores for the developmental periods and individual-level scores for the free-running periods. We checked the normality of the distribution of the scores for each genotype separately using the Kolmogorov–Smirnov test, and equality of variance of the data sets using F test. To correct for multiple tests with different genotypes, we applied the Benjamini and Hochberg (1995) procedure to control the false discovery rate. Deviation from the normal distribution was considered significant if the adjusted false discovery rate P-value was <0.05. As a result, no significant deviations from the normal distribution were detected in any of the cases in our study. For the ANOVA, we used the average vial-level developmental period or individual-level free-running period as the dependent variable, whereas the genotype (+/+ or Df/+) as the independent variable. Correction for multiple tests was performed using the Benjamini–Hochberg procedure, as in the normality test described above. In addition, we calculated the effect size (Cohen's d) of each deficiency to draw a robust conclusion, regardless of the sample size variation and the existence of outliers, and to make the results of different tests comparable. For the developmental periods, we performed separate analyses of sexes and tested correlation of the effect sizes of the developmental periods between males and females to determine any sex-specific effect of the deletions. We also tested the correlation between the effect sizes of deletions on the developmental and free-running periods to determine any genetic correlation. All statistical analyses were performed using the statistical software R 2.8.1 (R Development Core Team 2005).
Results
Effects of artificial selection on the developmental periods
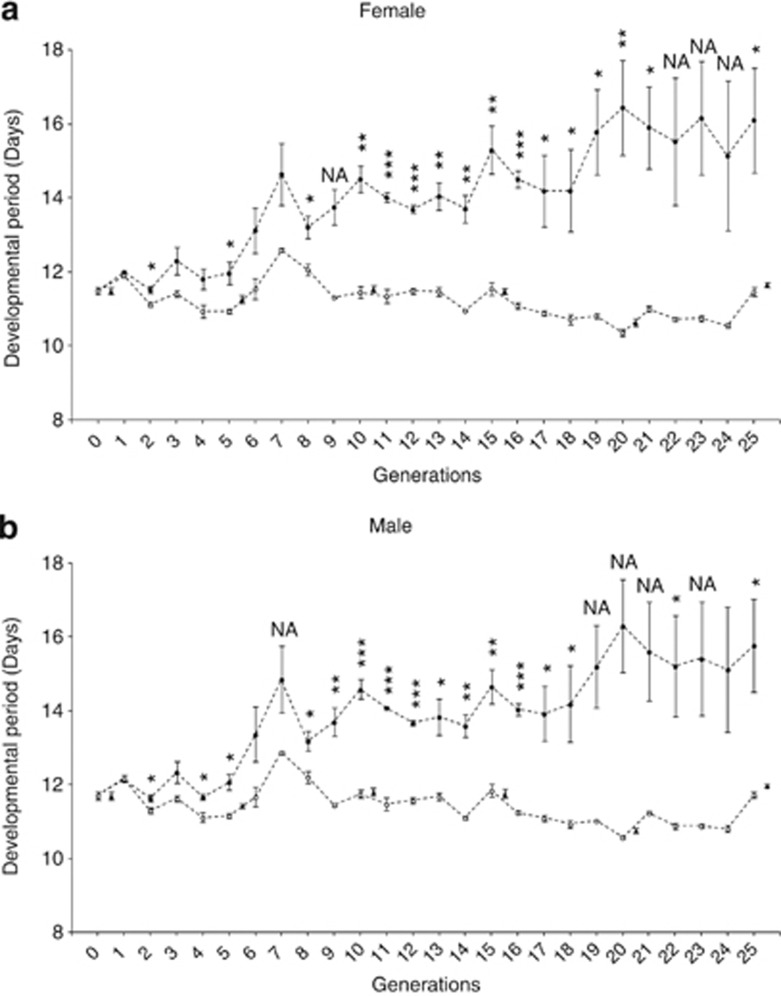
As a result of artificial selection, the developmental periods of long and short lines diverged significantly in both females and males where there were a few cases that violated the requirements for ANOVA and were not analysed (Figure 1). The mean developmental periods of the short lines remained at the same level as the control lines throughout selection, whereas the mean developmental periods of the long lines increased continuously until the 20th generation (Figure 1).
Figure 1.
Selection responses of the female flies (a) and the male flies (b). Short lines (○) were selected for a shorter developmental period, long lines ( ) were selected for a longer developmental period, whereas control lines (
) were selected for a longer developmental period, whereas control lines ( ) were not subjected to any selection. Error bars represent s.e's. Asterisks represent statistically significant differences between short and long lines: *P<0.05, **P<0.001, ***P<0.0001. NA indicates cases where the data sets violated the requirements of ANOVA and the test was not applied.
) were not subjected to any selection. Error bars represent s.e's. Asterisks represent statistically significant differences between short and long lines: *P<0.05, **P<0.001, ***P<0.0001. NA indicates cases where the data sets violated the requirements of ANOVA and the test was not applied.
Locomotor activity rhythms of the selected lines
The free-running periods of the long lines (average score±s.e.: 24.25±0.09) were significantly increased (P=0.016) compared with the control lines (23.82±0.06), whereas those of the short lines (23.96±0.08) were not significantly different from the control lines.
Effects of deficiencies on the developmental periods
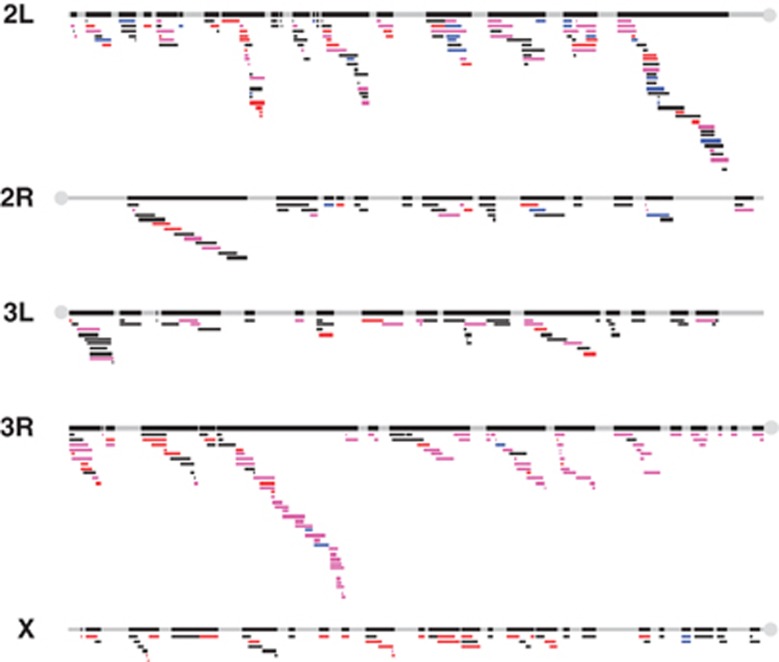
As a result of screening, we found 81 genomic regions with significant effects on the development periods in females only, 27 genomic regions with significant effects in males only and 145 genomic regions with significant effects in both females and males (Figure 2, Appendix 1).
Figure 2.
Distribution of deficiencies on the second, third and X chromosomes. Genomic regions covered by deficiencies are filled with black, while bars below each chromosome represent the location of each deficiency. Bars representing deficiencies with significant effects on the developmental periods are filled with different colours based on sex specificity, that is, a significant effect only in female flies is shown in red; a significant effect only in male flies is shown in blue; and a significant effect in both female and male flies is shown in purple. A full color version of this figure is available at the Heredity journal online.
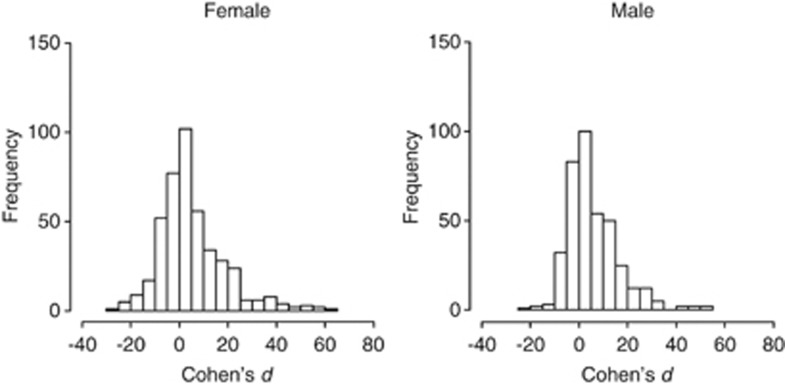
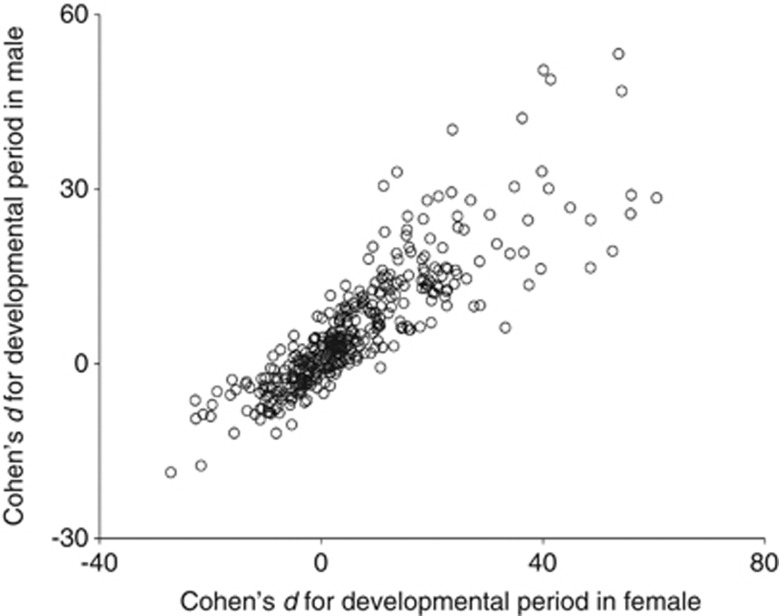
Compared with the developmental period of +/+ (13.51 days in female and 13.45 days in male on average), developmental period of Df/+ deviated positively in both females and males (0.39 on average ranging from −1.35 to 4.89 days in females and 0.53 on average ranging from −1.45 to 4.71 days in males). The frequency distribution of the effect size of deficiencies on the developmental periods was assessed using Cohen's d for the term ‘genotype' in the ANOVA model as shown in Figure 3. The effect sizes were centred around zero, indicating that most deficiencies had little effect on the developmental periods. Longer tails of the effect size distributions on the positive side indicated that deficiencies tended to prolong the developmental periods in females and males (Figure 3). We found a positive correlation between the effect sizes in females and males (correlation coefficient: 0.863, P<0.0001; Figure 4), suggesting that a large number of deficiencies had consistent effects on the developmental periods in females and males.
