Abstract
DNAzymes were used as inhibitory agents in a variety of experimental disease settings, such as cancer, viral infections and even HIV. Drugs that become active only upon the presence of preprogrammed abnormal environmental conditions may enable selective molecular therapy by targeting abnormal cells without injuring normal cells. Here we show a novel programmable DNAzyme library composed of variety of Boolean logic gates, including YES, AND, NOT, OR, NAND, ANDNOT, XOR, NOR and 3-input-AND gate, that uses both miRNAs and mRNAs as inputs. Each gate is based on the c-jun cleaving Dz13 DNAzyme and active only in the presence of specific input combinations. The library is modular, supports arbitrary inputs and outputs, cascadable, highly specific and robust. We demonstrate the library's potential diagnostic abilities on miRNA and mRNA combinations in cell lysate and its ability to operate in a cellular environment by using beacon-like c-jun mimicking substrate in living mammalian cells.
‘DNAzymes’ are a catalytically active class of antisense reagents discovered in 19941. In the past decade, DNAzymes (and mostly ‘10–23’) have been used as inhibitory agents in a variety of experimental disease settings, suggesting their possible clinical utility2,3,4,5,6.
Autonomous bio-molecular systems that can interact with naturally occurring biomolecules (such as indicators of particular diseases) and analyze their presence7,8,9,10,11,12,13, may be the basis of ‘programmable drugs’, potent drugs that become active only if preprogrammed abnormal environmental conditions hold.
Results
We modified the Dz13 DNAzyme (which showed promise of a therapeutic agent in preclinical studies by targeting the c-jun mRNA14) and preprogrammed it into a library of Boolean logic gates where only upon specific conditions, Dz13 regains its catalytic activity. These conditions are determined by the presence of predefined input molecules, in the form of miRNA or mRNA molecules, and the Boolean logic gate rules.
Our system is based on multi-component units in which computations are performed by three mechanisms: (a) splitting the DNAzyme at the core catalytic region in a way that only when an appropriate input molecule exists the complete DNAzyme complex is formed15 (b) caging the DNAzyme arms using a stem-loop structure, which can be un-caged when an appropriate input exits16 and (c) toehold exchange in which a longer hybridization is favored, where the presence of an input molecule changes the components' conformation.
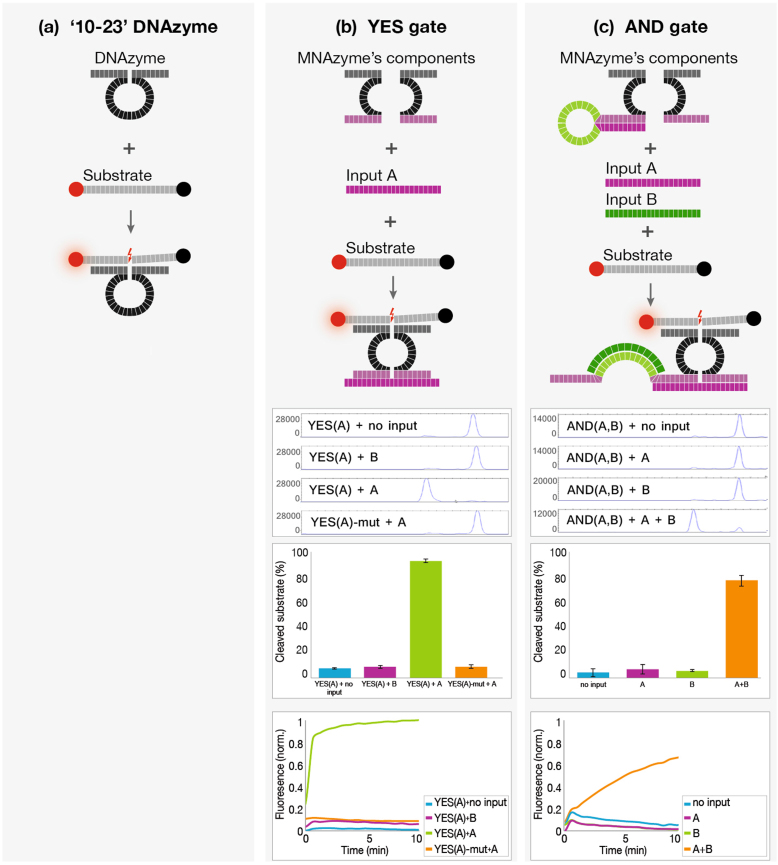
To construct the simple YES gate, which is active only when a single input molecule is present, we used the ‘10–23’ DNAzyme Dz13 (Fig. 1a). By following a previous design17 the DNAzyme's catalytic core was split into two parts between T8 and A9. Only upon the presence of an appropriate input molecule, the two parts are joined, the DNAzyme complex is formed and the cleavage of RNA occurs (‘True’ output) (Fig. 1b).
