Abstract
Expression in the adult heart of a number of cardiac genes, including the two genes comprising the cardiac myosin heavy chain locus (Myh), is controlled by thyroid hormone (T3) levels, but there is minimal information concerning the epigenetic status of the genes when their expressions change. We fed mice normal chow or a propyl thio uracil (PTU, an inhibitor of T3 production) diet for 6 weeks, or the PTU diet for 6 weeks followed by normal chow for a further 2 weeks. Heart ventricles from these groups were then used for ChIP-seq analyses with an antibody to H3K4me3, a well-documented epigenetic marker of gene activation. The resulting data show that, at the Myh7 locus, H3K4me3 modifications are induced primarily at 5′ transcribed region in parallel with increased expression of beta myosin heavy chain (MHC). At the Myh6 locus, decreases in H3K4me3 modifications occurred at the promoter and 5′ transcribed region. Extensive H3K4me3 modifications also occurred at the intergenic region between the two Myh genes, which extended into the 3′ transcribed region of Myh7. The PTU-induced changes in H3K4me3 levels are, for the most part, reversible but are not invariably complete. We found full restoration of Myh6 gene expression upon PTU withdrawal; however, the H3K4me3 pattern was only partially restored at Myh6, suggesting that full reexpression of Myh6 does not require that the H3K4me3 modifications return fully to the untreated conditions. Together, our data show that the H3K4me3 modification is an epigenetic marker closely associated with changes in Myh gene expression.
Key words: Myosin heavy chain, Epigenetic, ChIP-seq, Thyroid hormone, Heart, Histone methylation
INTRODUCTION
The amino-terminal tails of histones are the target of multiple posttranslational modifications: acetylation, methylation, phosphorylation, and ubiquitination (14,17). Multiple studies have revealed predictive correlations between histone modifications and gene expression (6,18). Trimethylation of lysine residue at position 4 of histone 3 (H3K4me3) is one such modification that is highly correlated with active gene expression (24). Genome-wide studies have found this modification to be present frequently within the promoters of genes that are actively transcribed, and it is now considered to be a well-conserved marker of gene activation (16,29). Several components of the transcriptional machinery contain domains (such as PHD, chromo, bromo, MCT, and Tudor) that can recognize H3K4me3 (12). The H3K4me3 modification may serve as a general means of recruiting transcriptional machinery. Determining how the distribution of H3K4me3 modification changes in response to defined stimuli is critical to understanding how genes are regulated.
Thyroid hormone (T3) is a fat-soluble hormone produced by the thyroid gland, which is released into the circulation and plays critical roles in differentiation, growth, and metabolism (30). The effects of T3 on heart function are well documented, and various studies have identified multiple genes whose expression levels are responsive to T3 levels, including the two cardiac myosin heavy chain (Myh) genes, sarcoplasmic reticulum Ca2+ ATPase, and several membrane-bound sodium and potassium channels (3,11,21). T3 binds to the thyroid hormone receptor (THR)—a member of nonsteroidal nuclear hormone receptor family—and exerts its effects by either inducing or silencing the expression of target genes (2). THR binds to its recognition site independently of T3, although the presence or absence of T3 controls whether THR is a transcriptional activator or transcriptional repressor.
The two genes of the Myh locus, Myh6 and Myh7, are some of the most T3-responsive genes in the heart. They code for the developmental stage-specific isoforms of the myosin heavy chain component of the sarcomere—the fundamental unit of contraction of the muscle cell. Myh7 (beta-MHC) is expressed at high levels during the fetal stage in the mouse ventricle, during which Myh6 (alpha-MHC) is expressed at very low levels. Immediately after birth, in response to a surge in the circulating levels of T3, the expression of Myh7 is silenced and that of Myh6 increases so that alpha-MHC becomes the predominant isoform of myosin heavy chain. Under conditions of hypothyroidism, heart failure, and cardiac hypertrophy, there is a reversal in the expression pattern of these two genes: Myh7 expression is induced and Myh6 is silenced.
The degree to which T3-mediated changes in Myh gene expression are associated with changes in H3K4me3 levels is of considerable interest, as is the degree to which H3K4me3 modification is reversible during reversible changes in gene expression. Previous work conducted by us using a chromatin immunoprecipitation (ChIP)-qPCR assay demonstrated that this modification occurred at the promoter of Myh6 but not Myh7 (20). However, this previous investigation did not interrogate regions outside the promoters. Consequently, in this report, we have comprehensively investigated the changes in H3K4me3 levels across the entire Myh loci using the more powerful ChIP-seq procedure. This combines ChIP with deep sequencing, which results in a sensitive and unbiased way of investigating the H3K4me3 status within individual genes across the whole genome. Here, we show that the H3K4me3 modification occurs at multiple regions within the Myh locus, including the 5′ transcribed portion of Myh7. We also show H3K4me3 modification at an intergenic region that coincides with a previously demonstrated antisense promoter. Finally, we show that the H3K4me3 modification occurs at many regions within the genome and that it is at least partially reversible in response to reversible changes in T3.
MATERIALS AND METHODS
Animals
Animals used in this study were 4-month-old 129/Svev. Animals were fed an iodine-deficient diet containing 0.15% PTU (Harlan Teklad, TD. 97061, Rx257896) for 6 weeks, and then either sacrificed (PTU group) or fed normal chow for another 2 weeks (reversal). Control mice were fed normal chow. The mice were maintained in a facility accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International according to Institutional Animal Care and Use Committee-approved guidelines.
Organ Preparation for Chromatin Immunoprecipitation (ChIP)
Hearts were excised immediately after euthanasia and cut into two pieces to remove residual blood. Then they were washed in PBS and snap frozen in liquid nitrogen. Hearts were treated with 1% formaldehdye for 20 min, followed by treatment with glycine to quench fixation. Nuclear extracts were prepared by crushing with Cryo-Press CP-100 (Microtech Nichion, Co. Ltd, Japan), as described previously (27). Briefly, approximately 300 mg of frozen heart tissue was physically crushed with Cryo-Press for 1 min four times, changing direction of liquid nitrogen-cooled sample container. Frozen powder of heart was immediately suspended in 2 ml of SDS lysis buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% SDS, 1 mM EDTA). Cell suspension including DNA–protein complex was applied to fragmentation using Sonifier (Branson); 10 min for highspeed sequencer. Samples were diluted by 2 ml of ChIP dilution buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) and applied to chromatin immunoprecipitation. Detailed experimental procedure for chromatin immunoprecipitation followed with high-speed sequencer was modified as described elsewhere (28). H3K4me3-bound genomic DNA was isolated from whole cell lysate using a mouse monoclonal antibody [16H10, a gift of Dr. Hiroshi Kimura (13)], in combination with magnetic beads (Dynal/Invitrogen). Prepared DNA was quantified using Qubit (Invitrogen) and more than 10 ng of DNA was processed as described below.
ChIP-seq Sample Preparation and Analysis
All protocols for Illumina/Solexa sequence preparation, sequencing, and quality control were provided by Illumina (http://www.illumina.com/pages.ilmn?ID=203) (25).
Microarray Analyses
Ventricles of the hearts from the different conditions were surgically isolated, frozen in liquid nitrogen, and RNA isolated. Total RNA was submitted for microarray analyses as previously described (27).
RESULTS
ChIP-seq Analyses for H3K4me3 Occupancy
The distribution of the H3K4me3 modification was determined by chromatin immunoprecipitation/high throughput sequencing (ChIP-seq) analyses on heart samples, which enables analysis of H3K4me3 distribution across the whole genome. We used cardiac ventricles from 4-month-old 129/Svev mice and conducted ChIP-seq assays with an antibody specific for the H3K4me3 modification. The sites and levels of H3K4me3 modification were determined by massive parallel sequencing on an Illumina deep sequencing platform. Distribution patterns of H3K4me3 were quantified and graphically visualized with an integrated genome browser, which depicts H3K4me3 levels on the y-axis and the locus map for each gene on the x-axis.