Figure 3.
Frequency distribution of the effect size (Cohen's d) of deletions on the developmental periods in female and male flies.
Figure 4.
Correlation between the effects of deficiencies on the developmental periods in female and male flies.
Effects of deficiencies on the locomotor activity rhythms
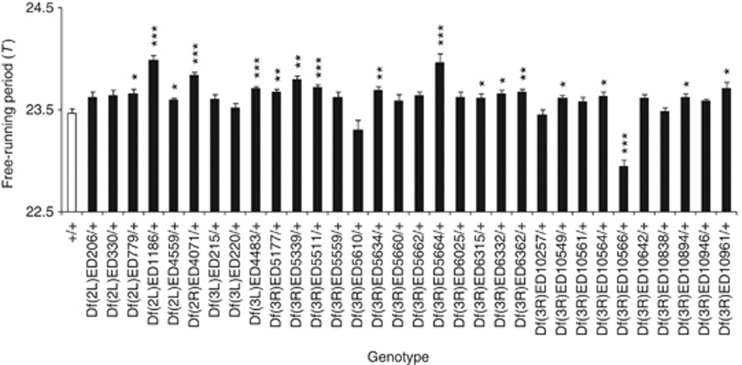
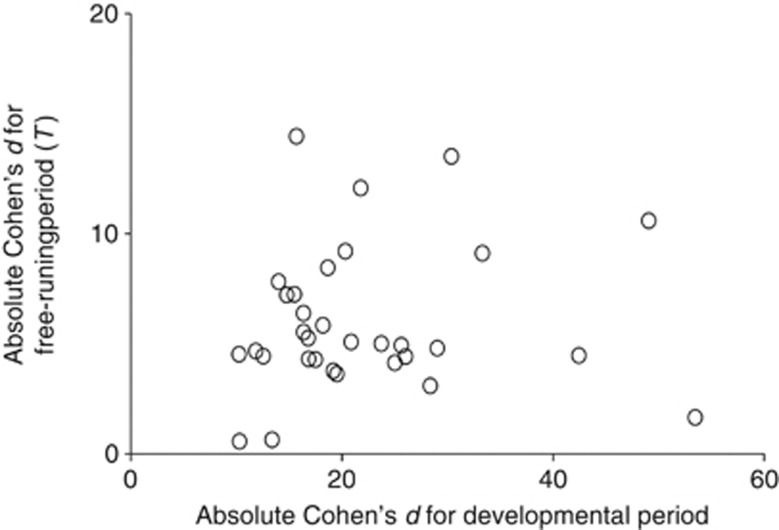
Of the 32 deficiencies with effects on developmental periods, 18 deficiencies had a significant effect on the free-running periods (Figure 5). The overall correlation between the effects of deficiencies on the developmental and free-running periods was not significant (correlation coefficient: 0.093, P>0.05; Figure 6).
Figure 5.
Free-running periods of the control homozygotes (+/+) and deficiency heterozygotes (Df/+). Error bars represent s.e's. Asterisks represent statistically significant differences between the +/+ and each Df/+ genotype: *P<0.05, **P<0.01, ***P<0.001.
Figure 6.
The overall correlation between the effects of deficiencies on the developmental and free-running periods.
Discussion
In our study, we observed a significant response to artificial selection for longer developmental periods, and this selection resulted in increased free-running periods in the selected lines, indicating a genetic correlation between the developmental period and circadian rhythm in D. melanogaster. We also found that 18 genomic deficiencies affected the developmental periods and circadian rhythms, suggesting that multiple genetic factors contribute to the genetic correlation between them.
A significant response to artificial selection for longer developmental periods and lack of response to selection for shorter developmental periods were observed in our study. This pattern of response to disruptive selection on the developmental period was similar to that observed by Zwaan et al. (1995) in D. melanogaster and by Miyatake (1995) in B. cucurbitae. The asymmetric response to disruptive selection might be attributable to a scarcity of natural genetic variations that shorten the developmental period. In Drosophila species, at least, natural selection seems to favour a shorter developmental period because most endoparasitic wasps attack the larval stage or feed externally on the pupae (Wertheim et al., 2005), and a shorter developmental period might reduce the risk of such parasitism. In addition, most Drosophila species utilize patchy and ephemeral resources such as mushrooms or fallen fruits (Takahashi et al., 2005; Mitsui et al., 2006), so rapidly completing their pre-adult development before the degradation of resource patches might be advantageous. Furthermore, for a species such as D. melanogaster whose small overwintering population increases in the absence of population pressure every spring, reduction in developmental period leads to the higher intrinsic rate of increase of the population (Lewontin, 1965). This demographic fitness effects is stronger in developmental period than in other life-history traits such as fecundity and longevity (Lewontin, 1965). If these selective advantages lead to a higher selection pressure that favours a shorter developmental period, natural genetic variations for a shorter developmental period will be more deficient than those for a longer developmental period. Selective advantage of shorter developmental period is not necessarily true for other organisms such as a comma butterfly Polygonia c-album, whose seasonal variation in developmental period is well known (Nylin, 1988, 1992). Under a variable environment, plasticity in a life-history trait such as developmental period can be adaptive (Nylin and Gotthard, 1998).
The pattern of genetic correlation between the developmental periods and circadian rhythms found in our selection experiments (a longer developmental period corresponded to a longer free-running period) was consistent with the pattern found in previous studies on D. melanogaster and B. cucurbitae (Kyriacou et al., 1990; Shimizu et al., 1997). Other than these fly species, a genetic correlation between the developmental period and circadian rhythm has only been examined in a seed beetle Callosobruchus chinensis; however, no significant genetic correlation was observed (Harano and Miyatake, 2011). Although the genetic architecture underlying this genetic correlation remains unclear, and it might be different among species, the pattern of genetic correlation might be broadly conserved across Dipteran insects. Further studies are needed to evaluate whether this genetic correlation is a widespread phenomenon in broader taxonomic groups.
In the deficiency screening for genomic regions with effects on the developmental periods, we found a large number of genomic deficiencies that had effects on the developmental periods in females and males. As the genomic deficiencies examined in our study were experimentally generated, the significant effect of these genomic regions does not necessarily mean that they contribute to natural genetic variations in the developmental periods in D. melanogaster. However, it does suggest that a large number of quantitative trait loci in the D. melanogaster genome are potentially involved in the developmental period. The effect size distributions of the deficiencies deviated positively from zero in females and males, indicating that a larger number of deficiencies prolonged the developmental period. The positively biased effect of deficiencies might support the hypothesis that flies have evolved to develop faster, which partially explains the asymmetric response to disruptive selection in the current and previous studies (Miyatake, 1995; Zwaan et al., 1995).
Although the speed of circadian clocks is known to correlate with developmental period (Paranjpe et al., 2005), how the deficiencies affected developmental period in this study is unclear. In fact, the deleterious effect of the deficiencies on pre-adult survival was shown in Takahashi et al. (2011b), and it might also impair normal developmental processes and slow down the pre-adult development. Such deleterious effect of deficiencies may obscure the general correlation of the deficiencies' effects on developmental period and circadian rhythm because the indirect fitness effect of the deficiencies on pre-adult period is not necessarily expected to affect circadian rhythm at adult stage. In our study, the correlation between deficiency effects on the developmental and free-running periods was not significant, indicating no general genetic correlation between them. However, we found 18 genomic deficiencies with significant effects on both the developmental and free-running periods that might mediate the genetic correlation between them. The general lack of correlation between developmental and free-running periods indicates that there are many genomic regions with little pleiotropic effects. On the contrary, only a limited number of the genomic regions showed such pleiotropic effects. This suggests that these genomic regions have the potential to mediate the genetic correlation between the developmental period and circadian rhythm that was found in the selection experiment in our study. As these deficiencies encompass 33.9 genes on an average, it remains unclear whether a single gene within these deficiencies had a pleiotropic effect that affected the developmental and free-running periods. MacDonald and Rosbash (2001) performed a microarray analysis to study global circadian gene expression in D. melanogaster and found 134 cycling genes under constant dark conditions. Ueda et al. (2002) also performed a microarray analysis using different strains of D. melanogaster from the ones used by MacDonald and Rosbash (2001) to profile gene expression patterns and found 455 periodically expressed genes under constant dark conditions. Among the 18 deficiencies that had effects on both the developmental and free-running periods, three of the deficiencies encompassed eight genes that were found to be expressed periodically by McDonald and Rosbash (2001), whereas 12 deficiencies encompassed 27 genes that were found to be expressed periodically by Ueda et al., 2002 (Table 1). In our study, whether a change in the expression level of these genes affected the free-running periods of the Df/+ flies was not clear, but they are primary candidate genes with potential effects on the free-running period. Six of the 18 deficiencies encompassed no periodically expressed genes that were found in the two expression profiling studies (Table 1). As these deficiencies encompassed a relatively small number of genes (4.3 on average), a further detailed examination of individual candidate genes might lead to the discovery of novel clock genes. In addition, future examination of the individual candidate genes using RNAi or mutation approaches might elucidate how the genetic correlation between the developmental period and circadian rhythm was mediated in these deficiencies.