Figure 1. YES or AND gates demonstration in vitro.
(a) Illustration of Dz13 cleaving its proper fluorescently labeled RNA substrate. (b and c) Each panel is an illustration of YES or AND gate's operation upon presence of its inputs (upper panel); Capillary-electrophoresis demonstrates cleavage or non-cleavage of the fluorescent substrate (upper middle panel); Quantitative cleavage results of 3 independent experiments (bottom-middle panel) and Plate Reader results showing reaction kinetics (bottom panel). Each gate's results match its truth table. For YES: input A = miR155, un-proper input B = miR31; For AND: input A = miR21, input B = miR125b. Standard deviation errors from three independent experiments are shown (bars).
To form the AND gate, an additional binding loop was added consisting of a complementary sequence for the second input, followed by a ‘caging’ sequence which is complementary to the DNAzyme arm to form a stem-loop structure (Fig. 1c). When the second input is present, the arm is un-caged (since the open conformation is favored) and the gate can accept the first input which joins the two sub-components. Only when both inputs are present, the complete DNAzyme complex is formed. Caging may be done on either one of the arms, as demonstrated in Supplementary information Fig. 2.
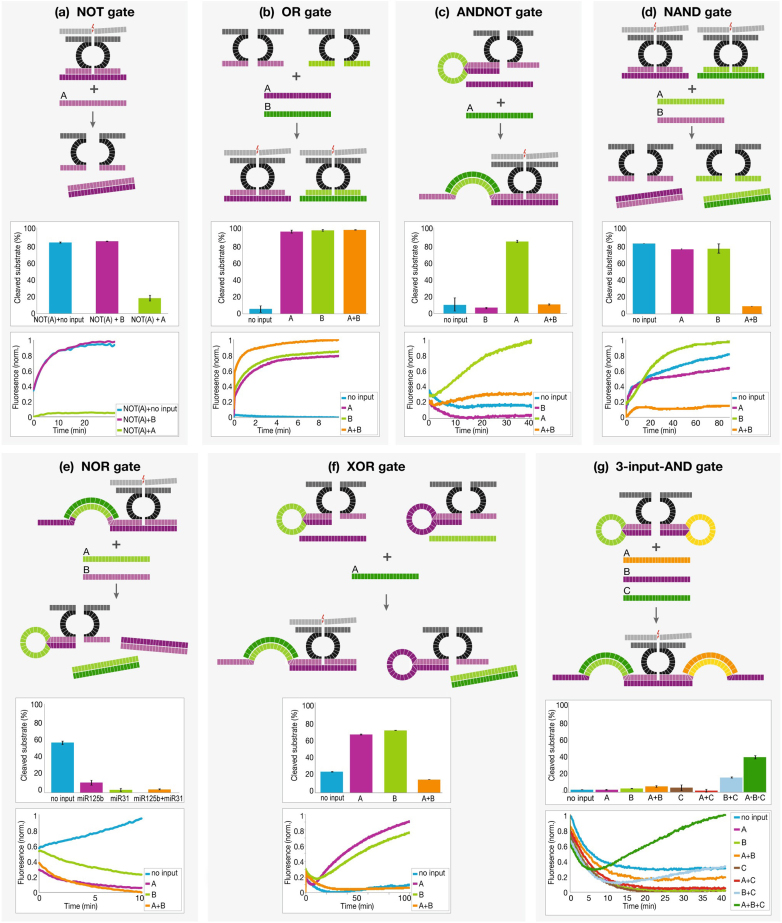
To complete the basic Boolean logic gates, we also implemented the NOT gate using an additional strand which contained the input's complementary sequence (‘anti-input’ molecule). When the input is present, it ‘cancels’ the anti-input molecule, the DNAzyme's components are separated, resulting in a ‘False’ output (Fig. 2a). The OR gate was implemented by using two YES gates operating in parallel on two different inputs (Fig. 2b).
