Abstract
Benzene, a tobacco constituent, is a leukemogen in humans and a carcinogen in rodents. Several benzene metabolites generate superoxide anion (O2•−) and induce nitric oxide synthase in the bone marrow of mice. We hypothesized that the reaction of nitric oxide (•NO) with O2•− leads to the formation of peroxynitrite as an intermediate during benzene metabolism. This hypothesis was supported by demonstrating that the exposure of mice to benzene produced nitrated metabolites and enhanced the levels of protein-bound 3-nitrotyrosine in the bone marrow of mice in vivo. In the current study, we investigated the influence of nitric oxide, generated from sodium 1-(N,N-diethylamino)diazen-1-ium-1,2-diolate, on DNA strand breaks induced by each single or binary benzene metabolite at different doses and compared the levels of the DNA damage induced by each benzene metabolite in the presence of nitric oxide with the levels of DNA strand breaks induced by peroxynitrite at similar doses in vitro. We found that among benzene metabolites only 1,2,4-trihydroxybenzene (BT) can induce significant DNA damage in the absence of nitric oxide. While 1,4-dihydroxybenzene (HQ), 1,4-benzo-quinone (BQ) and 1,2-dihydroxybenzene (CAT) require •NO to induce DNA strand breaks, hydroquinone was the most potent DNA-damaging benzene metabolite in the presence of •NO. The order of DNA breaks by benzene metabolites in the presence of •NO is: Peroxynitrite = HQ > BT > BQ > CAT. The •NO and O2•− scavengers inhibited DNA damage induced by [HQ+•NO]. Benzene, trans,trans-muconaldehyde, and phenol, do not induce DNA strand breaks either in the absence or presence of •NO. However, adding phenol to [HQ+•NO] leads to greater DNA damage than [HQ+•NO] alone. Collectively, these results suggest that nitric oxide is an important factor in DNA damage induced by certain benzene metabolites, probably via the formation of the peroxynitrite intermediate. Phenol, the major benzene metabolite that does not induce DNA damage alone and is inactive in vivo, synergistically enhances DNA damage induced by potent benzene metabolite in the presence of nitric oxide.
Keywords: benzene, peroxynitrite, nitric oxide, DNA strand breaks
Introduction
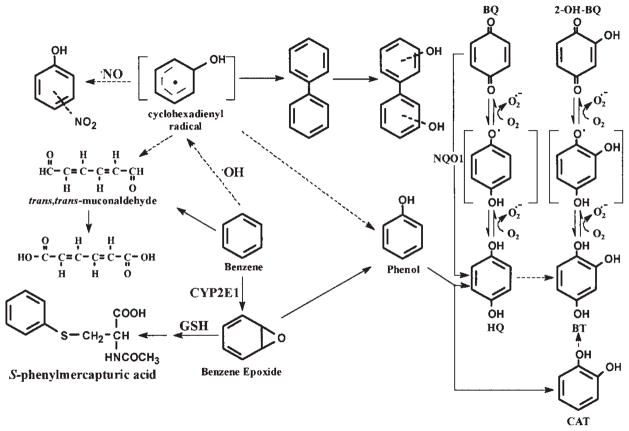
Benzene is known to depress bone marrow and to induce leukemia in humans and is a multisite carcinogen in rodents (1–3). It is a major industrial chemical, air pollutant and tobacco smoke constituent (4,5). Of the benzene-related leukemia cases in the USA, ~50% are attributed to tobacco smoking (6), and smokers have a two- to three-fold higher risk of myeloid leukemia than non-smokers (7). The molecular mechanism(s) responsible for benzene-induced bone marrow toxicity and leukemia have yet to be clearly defined. However, metabolism is essential for the myelotoxic effects of benzene (8–11). The cytochrome P450 2E1 which catalyzed the metabolic activation of benzene that primarily leads to ring-hydroxylated benzene metabolites (Fig. 1) cannot fully account for the benzene-induced toxicity because its primary metabolite (phenol) does not induce the same toxic reactions as benzene does (8–15). Thus, it is likely that other cellular processes participate in the bioactivation of benzene.
Figure 1.
The metabolic activation pathways of benzene.