Table 1. Deficiencies with significant effects on both developmental period and circadian rhythm, and cycling genes found in expression profling studies (McDonald and Rosbash, 2001; Ueda et al., 2002) encompassed in each deficiency.
| Chromosome | Deficiency | No. of genes deleted | McDonald and Rosbash, 2001 | Ueda et al., 2002 |
|---|---|---|---|---|
| 2L | Df(2L)ED779 | 16 | CG9934, CG16978 | |
| Df(2L)ED1186 | 61 | CG10283, CG10383 | ||
| Df(2L)ED4559 | 66 | CG3523, CG3605 | ||
| 2R | Df(2R)ED4071 | 103 | Eps-15, Tina-1, CG3511, CG3608 | |
| 3L | Df(3L)ED4483 | 39 | CG10616, CG10657 | sowah, CG10418, CG10638 |
| 3R | Df(3R)ED5177 | 7 | ||
| Df(3R)ED5339 | 22 | CG8861 | ||
| Df(3R)ED5511 | 47 | Ugt35b, Ugt86Da | Tctp, Ugt35b | |
| Df(3R)ED5634 | 40 | CG9631, CG9649, CG31326, CG33109 | Cyp6d5, CG9649 | |
| Df(3R)ED5664 | 53 | Art3, smp-30, Spn88Eb, CG12241 | ||
| Df(3R)ED6315 | 2 | |||
| Df(3R)ED6332 | 4 | |||
| Df(3R)ED6362 | 6 | |||
| Df(3R)ED10549 | 2 | |||
| Df(3R)ED10564 | 29 | Art3, Spn88Eb, CG12241 | ||
| Df(3R)ED10566 | 29 | Art3, Spn88Eb, CG12241 | ||
| Df(3R)ED10894 | 80 | Lsd-1, mbc, Rpn9, CG10208, CG10214 | ||
| Df(3R)ED10961 | 5 |
In our study, we performed disruptive selection on the developmental periods of D. melanogaster and found a genetic correlation between the developmental periods and circadian rhythms. We also identified 18 genomic deficiencies with effects on the developmental periods and circadian rhythms, and postulated that these genomic regions might potentially mediate the genetic correlation between them. The novel findings reported in our study might lead to a better understanding of how this correlation is regulated genetically in broader taxonomic groups.
Data archiving
There were no data to deposit.
Acknowledgments
This work was financially supported by Special Coordination Funds for Promoting Sciences and Technology of The Ministry of Education, Sport, Culture, Science and Technology of Japan, and a Grant-in-Aid for Scientific Research (KAKENHI 23770087) to KHT, and a Grant-in-Aid for Scientific Research (KAKENHI 23570027) to TM.
Appendix 1
Table A1. Deficiencies used for the screening, and their location, size, and mean developmental period and FDR from ANOVA.
| Chromosome | Deletion ID | Region | Deletion size (bp) |
Developmental period |
|
|---|---|---|---|---|---|
| Female | Male | ||||
| 2L | Df(2L)ED3 | 35B2–35D1 | 843185 | 13.797 (0.104) | 13.537 (0.760) |
| Df(2L)ED21 | 21B3–21B7 | 125158 | 14.736 (0.002) | 14.683 (0.001) | |
| Df(2L)ED40 | 21D1–21D2 | 9980 | 13.037 (0.015) | 13.139 (0.092) | |
| Df(2L)ED49 | 1A1–100E1 | 19888 | 13.712 (0.054) | 13.564 (0.607) | |
| Df(2L)ED87 | 21E2–21E2 | 284732 | 14.006 (0.034) | 13.894 (0.278) | |
| Df(2L)ED94 | 21E2–21E3 | 468874 | 14.482 (0.000) | 14.264 (0.002) | |
| Df(2L)ED105 | 21E2–22A1 | 567674 | 13.495 (0.977) | 14.396 (0.019) | |
| Df(2L)ED108 | 21F1–22A1 | 301394 | 12.945 (0.021) | 14.000 (0.467) | |
| Df(2L)ED122 | 22B1–22D4 | 494297 | 13.444 (0.631) | 13.812 (0.130) | |
| Df(2L)ED123 | 22B8–22D4 | 236161 | 13.396 (0.711) | 13.529 (0.832) | |
| Df(2L)ED124 | 22D3–22D4 | 23445 | 13.266 (0.086) | 13.814 (0.106) | |
| Df(2L)ED125 | 22B2–22D4 | 484626 | 13.582 (0.620) | 13.201 (0.221) | |
| Df(2L)ED132 | 23A3–23A3 | 106 | 12.804 (0.000) | 13.015 (0.035) | |
| Df(2L)ED136 | 22F4–23A3 | 260190 | 13.227 (0.034) | 13.299 (0.422) | |
| Df(2L)ED206 | 23B8–23C5 | 181763 | 12.283 (0.000) | 12.338 (0.000) | |
| Df(2L)ED216 | 23B8–23C5 | 181892 | 13.389 (0.674) | 13.329 (0.656) | |
| Df(2L)ED234 | 23C4–24A2 | 632936 | 15.142 (0.299) | 15.290 (0.111) | |
| Df(2L)ED243 | 24A2–24A4 | 24683 | 13.607 (0.209) | 13.777 (0.321) | |
| Df(2L)ED247 | 24A2–24C3 | 138959 | 13.791 (0.127) | 13.920 (0.027) | |
| Df(2L)ED250 | 24F4–25A7 | 344209 | 13.874 (0.226) | 13.505 (0.874) | |
| Df(2L)ED256 | 25B1–25B10 | 108097 | 13.853 (0.312) | 13.484 (0.943) | |
| Df(2L)ED270 | 25F2–25F5 | 141567 | 13.001 (0.012) | 13.029 (0.022) | |
| Df(2L)ED279 | 25F2–26A1 | 248827 | 13.165 (0.049) | 13.436 (0.955) | |
| Df(2L)ED280 | 25F5–26A1 | 105526 | 13.172 (0.031) | 13.385 (0.758) | |
| Df(2L)ED284 | 25F2–26A3 | 285333 | 13.050 (0.004) | 13.273 (0.398) | |
| Df(2L)ED285 | 25F5–26A3 | 142032 | 12.890 (0.001) | 12.979 (0.020) | |
| Df(2L)ED292 | 25F5–26B2 | 179079 | 13.801 (0.015) | 14.300 (0.008) | |
| Df(2L)ED299 | 26B1–26B2 | 2194 | 12.556 (0.060) | 12.893 (0.213) | |
| Df(2L)ED330 | 26A3–26B2 | 55750 | 14.834 (0.005) | 14.680 (0.007) | |
| Df(2L)ED331 | 26B2–26B2 | 18588 | 13.012 (0.141) | — | |
| Df(2L)ED334 | 25F2–26B2 | 341038 | 14.246 (0.032) | 14.813 (0.095) | |
| Df(2L)ED343 | 26B2–26B5 | 82250 | 13.396 (0.256) | 13.095 (0.046) | |
| Df(2L)ED347 | 25F5–26B5 | 280456 | 13.465 (0.923) | 13.613 (0.604) | |
| Df(2L)ED353 | 26B2–26B5 | 83109 | 13.258 (0.291) | 13.115 (0.234) | |
| Df(2L)ED354 | 26B1–26B5 | 102961 | 13.889 (0.571) | 13.875 (0.151) | |
| Df(2L)ED369 | 26C3–26D1 | 72246 | 12.853 (0.000) | 13.018 (0.055) | |
| Df(2L)ED371 | 26C3–26D1 | 73530 | 13.798 (0.005) | 13.416 (0.873) | |
| Df(2L)ED373 | 26B2–26D1 | 430254 | 12.900 (0.078) | 13.575 (0.691) | |
| Df(2L)ED374 | 26B10–26D1 | 232319 | 12.469 (0.000) | — | |
| Df(2L)ED384 | 26B2–26D7 | 465648 | 12.860 (0.043) | 13.214 (0.285) | |
| Df(2L)ED385 | 26B1–26D7 | 485500 | 14.746 (0.000) | 15.261 (0.000) | |
| Df(2L)ED438 | 27D1–27D4 | 52278 | 13.676 (0.572) | 13.973 (0.095) | |
| Df(2L)ED440 | 27D3–27E1 | 74563 | 13.582 (0.772) | 13.416 (0.893) | |
| Df(2L)ED463 | 27F4–27F7 | 661 | 13.681 (0.224) | 13.483 (0.905) | |
| Df(2L)ED478 | 27F7–28B1 | 139196 | 13.246 (0.072) | 13.306 (0.422) | |
| Df(2L)ED494 | 27F4–28B1 | 153371 | 14.267 (0.010) | 14.330 (0.006) | |
| Df(2L)ED496 | 28C4–28C4 | 9590 | 13.003 (0.001) | 13.154 (0.184) | |
| Df(2L)ED501 | 27F7–28C4 | 376256 | 13.658 (0.225) | 13.644 (0.347) | |
| Df(2L)ED502 | 28C1–28C4 | 122088 | 12.914 (0.013) | 12.623 (0.003) | |
| Df(2L)ED508 | 28B1–28C4 | 223552 | 13.159 (0.198) | 13.783 (0.377) | |
| Df(2L)ED517 | 27F7–28D2 | 447744 | 13.502 (0.993) | 13.845 (0.127) | |
| Df(2L)ED548 | 28E1–28E9 | 91467 | 13.766 (0.174) | 14.090 (0.007) | |
| Df(2L)ED573 | 28F1–29A2 | 95377 | 13.082 (0.089) | 13.437 (0.943) | |
| Df(2L)ED578 | 28F1–29A3 | 103066 | 13.735 (0.474) | 14.180 (0.142) | |
| Df(2L)ED611 | 29B4–29C3 | 36967 | 13.694 (0.529) | 13.916 (0.080) | |
| Df(2L)ED623 | 29C1–29E4 | 296560 | 13.052 (0.007) | 13.083 (0.091) | |
| Df(2L)ED629 | 29B4–29E4 | 317273 | 13.913 (0.097) | 13.602 (0.351) | |
| Df(2L)ED630 | 29C3–29E4 | 278827 | 13.028 (0.003) | 12.923 (0.009) | |
| Df(2L)ED632 | 29E1–29E4 | 156152 | 12.776 (0.000) | 13.167 (0.095) | |
