Abstract
Hybrid zones provide excellent opportunities to study processes and mechanisms underlying reproductive isolation and speciation. Here we investigated sex-specific clines of molecular markers in hybrid zones of morphologically cryptic yet genetically highly-diverged evolutionary lineages of the European common vole (Microtus arvalis). We analyzed the position and width of four secondary contact zones along three independent transects in the region of the Alps using maternally (mitochondrial DNA) and paternally (Y-chromosome) inherited genetic markers. Given male-biased dispersal in the common vole, a selectively neutral secondary contact would show broader paternal marker clines than maternal ones. In a selective case, for example, involving a form of Haldane's rule, Y-chromosomal clines would not be expected to be broader than maternal markers because they are transmitted by the heterogametic sex and thus gene flow would be restricted. Consistent with the selective case, paternal clines were significantly narrower or at most equal in width to maternal clines in all contact zones. In addition, analyses using maximum likelihood cline-fitting detected a shift of paternal relative to maternal clines in three of four contact zones. These patterns suggest that processes at the contact zones in the common vole are not selectively neutral, and that partial reproductive isolation is already established between these evolutionary lineages. We conclude that hybrid zone movement, sexual selection and/or genetic incompatibilities are likely associated with an unusual unidirectional manifestation of Haldane's rule in this common European mammal.
Keywords: cline analyses, Microtus arvalis, secondary contact zone, Haldane's rule, Y-chromosome, speciation
Introduction
Speciation involves the establishment of reproductive isolation between organisms. Reproductive isolation may be achieved through various mechanisms acting singly or together at the pre- and/or post-zygotic stage, such as assortative mating or genetic incompatibilities (Turelli and Begun, 1997; Coyne and Orr, 2004). Incomplete reproductive isolation is often characterized by sex-specific processes, and in particular intrinsic genetic incompatibilities often more strongly affect hybrids of the heterogametic sex, a pattern known as Haldane's rule (Haldane, 1922; Schilthuizen et al., 2011). One expectation from Haldane's rule is that the extent of gene flow between incipient species may strongly differ if a gene is paternally or maternally transmitted (Turelli and Begun, 1997; Coyne and Orr, 2004; Macholan et al., 2007).
Hybrid zones offer the possibility to study the extent and potential sex-specificity of gene flow between incipient species in their natural environment. Here the consistency of introgression of genetic markers may provide valuable information on the degree of reproductive isolation between organisms (Barton and Hewitt, 1989; Raufaste et al., 2005; Schilthuizen et al., 2011). For example, in a hybrid zone with a high degree of reproductive isolation, Haldane's rule might lead to very low gene flow for male-specific Y-chromosomal markers in mammals, whereas no impediments for gene flow might exist for maternally transmitted mitochondrial markers (for example, Beysard et al., 2012). Despite the recognition of the crucial information carried by the Y-chromosome in mammals (Petit et al., 2002), its investigation in hybrid zones of non-model organisms remains restricted to very few systems (for example, Jaarola et al., 1997; Hellborg et al., 2005; Yannic et al., 2008), most probably because of technical limitations.
The rich technical and genomic resources available for the laboratory mouse have eased the development of the secondary contact zone between the house mouse subspecies Mus musculus musculus and Mus musculus domesticus, running through Europe to a potential model system for speciation research in mammals. The exploration of this contact zone has shown, for example, that the subspecies may mate assortatively (Ganem et al., 2008), hybrids may differ from parentals in parasite load (see Baird et al., 2012) or male hybrids may be sterile (reviewed in Britton-Davidian et al, 2005; and also Mihola et al., 2009). The observation of male sterility combined with the detection of abrupt Y- and X-chromosome marker clines (Dod et al., 1993; Macholan et al., 2007, 2008, 2011) relative to other markers across the hybrid zone are consistent with the hypothesis that sex chromosomes and in particular factors on the X-chromosome have an important role in causing genetic incompatibilities in mouse hybrids (Coyne and Orr, 2004). Nevertheless, there is extensive geographical and individual variation in male sterility (Britton-Davidian et al., 2005; Good et al., 2008; Turner et al., 2012), and sex-chromosomal marker clines may be not coincident with other genetic markers in particular geographical regions, potentially due to asymmetrical processes (Macholan et al., 2008; Jones et al., 2010). For example, the Y-chromosome was found non-coincident with other markers in an area of about 330 km2 in the Czech–Bavarian part of the mouse hybrid zone (Macholan et al., 2008). This may be explained by two non-mutually exclusive scenarios: first, the hybrid zone may have moved and left the M. m. musculus Y-chromosome in the M. m. domesticus territory or second, the M. m. musculus Y-chromosome might have escaped from a static hybrid zone centre comprising autosomal and X-chromosomal markers and advanced up to 22 km into the M. m. domesticus territory (Macholan et al., 2008). It was proposed that a genetic conflict between the sexes may act together with a distortion of the sex-ratio to promote unidirectional sex-specific gene flow between the subspecies (Macholan et al., 2008). The variability in marker cline patterns stresses the importance of replicated analyses for hybrid zone analyses. In the present study, we collected information on multiple hybrid zones to assess the properties of sex-specific marker clines in a speciation-prone European small mammal.