Benzene induces iNOS and generates •NO and reactive oxygen species (ROS), such as superoxide anion (O2•−), hydroxyl radicals, hydrogen peroxide and singlet oxygen in the bone marrow of mice (16–23). iNOS is a cytochrome P-450 type of hemoprotein that contains a reductase and heme domain on the same polypeptide. The heme present in NOS is involved in the conversion of L-arginine to •NO and citrulline. The flavin-containing reductase domain is similar to the function of NADPH-dependent CYP reductase and is capable of producing O2•− with compounds such as quinones (24). The simultaneous production of nitric oxide and superoxide by iNOS may lead to the formation of peroxynitrite (ONOO−), a reactive intermediate that is known to cause protein damage through the nitration of tyrosine, tryptophan or cystein residue. It can also oxidize, nitrate and nitrosate biomolecules, such as DNA and lipids, thereby altering their function (25–29). Peroxynitrite can also nitrate aromatic compounds such as benzene and its metabolites, leading to the formation of toxic nitro-derivatives of benzene. Previously, we demonstrated the formation of nitro-derivatives of [14C] benzene metabolites in the bone marrow of mice treated with [14C] benzene (30) and found that benzene enhanced the levels of protein-bound 3-nitrotyrosine in the bone marrow of mice treated with benzene (31), suggesting the involvement of peroxynitrite in benzene metabolism. The present study initially aimed to investigate the influence of •NO on DNA damage induced by benzene and its metabolites and to compare them with DNA damage induced by peroxynitrite at the same doses. A second aim was to examine the synergistic interaction between benzene metabolites in the absence and presence of •NO on the induction of DNA nicks in vitro.
Materials and methods
Chemicals
Sodium phosphate, dimethyl sulfoxide (DMSO), diethylenetriamine penta-acetic acid (DTPA), xylene cyanol FF, glycerol, agarose, tris-borate/EDTA buffer, ethidium bromide and superoxide dismutase (SOD) were purchased from Sigma Chemical Co. (St. Louis, MO). Phenol, 1,4-dihydroxybenzene (hydroquinone, HQ), 1,4-benzoquinone (BQ), 1,2,4-trihydroxybenzene (benzenetriol, BT), 1,2-dihydroxybenzene (catechol, CAT), sodium azide, bromo-phenol blue, sodium periodate and cis-3,5-cyclohexadien-1,2-diol were procured from Aldrich Chemical Co. (Milwaukee, WI). The plasmid pBR322 DNA was obtained from Amersham Pharmacia Biotech (Piscataway, NJ). Sodium 1-(N,N-diethylamino)diazen-1-ium-1,2-diolate (DEA-NO) and 1H-imidazol-1-yloxy,2-(4-carboxyphenyl)-4,5-dihydro-4,4,5,5-tetramethyl-3-oxide potassium salt (carboxy-PTIO) were bought from Cayman Chemical Co. (Ann Arbor, MI).
The synthesis of peroxynitrite
Peroxynitrite was prepared by reacting ozone with sodium azide (32). Ozone was generated by passing dry oxygen (100 ml/min) through an ozonator (Fisher Model 500) at 120 V. The gas stream from the ozonator containing ~5% ozone in oxygen was bubbled through a glass frit into 100 ml of sodium azide solution at pH 12 and kept in an ice bath at 4°C (peroxynitrite is stable at pH 12). Peroxynitrite was quantified spectrophotometrically after dilution with 0.01 N NaOH (ε = 1670 M−1cm−1 for peroxy-nitrite at λ = 302 nm).
The synthesis of muconaldehydes
The cis,cis- and trans, trans-muconaldehydes were synthesized according to a previously reported procedure (33). The characteristic feature of the MS of synthesized muconaldehydes is the molecular ion at m/z 110 and a major fragment at m/z 81 (M-CHO)+. The UV spectrum in MeOH corresponded to the literature data at λmax 269 nm. The cis,cis- and trans,trans-muconaldehydes were analyzed by HPLC using a 300x3.9 mm, 10-μm Bone-clone column (Phenomenex, Torrance, CA). The column was eluted with a linear gradient from 100% H2O to 25:75 MeOH: H2O for 35 min at a flow rate of 1 ml/min. The cis,cis-muconaldehyde eluted at 22.6 min and the trans,trans-muconaldehyde at 24.2 min.