| Df(2L)ED647 | 29E1–29F5 | 414176 | 13.752 (0.043) | 13.574 (0.632) | |
| Df(2L)ED659 | 29E1–30A3 | 646785 | 14.150 (0.007) | 14.392 (0.012) | |
| Df(2L)ED673 | 30A4–30B3 | 226380 | 13.171 (0.069) | 12.912 (0.006) | |
| Df(2L)ED677 | 30B3–30B12 | 144271 | 13.416 (0.486) | 13.600 (0.617) | |
| Df(2L)ED678 | 29F5–30B12 | 623585 | 13.563 (0.753) | 13.552 (0.581) | |
| Df(2L)ED679 | 30B12–30B12 | 10552 | 13.644 (0.624) | 13.684 (0.604) | |
| Df(2L)ED680 | 30A4–30B12 | 376664 | 13.441 (0.606) | 13.622 (0.370) | |
| Df(2L)ED684 | 30B12–30C1 | 42145 | 13.926 (0.107) | 13.789 (0.110) | |
| Df(2L)ED690 | 30B3–30E4 | 480705 | 13.176 (0.312) | 13.080 (0.334) | |
| Df(2L)ED692 | 30B12–30E4 | 346986 | 14.350 (0.001) | 14.558 (0.001) | |
| Df(2L)ED695 | 30C5–30E4 | 218967 | 13.259 (0.078) | 13.295 (0.490) | |
| Df(2L)ED697 | 30C1–30E4 | 301348 | 13.554 (0.711) | 13.415 (0.836) | |
| Df(2L)ED700 | 30E1–30E4 | 20668 | 12.849 (0.158) | 12.904 (0.239) | |
| Df(2L)ED701 | 30C5–30F1 | 249119 | 14.252 (0.001) | 14.436 (0.001) | |
| Df(2L)ED729 | 31B1–31D7 | 100900 | 14.059 (0.005) | 14.485 (0.034) | |
| Df(2L)ED746 | 31F4–32A5 | 225931 | 13.774 (0.022) | 13.769 (0.170) | |
| Df(2L)ED748 | 31B1–32A5 | 485690 | 14.408 (0.000) | 14.345 (0.001) | |
| Df(2L)ED758 | 33C1–33E4 | 367471 | 13.591 (0.567) | 13.649 (0.406) | |
| Df(2L)ED760 | 33B8–33E5 | 426429 | 13.925 (0.007) | 13.922 (0.052) | |
| Df(2L)ED761 | 33A2–33E5 | 627604 | 13.596 (0.288) | 13.983 (0.190) | |
| Df(2L)ED769 | 33E9–34A1 | 277041 | 13.675 (0.155) | 14.165 (0.001) | |
| Df(2L)ED771 | 33E4–34A1 | 388303 | 14.323 (0.009) | 14.703 (0.012) | |
| Df(2L)ED773 | 33E9–34A3 | 429228 | 13.596 (0.689) | 13.751 (0.110) | |
| Df(2L)ED774 | 34A3–34A3 | 683 | 13.538 (0.869) | 13.257 (0.293) | |
| Df(2L)ED775 | 33B8–34A3 | 965018 | 13.862 (0.036) | 14.651 (0.002) | |
| Df(2L)ED776 | 33E4–34A3 | 540490 | 13.758 (0.260) | 14.181 (0.009) | |
| Df(2L)ED777 | 33E7–34A3 | 490576 | 13.444 (0.727) | 14.050 (0.016) | |
| Df(2L)ED778 | 33E9–34A7 | 619745 | 13.758 (0.079) | 14.243 (0.012) | |
| Df(2L)ED779 | 34A3–34A7 | 191200 | 15.034 (0.000) | 15.014 (0.001) | |
| Df(2L)ED780 | 33E4–34A7 | 731007 | 13.846 (0.029) | 14.300 (0.003) | |
| Df(2L)ED784 | 34A4–34B6 | 327612 | 14.388 (0.011) | 14.078 (0.325) | |
| Df(2L)ED791 | 34E1–35B4 | 811156 | 14.445 (0.024) | 14.367 (0.003) | |
| Df(2L)ED793 | 34E4–35B4 | 754489 | 14.832 (0.001) | 14.712 (0.027) | |
| Df(2L)ED796 | 35C1–35C4 | 152111 | 14.046 (0.070) | 13.890 (0.143) | |
| Df(2L)ED929 | 21B3–21B3 | 18484 | 13.965 (0.130) | 13.762 (0.300) | |
| Df(2L)ED1000 | 35B8–35D1 | 336213 | 13.490 (0.962) | 13.871 (0.180) | |
| Df(2L)ED1004 | 35B10–35D1 | 272692 | 14.107 (0.089) | 14.260 (0.070) | |
| Df(2L)ED1050 | 35B8–35D4 | 765231 | 13.524 (0.890) | 13.581 (0.679) | |
| Df(2L)ED1054 | 35B10–35D4 | 701710 | 14.509 (0.002) | 14.367 (0.007) | |
| Df(2L)ED1056 | 35D2–35D4 | 284125 | 13.834 (0.154) | 14.099 (0.009) | |
| Df(2L)ED1092 | 35F12–36A10 | 329835 | 13.632 (0.287) | 13.907 (0.022) | |
| Df(2L)ED1102 | 35F12–36A10 | 334647 | 13.739 (0.097) | 14.476 (0.009) | |
| Df(2L)ED1109 | 36A3–36A10 | 164790 | 13.649 (0.312) | 13.842 (0.124) | |
| Df(2L)ED1143 | 36A10–36B1 | 106247 | 13.369 (0.604) | 13.633 (0.440) | |
| Df(2L)ED1153 | 35F12–36B2 | 502171 | 14.371 (0.011) | 14.243 (0.012) | |
| Df(2L)ED1158 | 36B1–36C9 | 658852 | 14.270 (0.005) | 14.228 (0.040) | |
| Df(2L)ED1161 | 36A10–36C9 | 788014 | 13.920 (0.043) | 13.770 (0.180) | |
| Df(2L)ED1164 | 36A10–36C9 | 787566 | 14.424 (0.005) | 14.547 (0.001) | |
| Df(2L)ED1165 | 36C1–36C9 | 360447 | 14.071 (0.134) | 14.244 (0.010) | |
| Df(2L)ED1175 | 36C1–36C10 | 383436 | 13.023 (0.005) | 13.222 (0.227) | |
| Df(2L)ED1183 | 36E6–36F7 | 465553 | 14.700 (0.000) | 14.796 (0.000) | |
| Df(2L)ED1186 | 36E6–37A2 | 581220 | 15.539 (0.000) | 15.559 (0.001) | |
| Df(2L)ED1187 | 36F7–37A2 | 115693 | 12.594 (0.000) | — | |
| Df(2L)ED1196 | 36E6–37B1 | 671892 | 15.616 (0.000) | 16.005 (0.000) | |
| Df(2L)ED1198 | 36F7–37B1 | 206365 | 14.008 (0.150) | 14.359 (0.017) | |
| Df(2L)ED1200 | 44D8–45B4 | 155049 | 13.739 (0.504) | 13.700 (0.376) | |
| Df(2L)ED1202 | 37B1–37C5 | 334948 | 13.600 (0.638) | 13.449 (0.981) | |
| Df(2L)ED1226 | 37B9–37E3 | 460658 | 12.967 (0.008) | 13.407 (0.835) | |
| Df(2L)ED1231 | 37C5–37E3 | 305616 | 13.897 (0.267) | 14.071 (0.135) | |
| Df(2L)ED1236 | 37B9–37E4 | 483659 | 14.190 (0.001) | 14.054 (0.058) | |
| Df(2L)ED1238 | 37C1–37E4 | 357571 | 13.600 (0.765) | 13.671 (0.456) | |
| Df(2L)ED1242 | 37E5–37F1 | 15994 | 13.709 (0.460) | 13.820 (0.225) | |
| Df(2L)ED1243 | 37B9–37F1 | 524349 | 13.676 (0.281) | 13.787 (0.294) | |
| Df(2L)ED1245 | 37C1–37F1 | 398261 | 14.035 (0.055) | 14.701 (0.007) | |
| Df(2L)ED1250 | 37E5–37F1 | 24869 | 13.665 (0.264) | 14.193 (0.024) | |
| Df(2L)ED1251 | 37B9–37F1 | 533224 | 14.610 (0.000) | 14.334 (0.045) | |
| Df(2L)ED1272 | 37C5–38A2 | 594884 | 13.577 (0.611) | 14.381 (0.003) | |
| Df(2L)ED1303 | 37E5–38C6 | 864775 | 12.979 (0.314) | 12.853 (0.285) | |
| Df(2L)ED1305 | 38B4–38C6 | 296988 | 13.974 (0.009) | 13.832 (0.084) | |
| Df(2L)ED1315 | 38B4–38F5 | 832122 | 13.384 (0.690) | 13.298 (0.604) | |
| Df(2L)ED1317 | 38D1–38F5 | 278939 | 12.811 (0.001) | 13.185 (0.315) | |
| Df(2L)ED1375 | 38F5–39D2 | 457289 | 13.355 (0.650) | 13.295 (0.495) | |
| Df(2L)ED1378 | 38F1–39D2 | 574133 | 14.519 (0.002) | 14.539 (0.002) | |
| Df(2L)ED1382 | 39B4–39D2 | 159357 | 14.625 (0.000) | 14.723 (0.002) | |
| Df(2L)ED1384 | 38F5–39D2 | 474447 | 14.049 (0.072) | 13.999 (0.066) | |
| Df(2L)ED1451 | 38F5–39E2 | 666875 | 13.094 (0.059) | 12.919 (0.035) | |
| Df(2L)ED1454 | 39E3–39E6 | 28361 | 13.345 (0.267) | 13.498 (0.867) | |
| Df(2L)ED1455 | 39A1–39E6 | 605551 | 13.742 (0.146) | 13.468 (0.956) | |
| Df(2L)ED1462 | 39B4–39E6 | 406785 | 12.744 (0.299) | 12.889 (0.370) | |
| Df(2L)ED1466 | 39E3–40A5 | 199232 | 13.027 (0.104) | 13.609 (0.578) | |
| Df(2L)ED1473 | 39B4–40A5 | 577656 | 14.392 (0.000) | 14.710 (0.000) | |
| Df(2L)ED2809 | 21B1–21B1 | 5306 | 13.059 (0.018) | 12.925 (0.016) | |
| Df(2L)ED4330 | 23C4–23C5 | 56592 | 14.350 (0.012) | 14.173 (0.055) | |
| Df(2L)ED4559 | 23C4–23F6 | 479077 | 15.643 (0.000) | 15.024 (0.001) | |
| Df(2L)ED4651 | 23B8–23F6 | 604377 | 15.272 (0.000) | 14.978 (0.000) | |
| Df(2L)ED5878 | 21B1–21B3 | 93755 | 14.447 (0.001) | 14.133 (0.046) | |
| Df(2L)ED6569 | 27A1–27C4 | 212193 | 13.582 (0.526) | 13.574 (0.460) | |
| Df(2L)ED7007 | 27A1–27C7 | 254709 | 13.915 (0.167) | 13.509 (0.762) | |
| Df(2L)ED7666 | 22F4–22F4 | 1188 | 13.295 (0.460) | 13.150 (0.343) | |
| Df(2L)ED7733 | 21E2–21F1 | 266285 | 13.307 (0.545) | 13.516 (0.875) | |