Voles in the genus Microtus have been undergoing the fastest radiation known in mammals and diversified into at least 65 species across the world within <2 million years (Jaarola et al., 2004; Fink et al., 2010). Despite very little morphological differentiation, karyotypic variation and differentiation into deep genetic lineages is relatively common within recognized species, suggesting the presence of cryptic species or ongoing speciation processes (Jaarola et al., 2004; Heckel et al., 2005; Hellborg et al., 2005; Bastos-Silveira et al., 2012). In the common vole (Microtus arvalis), intraspecific divergence has resulted in four main, phenotypically cryptic evolutionary lineages in Europe (Western, Central, Italian and Eastern lineages; Heckel et al., 2005). Divergence between these lineages based on population samples of mitochondrial DNA (mtDNA) sequences and autosomal information was estimated to have coincided with or predated the last glacial maximum depending on the lineage, resulting, for example, in more than 50 000 generations of divergence between the Western and Central lineages (Heckel et al., 2005). An initial analysis of phylogeographic patterns in the region of secondary contact between lineages in the Alps demonstrated clear-cut parapatry of mtDNA lineages (Braaker and Heckel, 2009). Multi-locus nuclear microsatellite markers allow the allocation of individuals to genetic clusters whose distributions are highly consistent with mtDNA lineages—except for first evidence of cytonuclear discordance in a few individuals close to the potential contact zones (Braaker and Heckel, 2009). The detected patterns are consistent with male-biased dispersal in the species (Schweizer et al., 2007; Hahne et al., 2011) and relatively complex processes of genetic erosion after colonization of the region (Braaker and Heckel, 2009). However, they may also be explained by an ongoing speciation process and the effect of sex-specific gene flow associated with yet undocumented partial reproductive isolation between the highly divergent evolutionary lineages in M. arvalis. With male-biased dispersal in the species, a selectively neutral secondary contact would show broader paternal marker clines than maternal ones. In a case involving selection, for example, a form of Haldane's rule, paternal marker clines would not be expected to be broader than maternal ones because they are transmitted by the heterogametic sex, and thus gene flow would be expected to be restricted.
Here we fine-map the geographic location of the secondary contact zones between all three evolutionary lineages (Western, Central, Italian) of M. arvalis, occurring in the region of the central Alps. We performed cline analyses of Y-chromosomal and mtDNA markers across these contact zones to quantify the relative contribution of sex-specific processes and the importance of non-neutral evolution linked to potential incipient speciation in the probably most abundant wild European mammal.
Materials and methods
Sampling of transects
We investigated the different secondary contact zones in the region of the Alps between three evolutionary lineages of the common vole with a total number of 706 voles in transects of 50–160 km length consisting of 23–40 localities each (Figure 1, Table 2, Supplementary Information Table 1). The first transect (Jura) was sampled perpendicular to the Jura mountain ridge, where the Western and the Central lineages meet. The second transect (Grisons) was sampled along the Rhine valley, a contact zone of the Central and the Italian lineages. The third transect (Valais) comprises transitions from the Central to the Western and then to the Italian lineage along the Rhone valley. Samples were obtained as described in Braaker and Heckel (2009).
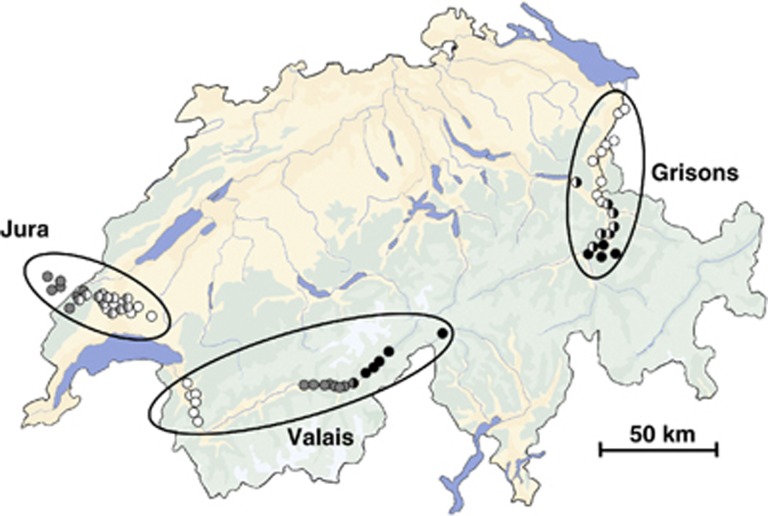
Figure 1.
Map of Switzerland showing the sampling localities of Microtus arvalis samples included in cline analyses of the three transects. Transects are described in Table 1. Localities are designated according to the cytb lineage they harbour (white=Central, grey=Western, black=Italian). The Valais transect along the Rhone valley contains a double transition from the Central through the Western to the Italian lineage. Mixed populations (Figure 2) are indicated by bi-coloured circles. Localities closer than two kilometres to each other were merged for this figure.
Table 1. Haplotypes and polymorphic positions in the Y-chromosomal marker (intron SMCY 11; 730 bp) in the studied transects across contact zones in M. arvalis.
| Haplotype | N | 133 | 177 | 295 | 336 | 405 |
|---|---|---|---|---|---|---|
| Western Y1 | 42 | C | T | C | G | T |
| Italian Y | 51 | T | A | C | G | T |
| Central Y1 | 192 | C | T | T | — | A |
The position of the substitutions and insertion/deletion are indicated in the first row. The number of individuals sequenced per haplotype is displayed under N.