Figure 2. Logic gate design and implementation.
Each panel shows: upper panel, schematic gate operation in the presence of its entire set of inputs; middle panel, quantitative cleavage results of 3 independent experiments; bottom panel, reaction kinetics. The results of each gate match its truth table. (a) for NOT: input A = miR31; (this is a single input gate, only A is shown) (b) for OR: input A = miR21, input B = miR125b; (c), (e) and (f) for ANDNOT, NOR & XOR: input A = miR31, input B = miR125b; (d) for NAND: input A = miR31, input B = miR155; (g) for 3-input-AND gate, input A = miR31, input B = miR21 and input C = miR125b. In (c) & (f) only input A is shown since this is the only (c) or one of (f) the input combinations that has a positive (‘True’) result in which the complex is formed. Standard deviation errors from three independent experiments are shown (bars).
After implementing the basic Boolean gates, we extended our library to more complex gates that combine two inputs. The ANDNOT gate is implemented by combining the AND and NOT gates' design. When both inputs are present, the second input cancels the anti-input molecule, resulting in a ‘False’ output (Fig. 2c). The NAND gate is formed by combining two NOT gates for the two different inputs. Only when both inputs are present, the two gates are inactive (Fig. 2d). The NOR gate is an inverse AND gate. When either is present, the complete DNAzyme complex is disassembled, resulting in a ‘False’ output (Fig. 2e). The XOR gate is formed by combining two ANDNOT gates. When either input is present, it activates one of the sub-gates, but also cancels the orthogonal sub-gate by binding to its anti-input molecule. Only when a single input (but not both) is present, the gate is active (Fig. 2f). As shown, the gates possess a ‘digital’, non-leaky behavior. This feature is a key for therapeutic agents in future diagnostic systems: only cells that meet a specific abnormal profile will be targeted, sparing healthy ones.
Finally, we extended our library to support an additional input. The 3-input-AND gate is formed using an AND gate in which both arms are caged. Only when all 3 inputs are present, the complete DNAzyme complex is formed resulting in a ‘True’ output (Fig. 2g). Theoretical truth tables of the gates shown in Fig. 1 and Fig. 2 are demonstrated in Supplementary information Fig. 3.
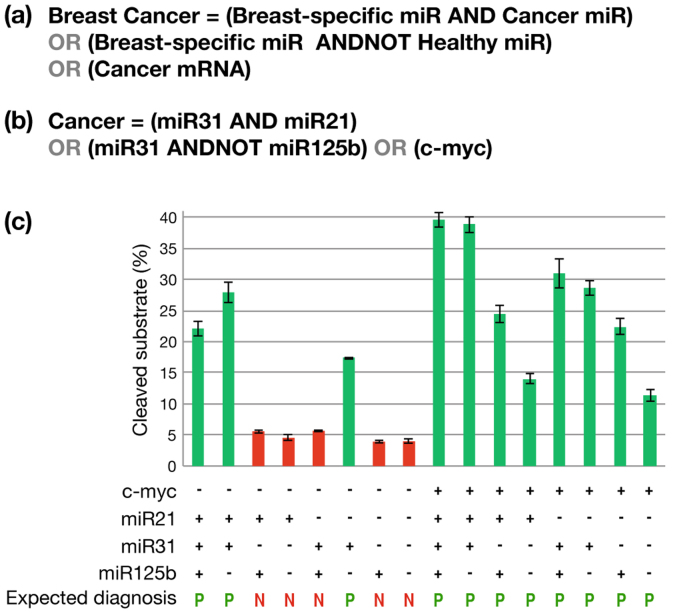
Abnormal cells can be diagnosed by using a complex Boolean expression, in which inputs such as mRNA and miRNA serve as ‘disease markers’. To demonstrate our library's ability to diagnose unhealthy cells by solving such expressions, the following expression was chosen: Breast Cancer = (Breast-specific miRNA AND Cancer miRNA) OR (Breast-specific miRNA AND NOT Healthy miRNA) OR (Cancer mRNA).
