Abstract
Maize protein EMB564 is a member of group 1 LEA (late embryogenesis abundant) proteins. Currently, the molecular functions of group 1 LEA proteins remain largely unclear. We here report on the functional assignment to EMB564 by computational analysis. EMB564 is predicted as nuclear localization by five different predictors including CELLO, Plant-mPLoc, WoLF PSORT, Predotar and TargetP. EMB564 is found to be remote homologous with DNA/RNA helicases and single-stranded DNA-binding proteins, and their sequences contains similar DNA/RNA binding sites. Furthermore, the three-dimensional (3D) model of EMB564 structurally resembles a variety of nuclear and DNA/RNA-binding proteins, especially those involving in the regulation of cell division, chromosomal replication and DNA unwinding or repairing. Our results reveal that EMB564 protein is most likely to function within the cell nucleus.
Keywords: Computational analysis, DNA-binding, EMB564 protein, Function assignment, Zea Mays
Background
Late embryogenesis abundant (LEA) proteins accumulate in seeds and pollen during maturation and in vegetative tissues in responses to drought, salinity, temperature stress and abscisic acid [1, 2]. After 32 years since the first LEA protein discovered [3], the molecular functions and mechanisms of most LEA proteins remain largely unknown [1]. LEA proteins are widely assumed to play a role in cellular dehydration tolerance and in controlling water uptake during imbibition [2]. According to the features of particular motifs, sequence similarity or biased amino acid composition, LEA proteins are classified into different groups. Group 1 LEA proteins are small (8-12 kD), acidic (pI 5.4-6.6) and highly hydrophilic, and contain 20-mer motif TVVPGGTGGKSVEAQEHLAE [3]. Based on consensus protein or oligonucleotide probability profile (POPP) analysis, group 1 is subdivided as la and 1b, and group 1a is hypothesized to match with chromosomal and nuclear proteins with a possible DNA binding function [4]. However, no further computational and experimental data supports this hypothesis. Recently, we have found that group 1a LEA protein EMB564 is greatly expressed in maize seeds and its abundance is associated with seed vigor [5]. This implies a relationship between EMB564 and seed vigor. Currently, there are still no clear annotations for group 1 LEA proteins (including EMB564) relating to their molecular functions and subcellular locations in the Universal Protein Resource KnowledgeBase (UniProtKB) (http:// www.uniprot.org). In the present paper, we have analyzed the subcellular localization, DNA/RNA-binding propensity and three-dimensional (3D) structure of EMB564 in order to elucidate the molecular basis of its functions. Our analysis highlights EMB564 functioning within the cell nucleus.
Methodology
Subcellular localization of EMB564 protein:
The protein sequences of EMB564 and other proteins examined in this paper were retrieved from UniProtKB. Subcellular localization of EMB564 was predicted with CELLO, PlantmPLoc, WoLF PSORT, Predotar and TargetP, respectively.
DNA/RNA-binding prediction:
DNA-binding propensity was predicted using Loctree (https://rostlab.org/owiki/index.php/Loctree) and DNAbinder (http://www.imtech.res.in/raghava/dnabinder). DNA and RNA binding residues were predicted using BindN (http://bioinfo.ggc.org/bindn) and DP-Bind (http://lcg.rit.albany.edu/dp-bind).
Homology search:
Standalone PSI-Blast ( http://www.ebi.ac.uk/Tools/ sss/psiblast/) was performed for 3 iterations (E-value: 0.001) using the protein sequence of EMB564 as a query to search remote homologues against the UniProtKB/TrEMBL.
Protein modeling and function assignments:
The 3D model of EMB564 was built using the on-line server ITASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER) in automatic model [6]. Function assignment was made by comparing the 3D model of EMB564 against those in the Protein Data Bank using the Dali server (DaliLite v.3 in -q mode) [7].
Discussion
Nucleus location of EMB564:
To date, despite the intensive characterization of many LEA proteins, their molecular functions are enigmatic and largely unknown [3]. Computational analysis will provide insights on the molecular basis of biological functions of these LEA proteins, especially on their subcellular locations, sequence binding sites, homology and modeling. According to our examination of the scientific literature, only 22 LEA proteins have been experimentally localized in the subcellular compartments, but no any group 1 LEA proteins are experimentally localized. In this study, we used five popular predictors (CELLO, Plant-mPLoc, WoLF PSORT, Predotar and TargetP), rather than relying on a single predictor, to predict the subcellular localization of EMB564 protein. These predictors use both defined characteristics and machine-learning techniques. All prediction results support the nucleus location of EMB564 Table 1 (see supplementary material). Therefore, the predicted nucleus location of EMB564 is highly reliable. After its synthesis, how EMB564 is targeted to the nucleus remains unclear, since it does not contain any known nuclear localization signals [8]. Our analysis indicated that EMB564 does contain many positively charged lysines or arginines exposed on the protein surface as nuclear (chromosomal) proteins. Due to its small size, EMB564 may diffuse through the nuclear pore into the nucleus [9]. It is also possible that small LEA proteins may be co-transported to the nucleus through protein-protein interactions [10].