Molecular markers, restriction fragment length polymorphism analysis and sequencing
We first determined a diagnostic Y-chromosome marker for the Italian, Western and Central lineages as follows: 730 bp of intron 11 of the SMCY gene were sequenced (see below for amplification and sequencing protocol) in 71 voles from across Europe, covering the range of the Western (Spain, France, and Belgium), Central (Switzerland, Germany and Austria) and Italian (Italy, Switzerland) lineages. Six haplotypes were found across Europe with a phylogeographic structure mirroring the parapatric distribution of mitochondrial lineages in M. arvalis (Heckel et al., 2005). These were termed Western Y (three haplotypes), Central Y (two haplotypes) and Italian Y (one haplotype) analogous to the mitochondrial lineages and genetic clusters in multi-locus nuclear markers (Heckel et al, 2005; Braaker and Heckel, 2009). Three of these lineage-diagnostic Y haplotypes defined by four substitutions and one insertion/deletion were found in the transects studied here (Tables 1 and 2).
Table 2. Summary of the three transects across contacts between evolutionary lineages in M. arvalis.
| Transect | Lineage transition | First/last locality | Length (km) | Localities | N (Cytb) | N (SMCY11) |
|---|---|---|---|---|---|---|
| Jura | Western–Central | Cerniébaud/Les Cullayes | 51 | 40 | 281 | 126 |
| (178 W–103 C) | (36 W–90 C) | |||||
| Grisons | Central–Italian | Diepoldsau/Jochalp | 71 | 26 | 287 | 104 |
| (133 C–154 It) | (90 C–14 It) | |||||
| Valais | Central–Western | Aigle/Villa | 159 | 23 | 138 | 55 |
| Western–Italian | (40 C—79 W—19 It) | (12 C—6 W—37 It) |
Positions of transects and sampling localities are shown in Figure 1. The Valais transect along the Rhone valley contains a double transition from the Central (C) through the Western (W) to the Italian (It) lineage. Sample sizes are given for mtDNA (cytb) and Y-chromosomal sequences.
SMCY11 fragments were amplified in a reaction volume of 25 μl using the primers SMCY11-f (5′-CTGCCCTGYRCCATGCAT-3′) and SMCY11-r (5′-TCCACCTGTTSMAGRACAT-3′) from Hellborg and Ellegren (2003) developed for M. agrestis. For PCR amplification, an initial step of denaturation at 95 °C for 2 min was followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 49 °C for 1 min and extension at 72 °C for 90 s. A final step at 72 °C for 10 min was added to complete primer extension. All PCR amplifications were performed in thermal cyclers PCR System 9700 (GeneAmp, Applied Biosystems, Rotkreuz, Switzerland) or PTC-100TM (MJ Research, Watertown, MA, USA). For sequencing, PCR products were cleaned with a GenElute PCR clean-up kit (Sigma-Aldrich, St Louis, MO, USA) and dissolved in 40 μl bi-distilled water. Cycle sequencing reactions were carried out using the Terminator Ready Reaction Mix ‘Big Dye' 3.1 from Applied Biosystems and the PCR primers. The reactions (10 μl) were performed with the following conditions: denaturation at 95 °C for 50 s, 30 cycles of 96 °C for 10 s, 57 °C for 10 s and 60 °C for 4.5 min. PCR products were cleaned by precipitation according to the Applied Biosystems manual, and separated and detected on an Applied Biosystems Prism 3100 Genetic Analyser.
The mitochondrial Cytochrome b (cytb) gene was amplified following Fink et al. (2004; 2006). The allocation of cytb amplification products to one of the three lineages present in the study region was performed by enzyme digestion with AFLIII and BsrI and restriction fragment length polymorphism analysis. Samples from the Jura transect were analyzed with AFLIII, which cuts the Central, Italian and Eastern lineage cytb but not the Western lineage. Samples from the Grisons transect were analyzed with BsrI, an enzyme cutting cytb fragments of the Western, Central and Eastern lineage twice, whereas Italian samples are only cut once. The samples from the Valais transect were analyzed subsequently with both enzymes to enable allocation to one of the three lineages present. As a control, 20–40 samples per transect were additionally sequenced (Fink et al., 2007; Braaker and Heckel, 2009) to verify lineage allocation by restriction fragment length polymorphism. No disagreements were found. For completeness, published cytb data of individuals from the study region (Fink et al., 2004; Heckel et al., 2005; Braaker and Heckel 2009) were integrated into our analyses. Sequences were aligned using the Clustal W algorithm (Thompson et al., 1997) implemented in BioEdit 7.0.5 (Hall, 1999) and revised manually.
Cline analyses
Contact zones can be described by a sigmoid cline in allele frequencies (p) with a function of cline centre (c) and width (w; defined as the inverse of the maximum slope) following
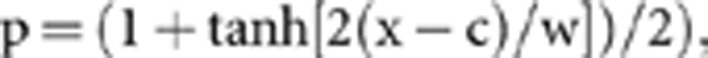 |
with x equal to the position of the population along the cline (Szymura and Barton, 1986). Cline-fitting analysis was performed by reducing the location data to one-dimensional transects. The frequency of a lineage in a population was plotted against distance (km) along the transects from the north-westernmost (Jura), the northernmost (Grisons) or the westernmost (Valais) locality. Tanh curves were then fitted for the cytb and SMCY11 data separately using the ‘Fit 1D cline' operation in the programme ANALYSE version 1.3 (Barton and Baird, 2002). Model parameters (centre and width) were computed using 2000 iterations from 20 different starting points. Likelihood profiles and two log-likelihood support limits (corresponding to 95% confidence intervals; Edwards, 1972) were explored using the ‘cross-section' option. Cline coincidence (centres at the same position) and concordance (equal cline widths) were explored with a likelihood ratio test (LRT) following the procedure used by Phillips et al. (2004) and adapted by Leache and Cole (2007).