Distribution of H3K4me3 Modification at the Myh Locus
Untreated Mice
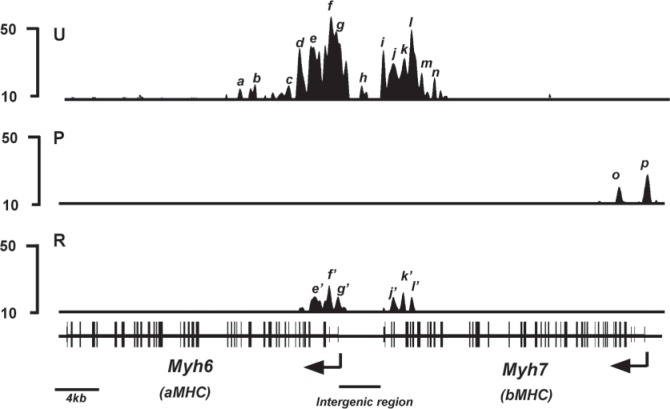
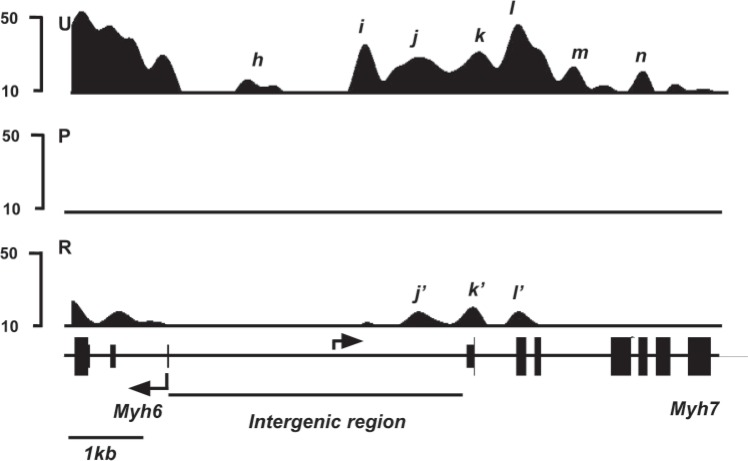
We first analyzed the distribution of H3K4me3 in untreated animals, which express Myh6 at high levels and Myh7 at very low levels. The top panel (U, untreated) of Figure 1 illustrates the distribution pattern of H3K4me3 signals at the Myh loci. A schematic of the locus is shown at the bottom of the figure. Under untreated conditions, Myh6 shows two regions of H3K4me3 enrichment. One region lies within the Myh6 promoter [peak h, located at −1.0 kb relative to the transcription start site (TSS)]. The other region is located within the 5′ transcribed portion (peaks a through g in Fig. 1, between TSS and +8.5 kb relative to TSS) of Myh6 gene. The presence of these H3K4me3 modifications within Myh6 is consistent with the high level of expression of Myh6. Myh7, on the other hand, shows essentially undetectable H3K4me3 signals within the promoter and most of the transcribed portion of the gene in the untreated mice. This lack of H3K4me3 modification at Myh7 is consistent with the very low expression of Myh7 under untreated conditions. Interestingly, a region of H3K4me3 enrichment is found at the intergenic region between the two Myh genes, which is seen as multiple peaks (peaks i through n in Fig. 1) beginning at approximately 2.5kb upstream of Myh6 TSS and extending approximately 4 kb into the 3′ transcribed portion of Myh7 [illustrated in greater detail in Fig. 2, top panel (U)].
Figure 1.
H3K4me3 enrichment profile of Myh loci. H3K4me3 signal intensities are shown on the y-axis and schematic of the Myh locus shown on the x-axis. Animals were fed either regular chow (untreated), PTU diet for 6 weeks (PTU), or PTU diet for 6 weeks followed by regular chow for a further 2 weeks (reversal), and hearts were isolated for analyses. ChIP-seq analyses were conducted using anti-H3K4me3 antibody from chromatin isolated from untreated (U, top panel), PTU-treated (P, middle panel), or reversal (R, bottom panel) animals. H3K4me3 signals are illustrated from the Integrated Genome Browser interface. Exons are represented as closed boxes; bent arrows show the transcriptional start site (TSS) and direction of transcription. Intergenic region between the two Myh genes is identified with a horizontal bar. Individual peaks are identified by letters in italics. Scale bar for the locus is shown at the bottom left.
Figure 2.
H3K4me3 enrichment profile of Myh intergenic region. H3K4me3 signal intensities are shown on the y-axis and schematic of the locus shown on the x-axis. Animals were fed either regular chow (untreated), PTU diet for 6 weeks (PTU), or PTU diet for 6 weeks followed by regular chow for a further 2 weeks (reversal), and hearts were isolated for analyses. ChIP-seq analyses were conducted using anti-H3K4me3 antibody from chromatin isolated from Untreated (U, top panel), PTU-treated (P, middle panel), or reversal (R, bottom panel) animals. Exons are represented as closed boxes; bent arrows show the transcriptional start site (TSS) and direction of transcription. Intergenic region between the two Myh genes is identified with a horizontal bar. Individual peaks are identified by letters in italics. Scale bar for the locus is shown at the bottom left.
PTU-Treated Mice
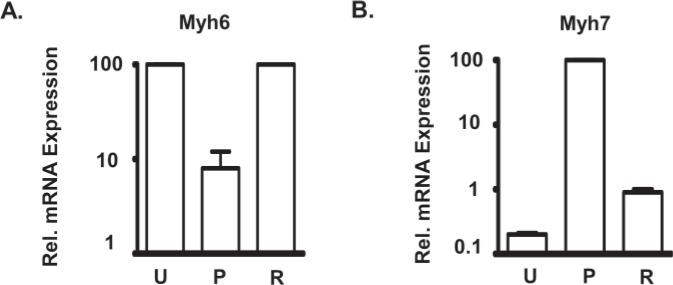
To change the expression levels of the two Myh genes, we induced a hypothyroid condition by feeding mice a diet containing 0.15% PTU for 6 weeks. PTU is a specific inhibitor of thyroid hormone synthesis, and it has been well established that this level of PTU in the diet is sufficient to induce changes in the heart that are typical of hypothyroid conditions (15,22,26). PTU treatment dramatically silences Myh6 expression and substantially increases Myh7 gene expression (22,26). In our mice, mRNA analyses using real-time PCR from the PTU-treated and untreated hearts show, as expected, a substantial silencing of Myh6 (approximately 10-fold decrease) and a dramatic increase in Myh7 gene expression (approximately 50-fold increase) in response to PTU (Fig. 3). The middle section (P) of Figure 1 illustrates the H3K4me3 distribution under PTU-treated conditions. At the Myh6 locus this treatment leads to a dramatic decrease of the H3K4me3 signals to undetectable levels (reduction to <20% of its maximal value, a level below which random noise predominates) at the two regions of the gene that showed high levels of H3K4me3 modification in the untreated mice (peaks a through n in Fig. 1). This loss of H3K4me3 modification is consistent with the greatly reduced level of gene expression. At the Myh7 locus, PTU treatment causes the development of two new peaks of H3K4me3 enrichment. One peak is at +0.5 kb (peak p in Fig. 1) and the other peak is located at +2.5kb (peak o in Fig. 1) relative to the Myh7 TSS. This gain of H3K4me3 modification is consistent with the greatly increased expression levels of Myh7. There are, however, no H3K4me3 peaks present within the Myh7 promoter, confirming our previous observation that H3K4me3 modifications are absent at the Myh7 promoter when it is expressed in the adult heart under hypothyroid condition (20). The intergenic region, noted above, completely loses the H3K4me3 signals during PTU treatment (note absence of peaks i though n in panel P, Figs. 1 and 2).
Figure 3.
Reversible changes in mRNA expression levels of Myh genes in response to PTU treatment. Comparisons of mRNA expressions for Myh6 (A) and Myh7 (B) from hearts of untreated (U), PTU-treated (P), and reversal (R) groups. The mean for untreated group (for Myh6) or PTU-treated group (for Myh7) was set at 100, and the values for the other groups are relative to this value (relative expression). Animals were fed either regular chow (U), PTU diet for 6 weeks (P), or PTU diet for 6 weeks followed by regular chow for a further 2 weeks (R), and hearts were isolated for analyses. Quantitative gene expressions were determined by Taqman real-time PCR. Individual values ± SD are: Myh6 (U): 100 ± 4; Myh6 (P): 8 ± 4; Myh6 (R): 115 ± 8; Myh7 (U): 0.2 ± 0.01; Myh7(P): 100 ± 5.7; Myh7(R): 0.9 ± 0.1.