Sequence similarity of EMB564 with DNA helicase:
LOCtree predicted EMB564 as a DNA-binding protein with 90% reliability. Likewise, DNAbinder predicted EMB564 as a DNAbinding protein. Furthermore, the prediction results of DP-Bind and BindN indicate that EMB564 contains three major DNAbinding segments: PGGTGGKS (residues 23-30), EGRSRGGQTRREQ (residues 39-51) and KKGGLSTT (residues 62-69) (Figure 1A). BindN predicted similar DNA-residues as DP-Bind but some more along the sequence of EMB564 (Figure 1A). The remote sequence similarities of EMB564 with DNAbinding proteins were searched using PSI-Blast. The first iteration of PSI-Blast retrieved many homologues with 23%-98% identities, such as group 1 LEA proteins, stress proteins and uncharacterized proteins. Strikingly, four DNA/RNA helicases with identities 33%-41% were retrieved from the bacteria Brachybacterium faecium (C7M9T8) and Fulvimarina pelagi (Q0G1N7) and the single celled green alga Chlamydomonas reinhardtii (Q9FPR8, A8JGB2). The second iteration also retrieved C7M9T8, Q0G1N7 and other three bacterial RNA helicase. Likewise, using the sequences of other group 1a LEA proteins as a query, the above-described helicases were also retrieved.
Figure 1.
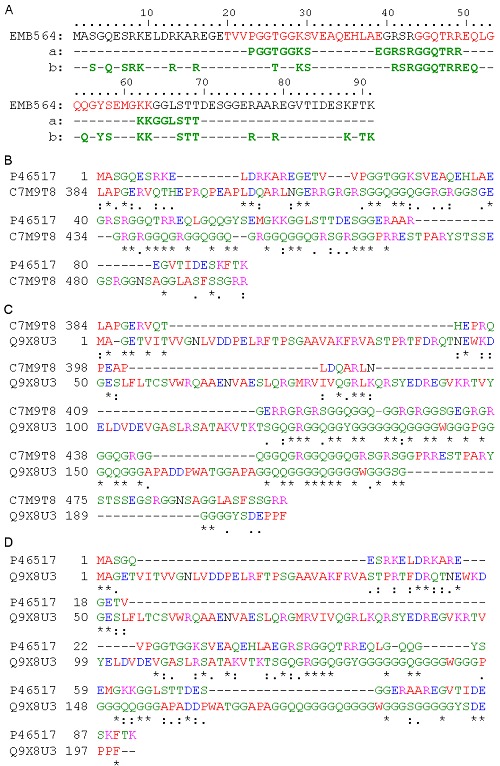
DNA-binding residues in EMB564 and its similarity with DNA-binding proteins. Two conserved motifs in EMb564 sequence, characteristic of group 1a LEA proteins, are marked in red color. A) DNA-binding residues predicted by DP-Bind (a) and BindN. (b), respectively; B), C) and D), sequence alignments by T-Coffee. * Single, fully conserved residue; : conservation of strong groups; conservation of weak groups; - no consensus. C7M9T8 = DNA/RNA helicase, Q9X8U3 = single-stranded DNA-binding protein 2.
C7M9T8 is chosen for further analysis due to its relatively high similarity with EMB564. Alignment analysis indicates that EMB564 is homologous with C7M9T8 at the C-terminal (Figure 1B). Moreover, the C-terminal sequence of C7M9T8 shares high identity with the well-characterized single-stranded DNAbinding protein 2 (SSB2, Q9X8U3) (Figure 1C). EMB564 share the similar DNA/RNA-binding sequence [E/S] G[Q/R][G/S]RGGQ as C7M9T8 and Q9X8U3 (Figure 1D). In addition, by the domain prediction protocol Ginzu in de novo 3D modeling server Robetta (http://robetta.bakerlab.org/), we found that EMB564 contains a domain homologous with the domain involved in branched DNA processing in ATPdependent RNA helicase (Q8TZH8; PDB: 1wp9-A) from Pyrococcus furiosus. Q8TZH8 is annotated in UniProtKB as DNA binding, ATP-dependent helicase activity and nuclease activity in DNA repair etc. In summary, the above-described analyses implied EMB564 protein having DNA/RNA-binding ability.