| Df(2L)ED7762 | 22A6–22D3 | 539713 | 13.730 (0.460) | 14.008 (0.028) | |
| Df(2L)ED7853 | 25A3–25B10 | 299273 | 13.981 (0.049) | 13.507 (0.794) | |
| Df(2L)ED8142 | 31E1–32A4 | 271218 | 13.185 (0.010) | 13.564 (0.608) | |
| Df(2L)ED8185 | 34E1–35A4 | 456009 | 14.205 (0.003) | 14.336 (0.001) | |
| Df(2L)ED8186 | 34E4–35A4 | 399342 | 13.715 (0.244) | 13.802 (0.247) | |
| Df(2L)ED8368 | 36B1–36B2 | 33618 | 13.052 (0.008) | 12.876 (0.015) | |
| Df(2L)ED8386 | 36C1–36C9 | 337487 | 14.427 (0.000) | 14.894 (0.000) | |
| Df(2L)ED12487 | 25C3–25F2 | 604135 | 14.180 (0.001) | 14.141 (0.058) | |
| Df(2L)ED12527 | 28C4–28D3 | 203671 | 13.660 (0.506) | 13.507 (0.812) | |
| Df(2L)ED13216 | 36A12–36B1 | 70720 | 13.694 (0.503) | 13.443 (0.958) | |
| 2R | Df(2R)ED1 | 53E9–53F8 | 70595 | 13.464 (0.766) | 13.625 (0.507) |
| Df(2R)ED1482 | 42A8–42A11 | 96487 | 12.641 (0.000) | 12.945 (0.010) | |
| Df(2R)ED1484 | 42A2–42A14 | 351791 | 13.136 (0.092) | 13.103 (0.129) | |
| Df(2R)ED1552 | 42A11–42C7 | 608682 | 12.923 (0.143) | 12.713 (0.130) | |
| Df(2R)ED1612 | 42A13–42E6 | 829140 | 13.294 (0.335) | 13.305 (0.495) | |
| Df(2R)ED1618 | 42C3–43A1 | 518138 | 13.074 (0.001) | 13.230 (0.258) | |
| Df(2R)ED1673 | 42E4–43D3 | 547751 | 13.013 (0.014) | 13.092 (0.337) | |
| Df(2R)ED1715 | 43A4–43F1 | 589972 | 13.633 (0.616) | 13.799 (0.276) | |
| Df(2R)ED1725 | 43E4–44B5 | 542121 | 13.943 (0.005) | 14.367 (0.002) | |
| Df(2R)ED1735 | 43F8–44D4 | 638302 | 13.364 (0.460) | 13.573 (0.655) | |
| Df(2R)ED1742 | 44B8–44E3 | 549961 | 14.059 (0.029) | 14.051 (0.005) | |
| Df(2R)ED1770 | 14A8–14C6 | 551912 | 13.569 (0.808) | 13.345 (0.832) | |
| Df(2R)ED1791 | 44F7–45F1 | 630522 | 13.504 (0.980) | 13.679 (0.258) | |
| Df(2R)ED2076 | 47A10–47C1 | 343202 | 13.206 (0.467) | 13.416 (0.939) | |
| Df(2R)ED2098 | 47A7–47C6 | 482345 | 13.727 (0.482) | 14.060 (0.167) | |
| Df(2R)ED2155 | 47C6–47F8 | 503346 | 13.121 (0.074) | 13.120 (0.073) | |
| Df(2R)ED2219 | 47D6–48B6 | 467979 | 13.925 (0.338) | 13.667 (0.579) | |
| Df(2R)ED2222 | 47F13–48B6 | 212411 | 13.921 (0.021) | 14.149 (0.004) | |
| Df(2R)ED2308 | 49D3–49E7 | 216614 | 13.517 (0.959) | 13.199 (0.110) | |
| Df(2R)ED2311 | 49E4–49F10 | 277450 | 13.861 (0.256) | 14.217 (0.065) | |
| Df(2R)ED2354 | 50E6–51B1 | 315512 | 13.361 (0.448) | 13.469 (0.949) | |
| Df(2R)ED2423 | 51C5–51F11 | 520185 | 13.747 (0.075) | 13.602 (0.484) | |
| Df(2R)ED2426 | 51E2–52B1 | 482016 | 13.436 (0.571) | 13.870 (0.151) | |
| Df(2R)ED2436 | 51F11–52D11 | 627239 | 13.832 (0.027) | 14.057 (0.008) | |
| Df(2R)ED2457 | 52D11–52E7 | 129848 | 14.078 (0.001) | 14.268 (0.001) | |
| Df(2R)ED2487 | 52E6–53C4 | 261478 | 13.026 (0.011) | 13.258 (0.428) | |
| Df(2R)ED2748 | 53D11–53F8 | 268682 | 13.251 (0.192) | 13.458 (0.988) | |
| Df(2R)ED2751 | 53D14–53F8 | 240132 | 13.229 (0.145) | 13.510 (0.874) | |
| Df(2R)ED3181 | 57F10–57F10 | 524868 | 13.682 (0.376) | 13.731 (0.169) | |
| Df(2R)ED3610 | 54F1–55C8 | 561128 | 14.107 (0.011) | 13.733 (0.490) | |
| Df(2R)ED3683 | 55C2–56C4 | 940122 | 13.139 (0.250) | 13.285 (0.391) | |
| Df(2R)ED3728 | 56D10–56E2 | 264297 | 13.827 (0.460) | 13.785 (0.266) | |
| Df(2R)ED3791 | 57B1–57D4 | 552570 | 13.507 (0.964) | 13.608 (0.347) | |
| Df(2R)ED3921 | 57F9–57F10 | 11246 | 13.063 (0.004) | 13.030 (0.024) | |
| Df(2R)ED3923 | 57F6–57F10 | 67570 | 13.897 (0.007) | 13.935 (0.018) | |
| Df(2R)ED3943 | 37B9–37C5 | 688723 | 14.051 (0.139) | 14.440 (0.034) | |
| Df(2R)ED3952 | 58B10–58E5 | 386674 | 13.111 (0.181) | 13.196 (0.151) | |
| Df(2R)ED4061 | 60C8–60D13 | 270614 | 13.308 (0.282) | 13.517 (0.794) | |
| Df(2R)ED4071 | 60C8–60E8 | 540173 | 15.150 (0.000) | 15.192 (0.000) | |
| Df(2R)ED9039 | 48C5–48E4 | 283867 | 13.695 (0.256) | 13.936 (0.016) | |
| Df(2R)ED9045 | 48F5–49A7 | 212898 | 12.561 (0.009) | 12.625 (0.051) | |
| 3L | Df(3L)ED201 | 91A5–91F1 | 224017 | 13.677 (0.338) | 13.463 (0.973) |
| Df(3L)ED202 | 61C9–61F7 | 597642 | 13.666 (0.314) | 13.989 (0.133) | |
| Df(3L)ED207 | 61C9–62A6 | 829369 | 14.917 (0.000) | 14.586 (0.001) | |
| Df(3L)ED208 | 63C1–63F5 | 644000 | 13.142 (0.059) | 13.155 (0.200) | |
| Df(3L)ED210 | 64B9–64C13 | 804208 | 13.650 (0.621) | 14.010 (0.135) | |
| Df(3L)ED211 | 65A9–65B4 | 334624 | 13.078 (0.179) | 12.973 (0.124) | |
| Df(3L)ED215 | 69B5–69C4 | 86745 | 14.816 (0.000) | 15.106 (0.001) | |
| Df(3L)ED217 | 70F4–71E1 | 831026 | 13.044 (0.397) | 13.512 (0.901) | |
| Df(3L)ED218 | 71B1–71E1 | 575028 | 13.824 (0.066) | 14.073 (0.201) | |
| Df(3L)ED220 | 72D4–72F1 | 324193 | 15.556 (0.000) | 16.338 (0.000) | |
| Df(3L)ED223 | 73A1–73D5 | 439052 | 14.400 (0.006) | 14.075 (0.181) | |
| Df(3L)ED224 | 75B1–75C6 | 429316 | 13.678 (0.398) | 13.670 (0.495) | |
| Df(3L)ED225 | 75C1–75D4 | 435192 | 12.806 (0.008) | 13.076 (0.185) | |
| Df(3L)ED228 | 76A1–76D2 | 701102 | 13.070 (0.251) | 13.189 (0.637) | |
| Df(3L)ED230 | 79C2–80A4 | 699720 | 14.182 (0.009) | 14.657 (0.000) | |
| Df(3L)ED231 | 80B1–80C1 | 73704 | 13.299 (0.448) | 13.176 (0.239) | |
| Df(3L)ED4079 | 61A5–61B1 | 91461 | 13.048 (0.005) | 13.082 (0.082) | |
| Df(3L)ED4177 | 61C1–61E2 | 715336 | 13.693 (0.603) | 13.784 (0.221) | |
| Df(3L)ED4191 | 61C3–62A2 | 934664 | 14.067 (0.134) | 14.225 (0.151) | |
| Df(3L)ED4196 | 61C7–62A2 | 839354 | 13.322 (0.630) | 13.183 (0.656) | |
| Df(3L)ED4238 | 61C9–62A4 | 808192 | 13.773 (0.231) | 14.281 (0.109) | |
| Df(3L)ED4256 | 62A3–62A6 | 40559 | 13.158 (0.104) | 13.148 (0.391) | |
| Df(3L)ED4284 | 62B4–62B12 | 168110 | 13.856 (0.276) | 13.657 (0.523) | |
| Df(3L)ED4287 | 62B4–62E5 | 756319 | 13.585 (0.600) | 13.356 (0.652) | |
| Df(3L)ED4288 | 63A6–63B7 | 78264 | 13.748 (0.411) | 13.577 (0.643) | |
| Df(3L)ED4293 | 63C1–63C1 | 24226 | 13.607 (0.624) | 13.391 (0.866) | |
| Df(3L)ED4341 | 63F6–64B9 | 637145 | 15.781 (0.000) | 15.684 (0.001) | |
| Df(3L)ED4342 | 64A12–64B12 | 347385 | 14.029 (0.001) | 14.242 (0.003) | |
| Df(3L)ED4408 | 66A22–66C5 | 320467 | 14.596 (0.001) | 15.186 (0.000) | |
| Df(3L)ED4414 | 66D12–66E6 | 233661 | 13.299 (0.146) | 13.386 (0.758) | |
| Df(3L)ED4415 | 66D12–66E6 | 213016 | 13.571 (0.690) | 14.124 (0.169) | |
| Df(3L)ED4416 | 66E1–67B1 | 522145 | 12.963 (0.049) | 12.969 (0.079) | |
| Df(3L)ED4421 | 66D12–67B3 | 638749 | 13.420 (0.869) | 13.409 (0.919) | |
| Df(3L)ED4457 | 67E2–68A7 | 761858 | 12.730 (0.025) | 12.902 (0.088) | |
| Df(3L)ED4470 | 68A6–68E1 | 736241 | 14.399 (0.000) | 14.616 (0.004) | |
| Df(3L)ED4483 | 69A5–69D3 | 415994 | 15.832 (0.000) | 15.876 (0.000) | |
| Df(3L)ED4486 | 69C4–69F6 | 518066 | 14.190 (0.061) | 14.047 (0.091) | |
| Df(3L)ED4502 | 70A3–70C10 | 765786 | 13.485 (0.959) | 13.674 (0.264) | |