Reversal Mice
We next determined the degree to which the PTU-induced changes in H3K4me3 distribution at the Myh locus reverse when the gene expression levels are reversed, which our previous work shows is complete by 2 weeks after withdrawal of PTU. Accordingly, mice were treated with PTU for 6 weeks, PTU treatment was then withdrawn, and 2 weeks later we sacrificed the mice. Real-time PCR analyses for mRNA expression show that Myh6 gene expression returns to its original high levels, while expression of Myh7 is reduced dramatically to its original low levels (Fig. 3). ChIP-seq analyses conducted on the hearts of these animals show that the PTU-induced changes in H3K4me3 enrichment within the Myh7 gene have likewise returned to the untreated levels. Thus, as illustrated in bottom panel (R) of Fig. 1, the two H3K4me3 peaks (peaks o and p) present at Myh7 gene under PTU conditions decrease dramatically during reversal to levels below the threshold and so indistinguishable from those of untreated animals. The Myh6 gene shows increases in the H3K4me3 levels within the 5′ transcribed portion during reversal, but neither the levels nor the patterns are restored to their untreated levels. The peaks a, b, and c present in the 5′ transcribed region are not reattained during reversal conditions. Nor is the peak in the promoter (peak h) reattained under reversal conditions. The heights of the H3K4me3 modification within the 5′ transcribed portion were also reduced under reversal condition compared with the untreated condition (compare peaks e, f, and g with peaks e′, f′, and g′, respectively, in Fig,. 1). The intergenic region also shows increases in the levels of H3K4me3 modification. However, similar to the situation for Myh6, some peaks (peaks i, m, and n in Figs. 1 and 2) are not reattained, and some peaks show decreased heights (compare peaks j, k, and l with peaks j′, k′, and l′, respectively, in Figs. 1 and 2). Thus, reversal of gene expression of Myh genes to untreated levels results in a full reversal of H3K4me3 modification at the Myh7 gene, but that at the Myh6 gene is only partial.
Distribution of H3K4me3 Modification in Other T3-Responsive Genes
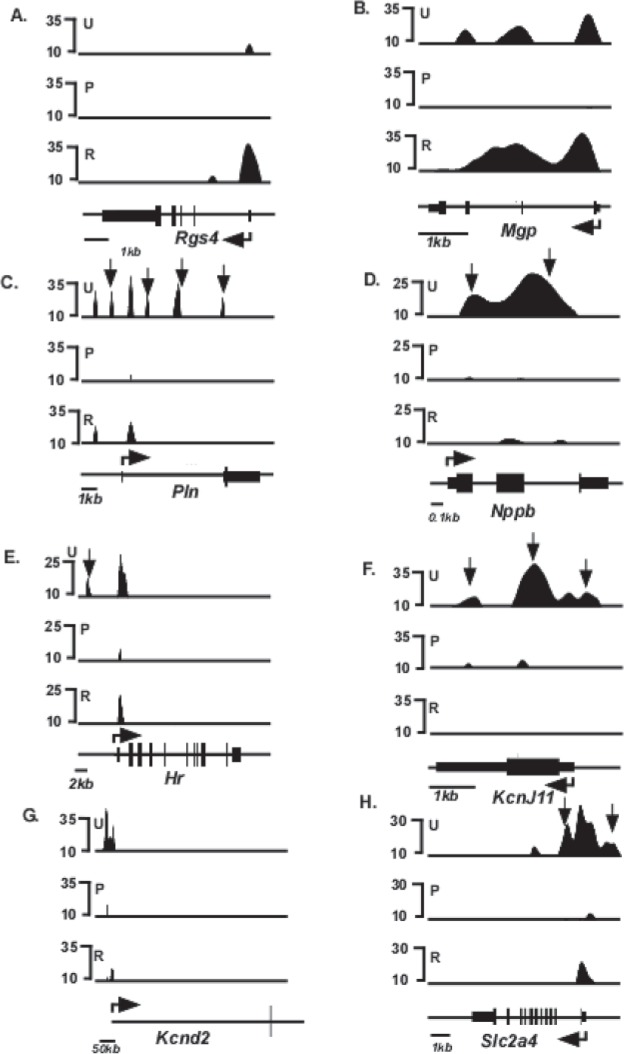
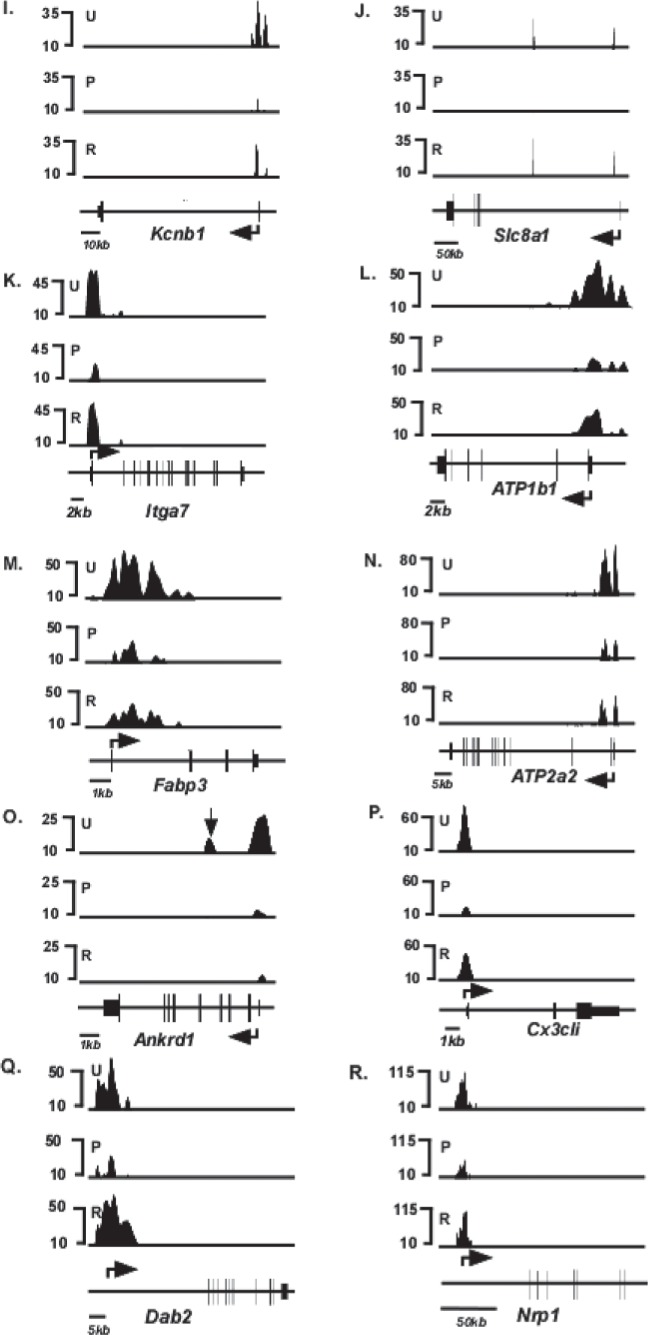
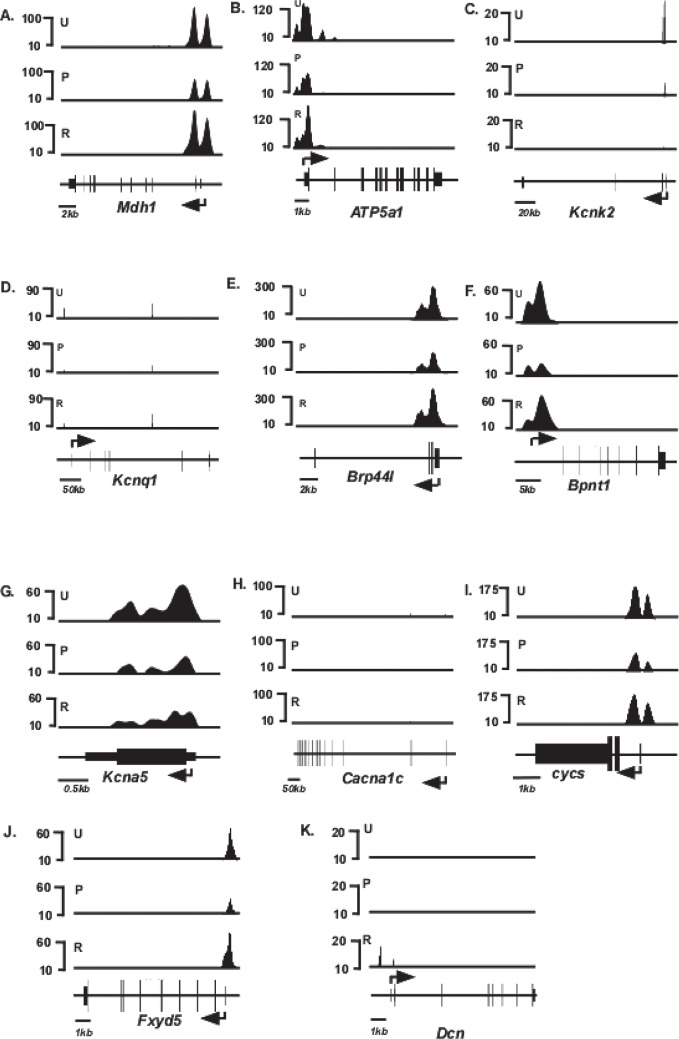
Forty genes, in addition to Myh6 and Myh7, have been shown by previous investigators to be responsive to T3 (1,15) (Table 1). Inspection of the H3K4me3 distribution patterns of these 40 genes in the untreated mice show that 28 have detectable H3K4me3 modifications indicative of expression [top panel (U) of Figs. 4 and 5] These H3K4me3 peaks are distributed primarily at the 5′ transcribed portions of these genes, as has been observed for other actively transcribed genes. PTU treatment changed the level of H3K4me3 modification significantly (defined as >2× changes in H3K4me3 levels in either direction) in 18 of these genes [middle panel (P) of Fig. 4], while the remaining 10 genes did not show a significant PTU-response [middle panel (P) of Fig. 5]. Under conditions of reversal, H3K4me3 levels within these 18 genes had a tendency to return to the original untreated conditions [bottom panel (R) of Fig. 4], although the levels did not always show full recovery. Six genes (Kcnj11, Hr, Pln, Nppb, Slc2a4, and Ankrd1) do not show full recovery. Kcnj11 loci shows dramatically reduced H3K4me3 levels under reversal conditions, compared to untreated conditions (Fig. 4F), while the H3K4me3 peaks present in the promoter of Hr and Slc2a4 (arrows, Fig. 4E and H, respectively), 5′ transcribed portion of Pln (arrows, Fig. 4C), and within the gene of Nppb and Anrd1 (arrows, Fig. 4D and O, respectively) are not reattained under reversal conditions. Interestingly, one gene (Dcn) that had previously showed undetected levels of H3K4me3 under untreated and PTU conditions shows detectable levels of H3K4me3 levels under conditions of reversal (Fig. 5K). Quantified values of H3K4me3 levels at the entire loci of all these genes are shown in Table 2, and the distributions within various regions of individual genes are shown in Table 3.