The reaction of plasmid DNA with benzene metabolites in the presence or absence of a •NO-releasing compound
These experiments were carried out as described by Yoshie and Ohshima (34) by incubating (at 37°C for 1 h) the plasmid pBR322 DNA (100 ng) in 100 mM sodium phosphate buffer, pH 7.4, containing 0.1 mM diethylenetriamine penta-acetic acid (DTPA) with the NO-releasing compound DEA-NO, (0.01–2 mM) alone or with benzene, or with one of its individual metabolites (0.01–1 mM), i.e., phenol, 1,4-dihydroxy-benzene (HQ), 1,4-benzoquinone (BQ), 1,2-dihydroxybenzene (CAT), 1,2,4-trihydroxybenzene (BT) or trans,trans-muconaldehyde with the volume of the final mixture at 10 μl. The reaction was terminated by adding 2 μl of the electrophoresis-loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol FF and 30% glycerol) followed by agarose gel electrophoresis. The pBR322 DNA incubated in buffer alone was used as the control. All experiments were carried out in triplicate. The effects of different concentrations of DEA-NO (0.01–5 mM) on DNA damage induced by HQ (0.2 mM) were also studied using plasmid DNA.
The reaction of plasmid DNA with peroxynitrite
The concentration of the synthetic peroxynitrite stock solution was determined by measuring the absorbance at λ = 302 nm of an aliquot diluted with 0.01 M aqueous NaOH versus that of an aliquot that was decomposed in 0.1 M sodium phosphate buffer (pH 7.4). The experiment was carried out by incubating plasmid pBR322 DNA (100 ng) in 100 mM sodium phosphate buffer, pH 7.4 containing 0.1 mM DTPA with the desired concentrations of peroxynitrite (0.01–1 mM). After adding peroxynitrite, the reaction mixture was immediately vortexed for 2 min, and then incubated at 37°C for 1 h. All reactions were carried out in 1 ml micro-centrifuge tubes allowing for large headspace.
The effects of various reactive oxygen and nitrogen scavengers on DNA strand breakage induced by hydroquinone in the presence of •NO
Incubation mixtures similar to that described above were prepared. In brief, the incubation included plasmid pBR322 DNA (100 ng) in 100 mM sodium phosphate buffer at pH 7.4, containing 0.1 mM DTPA, HQ (0.2 mM) and one of the scavengers (10 mM carboxy-PTIO, 10 mM DMSO or 5000 U/ml SOD). After adding DEA-NO (0.5 mM) to the above mixture, the reaction was incubated for 1 h at 37°C.
Agarose gel electrophoresis and measurements of pBR322 DNA strand breaks
A 10 μl sample out of 12 μl of each of the DNA incubation mixtures was loaded onto a 0.7% agarose gel prepared with 0.5X TBE buffer (45 mM tris-borate, 1 mM EDTA, pH 8.0), containing 0.5 μg/ml ethidium bromide. A Bio-Rad (Hercules, CA) sub-cell GT wide mini apparatus was used for gel electrophoresis at 8 V/cm for 80 min. The Bio-Rad molecular analyst software version 4 enabled us to measure the intensities of the supercoiled (I), relaxed (II, SSB) and linear form (III, double-strand break) DNA. The number of single-strand breaks per pBR322 DNA molecule was calculated according to equation 1, taking into account that a relaxed form of DNA (form II), when stained with ethidium bromide, gives a fluorescence intensity 1.4-fold greater than the supercoiled DNA (35).
| Equation (1) |
In Equation (1), SSB represents the number of single-strand breaks per DNA molecule induced directly by the damaging agent, while SC and OC are the fluorescence intensities of the supercoiled and relaxed circular forms of the DNA, respectively. The results were also expressed as the number of SSB per 104 bp (pBR322 consists of 4363 bp). The untreated pBR322 DNA contains ~3–10% SSB.