| Df(3L)ED4515 | 70C6–70C15 | 97860 | 13.398 (0.394) | 13.329 (0.604) | |
| Df(3L)ED4528 | 70C15–70D2 | 39982 | 13.417 (0.711) | 13.229 (0.378) | |
| Df(3L)ED4534 | 70C15–70D3 | 156653 | 13.228 (0.294) | 13.052 (0.245) | |
| Df(3L)ED4536 | 70C11–70D3 | 202563 | 13.211 (0.181) | 13.213 (0.318) | |
| Df(3L)ED4543 | 70C6–70F4 | 822815 | 15.457 (0.000) | 15.225 (0.000) | |
| Df(3L)ED4606 | 72D4–73C4 | 692639 | 14.750 (0.012) | 15.115 (0.012) | |
| Df(3L)ED4674 | 73B5–73E5 | 388134 | 13.679 (0.373) | 13.651 (0.337) | |
| Df(3L)ED4685 | 73D5–74E2 | 721094 | 13.645 (0.368) | 13.394 (0.790) | |
| Df(3L)ED4710 | 74D1–75B11 | 651836 | 14.658 (0.004) | 15.145 (0.000) | |
| Df(3L)ED4743 | 75D4–75D8 | 133616 | 13.261 (0.210) | 13.317 (0.423) | |
| Df(3L)ED4744 | 75D8–75E1 | 14368 | 13.572 (0.437) | 13.713 (0.351) | |
| Df(3L)ED4782 | 75F2–76A1 | 174808 | 13.128 (0.129) | 13.524 (0.812) | |
| Df(3L)ED4786 | 75F7–76A5 | 194711 | 13.210 (0.281) | 13.605 (0.490) | |
| Df(3L)ED4789 | 76A1–76A5 | 124956 | 13.455 (0.706) | 13.231 (0.180) | |
| Df(3L)ED4799 | 76A1–76B3 | 311466 | 13.130 (0.129) | 13.145 (0.105) | |
| Df(3L)ED4858 | 76D3–77C1 | 506447 | 13.685 (0.670) | 13.273 (0.624) | |
| Df(3L)ED4957 | 78C3–78F1 | 530381 | 14.150 (0.114) | 14.042 (0.151) | |
| Df(3L)ED4978 | 78D5–79A2 | 346878 | 13.709 (0.229) | 14.120 (0.143) | |
| Df(3L)ED5013 | 80A1–80B1 | 150650 | 13.438 (0.620) | 13.795 (0.142) | |
| Df(3L)ED5017 | 80A4–80C2 | 162804 | 15.020 (0.004) | 15.512 (0.000) | |
| 3R | Df(3R)ED2 | 21E2–21E2 | 697540 | 14.896 (0.000) | 15.026 (0.002) |
| Df(3R)ED5020 | 82A3–82B1 | 108705 | 13.058 (0.015) | 13.056 (0.127) | |
| Df(3R)ED5021 | 82A1–82B1 | 193118 | 13.114 (0.311) | 12.879 (0.055) | |
| Df(3R)ED5046 | 82A1–82D3 | 541858 | 13.517 (0.949) | 13.402 (0.809) | |
| Df(3R)ED5066 | 82D1–82E4 | 302797 | 13.947 (0.002) | 13.682 (0.280) | |
| Df(3R)ED5071 | 82A1–82E4 | 755409 | 14.519 (0.027) | 14.737 (0.005) | |
| Df(3R)ED5092 | 82A3–82E8 | 805399 | 16.192 (0.000) | 15.897 (0.001) | |
| Df(3R)ED5095 | 82D1–82E8 | 437200 | 13.993 (0.004) | 13.657 (0.312) | |
| Df(3R)ED5100 | 82A1–82E8 | 889812 | 15.927 (0.000) | 15.953 (0.000) | |
| Df(3R)ED5138 | 82D5–82F8 | 483811 | 13.700 (0.250) | 13.592 (0.607) | |
| Df(3R)ED5142 | 82B3–82F8 | 811587 | 14.413 (0.006) | 13.938 (0.035) | |
| Df(3R)ED5147 | 82E8–83A1 | 280684 | 14.583 (0.000) | 14.745 (0.001) | |
| Df(3R)ED5156 | 82F8–83A4 | 193919 | 13.179 (0.011) | 13.413 (0.791) | |
| Df(3R)ED5177 | 83B4–83B6 | 23466 | 14.994 (0.000) | 15.166 (0.001) | |
| Df(3R)ED5187 | 83B7–83B8 | 6020 | 13.840 (0.411) | 13.315 (0.604) | |
| Df(3R)ED5196 | 83B9–83D2 | 323565 | 14.812 (0.001) | 14.922 (0.000) | |
| Df(3R)ED5197 | 83B7–83D2 | 359362 | 13.095 (0.019) | 13.191 (0.127) | |
| Df(3R)ED5220 | 84E6–84E11 | 116309 | 13.035 (0.015) | 13.316 (0.424) | |
| Df(3R)ED5221 | 84C4–84E11 | 965801 | 12.803 (0.000) | 13.098 (0.130) | |
| Df(3R)ED5223 | 84D9–84E11 | 602379 | 12.537 (0.003) | 12.743 (0.118) | |
| Df(3R)ED5230 | 84E6–85A5 | 675360 | 12.900 (0.018) | 12.903 (0.068) | |
| Df(3R)ED5296 | 84F6–85C3 | 806270 | 13.612 (0.612) | 13.797 (0.169) | |
| Df(3R)ED5301 | 85C3–85C3 | 22497 | 13.007 (0.000) | 13.247 (0.370) | |
| Df(3R)ED5327 | 85D1–85D1 | 2719 | 13.251 (0.159) | 13.404 (0.821) | |
| Df(3R)ED5330 | 85A5–85D1 | 560209 | 13.350 (0.534) | 13.967 (0.310) | |
| Df(3R)ED5331 | 85C3–85D1 | 195601 | 13.296 (0.363) | 13.265 (0.523) | |
| Df(3R)ED5339 | 85D1–85D11 | 125299 | 14.819 (0.001) | 15.291 (0.000) | |
| Df(3R)ED5416 | 85D16–85E6 | 335297 | 13.279 (0.114) | 13.363 (0.744) | |
| Df(3R)ED5428 | 85E1–85F8 | 417820 | 14.345 (0.001) | 13.946 (0.051) | |
| Df(3R)ED5438 | 85E5–85F8 | 321934 | 13.401 (0.727) | 13.517 (0.779) | |
| Df(3R)ED5472 | 85F16–86B1 | 180223 | 12.976 (0.113) | 13.156 (0.221) | |
| Df(3R)ED5474 | 85F11–86B1 | 241312 | 12.819 (0.068) | 12.757 (0.055) | |
| Df(3R)ED5495 | 85F16–86C7 | 716259 | 13.103 (0.078) | 13.911 (0.296) | |
| Df(3R)ED5506 | 86C7–86D5 | 287750 | 13.012 (0.001) | 13.179 (0.151) | |
| Df(3R)ED5511 | 86C7–86D9 | 359178 | 12.267 (0.000) | 12.376 (0.000) | |
| Df(3R)ED5514 | 86C7–86E11 | 684255 | 14.812 (0.014) | 15.350 (0.000) | |
| Df(3R)ED5516 | 86D8–86E13 | 385730 | 13.222 (0.355) | 13.422 (0.938) | |
| Df(3R)ED5518 | 86C7–86E13 | 734902 | 16.708 (0.000) | 16.480 (0.000) | |
| Df(3R)ED5519 | 86E11–86E13 | 53930 | 13.442 (0.590) | 13.415 (0.815) | |
| Df(3R)ED5554 | 87B5–87B11 | 162903 | 13.172 (0.005) | 13.239 (0.420) | |
| Df(3R)ED5558 | 86F9–87B11 | 615275 | 13.967 (0.021) | 13.584 (0.495) | |
| Df(3R)ED5559 | 86E11–87B11 | 874834 | 16.437 (0.000) | 16.316 (0.000) | |
| Df(3R)ED5573 | 87B5–87B13 | 196465 | 12.413 (0.007) | 12.480 (0.016) | |
| Df(3R)ED5573 | 87B5–87B13 | 196465 | 12.413 (0.007) | 12.480 (0.016) | |
| Df(3R)ED5577 | 86F9–87B13 | 648837 | 15.059 (0.000) | 15.191 (0.000) | |
| Df(3R)ED5591 | 87B7–87C7 | 369479 | 14.148 (0.003) | 14.157 (0.012) | |
| Df(3R)ED5608 | 87C7–87D7 | 275690 | 14.633 (0.005) | 15.577 (0.003) | |
| Df(3R)ED5610 | 87B11–87D7 | 551659 | 15.055 (0.000) | 14.990 (0.000) | |
| Df(3R)ED5612 | 87C7–87F6 | 925149 | 14.381 (0.003) | 14.908 (0.001) | |
| Df(3R)ED5613 | 87E3–87F6 | 385385 | 14.031 (0.003) | 14.607 (0.001) | |
| Df(3R)ED5622 | 87F10–88A4 | 300090 | 14.410 (0.281) | 14.339 (0.030) | |
| Df(3R)ED5623 | 87E3–88A4 | 724163 | 15.169 (0.005) | 15.043 (0.020) | |
| Df(3R)ED5634 | 88A4–88B1 | 260040 | 15.023 (0.001) | 14.968 (0.001) | |
| Df(3R)ED5642 | 87F10–88C2 | 797952 | 14.295 (0.000) | 14.238 (0.001) | |
| Df(3R)ED5644 | 88A4–88C9 | 607806 | 13.849 (0.057) | 13.920 (0.024) | |
| Df(3R)ED5657 | 88D1–88D7 | 221350 | 15.628 (0.000) | 15.788 (0.000) | |
| Df(3R)ED5660 | 88D1–88E1 | 396848 | 15.593 (0.000) | 16.485 (0.000) | |
| Df(3R)ED5662 | 88D1–88E2 | 434545 | 16.315 (0.000) | 16.406 (0.000) | |
| Df(3R)ED5664 | 88D1–88E3 | 531540 | 16.487 (0.000) | 16.892 (0.000) | |
| Df(3R)ED5688 | 88E12–88F1 | 37068 | 12.838 (0.006) | 12.759 (0.018) | |
| Df(3R)ED5705 | 88E12–89A5 | 502138 | 14.695 (0.006) | 14.733 (0.035) | |
| Df(3R)ED5780 | 89E11–90C1 | 625324 | 13.339 (0.181) | 13.331 (0.437) | |
| Df(3R)ED5781 | 89E13–90C1 | 562695 | 12.786 (0.094) | 13.074 (0.377) | |
| Df(3R)ED5785 | 90C2–90D1 | 225960 | 13.234 (0.068) | 13.199 (0.377) | |
| Df(3R)ED5807 | 90C2–91A5 | 681121 | 13.654 (0.405) | 13.500 (0.875) | |
| Df(3R)ED5815 | 90F4–91B8 | 491112 | 16.000 (0.030) | — | |
| Df(3R)ED5911 | 91C5–91F8 | 422856 | 14.796 (0.000) | 15.143 (0.001) | |
| Df(3R)ED5938 | 91D4–92A11 | 735402 | 15.433 (0.004) | 15.173 (0.001) | |
| Df(3R)ED6025 | 92A11–92E2 | 666791 | 16.041 (0.000) | 16.167 (0.000) | |