TABLE 1.
T3-RESPONSIVE HEART GENES
| Actg2 | Actin, gamma 2, smooth muscle, enteric |
| Aldoc | Aldose C, fructose-bisphosphate |
| Ankrd1 | Ankyrin repeat domain 1 protein |
| Atp1b1 | Sodium/Potassium-transporting ATPase subunit beta-1 |
| Atp2a2 | ATPase, Ca2+ transporting, cardiac muscle, slow twitch2 |
| Atp5a1 | Mitochondrial H+-ATPase synthase alpha subunit |
| Brp44l | Brain protein 44-like |
| Bpnt1 | 3′(2′),5′-bisphosphate nucleotidase |
| Cacna1c | Ca2+ channel |
| Ctrb1 | Chymotrypsin B |
| Cycs | Cytochrome C |
| Cx3c | Chemokin Cx3c |
| Dab2 | DOC-2 p82 isoform |
| Dcn | Decorin |
| Fabp3 | Adipocyte lipid-binding protein |
| Fbp1 | Fructose 1,6-biphosphate |
| Fxyd5 | FXYD domain-containing ion transport regulator-1 |
| Hr | Hairless |
| Itga7 | Alpha7A integrin |
| Kcna5 | Potassium channel a5 |
| Kcnb1 | Potassium channel b1 |
| Kcnd2 | Potassium channel d2 |
| Kcne1 | Potassium channel e1 |
| Kcnj11 | Sulfonyl urea receptor |
| Kcnk2 | Potassium channel k2 |
| Kcnq1 | Potassium channel q1 |
| Mdh1 | Malate dehydrogenase-like enzyme |
| Mgp | Matrix G1a protein |
| Myh6 | Cardiac myosin heavy chain, alpha |
| Myh7 | Cardiac nyosin heavy chain, beta |
| Myh13 | Myosin, heavy chian polypeptide, 13, skeletal muscle |
| Nppa | Natriuretic peptide precursor A |
| Nppb | Brain natriuretic factor |
| Nrp1 | Neuropilin |
| Pln | Phospholamban |
| Rgs4 | Regulator of G-protein signaling 4 |
| S100a4 | S100 calcium binding protein A4 |
| Slc8a1 | Ca2+ regulatory channel |
| Slc2a4 | Glucose transporter 4, insulin responsive |
| Tmprss13 | Protease, serine, 11 (IgF binding) |
| Tnp2 | Transition protein 2 |
| Wfdc8 | WAP four-disulfide core domain 8 |
Putative T3-resposnive genes whose expression was previously shown to change >2× in response to changes in T3 levels (1,15).
Figure 4.
Schematic of H3K4me3 enrichment profile of T3-responsive genes that respond to PTU treatment. Relative H3K4me3 signal intensities are shown on the y-axis and schematics of individual gene loci shown on the x-axis. ChIP-seq analyses were conducted using anti-H3K4me3 antibody from chromatin isolated from untreated (U, top panel), PTU-treated (P, middle panel), or reversal (R, bottom panel) animals. Exons are represented as closed boxes; bent arrows show the transcriptional start site (TSS) and direction of transcription. Quantitation of these data is presented in Tables 2 and 3. Vertical arrows in (D, F, G) (Hr, Kcnj11, and Pln) identify H3K4me3 peaks that are present in untreated (U) but not in reversal (R) conditions, depicting incomplete reversal. Genes whose loci showed >2× or <2× changes in H3K4me3 levels in response to PTU were considered PTU responsive. Scale bars for each locus are shown at the bottom left of each panel.
Figure 5.
Schematic of H3K4me3 enrichment profile of T3-responsive genes that do not respond to PTU. Relative H3K4me3 signal intensities are shown on the y-axis and schematic of individual gene loci shown on the x-axis. ChIP-seq analyses were conducted using anti-H3K4me3 antibody from chromatin isolated from untreated (U, top panel), PTU-treated (P, middle panel), or reversal (R, bottom panel) animals. Exons are represented as closed boxes; bent arrows show the transcriptional start site (TSS) and direction of transcription. Quantitation of these data is presented in Tables 2 and 3. Scale bars for each locus are shown at the bottom left of each panel.
TABLE 2.