Results
Lineage assignment and marker mismatches
Dedicated sampling along transects allowed the precise localization of the intraspecific contact zones in M. arvalis and a general refinement of the distribution of mtDNA lineages compared with a previous study (Braaker and Heckel, 2009). The distribution of Y-chromosomal lineages was in general agreement with the mtDNA contact zones but with highly significant differences in each transect (Figure 2; Table 3).
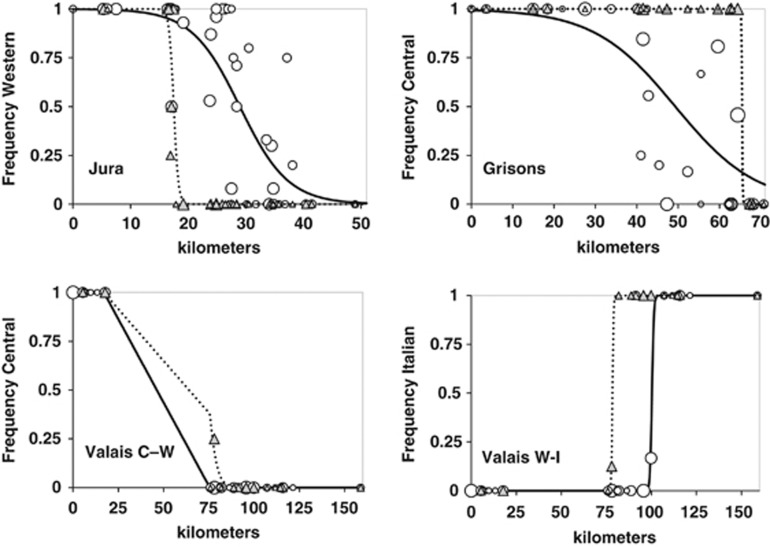
Figure 2.
Plots of lineage frequencies in populations versus position (km) along the transects Jura, Grisons and Valais for mtDNA (white circles) and Y-chromosomal (grey triangles) markers. Sizes of symbols are proportional to the log number of samples. Bold lines are fitted clines for mtDNA and dashed lines for Y-chromosomal markers. Despite a sampling gap in the Valais transect, an admixed population was detected at km 78, pointing to at least partial contact between the Western and the Central lineage in this transect. See Table 2 for cline properties and statistical comparisons.
Table 3. Contact zone cline parameters and LRT results of the null hypothesis that cline centres and widths do not differ significantly between the maternal and parental marker for a given transect.
| Parameter | cytb | SMCY11 | ∑lnLu | ∑lnLc | Δ=∑lnLc-∑lnLu | X 2=2 Δ |
|---|---|---|---|---|---|---|
| Jura | ||||||
| Cline centre | 28.98 (27.68–30.38) | 17.5 (17.15–18.05) | −63.7 | −128.399 | 64.699 | *129.398 |
| Cline width | 16.81 (13.31–21.91) | 1.59 (0.89–3.34) | −63.7 | −80.871 | 17.171 | *34.342 |
| lnLu | −53.973 | −9.727 | ||||
| Grisons | ||||||
| Cline centre | 49.32 (46.12–52.22) | 65.47 (64.47–67.37) | −76.6 | −111.595 | 34.995 | *69.990 |
| Cline width | 39.47 (31.00–50.97) | 0.25 (0.0–2.0) | −76.6 | −104.987 | 28.387 | *56.774 |
| lnLu | −76.6 | −0.000 | ||||
| Valais (C–W) | ||||||
| Cline centre | 28.4–70 (17.7–75.7) | 74.23 (45.8–77.9) | −0.701 | −0.702 | 0.001 | 0.002 |
| Cline width | 2.1–10.8 (0.01–37.7) | 10.19 (2.2–56.6) | −0.701 | −0.701 | 0.000 | 0.000 |
| lnLu | −0.000 | −0.701 | ||||
| Valais (W–I) | ||||||
| Cline centre | 100.3 (99.9–105.2) | 78.4 (78.1–81.6) | −0.001 | −26.446 | 26.445 | *52.890 |
| Cline width | 0.1–2 (0.01–10.9) | 0.1–1.1 (0.01–9.2) | −0.001 | −0.001 | 0.000 | 0.000 |
| lnLu | −0.001 | −0.000 | ||||
Maximum likelihood estimates of the positions of cline centres and their widths are given in kilometres and confidence intervals (two log-likelihood units≈95% confidence interval) are shown in parentheses. lnLu is the unconstrained log-likelihood support. lnLc is the maximum constrained likelihood in an analysis where cline centre or width was held constant at intervals while allowing the other parameter (width or centre) to vary freely. An asterisk indicates rejection of the null hypothesis at α=0.01 (critical χ2 value=6.635; one degree of freedom). Estimates for centres and widths in the Valais transect are given separately for the two different transition zones (Central–Western and Western–Italian).