| Df(3R)ED6027 | 92B3–92E2 | 472646 | 13.051 (0.015) | 13.011 (0.027) | |
| Df(3R)ED6052 | 93D4–93D8 | 68869 | 12.421 (0.002) | 12.520 (0.014) | |
| Df(3R)ED6058 | 93D4–93F6 | 423105 | 13.936 (0.072) | 14.315 (0.005) | |
| Df(3R)ED6076 | 93E10–94A1 | 409323 | 15.137 (0.000) | 14.307 (0.002) | |
| Df(3R)ED6079 | 94A1–94A2 | 91507 | 12.831 (0.000) | 12.724 (0.003) | |
| Df(3R)ED6085 | 93F14–94B5 | 706744 | 12.909 (0.220) | 13.009 (0.262) | |
| Df(3R)ED6090 | 94A1–94C1 | 656195 | 15.311 (0.002) | 14.784 (0.003) | |
| Df(3R)ED6091 | 94B5–94C4 | 138626 | 12.950 (0.024) | 12.879 (0.057) | |
| Df(3R)ED6093 | 94A2–94C4 | 592519 | 15.693 (0.000) | 15.445 (0.000) | |
| Df(3R)ED6096 | 94B5–94E7 | 634288 | 15.238 (0.000) | 15.042 (0.002) | |
| Df(3R)ED6103 | 94D3–94E9 | 359862 | 14.967 (0.000) | 14.465 (0.006) | |
| Df(3R)ED6105 | 94E9–94E11 | 37549 | 15.042 (0.000) | 14.990 (0.000) | |
| Df(3R)ED6116 | 95B4–95C1 | 46434 | 12.850 (0.004) | 13.036 (0.152) | |
| Df(3R)ED6119 | 95C8–95C12 | 35707 | 15.143 (0.003) | 15.516 (0.002) | |
| Df(3R)ED6144 | 95C8–95D1 | 55833 | 12.513 (0.003) | 12.644 (0.024) | |
| Df(3R)ED6150 | 95D1–95D11 | 114310 | 12.696 (0.005) | 12.839 (0.054) | |
| Df(3R)ED6155 | 95B4–95D11 | 283236 | 14.231 (0.010) | 14.395 (0.002) | |
| Df(3R)ED6168 | 95D1–95F8 | 328141 | 12.396 (0.000) | 12.499 (0.012) | |
| Df(3R)ED6187 | 95D10–96A7 | 492295 | 15.640 (0.004) | 15.140 (0.001) | |
| Df(3R)ED6220 | 96A7–96C3 | 639975 | 14.415 (0.000) | 14.521 (0.001) | |
| Df(3R)ED6232 | 96F10–97D2 | 762106 | 16.057 (0.000) | 16.125 (0.000) | |
| Df(3R)ED6235 | 97B9–97D12 | 445273 | 13.148 (0.092) | 13.285 (0.299) | |
| Df(3R)ED6242 | 97E4–97E11 | 123525 | 14.735 (0.000) | 14.669 (0.001) | |
| Df(3R)ED6255 | 97D2–97F1 | 482865 | 15.119 (0.001) | 14.706 (0.000) | |
| Df(3R)ED6265 | 97E2–98A7 | 467511 | 12.530 (0.000) | 12.568 (0.014) | |
| Df(3R)ED6277 | 98B6–98B6 | 10924 | 12.711 (0.006) | 12.918 (0.179) | |
| Df(3R)ED6290 | 98C3–98E5 | 485726 | 14.406 (0.000) | 14.603 (0.001) | |
| Df(3R)ED6310 | 98F12–99B2 | 373258 | 14.381 (0.000) | 14.540 (0.001) | |
| Df(3R)ED6315 | 99B10–99C1 | 17077 | 15.210 (0.000) | 14.863 (0.003) | |
| Df(3R)ED6316 | 99A5–99C1 | 527344 | 15.444 (0.000) | 15.342 (0.000) | |
| Df(3R)ED6332 | 99E4–99F2 | 111366 | 15.221 (0.000) | 14.913 (0.000) | |
| Df(3R)ED6346 | 100A5–100B1 | 265322 | 18.383 (0.000) | 18.167 (0.000) | |
| Df(3R)ED6361 | 100C7–100E3 | 469313 | 15.383 (0.000) | 14.575 (0.001) | |
| Df(3R)ED6362 | 100E1–100E3 | 141893 | 14.821 (0.000) | 14.861 (0.001) | |
| Df(3R)ED7665 | 84B4–84E11 | 1003556 | 14.300 (0.161) | 14.200 (0.337) | |
| Df(3R)ED10257 | 83A7–83B4 | 81922 | 15.026 (0.000) | 15.102 (0.002) | |
| Df(3R)ED10549 | 88D6–88D7 | 17163 | 15.191 (0.000) | 15.051 (0.000) | |
| Df(3R)ED10555 | 88C9–88D8 | 361038 | 14.177 (0.000) | 14.170 (0.030) | |
| Df(3R)ED10556 | 88D6–88E1 | 192661 | 15.288 (0.000) | 15.723 (0.001) | |
| Df(3R)ED10557 | 88D6–88E2 | 230358 | 15.780 (0.001) | 15.707 (0.000) | |
| Df(3R)ED10561 | 88E2–88E2 | 25799 | 14.868 (0.000) | 14.829 (0.000) | |
| Df(3R)ED10564 | 88D6–88E3 | 327353 | 14.923 (0.001) | 15.267 (0.000) | |
| Df(3R)ED10566 | 88E2–88E5 | 118112 | 14.942 (0.000) | 15.751 (0.000) | |
| Df(3R)ED10639 | 89B7–89D2 | 268307 | 15.219 (0.000) | 14.799 (0.007) | |
| Df(3R)ED10642 | 89C7–89D5 | 171514 | 15.051 (0.000) | 14.778 (0.002) | |
| Df(3R)ED10811 | 93A4–93B8 | 111808 | 15.364 (0.001) | 14.993 (0.000) | |
| Df(3R)ED10820 | 93A4–93B12 | 162720 | 14.892 (0.001) | 14.455 (0.043) | |
| Df(3R)ED10838 | 93C1–93D4 | 162185 | 15.819 (0.000) | 15.639 (0.006) | |
| Df(3R)ED10893 | 95C8–95E1 | 217754 | 14.634 (0.000) | 14.210 (0.049) | |
| Df(3R)ED10894 | 95A7–95E1 | 435407 | 15.191 (0.000) | 15.194 (0.000) | |
| Df(3R)ED10946 | 96B20–96D1 | 221386 | 15.604 (0.000) | 15.305 (0.000) | |
| Df(3R)ED10953 | 96C6–96D1 | 70912 | 16.225 (0.000) | 16.207 (0.000) | |
| Df(3R)ED10961 | 97E11–97F1 | 19770 | 14.822 (0.000) | 15.015 (0.001) | |
| Df(3R)ED10966 | 97E11–97F1 | 28417 | 14.737 (0.000) | 14.581 (0.002) | |
| Df(3R)ED10970 | 97E11–98B5 | 652492 | 14.692 (0.008) | 14.530 (0.001) | |
| Df(3R)ED10993 | 99B10–99C2 | 41267 | 15.588 (0.000) | 15.175 (0.000) | |
| Df(3R)ED13102 | 99B1–99B10 | 279997 | 15.610 (0.000) | 15.324 (0.000) | |
| X | Df(1)ED404 | 1D2–1E3 | 200503 | 13.672 (0.321) | — |
| Df(1)ED409 | 2C7–2F5 | 275404 | 13.634 (0.869) | — | |
| Df(1)ED411 | 3A3–3A8 | 172827 | 13.592 (0.894) | — | |
| Df(1)ED418 | 5C7–5E4 | 377712 | 13.837 (0.044) | — | |
| Df(1)ED429 | 9D3–9D3 | 38567 | 14.373 (0.001) | 14.302 (0.005) | |
| Df(1)ED447 | 17C1–17F1 | 356796 | 14.914 (0.000) | — | |
| Df(1)ED6396 | 1B5–1B8 | 30101 | 14.692 (0.000) | — | |
| Df(1)ED6443 | 1B14–1E1 | 370684 | 14.746 (0.000) | — | |
| Df(1)ED6574 | 2E1–3A2 | 203136 | 13.271 (0.597) | — | |
| Df(1)ED6579 | 3A6–3A8 | 53476 | 15.794 (0.000) | — | |
| Df(1)ED6584 | 3A8–3B1 | 49222 | 12.961 (0.008) | — | |
| Df(1)ED6630 | 3B1–3C5 | 351370 | 14.868 (0.000) | — | |
| Df(1)ED6712 | 3D3–3F1 | 357080 | 14.100 (0.078) | — | |
| Df(1)ED6727 | 4B6–4D5 | 585887 | 14.266 (0.011) | — | |
| Df(1)ED6802 | 5A12–5D1 | 285900 | 13.346 (0.265) | — | |
| Df(1)ED6829 | 5C7–5F3 | 451119 | 13.048 (0.117) | — | |
| Df(1)ED6849 | 5F3–6D3 | 452200 | 13.862 (0.080) | — | |
| Df(1)ED6878 | 6C12–6D8 | 103655 | 13.514 (0.964) | — | |
| Df(1)ED6906 | 7A3–7B2 | 210722 | 13.191 (0.181) | — | |
| Df(1)ED6940 | 36A10–36B1 | 297221 | 12.454 (0.001) | — | |
| Df(1)ED6957 | 8B6–8C13 | 243242 | 13.179 (0.021) | — | |
| Df(1)ED6989 | 8F9–9B1 | 383820 | 13.428 (0.694) | — | |
| Df(1)ED6991 | 8F9–9B4 | 524871 | 12.155 (0.002) | — | |
| Df(1)ED7005 | 9B1–9D3 | 513509 | 15.667 (0.007) | — | |
| Df(1)ED7010 | 9D3–9D4 | 82437 | 14.383 (0.007) | — | |
| Df(1)ED7067 | 10B8–10C10 | 210959 | 13.265 (0.048) | — | |
| Df(1)ED7147 | 10D7–11A1 | 290417 | 13.373 (0.610) | — | |
| Df(1)ED7153 | 11A1–11B1 | 560373 | 13.976 (0.025) | — | |
| Df(1)ED7161 | 11A1–11B14 | 743779 | 14.781 (0.010) | — | |
| Df(1)ED7165 | 11B15–11E1 | 386346 | 14.032 (0.014) | — | |
| Df(1)ED7170 | 11B15–11E8 | 524724 | 13.839 (0.101) | — | |
| Df(1)ED7173 | 11B15–11F1 | 621133 | 14.467 (0.002) | — | |
| Df(1)ED7217 | 12A9–12C6 | 180238 | 13.205 (0.077) | — | |
| Df(1)ED7229 | 12E5–12F2 | 431710 | 12.961 (0.000) | — | |
| Df(1)ED7261 | 12F2–12F5 | 185603 | 13.074 (0.376) | — | |
| Df(1)ED7265 | 12F4–13A5 | 181838 | 13.986 (0.144) | — | |
| Df(1)ED7289 | 13A5–13A12 | 100973 | 13.941 (0.044) | — | |
| Df(1)ED7294 | 13B1–13C3 | 274883 | 13.860 (0.296) | — | |
| Df(1)ED7331 | 13C3–13F1 | 363268 | 12.469 (0.044) | — | |