CHANGES IN H3K4me3 LEVELS AMONG PUTATIVE T3-RESPONSIVE GENES
| Gene Symbol | Ctl Levels | PTU Levels | Rev Levels | PTU/Ctrl Ratio | Rev/PTU Ratio |
|---|---|---|---|---|---|
| Myh6 | 68026 | 0 | 21573 | 0.0 | — |
| Rgs4 | 2000 | 0 | 8942 | 0.0 | — |
| Mgp | 20267 | 602 | 42947 | 0.0 | 71.4 |
| Pln | 20090 | 1134 | \N7021 | 0.1 | 6.2 |
| Nppb | 18315 | 2351 | 3857 | 0.1 | 1.6 |
| Hr | 18641 | 2638 | 7858 | 0.1 | 3.0 |
| Kcnj11 | 34836 | 6006 | 0 | 0.2 | 0 |
| Kcnd2 | 19304 | 3331 | 1535 | 0.2 | 0.5 |
| Slc2a4 | 32417 | 6319 | 8694 | 0.2 | 1.4 |
| Kcnb1 | 14129 | 2766 | 3523 | 0.2 | 1.3 |
| Slc8a1 | 49337 | 11331 | 40608 | 0.2 | 3.6 |
| Itga7 | 51115 | 14827 | 33826 | 0.3 | 2.3 |
| Atp1b1 | 57529 | 16998 | 16922 | 0.3 | 1.6 |
| Fabp3 | 95984 | 31582 | 44893 | 0.3 | 1.4 |
| Atp2a2 | 84817 | 29090 | 47822 | 0.3 | 1.6 |
| Ankrd1 | 17212 | 5936 | 3077 | 0.3 | 0.5 |
| Cx3cl1 | 29320 | 10314 | 21145 | 0.4 | 2.1 |
| Dab2 | 42588 | 17915 | 69258 | 0.4 | 3.9 |
| Nrp1 | 171188 | 74166 | 179050 | 0.4 | 2.4 |
| Mdh1 | 47128 | 22051 | 49320 | 0.5 | 2.2 |
| Atp5a1 | 105564 | 49696 | 84171 | 0.5 | 1.7 |
| Kcnk2 | 17021 | 8040 | 1352 | 0.5 | 0.2 |
| Kcnq1 | 105988 | 50258 | 80012 | 0.5 | 1.6 |
| Brp44l | 101490 | 50152 | 88572 | 0.5 | 1.8 |
| Bpnt1 | 74190 | 38480 | 57761 | 0.5 | 1.5 |
| Kcna5 | 66864 | 35810 | 44242 | 0.5 | 1.2 |
| Cacna1c | 8782 | 4837 | 6618 | 0.6 | 1.4 |
| Cycs | 47220 | 28252 | 43321 | 0.6 | 1.5 |
| Fxyd5 | 26130 | 17449 | 33009 | 0.7 | 1.9 |
| Myh7 | 1160 | 12730 | 0 | 11.0 | 0 |
| Actg2 | ND | ND | ND | — | — |
| Aldoc | ND | ND | ND | — | — |
| Ctrb1 | ND | ND | ND | — | — |
| Dcn | ND | ND | 10456 | — | — |
| Fbp1 | ND | ND | ND | — | — |
| Kcne1 | ND | ND | ND | — | — |
| Myh13 | ND | ND | ND | — | — |
| Nppa | ND | ND | ND | — | — |
| S100a4 | ND | ND | ND | — | — |
| Tmprss13 | ND | ND | ND | — | — |
| Tnp2 | ND | ND | ND | — | — |
| Wfdc8 | ND | ND | ND | — | — |
Quantification of H3K4me3 levels within putative T3-reponsive genes under untreated (Ctrl), PTU-treated (PTU), and reversal (Rev) conditions. ND, not detected. Changes in H3K4me3 levels were quantified based on values for ChIP-seq.
TABLE 3.
QUANTITATION OF H3K4me3 LEVELS AT VARIOUS REGIONS IN INDIVIDUAL GENES
| Gene | Condition | Promoter | 5′ UTR | First Intron | Other Introns | All Exons | 3′ UTR |
|---|---|---|---|---|---|---|---|
| Myh6 | Control | 48029 | 19997 | 0 | 0 | 0 | 0 |
| PTU | 0 | 0 | 0 | 0 | 0 | 0 | |
| Reversal | 14496 | 7077 | 0 | 0 | 0 | 0 | |
| Rgs4 | Control | 0 | 0 | 2000 | 0 | 0 | 0 |
| PTU | 0 | 0 | 0 | 0 | 0 | 0 | |
| Reversal | 0 | 280 | 7806 | 0 | 857 | 0 | |
| Mgp | Control | 0 | 568 | 13329 | 4456 | 1915 | 0 |
| PTU | 0 | 0 | 521 | 0 | 81 | 0 | |
| Reversal | 0 | 451 | 29793 | 10286 | 2417 | 0 | |
| Pln | Control | 0 | 8643 | 6843 | 0 | 1956 | 848 |
| PTU | 0 | 567 | 567 | 0 | 0 | 0 | |
| Reversal | 0 | 3511 | 3511 | 0 | 0 | 0 | |
| Npph | Control | 0 | 646 | 2817 | 7903 | 6950 | 0 |
| PTU | 0 | 248 | 0 | 0 | 2103 | 0 | |
| Reversal | 0 | 0 | 558 | 1344 | 1955 | 0 | |
| Hr | Control | 5697 | 6496 | 822 | 211 | 5434 | 0 |
| PTU | 0 | 1325 | 185 | 0 | 1128 | 0 | |
| Reversal | 0 | 3938 | 746 | 0 | 3174 | 0 | |
| Kcnj11 | Control | 0 | 2688 | 0 | 0 | 28240 | 3809 |
| PTU | 0 | 355 | 0 | 0 | 3665 | 986 | |
| Reversal | 0 | 0 | 0 | 0 | 0 | 0 | |
| Kcnd2 | Control | 19304 | 0 | 0 | 0 | 0 | 0 |
| PTU | 3331 | 0 | 0 | 0 | 0 | 0 | |
| Revesal | 1535 | 0 | 0 | 0 | 0 | 0 | |
| Slc2a4 | Control | 0 | 2239 | 20953 | 2367 | 6858 | 0 |
| PTU | 0 | 1142 | 3625 | 0 | 1552 | 0 | |
| Reversal | 0 | 975 | 6325 | 0 | 1395 | 0 | |
| Kcnb1 | Control | 12711 | 709 | 0 | 0 | 709 | 0 |
| PTU | 2231 | 268 | 0 | 0 | 268 | 0 | |
| Reversal | 2523 | 0 | 0 | 0 | 0 | 0 | |
| Slc8a1 | Control | 0 | 24675 | 24293 | 0 | 369 | 0 |
| PTU | 0 | 5665 | 5665 | 0 | 0 | 0 | |
| Reversal | 0 | 20304 | 20304 | 0 | 0 | 0 | |
| Itga7 | Control | 0 | 2936 | 36801 | 0 | 11378 | 0 |
| PTU | 0 | 364 | 11509 | 0 | 2954 | 0 | |
| Reversal | 0 | 2067 | 22184 | 0 | 9575 | 0 | |
| Atp1b1 | Control | 0 | 7670 | 37385 | 0 | 12474 | 0 |
| PTU | 0 | 2245 | 10793 | 0 | 3959 | 0 | |
| Reversal | 0 | 1765 | 20615 | 0 | 4543 | 0 | |
| Fabp3 | Control | 4334 | 0 | 37385 | 0 | 2997 | 0 |
| PTU | 0 | 0 | 1073 | 0 | 1083 | 0 | |
| Reversal | 0 | 0 | 20615 | 0 | 1320 | 0 | |
| Atp2a2 | Control | 0 | 11767 | 30530 | 20653 | 20866 | 0 |
| PTU | 0 | 5172 | 14109 | 0 | 9808 | 0 | |
| Reversal | 0 | 1002 | 22140 | 19106 | 5575 | 0 | |
| Ankrd1 | Control | 0 | 0 | 5317 | 6689 | 5206 | 0 |
| PTU | 0 | 0 | 1614 | 1416 | 2906 | 0 | |
| Reversal | 0 | 0 | 1527 | 0 | 1550 | 0 | |
| Cx3cl1 | Control | 0 | 1848 | 21708 | 0 | 5764 | 0 |
| PTU | 0 | 505 | 8164 | 0 | 1644 | 0 | |
| Reversal | 0 | 1046 | 16856 | 0 | 3243 | 0 | |
| Dab2 | Control | 0 | 21308 | 18574 | 0 | 2706 | 0 |
| PTU | 0 | 8964 | 7828 | 0 | 1123 | 0 | |
| Reversal | 0 | 34650 | 31026 | 0 | 3581 | 0 | |
| Nrp1 | Control | 41277 | 13383 | 92480 | 2449 | 21600 | 0 |