Structure similarity of EMB564 with DNA-binding proteins:
Currently, no information on LEA proteins is available in the protein structural databases. The 3D model of EMB564 built using the I-TASSER server is given in (Figure 2A). It consists of α-helices and random coils: an N-terminal α-helix and a long coil, followed by three α-helices which are linked by short coils and parallel to each other in space, and a C-terminal coil. DNAbinding residues locate at the surface of the 3D model (indicated by vertical box). 3D structure comparison may reveal biologically interesting similarities that are not detectable by comparing sequences, since even remotely homologous proteins generally have quite similar 3D structure [11]. DALI comparison indicated that EMB564 structurally resembles a variety of nuclear (chromosomal) proteins involving in the regulation of cell division, chromosomal replication, DNA/RNA polymerase and helicase (Figure 2B).
Figure 2.
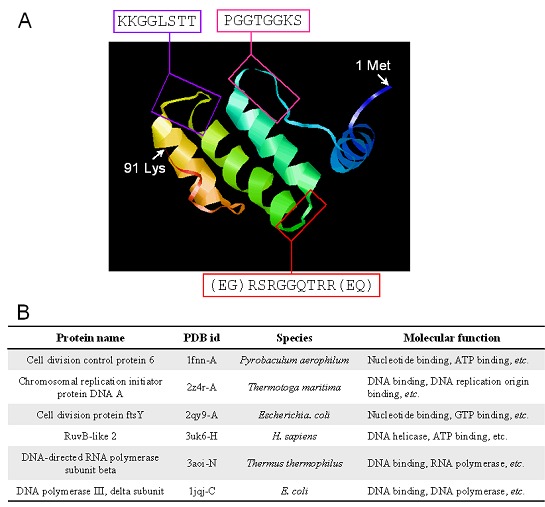
The 3D model of EMB564 and its structure homologues. A) The 3D model of EMB564 was built using the ITASSER server and deposited in the PMDB database (http://www.caspur.it/PMDB, id: PM0078754). Predicted DNA-binding residues are indicated by boxes; B) Six representative structure homologues with EMB564 determined in the DALI server using the 3D model of EMB564 as a query against those in PDB.
Conclusion
Our results indicate that EMB564 is localized in the cell nucleus, it has a high propensity of binding DNA, its sequence is homologous with DNA helicases, and finally, its 3D model structurally resembles a variety of DNA-binding and nuclear proteins. Therefore, we propose that EMB564 is most likely to function within the nucleus by binding DNA. This paper provides new data to support the hypothesis by Wise et al. [4] that group 1a LEA proteins may act as nucleus proteins that unwind or repair DNA and regulate transcription. It will help to understand the mechanism underlying the biological function of group 1 LEA proteins.
Supplementary material
Acknowledgments
This work was supported by State Key Laboratory of Crop Biology (Grant no. 2012KF01) at Shandong Agricultural University, China.
Footnotes
Citation:Wu et al, Bioinformation 9(6): 276-280 (2013)
References
- 1.Battaglia M, et al. Plant Physiol. 2008;148:6. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray EA, et al. Plant Physiol. 1993;103:1035. doi: 10.1104/pp.103.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dure L 3rd, et al. Biochemistry. 1981;7:4162. doi: 10.1021/bi00517a033. [DOI] [PubMed] [Google Scholar]
- 4.Wise MJ, Tunnacliffe A. Trends Plant Sci. 2004;9:13. doi: 10.1016/j.tplants.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Wu XL, et al. Acta Physiol Plant. 2011;33:181. DOI 10.1007/s11738-010-0536-4. [Google Scholar]
- 6.Roy A, et al. Nat Protoc. 2010;5:725. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holm L, et al. Nucl Acids Res. 2010;38:W545. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cokol M, et al. EMBO Rep. 2000;1:411. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaltsman A, et al. Plant Physiol. 2007;145:1264. doi: 10.1104/pp.107.105973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godey A, et al. Plant Cell. 1984;6:351. [Google Scholar]
- 11.Kinch LN, Grishin NV. Curr Opin Struct Biol. 2002;12:400. doi: 10.1016/s0959-440x(02)00338-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