The oxidation of dihydrorhodamine to rhodamine as an indicator of peroxynitrite formation
Peroxynitrite readily oxidizes dihydrorhodamine (DHR) to form the fluorescent rhodamine (36). To determine the formation of peroxynitrite, the incubation mixtures consisting of DHR (50 μM) and DTPA (0.1 mM), reacted either with synthetic peroxynitrite (0.5 mM) or with a benzene metabolite (0.5 mM) plus DEA-NO (0.5 mM) in 50 mM potassium phosphate buffer (pH 7.4). The formation of peroxynitrite was determined based on the formation of rhodamine by UV absorbance at λ = 500 nm. DHR has no absorbance at 500 nm, whereas rhodamine has a high molar extinction coefficient (ε500 nm = 74500 M−1cm−1) at this wavelength.
Statistical analysis
The percentage of DNA breaks induced by peroxynitrite alone was compared with the DNA damage induced by each benzene metabolite in the absence or presence of •NO, using one-way analysis of variance (ANOVA), followed by Dunnett’s multiple comparison procedure (37). This comparison was made separately for each concentration (0.1, 0.5 and 1.0 mM). Alternatively, an area under the curve (AUC) of the percentage of DNA breaks versus the concentration of individual benzene metabolite or peroxynitrite (0.0–1 mM) was estimated separately for each of the three replicate experiments for each metabolite, using WinNonlin statistical software (38). The AUC values obtained for each metabolite were then compared to the AUC for peroxynitrite, using one-way ANOVA and Dunnett’s procedure.
Results
The pBR322 DNA SSB induced by a single benzene metabolite or binary mixture of benzene metabolites in the absence and presence of nitric oxide
Table I summarizes the levels of DNA SSB induced by various benzene metabolites at 0.25 mM. The metabolites tested were BT, HQ, BQ, CAT, phenol, trans,trans-muconaldehyde, as well as binary combinations of metabolites in the absence or presence of the •NO-releasing compound, DEA-NO (1 mM). Except for BT, incubations of the plasmid DNA with benzene or with one of its metabolites resulted in SSB that were not significantly different from the untreated control DNA. BT was the only metabolite that induced a significant level of SSB in supercoiled DNA in the absence of DEA-NO (3.48 SSB per 104 DNA base pairs). However, HQ, CAT and BQ, in the presence of DEA-NO, induced 3.7, 1.15 and 1.13 SSB per 104 base pairs, respectively. No significant DNA damage was observed with benzene, phenol or trans, trans-muconaldehyde in the presence of DEA-NO (0.07–0.16 SSB per 104 base pairs).
Table I.
The pBR322 plasmid DNA single-strand breaks induced by benzene metabolites in the absence or presence of the NO-releasing compound (DEA-NO).a
| Benzene metabolites | SSB per 104 bp DNAb Absence of DEA-NO |
SSB per 104 bp DNAb Presence of DEA-NO |
Ratio of DNA single-strand breakage Presence/absence of DEA-NO |
|
|---|---|---|---|---|
| Control | - | 0.07±0.009 | 0.07±0.009 | 1.0 |
| HQ | - | 0.14±0.028 | 3.70±0.160 | 26.4 |
| BT | - | 3.48±0.038 | 3.96±0.100 | 1.1 |
| CAT | - | 0.07±0.008 | 1.15±0.200 | 16.4 |
| BQ | - | 0.08±0.026 | 1.13±0.079 | 14.1 |
| Phenol | - | 0.07±0.028 | 0.16±0.032 | 2.3 |
| Benzene | - | 0.07±0.009 | 0.07±0.007 | 1.0 |
| trans, trans-MA | - | 0.07±0.040 | 0.14 ±0.050 | 2.0 |
| HQ | CAT | 0.12±0.060 | 4.88±0.027 | 40.6 |
| HQ | BT | 1.14±0.120 | 3.38±0.080 | 3.0 |
| HQ | Phenol | 0.12±0.048 | 6.44 ±0.041 | 53.6 |
| HQ | BQ | 0.25±0.068 | 3.36±0.580 | 13.4 |
| CAT | BT | 0.87±0.210 | 2.58±0.095 | 3.0 |
| CAT | Phenol | 0.10±0.030 | 1.34±0.039 | 13.4 |
| BT | Phenol | 1.47±0.480 | 3.27±0.081 | 2.2 |
Incubations included pBR322 DNA (100 ng), DEA-NO (1 mM) and benzene metabolites (0.25 mM). Data are the means of at least three different experiments.