| PTU | 20895 | 7504 | 34309 | 0 | 11458 | 0 | |
| Reversal | 31449 | 13472 | 111118 | 637 | 22375 | 0 | |
| Mdh1 | Control | 0 | 4922 | 37028 | 0 | 5179 | 0 |
| PTU | 0 | 3587 | 14715 | 0 | 3750 | 0 | |
| Reversal | 0 | 3735 | 35533 | 0 | 7051 | 0 | |
| Atp5a1 | Control | 0 | 6115 | 79019 | 7882 | 12548 | 0 |
| PTU | 0 | 3650 | 39448 | 0 | 6598 | 0 | |
| Revesal | 0 | 4569 | 68859 | 2682 | 8061 | 0 | |
| Kcnk2 | Control | 0 | 0 | 7909 | 0 | 596 | 8517 |
| PTU | 0 | 0 | 3356 | 0 | 658 | 4026 | |
| Reversal | 0 | 0 | 676 | 0 | 0 | 676 | |
| Kcnq1 | Control | 0 | 1330 | 16363 | 76913 | 11382 | 0 |
| PTU | 0 | 470 | 8176 | 35882 | 5731 | 0 | |
| Reversal | 0 | 276 | 2575 | 71598 | 5563 | 0 | |
| Brp44I | Control | 3623 | 1955 | 88258 | 0 | 7654 | 0 |
| PTU | 547 | 944 | 44962 | 0 | 3699 | 0 | |
| Reversal | 2989 | 2044 | 76059 | 0 | 7522 | 0 | |
| Bpnt1 | Control | 53583 | 10332 | 7823 | 0 | 2452 | 0 |
| PTU | 28490 | 5006 | 4043 | 0 | 941 | 0 | |
| Reversal | 34568 | 10117 | 8414 | 0 | 1662 | 0 | |
| Kcna5 | Control | 0 | 4902 | 0 | 0 | 60396 | 1566 |
| PTU | 0 | 2298 | 0 | 0 | 33471 | 41 | |
| Reversal | 0 | 3248 | 0 | 0 | 39730 | 1264 | |
| Cacna1c | Control | 3540 | 5242 | 0 | 0 | 0 | 0 |
| PTU | 1976 | 2861 | 0 | 0 | 0 | 0 | |
| Reversal | 2456 | 4162 | 0 | 0 | 0 | 0 | |
| Cycs | Control | 0 | 23373 | 21723 | 0 | 2124 | 0 |
| PTU | 0 | 14144 | 13296 | 0 | 813 | 0 | |
| Reversal | 0 | 21429 | 20621 | 0 | 1272 | 0 | |
| Fxyd5 | Control | 0 | 12495 | 9107 | 476 | 4051 | 0 |
| PTU | 0 | 7831 | 5846 | 1067 | 2706 | 0 | |
| Reversal | 0 | 14289 | 11825 | 3119 | 3776 | 0 | |
| Myh7 | Control | 0 | 0 | 0 | 0 | 0 | 1160 |
| PTU | 0 | 0 | 12730 | 0 | 0 | 0 | |
| Reversal | 0 | 0 | 0 | 0 | 0 | 0 | |
| Actg2 | Control | 0 | 0 | 0 | 0 | 0 | 0 |
| PTU | 0 | 0 | 0 | 0 | 0 | 0 | |
| Reversal | 0 | 0 | 0 | 0 | 0 | 0 | |
| Aldoc | Control | 0 | 0 | 0 | 0 | 0 | 0 |
| PTU | 0 | 0 | 0 | 0 | 0 | 0 | |
| Reversal | 0 | 0 | 0 | 0 | 0 | 0 | |
| Ctrb1 | Control | 0 | 0 | 0 | 0 | 0 | 0 |
| PTU | 0 | 0 | 0 | 0 | 0 | 0 | |
| Reversal | 0 | 0 | 0 | 0 | 0 | 0 | |
| Dcn | Control | 0 | 0 | 0 | 0 | 0 | 0 |
| PTU | 0 | 0 | 0 | 0 | 0 | 0 | |
| Reversal | 7907 | 0 | 1279 | 108 | 1161 | 0 | |
| Fbp1 | Control | 0 | 0 | 0 | 0 | 0 | 0 |
| PTU | 0 | 0 | 0 | 0 | 0 | 0 | |
| Reversal | 0 | 0 | 0 | 0 | 0 | 0 | |
| Kcne1 | Control | 0 | 0 | 0 | 0 | 0 | 0 |
| PTU | 0 | 0 | 0 | 0 | 0 | 0 | |
| Reversal | 0 | 0 | 0 | 0 | 0 | 0 | |
| Myh13 | Control | 0 | 0 | 0 | 0 | 0 | 0 |
| PTU | 0 | 0 | 0 | 0 | 0 | 0 | |
| Reversal | 0 | 0 | 0 | 0 | 0 | 0 | |
| Nppa | Control | 0 | 0 | 0 | 0 | 0 | 0 |
| PTU | 0 | 0 | 0 | 0 | 0 | 0 | |
| Reversal | 0 | 0 | 0 | 0 | 0 | 0 | |
| S100a4 | Control | 0 | 0 | 0 | 0 | 0 | 0 |
| PTU | 0 | 0 | 0 | 0 | 0 | 0 | |
| Reversal | 0 | 0 | 0 | 0 | 0 | 0 | |
| Tmprss13 | Control | 0 | 0 | 0 | 0 | 0 | 0 |
| PTU | 0 | 0 | 0 | 0 | 0 | 0 | |
| Reversal | 0 | 0 | 0 | 0 | 0 | 0 | |
| Tnp2 | Control | 0 | 0 | 0 | 0 | 0 | 0 |
| PTU | 0 | 0 | 0 | 0 | 0 | 0 | |
| Reversal | 0 | 0 | 0 | 0 | 0 | 0 | |
| Wfdc8 | Control | 0 | 0 | 0 | 0 | 0 | 0 |
| PTU | 0 | 0 | 0 | 0 | 0 | 0 | |
| Reversal | 0 | 0 | 0 | 0 | 0 | 0 |
Quantification of H3K4me3 levels at different regions within putative T3-reponsive genes under untreated (Ctrl), PTU-treated (PTU), and reversal (Rev) conditions. ND, not detected. Changes in H3K4me3 levels were quantified based on values for ChIP-seq.
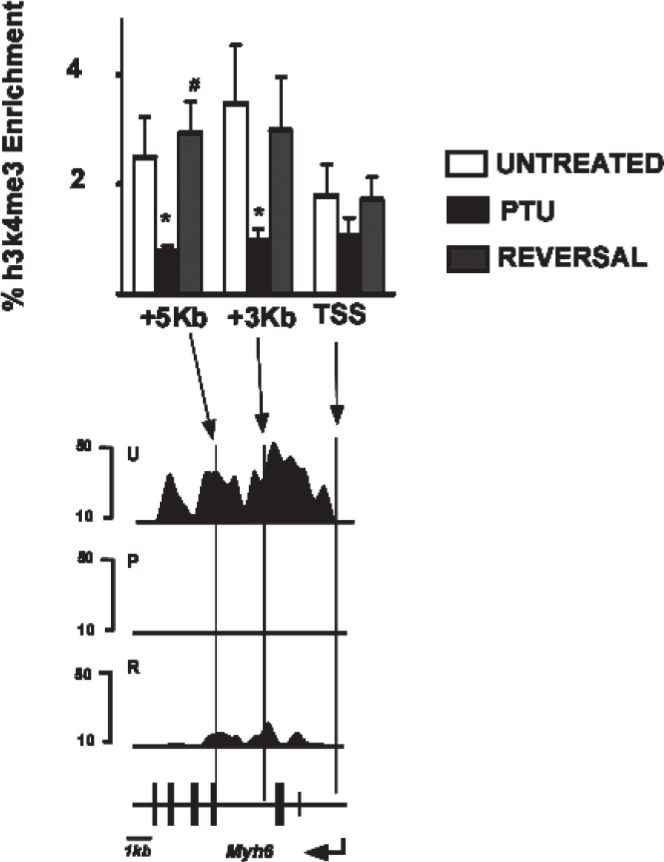
We compared the ChIP-seq data for Myh6 described in this work with ChIP-qPCR analyses of Myh6 carried out previously by us (20) in response to PTU treatment and PTU reversal. The previous ChIP-qPCR analyses showed that the +3 kb and +5 kb regions of Myh6 gene had significant reversible decreases in H3K4me3 signals in response to PTU treatment (Fig. 6, black bars), while the TSS region of Myh6 gene showed no changes in H3K4me3 levels in response to these treatments. These ChIP-qPCR data are in complete agreement with the ChIP-seq data for Myh6 at these locations presented in this report (Fig. 6, bottom panel).
Figure 6.
Validation of ChIP-seq data. Top: ChIP-qPCR analyses for H3K4me3 at the Myh6 transcribed region. % enrichment (relative to 100% input) of immunoprecipitated DNA is shown on y-axis and the various locations (TSS, transcription start site, +3 kb, +5 kb) of Myh6 are shown on the x-axis. ChIP-qPCRs were conducted using anti-H3K4me3 antibody form chromatin isolated from untreated (U, open bars), PTU-treated (P, black bars), or reversal (R, gray bars) animals. Data represent mean of seven independent ChIP experiments from four individual animals within each group. Error bars represent SEM. *p < 0.05 (relative to untreated group), #p < 0.005 (relative to PTU group). These data are reproduced from a previously published work (20). Bottom: Chip-seq analyses of Myh6 gene. Part of Figure 1 is shown below the graph for ease of comparison. Scale bar for the locus is shown at the bottom left.