In the Jura transect, 34 of 126 males showed a mismatch between the maternal and the paternal marker of which all but one had a Central Y-chromosome and Western mtDNA. In the Grisons transect, a mismatch between the maternal and paternal marker was detected in 41 of 104 males, all with Italian mtDNA and a Central Y-chromosome. In the Valais transect, our analyses showed a double transition between lineages along the Rhone valley for both markers. Despite extensive sampling attempts, no samples or evidence of the presence of M. arvalis could be obtained between kilometres 18 and 75 of the transect, separating the Central and Western mtDNA (Figure 1). Suitable habitats for the common vole are very scarce in this region due to intensive human usage. A mismatch between the maternal and paternal marker was detected in 28 out of 55 males sampled along the Valais transect. Two males had Western mtDNA and a Central Y-chromosome, whereas 26 had Western mtDNA and an Italian Y-chromosome. In the Valais transect, the Western Y-chromosome was found exclusively in combination with Western mtDNA.
Cline analyses
Cline shape analyses revealed significant cline centre shifts (non-coincidence) in all three transects and differences in cline widths (non-concordance) in two of the three transects (Figure 2; Table 3). In the Jura transect, the LRT revealed a significant north-westward shift of the Y-chromosomal cline relative to the mtDNA cline (χ12=129.398; for df=1 the critical value at P=0.01 is 6.635). Moreover, the mtDNA cline was significantly wider (χ12=34.342). The Grisons transect also displayed non-coincident (χ12=69.990) and non-concordant (χ12=56.774) clines. The Y-chromosomal cline was narrower than the mtDNA cline and the centre shifted to the South. In the Valais transect, analyses were performed separately for the two transition zones (Central–Western and Western–Italian). Centres and widths of the paternal and maternal cline of the Central–Western contact zone did not differ significantly in this part of the transect containing mostly unsuitable vole habitats. However, several males with a Central Y-chromosome were detected higher up in the Rhone valley at kilometre 78 in a population with only Western mtDNA, which was several dozens of kilometres away from the nearest known Central Y males along the transect (Figure 2). Coincidence of the Western–Italian contact zone was highly significantly rejected (χ12=52.890), whereas there was no evidence that the widths of the clines differed (χ12=0.000). Further analyses according to age classes of voles provided no evidence of differences in the position of centres or widths of clines between adults and juveniles in any transect (details not shown).
Discussion
Our analyses of sex-specific genetic markers across multiple contact zones revealed strong evidence for partial reproductive isolation between highly divergent evolutionary lineages within the common vole, M. arvalis. Cline analyses strongly suggest sex-specific processes acting in the contact zones consistent with the effect of a unidirectional manifestation of Haldane's rule.
Evidence for selection
Very narrow clines for the Y-chromosome in comparison to mtDNA suggest the action of selection on common vole males. The width of a cline across a hybrid zone is theoretically shaped by dispersal, which tends to widen it, and selection, which may narrow it (Barton and Hewitt, 1985). In M. arvalis, dispersal is male-biased like in most mammals (Hamilton et al., 2005; Schweizer et al., 2007; Hahne et al., 2011). Thus, without the action of selection, paternally inherited markers are expected to show wider clines across the contact zones than maternally inherited ones. At odds with this neutral expectation, Y-chromosome clines were significantly narrower or at most equal to mtDNA clines, suggesting selective forces acting on males. At present, the mechanisms leading to this pattern are unknown (see below), but it is compatible with Haldane's rule predicting negative fitness effects for the heterogametic sex due to recessive X-linked mutations or epistatic interactions between the Y-chromosome and another part of the genome (Haldane, 1922; Turelli and Orr, 2000; Coyne and Orr, 1998, 2004).
These patterns in M. arvalis are similar to other small mammal systems where speciation is ongoing or was recently completed, but where evolutionary divergence is much older and phenotypic differentiation between the taxa has been established. For example, in the European hybrid zone between M. m. musculus and M. m. domesticus (diverged >700 000 generations ago; She et al., 1990) the transition of sex chromosomes is similar in width to M. arvalis (for example, <4 km) and mtDNA clines are also wider (for example, Dod et al., 1993; Raufaste et al., 2005; Macholan et al., 2007, 2008). This is likely a consequence of Haldane's rule in place in the hybrid zone, which may—among other factors—prevent the mouse Y-chromosome cline width to increase (Britton-Davidian et al., 2005; but see Turner et al., 2012). The rapidly speciating Microtus genus harbours several other examples of restricted hybridization and evidence for selection acting prevalently on males—typically associated with morphological and karyotypic differences between the involved sibling species or subspecies (Meier et al., 1996; Jaarola et al., 1997; Bulatova et al., 2010; Bastos-Silveira et al., 2012; Beysard et al., 2012). Among the cryptic evolutionary lineages in M. arvalis however, partial reproductive isolation appears to be additionally acting in an unusual unidirectional manner.
Asymmetrical sex-specific processes
The second sex-specific process across the contact zones is the shift between maternal and paternal clines, leading to an asymmetrical pattern of mismatches. These mismatches may be the result of two main processes: introgression of mtDNA and/or introgression of the Y-chromosome. First, a range expansion of one lineage may have left behind a trail of introgressed mtDNA in the other lineage (Buggs, 2007 for a review). Simulation studies have shown that a replacement of one taxon by another may result in the introgression of selectively neutral local genes into the invading taxon, and that markers transmitted by the least dispersing sex would be most introgressed (Currat et al., 2008; Petit and Excoffier, 2009). In our case, this scenario would have resulted in asymmetrical introgression of mtDNA. However, this scenario would require a general fitness advantage of one lineage over the other, for which there is currently no support given the apparent absence of phenotypic differences between the lineages. mtDNA might also introgress owing to a selective advantage into other taxa (examples in Currat et al., 2008 and Petit and Excoffier, 2009), but there is no evidence of mtDNA introgression or of a selective advantage of particular mtDNA lineages (Fink et al., 2004; Hamilton et al., 2005; Braaker and Heckel, 2009; Fischer et al., 2011). In addition, very high diversity of mtDNA haplotypes in populations and lineages (Heckel et al., 2005; Braaker and Heckel, 2009) argues against a recent mtDNA sweep, but additional analyses together with the nuclear genome will be necessary for dedicated testing of mtDNA introgression in the hybrid zones.