Single-strand DNA breakage is calculated based on the Possion formula as described in Material and methods (35).
In assays that employed the binary combinations of metabolites, phenol, in the presence of •NO, increased the levels of HQ-induced SSB from 3.7 to 6.44 SSB per 104 DNA base pairs (Table I). In contrast, phenol, HQ or CAT, in the absence of DEA-NO, decreased BT-induced SSB from 3.48 to 1.47, 1.14 and 0.87 SSB per 104 base pairs, respectively (Table I).
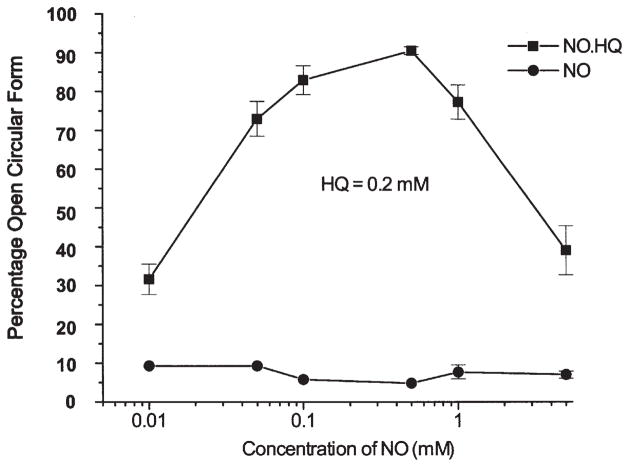
The effects of •NO concentrations on HQ-induced pBR322 plasmid DNA single-strand breaks
We used HQ in this study because, as described above, it is a potent DNA-damaging metabolite in the presence of •NO. Fig. 3 shows the levels of SSB induced in the plasmid DNA following incubation with 0.2 mM HQ while the concentrations of •NO varied from 0.01 to 5 mM. The DNA SSB reached a maximum and at concentrations >0.5 mM DEA-NO the number of SSB declined (Fig. 3).
Figure 3.
The effects of various concentrations of the NO-releasing compound (DEA-NO, 0.01–5 mM) on DNA single-strand breakage induced by HQ (0.2 mM).
The dose-response of DNA strand-breakage induced by benzene metabolites in the presence of •NO and the dose-response of DNA damage caused by peroxynitrite alone
Fig. 4 compares the levels of SSB induced by peroxynitrite alone, in the range of 0.01–1 mM, with those induced by the equimolar doses of benzene or its individual metabolites in the absence or presence of 0.5 mM DEA-NO. The mean of the SSB levels at 0.1, 0.5 and 1 mM and the areas under curve (AUCs) (0–1 mM) are shown in Table II. At 0.1, 0.5 or 1 mM, the mean of the SSB levels induced by peroxynitrite was not significantly different from that induced by [HQ+•NO] (Table II). AUCs (0–1 mM) for the SSB of DNA induced by peroxynitrite were ~1.4-, 1.8- and 2.1-fold greater than those obtained with [BT+•NO], [BQ+•NO] and [CAT+•NO], respectively (p=0.01). SSB were not produced by benzene, individual metabolites alone (except BT), [phenol+•NO] or [benzene+•NO], at any concentration tested (p<0.01, compared to peroxynitrite). The pattern of DNA strand breaks induced by [HQ+•NO], [BQ+•NO] or [CAT+•NO] was similar to that caused by peroxynitrite alone (Fig. 4).
Figure 4.
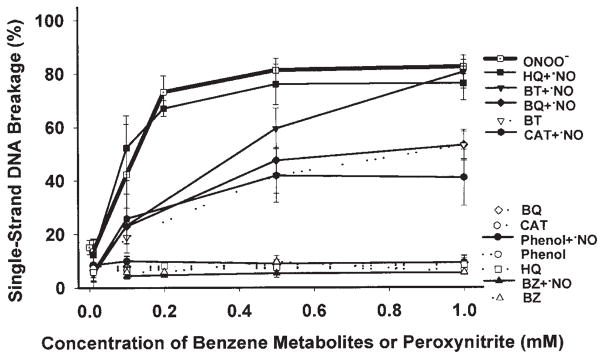
The percentage of DNA single-strand breaks induced by various concentrations of peroxynitrite (0.01–1 mM) or benzene metabolites in the absence or presence of 0.5 mM DEA-NO.