A breast cell is considered ‘cancerous’ if it either possesses: (1) combinations of breast-specific microRNA and a ‘cancerous’ microRNA or (2) breast-specific miRNA but lacks the ‘health indicative’ miRNA or (3) cancerous mRNA. The inputs demonstrated are miR31 as a breast-specific miRNA18, miR21 as a strong markers of breast cancer19,20 and miR125b as health indicative miRNA18. The c-myc oncogene serves as a cancerous mRNA marker20. Therefore, the expression can be further described as: Breast cancer = (miR31 AND miR21) OR (miR31 AND NOT miR125b) OR (c-myc).
To indicate the potential of our library to both ‘diagnose’ and ‘treat’ cancerous cells in the future, we demonstrated its ability to solve the above expression in a cellular environment (cancer cells lysate). As the gates are based on Dz13, a ‘true’ output will result in the active state of the DNAzyme, which can potentially cleave the c-jun mRNA, potentially a key step of an apoptosis-based ‘treatment’, as shown in Fig. 3.
Figure 3. Expression profile of breast cancer in cell lysates.
(a) and (b) Definition of Boolean expressions representing positive breast cancer diagnosis. (c) capillary electrophoresis results for the complex Boolean expression. Experiments were performed using the same conditions as the in vitro experiments in previous figures, by replacing DDW with cell lysate (1.8 mg/ml by BCA assay), and with addition of miRNAs & myc RNA to the reaction. Only upon fulfillment of conditions which meet the requirements defined for ‘breast cancer’ the DNAzyme became active and its substrate cleaved. The design of each basic logic gate (AND, ANDNOT, YES, OR) is as shown in previous figures. Variance in the cleavage efficiency of positive conditions may be explained by the different reaction kinetics underlying each ‘active’ complex (as seen in Fig. 1 & 2). Standard deviation errors from three independent experiments are shown (bars).
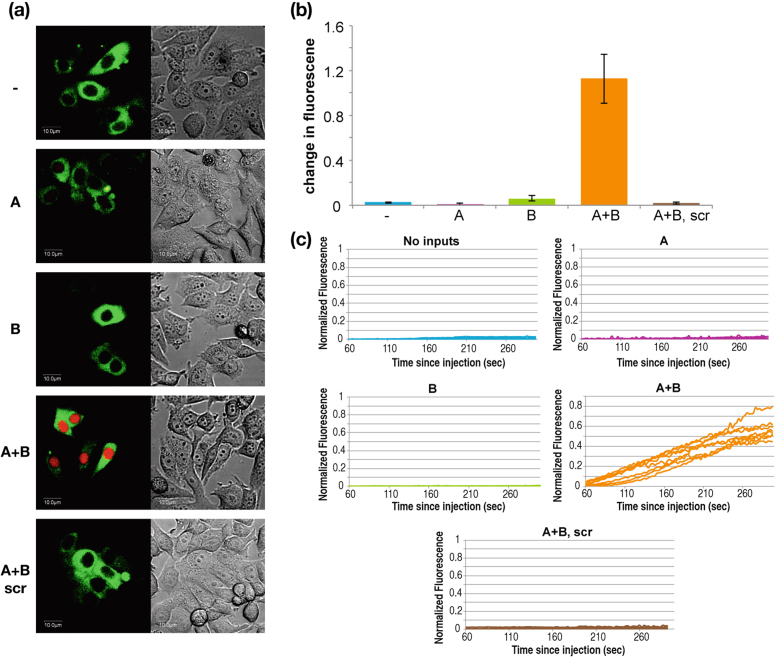
Finally, we turned to test the feasibility of our library's design principles to operate within living human cancer cells. For this purpose, we used a beacon-like substrate which contains a fluorophore and a quencher (Supplementary information, Materials & Methods section). Only when the substrate is cleaved by a DNAzyme, the fluorophore and quencher are separated resulting in light emission. To demonstrate this ability, we used the AND gate (which contains both core design elements our system is built upon – splitting the catalytic core & caging the DNAzyme arm). The gate's components, along with the substrate and exogenous inputs were introduced into cells by microinjection using a Confocal microscope (Supplementary information). Although microinjection might be harmful to living cells, it provides the best control over time, enabling demonstration of computation in its first several minutes. To protect our libraries' components from nucleases, an inverted thymidine modification was added to the 3′ end. As shown in Fig. 4, the AND gate, representing the library's design, operates within living cells, ex vivo, and computations are very rapid (less than 5 minutes). As clearly seen, the result of the cleavage (i.e.: red fluorescent signal) localizes entirely in the cell nucleus. This strengthens the fact that computations were performed inside the cells (as also demonstrated in "Video AB" in the Supplementary Information).