| Df(1)ED7344 | 13E1–13F17 | 241694 | 14.022 (0.022) | — | |
| Df(1)ED7355 | 14A8–14B7 | 186930 | 13.662 (0.583) | 13.600 (0.679) | |
| Df(1)ED7374 | 15A1–15E3 | 412445 | 12.860 (0.071) | — | |
| Df(1)ED7413 | 17D1–17F1 | 206484 | 13.793 (0.269) | — | |
| Df(1)ED7441 | 18A3–18C2 | 168474 | 13.211 (0.119) | — | |
| Df(1)ED7635 | 19A2–19C1 | 278714 | 13.568 (0.656) | 14.125 (0.045) | |
| Df(1)ED7664 | 19F1–19F6 | 250376 | 13.396 (0.576) | — | |
| Df(1)ED11354 | 61B1–61C1 | 191859 | 12.944 (0.094) | — | |
| Df(1)ED11437 | 2F6–3A4 | 518880 | 13.137 (0.198) | — | |
| Df(1)ED12405 | 19C4–19E5 | 594760 | 13.802 (0.224) | — | |
| Df(1)ED12425 | 19E7–19F3 | 216238 | 13.645 (0.473) | — | |
| Df(1)ED12432 | 20C1–20C1 | 97858 | 13.842 (0.287) | 13.716 (0.366) | |
| Df(1)ED13157 | 18F4–19C1 | 288549 | 13.747 (0.114) | 14.750 (0.005) | |
| Df(1)ED13478 | 16F6–16F7 | 16605 | 13.712 (0.219) | — | |
| Df(1)ED14021 | 20C1–20E1 | 320915 | 13.465 (0.803) | — | |
The authors declare no conflict of interest.
References
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Chiu JC, Ko HW, Edery I. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell. 2011;145:357–370. doi: 10.1016/j.cell.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda T, Mirowska K, Currie J, Kim MH, Rao NV, Bonilla G, et al. Adult circadian behavior in Drosophila requires developmental expression of cycle, but not period. PLoS Genet. 2011;7:e1002167. doi: 10.1371/journal.pgen.1002167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Harano T, Miyatake T. Independence of genetic variation between circadian rhythm and development time in the seed beetle, Callosobruchus chinensis. J Insect Physiol. 2011;57:415–420. doi: 10.1016/j.jinsphys.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Kumar S, Kumar D, Paranjpe DA, R AC, Sharma VK. Selection on the timing of adult emergence results in altered circadian clocks in fruit flies Drosophila melanogaster. J Exp Biol. 2007;210:906–918. doi: 10.1242/jeb.001354. [DOI] [PubMed] [Google Scholar]
- Kyriacou CP, Oldroyd M, Wood J, Sharp M, Hill M. Clock mutations alter developmental timing in Drosophila. Heredity. 1990;64:395–401. doi: 10.1038/hdy.1990.50. [DOI] [PubMed] [Google Scholar]
- Lewontin RC.1965Selection for colonizing abilityIn: Baker HG, Stebbins GL, (eds)The Genetics of Colonizing Species Academic Press New York; 79–94. [Google Scholar]
- Mazzoni EO, Desplan C, Blau J. Circadian pacemaker neurons transmit and modulate visual information to control a rapid behavioral response. Neuron. 2005;45:293–300. doi: 10.1016/j.neuron.2004.12.038. [DOI] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- Mitsui H, Takahashi KH, Kimura MT. Spatial distributions and clutch sizes of Drosophila species ovipositing on cherry fruits of different stages. Popul Ecol. 2006;48:233–237. [Google Scholar]
- Miyatake T. Two-way artificial selection for developmental period in Bactrocera cucurbitae (Diptera: Tephritidae) Ann Entomol Soc Am. 1995;88:848–855. [Google Scholar]
- Miyatake T. Pleiotropic effect, clock genes, and reproductive isolation. Popul Ecol. 2002;44:201–207. [Google Scholar]
- Miyatake T, Matsumoto A, Matsuyama T, Ueda HR, Toyosato T, Tanimura T. The period gene and allochronic reproductive isolation in Bactrocera cucurbitae. Proc R Soc Lond, Ser B: Biol Sci. 2002;269:2467–2472. doi: 10.1098/rspb.2002.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylin S. Host plant specialization and seasonality in a polyphagous butterfly, Polygonia C-Album (Nymphalidae) Oikos. 1988;53:381–386. [Google Scholar]
- Nylin S. Seasonal plasticity in life history traits: growth and development in Polygonia c-album (Lepidoptera: Nymphalidae) Biol J Linn Soc. 1992;47:301–323. [Google Scholar]
- Nylin S, Gotthard K. Plasticity in life-history traits. Annu Rev Entomol. 1998;43:63–83. doi: 10.1146/annurev.ento.43.1.63. [DOI] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Paranjpe DA, Anitha D, Chandrashekaran MK, Joshi A, Sharma VK. Possible role of eclosion rhythm in mediating the effects of light-dark environments on pre-adult development in Drosophila melanogaster. BMC Dev Biol. 2005;5:5. doi: 10.1186/1471-213X-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjpe DA, Anitha D, Sharma VK, Joshi A. Circadian clocks and life-history related traits: is pupation height affected by circadian organization in Drosophila melanogaster. J Genet. 2004;83:73–77. doi: 10.1007/BF02715831. [DOI] [PubMed] [Google Scholar]
- Ryder E, Ashburner M, Bautista-Llacer R, Drummond J, Webster J, Johnson G, et al. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics. 2007;177:615–629. doi: 10.1534/genetics.107.076216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E, Fiona B, Michael A, Rosa B-L, Darin C, Jenny D, et al. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004;167:797–813. doi: 10.1534/genetics.104.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Miyatake T, Watari Y, Arai T. A gene pleiotropically controlling developmental and circadian periods in the melon fly, Bactrocera cucurbitae (Diptera:Tephritidae) Heredity. 1997;79:600–605. [Google Scholar]
- Takahashi KH, Okada Y, Teramura K. Genome-wide deficiency mapping of the regions responsible for temporal canalization of the developmental processes of Drosophila melanogaster. J Hered. 2011a;102:448–457. doi: 10.1093/jhered/esr026. [DOI] [PubMed] [Google Scholar]
- Takahashi KH, Okada Y, Teramura K, Tsujino M. Deficiency mapping of the genomic regions associated with effects on developmental stability in Drosophila melanogaster. Evolution. 2011b;65:3565–3577. doi: 10.1111/j.1558-5646.2011.01400.x. [DOI] [PubMed] [Google Scholar]
- Takahashi KH, Tuno N, Kagaya T. The relative importance of spatial aggregation and resource partitioning on the coexistence of mycophagous insects. Oikos. 2005;109:125–134. [Google Scholar]
- Tsujino M, Takahashi KH. Natural genetic variation in fluctuating asymmetry of wing shape in Drosophila melanogaster. Ecol Res. 2012;27:133–143. [Google Scholar]
- Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- Wertheim B, Kraaijeveld AR, Schuster E, Blanc E, Hopkins M, Pletcher SD, et al. Genome-wide gene expression in response to parasitoid attack in Drosophila. Genome Biol. 2005;6:R94. doi: 10.1186/gb-2005-6-11-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaan B, Bijlsma R, Hoekstra RF. Artificial selection for developmental time in Drosophila melanogaster in relation to the evolution of aging: direct and correlated responses. Evolution. 1995;49:635–648. doi: 10.1111/j.1558-5646.1995.tb02300.x. [DOI] [PubMed] [Google Scholar]