The alternative main explanation for asymmetrical mismatches is the potential existence of asymmetric pre- or post-zygotic mechanisms, leading to an introgression of the Y-chromosome in the opposite direction. Pre-zygotic mechanisms might consist for example, in mating preferences of females from both lineages for a certain Y-chromosomal lineage or in specific male–male interactions (for example, aggressive behaviour; Roubertoux et al., 1994). A potential post-zygotic mechanism in M. arvalis would be a unidirectional manifestation of Haldane's rule with incompatibilities occurring only in backcrosses, involving hybrid males with one sex chromosome type but not the other. Mating and breeding experiments involving these M. arvalis lineages will allow to test for behavioural mechanisms or genetic incompatibilities acting as selective forces.
A possible unidirectional manifestation of Haldane's rule was suggested by Gavrilets (1997) based on theoretical work. Non-coincident marker clines similar to the ones in M. arvalis in a particular region of the European house mouse hybrid zone are compatible with such a process as well (Macholan et al., 2008). The M. m. musculus Y-chromosome and to a lesser extent X-chromosomal markers and mtDNA are here more introgressed into the M. m. domesticus territory than the rest of the genome (Macholan et al., 2008; 2011). The authors suggested that this unusual pattern resulted from genetic conflict between the sexes, which may promote unidirectional sex-specific gene flow between subspecies. To date, this pattern has only been detected in one of the studied transects and the general importance of stochastic vs deterministic effects in the house mouse hybrid zone deserves further investigation (Macholan et al., 2011; Turner et al., 2012). A potentially promising similar case was found in Scandinavia, where 15 mice from seven M. m. domesticus populations were introgressed with the M. m. musculus Y-chromosome (Jones et al., 2010). Our analyses, however, display a highly consistent pattern of very sharp Y-chromosomal clines shifted relative to mtDNA clines in several hybrid zones between the morphologically cryptic lineages of M. arvalis. At present, it is unclear if similar forces are acting in the different contact zones, but additional X-chromosomal and autosomal markers will allow a more detailed characterization of introgression patterns in the future. This may also reveal particular differences between hybrid zones, potentially depending on the evolutionary lineages involved and their level of divergence.
Conclusion
Our analyses of multiple hybrid zones provide evidence for sex-specific processes, which are compatible with strong selection acting at the contact between evolutionary lineages in the common vole. The mechanisms contributing to partial reproductive isolation and the extent of actual hybridization in natural populations remain currently unknown. However, more detailed population genetic analyses covering the nuclear genome widely combined with classical experimental crosses and mate-choice experiments, and the analysis of less-diverged lineages (for example, Eastern) have the potential to inform us about causes and consequences of asymmetric gene flow in hybrid zones of the probably most abundant wild European mammal.
Data archiving
The SMCY11 sequences as well as novel cytb haplotypes have been deposited in GenBank with accession numbers KC283238–KC283240 and KC349131- KC349136, respectively.
Acknowledgments
We are grateful to Laurent Excoffier and the University of Bern for ongoing support of the vole projects. We thank the editor and three reviewers for helpful suggestions, Irene Keller and Mathieu Foll for comments on the manuscript, Susanne Tellenbach for technical assistance, Joana Meier for unpublished data on Y-chromosomal variability, and Sonja Braaker, Martin Fischer and Birgit Thorenz for kindly providing access to specimens. This study was supported in part by grants from the Berne University Research Foundation and by the Swiss National Science Foundation number 31003A-127377 to GH.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Baird SJE, Ribas A, Macholán M, Albrecht T, Piálek J, Goüy de Bellocq J. Where are the wormy mice? A reexamination of hybrid parasitism in the European House mouse hybrid zone. Evolution. 2012;66:2757–2772. doi: 10.1111/j.1558-5646.2012.01633.x. [DOI] [PubMed] [Google Scholar]
- Barton NH, Baird SJE.2002. ANALYSE, Version 1.3: Freeware: available from http://helios.bto.edsac.uk/evolgen/Mac/Analyse .