Figure 4. DNAzyme-based AND gate operating within cancerous living cells.
Left panel, fluorescent view of the injected cells; Right panel, phase view of the cells during and after injection. For each inputs combination 15 cells were microinjected with the following mixtures: (1) AND gate components; (2) combination of miRNA inputs (A = miR21, B = mir125b), (3) a fluorescent-quenched substrate (red) and (4) green Dextran (70 KDa), which was used to normalize and mark injected cells. (a) Representative injected cells were imaged for 5 minutes after injection. Only when both inputs were present (A + B), the red substrate was cleaved and therefore visible. ‘scr’ represent injections of a scrambled DNAzyme's component sequence, which were otherwise identical to the normal injections; (b) Average of relative change in red fluorescence in 15 cells, 5 minutes after microinjection (change was calculated as: normalized fluorescence 5 minutes post injection minus normalized fluorescence 1 minute post injection). The design of the AND gate is as shown in Fig. 1. Standard deviation errors are shown (bars). (c) Kinetic results in living cells.
Discussion
The results of this experiment demonstrate that the principles underlying our library's design can generally operate in a cellular environment as a proof-of concept. The feasibility of applying these principles for real diagnosis has yet to be demonstrated. We chose to demonstrate these principles in living cells using the AND gate, which contains both core design elements our system is built upon – splitting the catalytic core & caging the DNAzyme arm. All other gates are based on similar principles. We demonstrated the ability of multiple gates (AND, ANDNOT, OR, YES) to operate in a cellular environment in the lysate experiment in lieu of demonstrations of additional gates ex vivo.
Our system can in principle receive any RNA (or DNA) sequence as its input, and thus can sense for both miRNA and mRNA, as demonstrated. Unlike previous systems which implemented logic libraries based on DNAzymes15 where the computational layer was dependent on the relation between inputs (and thus inputs depended on each other), our system does not require any dependence between inputs. Each input can be completely arbitrary, allowing integration of any input combinations. In addition, unlike previous implementations17, in which the input specificity was determined by a maximal amount of 7 nucleotides, our system is not restricted to short input sequences. As demonstrated, most of our logic gate input-binding sequences are 18–22 nucleotides long, allowing great specificity and minimizing identification of inappropriate inputs. This is a key feature for using cellular markers as inputs, since a cell contains a large amount of miRNA/mRNA populations, some with common sequences.