We also examined whether the T3-responsive genes that showed changes in H3K4me3 levels also showed changes in gene expression. To do this, we conducted microarray analyses from hearts of mice from the three groups, and the results are shown in Table 4. Eight out of 42 genes show significant changes in mRNA levels (determined as >2× change in either direction). Six of these genes (Myh6, Rgs4, Hr, Kcnb1, Kcnq1, and Myh7) also showed changes in H3K4me3 levels, while two genes (Tmprss13 and Kcne1) did not show any detectable H3K4me3 signals at their respective loci.
TABLE 4.
CHANGES IN mRNA LEVELS AMONG PUTATIVE T3-RESPONSIVE GENES
| Gene Symbol | PTU/Ctrl Ratio | Rev/PTU Ratio |
|---|---|---|
| Tmprss13 | 0.2 | 2.6 |
| Hr | 0.3 | 2.5 |
| Myh6 | 0.3 | 3.1 |
| Rgs4 | 0.4 | 2.5 |
| Kcnb1 | 0.5 | 2.9 |
| Kcnq1 | 2.1 | 0.4 |
| Kcne1 | 3.6 | 0.4 |
| Myh7 | 3.0 | 0.1 |
| Actg2 | — | — |
| Atp2a2 | — | — |
| Atp5a1 | — | — |
| Bpnt1 | — | — |
| Cx3cl1 | — | — |
| Cycs | — | — |
| Dab2 | — | — |
| Dcn | — | — |
| Fabp3 | — | — |
| Fxyd5 | — | — |
| Itga7 | — | — |
| Kcna5 | — | — |
| Kcnd2 | — | — |
| Kcnj11 | — | — |
| Mdh1 | — | — |
| Mgp | — | — |
| Nppa | — | — |
| Nppb | — | — |
| Nrp1 | — | — |
| Pln | — | — |
| S100a4 | — | — |
| Slc2a4 | — | — |
| Slc8a1 | — | — |
| Aldoc | ND | ND |
| Ankrd1 | ND | ND |
| Atp1b1 | ND | ND |
| Brp44l | ND | ND |
| Cacna1c | ND | ND |
| Ctrb1 | ND | ND |
| Fbp1 | ND | ND |
| Kcnk2 | ND | ND |
| Myh13 | ND | ND |
| Tnp2 | ND | ND |
| Wfdc8 | ND | ND |
Changes in mRNA levels of putative T3-reponsive genes under untreated (Ctrl), PTU-treated (PTU), and reversal (Rev) conditions. Genes whose expression changed <2× are denoted by —. ND, not detected. Changes in mRNA levels were quantified based on microarray analyses.
DISCUSSION
The ChIP-seq procedure combines chromatin immunoprecipitation with high-throughput direct sequencing of short fragments of DNA (19,31). The method is devoid of sequence bias and has great sensitivity and consequently provides a way of mapping a wide variety of histone modifications across the whole genome (10,23). In this study we have applied this procedure to investigate the distribution of H3K4me3 marks at the Myh locus in response to reversible changes in gene expression triggered by chemical inhibition of T3 synthesis.
Our ChIP-seq analyses show a correlation between H3K4me3 signals and the gene expression status of Myh6 and Myh7 genes. When Myh6 gene expression is high, as under untreated and reversal conditions, we find detectable levels of H3K4me3 at the Myh6 locus. Similarly, when Myh7 gene expression is high, as under conditions of PTU treatment, we find detectable levels of H3K4me3 modifications at the Myh7 locus. When Myh6 gene expression is silenced, as under conditions of PTU treatment, H3K4me3 levels also drops to low levels at the Myh6 locus. Similarly, when Myh7 gene expression is silenced, under untreated and reversal conditions, H3K4me3 modifications likewise drop to low levels.
Our previous work using ChIP-qPCR showed that the promoter of Myh7 did not show detectable changes in H3K4me3 modifications in response to PTU-induced changes in gene expression (20). Consistent with these previous ChIP-qPCR data, our current ChIP-seq data do not show PTU-induced changes in the levels of H3K4me3 levels in the promoter region of Myh7. Rather, the distribution of this modification occurs at a region downstream of the TSS. Thus, our new ChIP-seq data imply that if H3K4me3 modifications play a role in the expression of the Myh7 gene, it is not by changing the modification levels in the promoter region. The significance of the differences in H3K4me3 modification at the two Myh promoters remains to be determined.
Interestingly, our data show the presence of a region of H3K4me3 modification in the intergenic region between Myh6 and Myh7 under untreated conditions, beginning approximately 2.5 kb upstream of Myh6 TSS and extending for 6.5 kb into the 3′ region of Myh7. This observation is a strong indicator that there is transcriptional activity in this intergenic region of the cardiac MHC locus, in confirmation of several previous reports demonstrating the presence of an antisense intergenic promoter at this location (4,5,7,9). This antisense promoter is thought to direct a transcript across the Myh7 gene and may play a role in controlling the switch between Myh6 and Myh7 gene expression. Multiple binding sites for the THR are present within this intergenic region, and previous work suggests that this promoter is under the control of T3, and has documented the presence of a PTU-responsive change in the levels of h3k9/14 ac and h3k4/k9 me3 at this intergenic promoter (8). Consistent with this our data show that PTU treatment leads to a dramatic reduction of H3K4me3 modification in this region, which is reversed when the PTU treatment is ceased, and the intergenic changes correlates with changes occurring within the Myh6 gene. Together, these observations of T3-mediated epigenetic changes in H3K4me3 signals at this location make a strong case for the presence of a functional antisense promoter in this intergenic region.
Our analyses demonstrate that 18 other putative T3-responsive genes show very modest changes in H3K4me3 levels in response to PTU. All of these are reductions in the levels of H3K4me3, consistent with previous observations that the vast majority of T3-responsive genes are positively regulated by T3. Since cardiac myocytes do not divide in the adult mouse heart to any appreciable degree, any decreases in H3K4me3 levels must be a direct result of active removal of the modification via demethylase rather than being the result of dilution due to cell division.
Even though Myh6 gene expression levels fully returned to their original state, the H3K4me3 modification within the Myh6 locus only partially returned to the original state. Thus, some peaks that were present under untreated conditions were absent under reversal conditions, while other peaks had a marked decrease in amplitude, although the expression levels were comparable between the two conditions. The achievement of full restoration of Myh6 gene expression, even though the H3K4me3 pattern is only partially restored, shows that full reexpression of Myh6 does not require that the H3K4me3 modifications return fully to the untreated conditions. The degree of reversal also varied at the non-Myh loci. Thus, one gene (Kcnj11) showed no reversal while five genes (Hr, Pln, Nppb, Slc2a4, and Ankrd1) showed partial reversal of the H3K4me3 distribution when PTU was withdrawn. Since each nucleosome contains two h3 molecules, a single nucleosome can have at least three discrete H3K4me3 modification states: zero (both h3k4 unmodified), one (only one H3K4me3 modified), or two (both h3k4 modified). The partial H3K4me3 levels seen at a particular position may represent either partial modification of all nucleosomes at that position, full modification of a fraction of DNA molecules in the population, or a combination of both. More work is required to distinguish between these various possibilities.
Our results also demonstrate that genes that show changes in mRNA levels, in general, also show changes in H3K4me3 levels but not vice versa. Thus, 75% (6/8) of genes that showed changes in mRNA levels showed changes in H3K4me3 levels, but only 30% (6/20) genes that showed changes in H3K4me3 levels also showed changes in mRNA levels. It is possible that the two genes that showed changes in mRNA levels in absence of H4K4me3 may be regulated in a posttranscriptional manner, which generally does not involve changes in histone modifications. It has also been demonstrated that some genes show changes in H3K4me3 levels in the absence of changes in mRNA levels—a phenomenon termed “poised”—as a prelude to subsequent increases in transcription. It is possible that several of the 14 genes that showed changes in H3K4me3 levels in the absence of changes in mRNA levels may belong to this category.