Table II.
The pBR322 DNA damage induced at different concentrations of peroxynitrite or by benzene metabolites in the presence of the NO-releasing compound (DEA-NO).a
| Compounds | Percent of single-strand DNA breaks at different concentrations of peroxynitrite or benzene metabolites (mean ± SD) | AUCb 0.0–1.0 mM (mean ± SD) |
||
|---|---|---|---|---|
| Concentration of peroxynitrite or benzene metabolites | 0.1 mM | 0.5 mM | 1.0 mM | |
| Peroxynitrite | 42.3±18.9 | 81.2±4.5 | 82.4±2.7 | 72.4±5.9 |
| HQ+•NO | 54.0±12.2 | 77.7±7.6 | 78.0±6.3 | 70.2±6.2 |
| BQ+•NO | 22.8±3.3c | 47.5±4.9d | 53.2±5.8d | 40.6±4.3d |
| CAT+•NO | 24.3±6.9c | 39.6±6.7d | 38.1±6.0d | 33.7±6.3d |
| BT+•NO | 23.3±5.5c | 57.4±2.4d | 80.7±8.9 | 52.0±2.7d |
| Phenol+•NO | 4.3±3.3d | 3.3±2.3d | 3.6±1.3d | 3.6±2.3d |
| Benzene+•NO | 0.1±0.1d | 2.8±1.6d | 0.1±0.1d | 0.7±0.4d |
| HQ | 1.1±0.8d | 1.9±0.7d | 2.8±2.0d | 1.9±0.7d |
| BQ | 0.1±0.1d | 0.7±1.2d | 1.1±1.0d | 0.6±0.7d |
| CAT | 2.4±2.2d | 1.7±1.6d | 2.3±1.3d | 2.0±1.6d |
| BT | 20.2±5.3d | 42.6±5.5d | 53.3±3.6d | 37.7±4.5d |
| Phenol | 1.1±1.7d | 1.6±1.5d | 1.4 ±1.4d | 1.4±1.3d |
| Benzene | 0.3±0.4d | 1.3±0.5d | 1.3±0.8d | 1.0±0.9d |
Incubations included pBR322 DNA (100 ng), DEA-NO (0.5 mM), ring-hydroxylated benzene metabolites or peroxynitrite (0.01–1mM).
AUC, area under curve. The values shown in the column AUC include concentrations of 0.01, 0.1, 0.2, 0.5 and 1.0 mM that are shown in Fig. 4.
Indicates a significant difference from peroxynitrite alone at p=0.05.
Indicates a significant difference from peroxynitrite alone at p=0.01.
The oxidation of dihydrorhodamine to rhodamine in the presence of peroxynitrite or of benzene metabolites plus •NO
Incubations of dihydrorhodamine with 0.5 mM peroxynitrite, or 0.5 mM [HQ+•NO] or [CAT+•NO] resulted in the formation of rhodamine at levels of 0.14, 0.07 and 0.007 mM, respectively, as quantified by UV absorption at λ = 500 nm. The levels of rhodamine formed from the oxidation of dihydrorhodamine could not be determined for BQ and BT because of color interference.
The effects of reactive oxygen and nitrogen scavengers on plasmid DNA nicks induced by •NO+HQ
To identify the intermediate(s) that may be responsible for the synergistic effect of •NO with HQ in inducing SSB, we examined the effects of the various scavengers (at 10 mM) and SOD (at 5000 U/ml) on DNA nicks induced by HQ. We observed a 90% inhibition of DNA damage by PTIO (a •NO-trapping agent), 65% by SOD (an O2•− scavenger) and 45% by DMSO (an •OH trapping agent). Ascorbic acid did not inhibit DNA damage induced by HQ, but increased DNA strand breaks by 20%.