The output of the system (the target gene) is programmable and is selected by the RNA-binding arms' sequence. Since both output and inputs are RNA molecules, unlike other miRNA based systems7, simple reactions can be composed, in principle, to form cascaded (compositional) ones, where the output RNA of one gate can serve as the input for a downstream gate. In addition, our design supports more than 2 inputs for an AND gate (and for all other gates). As we have demonstrated, by ‘locking’ both input-binding arms, we have created a 3-input-AND gate, illustrated in Fig. 2g. By ‘locking’ also the substrate-binding-arms, the system can be extended to create a 5-input-AND gate. In previous designs, implementation of such a gate would require cascaded gates, each adding multiple molecular interactions. In our system, any additional input (beyond 2 inputs) requires the addition of a single molecular interaction, keeping the total chemical complexity relatively low. Demonstration of several AND gates also shows the programmability of the gate's inputs (Supplementary information 2).
Unlike previous systems based on DNAzymes15,21, we focused on the ability to interact with physiologic components (inputs and outputs), which is a necessary step towards clinical utilization. In a top-down approach, we modified the ‘Dz13’ drug into a ‘programmable drug’, which operates only when certain pre-programmed conditions are met. For this purpose, we preferred to utilize a member of the ‘10–23’ DNAzyme family as our hardware, over other families such as ‘E6’, ‘8–17’ (that were already shown as the basis of Boolean logic gates and automata16,21,22) mainly due to its powerful clinical antisense activity in vivo23,24 and its ability to operate in physiological conditions, such as low MgCl2 concentrations and 37°C25. Unlike previous systems13 which rely on gene transcription to perform computation, our computing element directly binds to both input and target RNA, keeping chemical interactions to a minimum and reducing the number of components which should be delivered to the cell.
Our current demonstrations are based on well-controlled concentrations of microRNAs and mRNAs, which are higher than endogenous levels26, and delivery of these exogenous inputs was performed using microinjection. This is a demonstration that our design is not adversely affected by the cellular environment, a first step towards practical diagnosis. In order to utilize the system for clinical diagnosis, its sensitivity should be increased, for example by integration of input amplifiers27. Applicability also depends on efficient delivery. As both programmable DNAzymes and simple DNAzymes are composed of DNA strands, a therapeutically feasible delivery mechanism of known DNAzymes may also be efficient for programmable DNAzymes. Finally, the library should be further be optimized to reduce false-positive detection and signal leakage, especially in the cellular environment. This may be achieved by further optimizing the length of the caged arms as well as adding a "suppressor" antisense molecule (as previously shown10).
In conclusion, we presented a library of programmable DNAzymes that operates in a cellular environment and can potentially operate in living cancer cells. The library is modular, supports arbitrary inputs and outputs, potentially cascadable, highly specific and robust. Unlike single-marker diagnosis, our library allows integration of multiple markers according to predefined rules, thus may be the first step towards a future of ‘smart’ medical diagnosis and therapy in the form of ‘programmable’ therapeutics.
Methods
DNAzyme design and synthesis
Programmable DNAzymes are based on the ‘10–23’ DNAzyme, with the catalytic core split into two parts between T8 and A9 (based on previous MNAzyme design15) so that each part contains a 9 nt substrate binding region and a 10–11 nt input binding region (input is split across the two parts). Prior to any experiment, gate components were warmed to 99°C and slowly brought down to 10°C in a 50 mM NaCl solution to allow hybridization and stored in −20°C. Synthetic single-stranded RNA substrate was labeled by a FAM fluorophore in its 3′ end. Unmodified DNA sequences were obtained from Sigma-Alderich (Standard Desalted). Modified DNA sequences and RNA sequences were obtained from Integrated DNA Technologies (HPLC Purified). Oligos were stored in −20°C in TE buffer. All sequences can be found in Supplementary Table 1.