ACKNOWLEDGMENTS
The work was supported by grants from NIH (HL49277), Grant-in-Aid for Scientific Research (B) 22310117, Grant-in-Aid for Exploratory Research 23659050, from the Japan Society for the Promotion of Science, and Grant-in-Aid for Scientific Research on Innovative Areas 23125503 from the Ministry of Education, Culture, Sports, Science and Technology, Japan. We would like to thank Akashi Izumi and Kaori Shiina for Solexa sequencing, and Hiroshi Kimura for monoclonal antibody for H3K4me3, and Feng Li for the critical reading of the manuscript.
REFERENCES
- 1. Adamson C.; Maitra N.; Bahl J.; Greer K.; Klewer S.; Hoying J.; Morkin E. Regulation of gene expression in cardiomyocytes by thyroid hormone and thyroid hormone analogs 3,5-diiodothyropropionic acid and CGS 23425 [N-[3,5-dimethyl-4-(4′-hydroxy-3′-isopropylphenoxy)-phenyl]-oxamic acid]. J. Pharmacol. Exp. Ther. 311:164–171; 2004. [DOI] [PubMed] [Google Scholar]
- 2. Aranda A.; Pascual A. Nuclear hormone receptors and gene expression. Physiol. Rev. 81:1269–1304; 2001. [DOI] [PubMed] [Google Scholar]
- 3. Dillmann W. H. Cellular action of thyroid hormone on the heart. Thyroid 12:447–452; 2002. [DOI] [PubMed] [Google Scholar]
- 4. Giger J.; Qin A. X.; Bodell P. W.; Baldwin K. M.; Haddad F. Activity of the beta-myosin heavy chain antisense promoter responds to diabetes and hypothyroidism. Am. J. Physiol. Heart Circ. Physiol. 292: H3065–3071; 2007. [DOI] [PubMed] [Google Scholar]
- 5. Giger J. M.; Bodell P. W.; Baldwin K. M.; Haddad F. The CAAT-binding transcription factor 1/nuclear factor 1 binding site is important in beta-myosin heavy chain antisense promoter regulation in rats. Exp. Physiol. 94:1163–1173; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guenther M. G.; Levine S. S.; Boyer L. A.; Jaenisch R.; Young R. A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130:77–88; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haddad F.; Bodell P. W.; Qin A. X.; Giger J. M.; Baldwin K. M. Role of antisense RNA in coordinating cardiac myosin heavy chain gene switching. J. Biol. Chem. 278:37132–37138; 2003. [DOI] [PubMed] [Google Scholar]
- 8. Haddad F.; Jiang W.; Bodell P. W.; Qin A. X.; Baldwin K. M. Cardiac myosin heavy chain gene regulation by thyroid hormone involves altered histone modifications. Am. J. Physiol. Heart Circ. Physiol. 299:H1968–1980; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haddad F.; Qin A. X.; Bodell P. W.; Zhang L. Y.; Guo H.; Giger J. M.; Baldwin K. M. Regulation of antisense RNA expression during cardiac MHC gene switching in response to pressure overload. Am. J. Physiol. Heart Circ. Physiol. 290:H2351–2361; 2006. [DOI] [PubMed] [Google Scholar]
- 10. Ishihara K.; Oshimura M.; Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol. Cell 23:733–42; 2006. [DOI] [PubMed] [Google Scholar]
- 11. Kahaly G. J.; Dillmann W. H. Thyroid hormone action in the heart. Endocr. Rev. 26:704–728; 2005. [DOI] [PubMed] [Google Scholar]
- 12. Kim J.; Daniel J.; Espejo A.; Lake A.; Krishna M.; Xia L.; Zhang Y.; Bedford M. T. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 7:397–403; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kimura H.; Hayashi-Takanaka Y.; Goto Y.; Takizawa N.; Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 33:61–73; 2008. [DOI] [PubMed] [Google Scholar]
- 14. Kouzarides T. Chromatin modifications and their function. Cell 12:693–705; 2007. [DOI] [PubMed] [Google Scholar]
- 15. Le Bouter S.; Demolombe S.; Chambellan A.; Bellocq C.; Aimond F.; Toumaniantz G.; Lande G.; Siavoshian S.; Baro I.; Pond A. L.; Nerbonne J. M.; Leger J. J.; Escande D.; Charpentier F. Microarray analysis reveals complex remodeling of cardiac ion channel expression with altered thyroid status relation to cellular and integrated electrophysiology. Circ. Res. 92:234–242; 2003. [DOI] [PubMed] [Google Scholar]
- 16. Lee B. M.; Mahadevan L. C. Stability of histone modifications across mammalian genomes implications for ‘epigenetic’ marking. J. Cell. Biochem. 108:22–34; 2009. [DOI] [PubMed] [Google Scholar]
- 17. Li B.; Carey M.; Workman J. L. The role of chromatin during transcription. Cell 128:707–719; 2007. [DOI] [PubMed] [Google Scholar]
- 18. Mellor J.; Dudek P.; Clynes D. A glimpse into the epigenetic landscape of gene regulation. Curr. Opin. Genet. Dev. 18:116–122; 2008. [DOI] [PubMed] [Google Scholar]
- 19. Mendenhall E. M.; Bernstein B. E. Chromatin state maps new technologies, new insights. Curr. Opin. Genet. Dev. 18:109–115; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pandya K.; Pulli B.; Bultman S.; Smithies O. Reversible epigenetic modifications of the two cardiac myosin heavy chain genes during changes in expression. Gene Expr. 15:51–59; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Portman M. A. Thyroid hormone regulation of heart metabolism. Thyroid 18:217–225; 2008. [DOI] [PubMed] [Google Scholar]
- 22. Rundell V. L.; Manaves V.; Martin A. F.; de Tombe P. P. Impact of beta-myosin heavy chain isoform expression on cross-bridge cycling kinetics. Am. J. Physiol. Heart Circ. Physiol. 288:H896–903; 2005. [DOI] [PubMed] [Google Scholar]
- 23. Saitoh N.; Bell A. C.; Recillas-Targa F.; West A. G.; Simpson M.; Pikaart M.; Felsenfeld G. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 19:2315–2322; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Santos-Rosa H.; Schneider R.; Bannister A. J.; Sherriff J.; Bernstein B. E.; Emre N. C.; Schreiber S. L.; Mellor J.; Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature 419:407–411; 2002. [DOI] [PubMed] [Google Scholar]
- 25. Seila A. C.; Calabrese J. M.; Levine S. S.; Yeo G. W.; Rahl P. B.; Flynn R. A.; Young R. A.; Sharp P. A. Divergent transcription from active promoters. Science 322:1849–1851; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Rooij E.; Sutherland L. B.; Qi X.; Richardson J. A.; Hill J.; Olson E. N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316:575–579; 2007. [DOI] [PubMed] [Google Scholar]
- 27. Wada H.; Hashimoto K.; Wada Y.; Kobayashi M.; Izumi A.; Sugiyama A.; Kohro T.; Hamakubo T.; Kodama T. Extensive oligonucleotide microarray transcriptome analysis of the rat cerebral artery and arachnoid tissue. J. Atheroscler. Thromb. 9:224–232; 2002. [DOI] [PubMed] [Google Scholar]
- 28. Wada Y.; Ohta Y.; Xu M.; Tsutsumi S.; Minami T.; Inoue K.; Komura D.; Kitakami J.; Oshida N.; Papantonis A.; Izumi A.; Kobayashi M.; Meguro H.; Kanki Y.; Mimura I.; Yamamoto K.; Mataki C.; Hamakubo T.; Shirahige K.; Aburatani H.; Kimura H.; Kodama T.; Cook P. R.; Ihara S. A wave of nascent transcription on activated human genes. Proc. Natl. Acad. Sci. USA 106:18357–18361; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Z.; Zang C.; Rosenfeld J. A.; Schones D. E.; Barski A.; Cuddapah S.; Cui K.; Roh T. Y.; Peng W.; Zhang M. Q.; Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40:897–903; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yen P. M. Physiological and molecular basis of thyroid hormone action. Physiol. Rev. 81:1097–1142; 2001. [DOI] [PubMed] [Google Scholar]
- 31. Zecchini V.; Mills I. G. Putting chromatin immunoprecipitation into context. J. Cell. Biochem. 107:19–29; 2009. [DOI] [PubMed] [Google Scholar]