- Barton NH, Hewitt GM. Analysis of hybrid zones. Annu Rev Ecol Syst. 1985;16:113–148. [Google Scholar]
- Barton NH, Hewitt GM. Adaptation, speciation and hybrid zones. Nature. 1989;341:497–503. doi: 10.1038/341497a0. [DOI] [PubMed] [Google Scholar]
- Bastos-Silveira C, Santos SM, Monarca R, Mathias ML, Heckel G. Deep mitochondrial introgression and hybridization between ecologically divergent vole species. Mol Ecol. 2012;21:5309–5323. doi: 10.1111/mec.12018. [DOI] [PubMed] [Google Scholar]
- Beysard M, Perrin N, Jaarola M, Heckel G, Vogel P. Asymmetric and differential gene introgression at a contact zone between two highly divergent lineages of field voles (Microtus agrestis) J Evol Biol. 2012;25:400–408. doi: 10.1111/j.1420-9101.2011.02432.x. [DOI] [PubMed] [Google Scholar]
- Braaker S, Heckel G. Transalpine colonisation and partial phylogeographic erosion by dispersal in the common vole (Microtus arvalis) Mol Ecol. 2009;18:2518–2531. doi: 10.1111/j.1365-294X.2009.04189.x. [DOI] [PubMed] [Google Scholar]
- Britton-Davidian J, Fel-Clair F, Lopez J, Alibert P, Boursot P. Postzygotic isolation between the two European subspecies of the house mouse: estimates from fertility patterns in wild and laboratory-bred hybrids. Biol J Linn Soc. 2005;84:379–393. [Google Scholar]
- Buggs RJA. Empirical study of hybrid zone movement. Heredity. 2007;99:301–312. doi: 10.1038/sj.hdy.6800997. [DOI] [PubMed] [Google Scholar]
- Bulatova NS, Potapov SG, Lavrenchenko LA. Genomic versus chromosomal polytypy in studies of mitochondrial and nuclear DNA markers in the Microtus arvalis group. Russ J Genet. 2010;46:586–594. [PubMed] [Google Scholar]
- Coyne JA, Orr HA. The evolutionary genetics of speciation. Phil Trans R Soc B Biol Sci. 1998;353:287–305. doi: 10.1098/rstb.1998.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sinauer Sunderland; 2004. [Google Scholar]
- Currat M, Ruedi M, Petit RJ, Excoffier L. The hidden side of invasions: massive introgression by local genes. Evolution. 2008;62:1908–1920. doi: 10.1111/j.1558-5646.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- Dod B, Jermiin LS, Boursot P, Chapman VH, Nielsen JT, Bonhomme F. Counterselection on sex-chromosomes in the Mus musculus European hybrid zone. J Evol Biol. 1993;6:529–546. [Google Scholar]
- Edwards AWF. Likelihood. Cambridge University Press Cambridge, UK; 1972. [Google Scholar]
- Fink S, Excoffier L, Heckel G. Mitochondrial gene diversity in the common vole Microtus arvalis shaped by historical divergence and local adaptations. Mol Ecol. 2004;13:3501–3514. doi: 10.1111/j.1365-294X.2004.02351.x. [DOI] [PubMed] [Google Scholar]
- Fink S, Excoffier L, Heckel G. Mammalian monogamy is not controlled by a single gene. Proc Natl Acad Sci USA. 2006;103:10956–10960. doi: 10.1073/pnas.0602380103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink S, Excoffier L, Heckel G. High variability and non-neutral evolution in the mammalian avpr1a gene. BMC Evol Biol. 2007;7:176. doi: 10.1186/1471-2148-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink S, Fischer MC, Excoffier L, Heckel G. Genomic scans support repetitive continental colonization events during the rapid radiation of voles (Rodentia: Microtus): the utility of AFLPs versus mitochondrial and nuclear sequence markers. Syst Biol. 2010;59:548–572. doi: 10.1093/sysbio/syq042. [DOI] [PubMed] [Google Scholar]
- Fischer MC, Foll M, Excoffier L, Heckel G. Enhanced AFLP genome scans detect local adaptation in high-altitude populations of a small rodent (Microtus arvalis) Mol Ecol. 2011;20:1450–1462. doi: 10.1111/j.1365-294X.2011.05015.x. [DOI] [PubMed] [Google Scholar]
- Gavrilets S. Hybrid zones with Dobzhansky-type epistatic selection. Evolution. 1997;51:1027–1035. doi: 10.1111/j.1558-5646.1997.tb03949.x. [DOI] [PubMed] [Google Scholar]
- Ganem G, Litel C, Lenormand T. Variation in mate preference across a house mouse hybrid zone. Heredity. 2008;100:594–601. doi: 10.1038/hdy.2008.20. [DOI] [PubMed] [Google Scholar]
- Good JM, Handel MA, Nachman MW. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution. 2008;62:50–65. doi: 10.1111/j.1558-5646.2007.00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne J, Jenkins T, Halle S, Heckel G. Establishment success and resulting fitness consequences for vole dispersers. Oikos. 2011;120:95–105. [Google Scholar]
- Haldane J. Sex ratio and unisexual sterility in hybrid animals. J Genet. 1922;12:101–109. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nuc Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hamilton G, Currat M, Ray N, Heckel G, Beaumont M, Excoffier L. Bayesian estimation of recent migration rates after a spatial expansion. Genetics. 2005;170:409–417. doi: 10.1534/genetics.104.034199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel G, Burri R, Fink S, Desmet JF, Excoffier L. Genetic structure and colonization processes in European populations of the common vole, Microtus arvalis. Evolution. 2005;59:2231–2242. [PubMed] [Google Scholar]
- Hellborg L, Ellegren H. Y chromosome conserved anchored tagged sequences (YCATS) for the analysis of mammalian male-specific DNA. Mol Ecol. 2003;12:283–291. doi: 10.1046/j.1365-294x.2003.01702.x. [DOI] [PubMed] [Google Scholar]