In vitro cleavage experiments
Experiments were performed by preparing a reaction mixture that contained the DNAzymes fluorogenic RNA substrate (0.1 μM final) in 10 μl reaction buffer (Tris-HCl 50 mM, pH 7.5) containing 150 mM NaCl and 10 mM MgCl2 and 1 μl of each input (2 μM final, or 10 uM final for the NAND & XOR gates). Computation was initiated by the addition of 1 μl of each of the DNAzyme components (2 μM final) followed by further incubation for 20 minutes (40 minutes for the NOT, ANDNOT & NAND gates; 120 minutes for the XOR gate) at 37°C. The reaction was terminated by passing 0.5 μl of each sample to 22 μl Formamide containing GeneScan LIZ120 size standards which were diluted 1:40 in the Formamide (Applied Biosystems). Samples were run on a capillary electrophoresis machine (ABI Prism, Avant-3100, Applied Biosystems) and analyzed using the GeneMapper software.
In vitro kinetic experiments
Experiments were performed by preparing a reaction mixture that contained the DNAzymes quenched/fluorogenic RNA substrate. The quencher used was 3BHQ (3′ end) and fluorophore was i6-FAMK (positioned internally between C4 and G5 (Sequence: CAAC/i6-FAMK/GCCTCguTCCTCCCG/3BHQ_1/, lowercase letters designate RNA nucleotides). Kinetics measurements were done using a Tecan Infinite® 200 microplate-reader. Fluorescence based experiments were done in a total volume of 30 μl in the same conditions as above, in x10 concentrations (1 uM substrate, 20 μM inputs and DNAzyme components). Excitation and emission values for FAM fluorophore were set to 485 nm and 518 nm, respectively. Initial fluorescence was measured for 3 seconds prior to reaction initialization by addition of MgCl2, to establish the fluorescence baseline.
Cell culture
Human MCF7 breast carcinoma cells were obtained from American Type Culture Collection (ATCC) and cultured in RPMI (Gibco-BRL) pH 7.4, supplemented with 10% fetal calf serum (FCS). All media were supplemented with 10 mg/ml streptomycin and 10 IU/ml penicillin and all cultures were incubated in a humidifed atmosphere of 5% CO2 at 37°C. Cells were passaged routinely by trypsinization. Subconfluent MCF7 cells (70%) were incubated overnight prior to microinjections.
Author Contributions
E.S. led the project. M.K.H. and Y.D. developed the concept, designed the study and performed experiments. All authors discussed the results and implications, commented on the manuscript at all stages and wrote the paper.
Supplementary Material
Supplementary Information
Video 1
Video 2
Video 3
Video 4
Video 5
Acknowledgments
We thank Dr. Tom Ran, Dr. Binyamin Gil, Dr. Michal Golan-Mashiach and Dr. Shai Kaplan for helpful discussions. We thank Vladimir Kiss for his excellent technical assistance with the Confocal microscopy experiments. We thank Keren Katzav for the excellent preparation and design of figures. The research was supported by the European Union FP7-ERC-AdG. M.K was supported by The ISF Converging Technologies (Grant No 1694/07). Ehud Shapiro is the Incumbent of The Harry Weinrebe Professorial Chair of Computer Science and Biology.