- Hellborg L, Gunduz I, Jaarola M. Analysis of sex-linked sequences supports a new mammal species in Europe. Mol Ecol. 2005;14:2025–2031. doi: 10.1111/j.1365-294X.2005.02559.x. [DOI] [PubMed] [Google Scholar]
- Jaarola M, Martinkova N, Gunduz I, Brunhoff C, Zima J, Nadachowski A, et al. Molecular phylogeny of the speciose vole genus Microtus (Arvicolinae, Rodentia) inferred from mitochondrial DNA sequences. Mol Phylogenet Evol. 2004;33:647–663. doi: 10.1016/j.ympev.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Jaarola M, Tegelström H, Fredga K. A contact zone with noncoincident clines for sex-specific markers in the fied vole (Microtus agrestis) Evolution. 1997;51:241–249. doi: 10.1111/j.1558-5646.1997.tb02405.x. [DOI] [PubMed] [Google Scholar]
- Jones EP, van der Kooij J, Solheim R, Searle JB. Norwegian house mice (Mus musculus musculus/domesticus): distributions, routes of colonization and patterns of hybridization. Mol Ecol. 2010;19:5252–5264. doi: 10.1111/j.1365-294X.2010.04874.x. [DOI] [PubMed] [Google Scholar]
- Leache AD, Cole CJ. Hybridization between multiple fence lizard lineages in an ecotone: locally discordant variation in mitochondrial DNA, chromosomes, and morphology. Mol Ecol. 2007;16:1035–1054. doi: 10.1111/j.1365-294X.2006.03194.x. [DOI] [PubMed] [Google Scholar]
- Macholan M, Baird SJE, Dufkova P, Munclinger P, Bimova BV, Pialek J. Assessing multilocus introgression patterns: a case study on the mouse X chromosome in Central Europe. Evolution. 2011;65:1428–1446. doi: 10.1111/j.1558-5646.2011.01228.x. [DOI] [PubMed] [Google Scholar]
- Macholan M, Baird SJE, Munclinger P, Dufkova P, Bimova B, Pialek J. Genetic conflict outweighs heterogametic incompatibility in the mouse hybrid zone. BMC Evol Biol. 2008;8:e14. doi: 10.1186/1471-2148-8-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macholan M, Munclinger P, Sugerkova M, Dufkova P, Bimova B, Bozikova E, et al. Genetic analysis of autosomal and X-linked markers across a mouse hybrid zone. Evolution. 2007;61:746–771. doi: 10.1111/j.1558-5646.2007.00065.x. [DOI] [PubMed] [Google Scholar]
- Meier MN, Golenishchev FN, Radzhabli SI, Sablina OL. Gray voles (subgenus Microtus) of the fauna of Russia and adjacent territories. Trudy Zoolog icheskogo insituta (Collection of Scientific Papers of Zoological Institute) St Petersburg: Ross Akad Nauk; 1996. [Google Scholar]
- Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- Petit E, Balloux F, Excoffier L. Mammalian population genetics: why not Y. Trends Ecol Evol. 2002;17:28–33. [Google Scholar]
- Petit RJ, Excoffier L. Gene flow and species delimitation. Trends Ecol Evol. 2009;24:386–393. doi: 10.1016/j.tree.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Phillips BL, Baird SJE, Moritz C. When vicars meet: A narrow contact zone between morphologically cryptic phylogeographic lineages of the rainforest skink, Carlia rubrigularis. Evolution. 2004;58:1536–1548. doi: 10.1111/j.0014-3820.2004.tb01734.x. [DOI] [PubMed] [Google Scholar]
- Raufaste N, Orth A, Belkhir K, Senet D, Smadja C, Baird SJE, et al. Inferences of selection and migration in the Danish house mouse hybrid zone. Biol J Linn Soc. 2005;84:593–616. [Google Scholar]
- Roubertoux PL, Carlier M, Degrelle H, Haasdupertuis MC, Phillips J, Moutier R. Co-segregation of intermale aggression with the pseudoautosomal region of the Y-chromosome in mice. Genetics. 1994;136:225–230. doi: 10.1093/genetics/136.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilthuizen M, Giesbers MCWG, Beukeboom LW. Haldane's rule in the 21st century. Heredity. 2011;107:95–102. doi: 10.1038/hdy.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer M, Excoffier L, Heckel G. Fine-scale genetic structure and dispersal patterns in the common vole (Microtus arvalis) Mol Ecol. 2007;16:2463–2473. doi: 10.1111/j.1365-294X.2007.03284.x. [DOI] [PubMed] [Google Scholar]
- She JX, Bonhomme F, Boursot P, Thaler L, Catzeflis F. Molecular phylogenies in the genus Mus–comparative analysis of electrophoretic, scnDNA hybridization, and mtDNA RFLP data. Biol J Linn Soc. 1990;41:83–103. [Google Scholar]
- Szymura JM, Barton NH. Genetic-analysis of a hybrid zone between the fire-bellied toads, Bombina bombina and Bombina variegata, near Cracow in southern Poland. Evolution. 1986;40:1141–1159. doi: 10.1111/j.1558-5646.1986.tb05740.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Begun DJ. Haldane's rule and X-chromosome size in Drosophila. Genetics. 1997;147:1799–1815. doi: 10.1093/genetics/147.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Orr HA. Dominance, epistasis and the genetics of postzygotic isolation. Genetics. 2000;154:1663–1679. doi: 10.1093/genetics/154.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner LM, Schwahn DJ, Harr B. Reduced male fertility is common but highly variable in form and severity in a natural house mouse hybrid zone. Evolution. 2012;66:443–458. doi: 10.1111/j.1558-5646.2011.01445.x. [DOI] [PubMed] [Google Scholar]
- Yannic G, Basset P, Hausser J. A hybrid zone with coincident clines for autosomal and sex-specific markers in the Sorex araneus group. J Evol Biol. 2008;21:658–667. doi: 10.1111/j.1420-9101.2008.01526.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.