References
- Breaker R. R. & Joyce G. F. A DNA enzyme that cleaves RNA. Chem Biol 1, 223–229 (1994). [DOI] [PubMed] [Google Scholar]
- Dass C. R., Galloway S. J. & Choong P. F. Dz13, a c-jun DNAzyme, is a potent inducer of caspase-2 activation. Oligonucleotides 20, 137–146 (2010). [DOI] [PubMed] [Google Scholar]
- Wu Y. et al. Inhibition of bcr-abl oncogene expression by novel deoxyribozymes (DNAzymes). Hum Gene Ther 10, 2847–2857 (1999). [DOI] [PubMed] [Google Scholar]
- Dass C. R., Choong P. F. & Khachigian L. M. DNAzyme technology and cancer therapy: cleave and let die. Mol Cancer Ther 7, 243–251 (2008). [DOI] [PubMed] [Google Scholar]
- Reyes-Gutiérrez P. & Alvarez-Salas L. M. Cleavage of HPV-16 E6/E7 mRNA mediated by modified 10-23 deoxyribozymes. Oligonucleotides 19, 233–242 (2009). [DOI] [PubMed] [Google Scholar]
- Jakobsen M. R., Haasnoot J., Wengel J., Berkhout B. & Kjems J. Efficient inhibition of HIV-1 expression by LNA modified antisense oligonucleotides and DNAzymes targeted to functionally selected binding sites. Retrovirology 4, 29 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Liu S., Bleris L. & Benenson Y. Logic integration of mRNA signals by an RNAi-based molecular computer. Nucleic Acids Res 38, 2692–2701 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. Y., Jensen M. C. & Smolke C. D. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc Natl Acad Sci U S A 107, 8531–8536 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culler S. J., Hoff K. G. & Smolke C. D. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science 330, 1251–1255 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benenson Y., Gil B., Ben-Dor U., Adar R. & Shapiro E. An autonomous molecular computer for logical control of gene expression. Nature 429, 423–429 (2004). [DOI] [PubMed] [Google Scholar]
- Elbaz J., Shimron S. & Willner I. pH-triggered switchable Mg2+-dependent DNAzymes. Chem Commun (Camb) 46, 1209–1211 (2010). [DOI] [PubMed] [Google Scholar]
- Nissim L. & Bar-Ziv R. H. A tunable dual-promoter integrator for targeting of cancer cells. Mol Syst Biol 6, 444 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Wroblewska L., Prochazka L., Weiss R. & Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333, 1307–1311 (2011). [DOI] [PubMed] [Google Scholar]
- Dass C. R. & Choong P. F. C-jun: pharmaceutical target for DNAzyme therapy of multiple pathologies. Pharmazie 63, 411–414 (2008). [PubMed] [Google Scholar]
- Elbaz J. et al. DNA computing circuits using libraries of DNAzyme subunits. Nat Nanotechnol 5, 417–422 (2010). [DOI] [PubMed] [Google Scholar]
- Stojanovic M. N. & Stefanovic D. A deoxyribozyme-based molecular automaton. Nat Biotechnol 21, 1069–1074 (2003). [DOI] [PubMed] [Google Scholar]
- Mokany E., Bone S. M., Young P. E., Doan T. B. & Todd A. V. MNAzymes, a versatile new class of nucleic acid enzymes that can function as biosensors and molecular switches. J Am Chem Soc 132, 1051–1059 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Ridzon D., Wong L. & Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8, 166 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan H., Miller N., Lowery A., Sweeney K. & Kerin M. MicroRNAs as Novel Biomarkers for Breast Cancer. J Oncol 2009, 950201 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N. & Penn L. Z. Reflecting on 25 years with MYC. Nat Rev Cancer 8, 976–990 (2008). [DOI] [PubMed] [Google Scholar]
- Macdonald J. et al. Medium scale integration of molecular logic gates in an automaton. Nano Lett 6, 2598–2603 (2006). [DOI] [PubMed] [Google Scholar]
- Stojanovic M., Mitchell T. & Stefanovic D. Deoxyribozyme-based logic gates. J Am Chem Soc 124, 3555–3561 (2002). [DOI] [PubMed] [Google Scholar]
- Zhang L. et al. Angiogenic inhibition mediated by a DNAzyme that targets vascular endothelial growth factor receptor 2. Cancer Res 62, 5463–5469 (2002). [PubMed] [Google Scholar]
- Fahmy R. G. et al. Suppression of vascular permeability and inflammation by targeting of the transcription factor c-Jun. Nat Biotechnol 24, 856–863 (2006). [DOI] [PubMed] [Google Scholar]
- Santoro S. W. & Joyce G. F. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry 37, 13330–13342 (1998). [DOI] [PubMed] [Google Scholar]
- Cissell K. & Deo S. Trends in microRNA detection. Anal Bioanal Chem 394, 1109–1116 (2009). [DOI] [PubMed] [Google Scholar]
- Seelig G., Soloveichik D., Zhang D. & Winfree E. Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Video 1
Video 2
Video 3
Video 4
Video 5