Discussion
It has been shown that benzene can induce iNOS in the bone marrow of mice (22). The objective of the current study was to investigate which benzene metabolite(s) is (are) the major contributor to the observed DNA damage in the presence of •NO. Our results indicate that benzene and its metabolites, except BT, do not induce any remarkable damage to the supercoiled DNA. While in the presence of the •NO-releasing compound, several benzene metabolites produce DNA strand breaks in supercoiled DNA. Furthermore, similar effects were also observed in calf thymus DNA by alkaline gel electrophoresis (data not shown). HQ is the most potent DNA-damaging metabolite in the presence of •NO, followed by BT, BQ and CAT (Fig. 4 and Table II). Benzene, phenol and trans, trans-muconaldehyde do not produce superoxide anion, and thus do not induce DNA damage in the absence or presence of •NO.
The comparison of DNA damage induced by peroxynitrite or by benzene metabolites with •NO in a dose response assay indicates that the patterns of DNA damage produced by [HQ+•NO], [CAT+•NO] and [BQ+•NO] are similar to the biphasic nature of DNA strand breaks induced by peroxynitrite (39) (Fig. 4). Moreover, the incubation of HQ or CAT in combination with •NO oxidizes dihydrorhodamine to rhodamine. Previous reports indicate that peroxynitrite is capable of oxidizing dihydrorhodamine, while H2O2 does not (36). Carboxy-PTIO (a NO-trapping agent), SOD (an O2•− scavenger) and, to a lesser extent, DMSO (an •OH scavenger) blocked DNA damage induction by HQ in the presence of •NO. This suggests that •NO and O2•− are needed to exert the effect on DNA breakage. Quantitatively, there is no significant difference between plasmid DNA damage induced by peroxynitrite or by [HQ+•NO] (Table II). Similar results were obtained with calf thymus DNA by alkaline gel electrophoresis (data not shown). It is possible that nitric oxide can mediate the conversion of HQ, CAT and BT to their respective semiquinone free radical forms, as shown in Fig. 2, resulting in a one-electron reduction of •NO to the nitroxyl anion (NO−). NO− (in its triplet state) can react with O2 to give ONOO−. Semiquinones can also react with •NO to form NO−, which subsequently leads to ONOO−. Alternatively, •NO can rapidly react with O2•−, generated during redox cycling, to form ONOO−. Thus, nitric oxide can have a dual role in the production of DNA strand breaks: 1) it can increase the oxidation of ring-hydroxylated benzene metabolites to corresponding quinones, and 2) it can lead to the formation of peroxynitrite. It has been demonstrated that ONOO−, as well as NO− can cause strand breaks in plasmid DNA (40).
Figure 2.
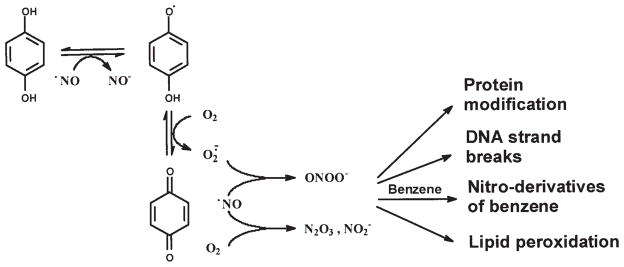
The proposed pathways of peroxynitrite generation by a representative benzene metabolite.
In assays that involved binary mixtures, the inclusion of phenol resulted in a two-fold enhancement of the DNA damage induced by [HQ+•NO] (Table I). This observation appears to be in agreement with in vivo genotoxicity studies in which HQ, administered separately to the mice, induced micronuclei in the mouse bone marrow and phenol alone had no genotoxic effects while the binary mixture that consisted of phenol and HQ led to a greater micronuclei induction in the bone marrow of mice than the exposure of the mice to HQ alone (41). Similarly, the co-administration of phenol with HQ resulted in a significant loss of bone marrow cellularity and an increased chromosome loss and breakage in the mice (42). The mechanism by which phenol increased HQ-induced toxicity is unknown. It has been demonstrated that the formation of peroxynitrite is maximal when •NO and O2•− are at equimolar levels, whereas by increasing the concentration of one relative to the other results in a decreased oxidation reaction (43). A similar phenomenon is observed in the current study as well (Fig. 3). On the basis of this observation, the mechanism by which phenol enhances DNA damage induced by HQ in the presence of •NO, occurs via a phenoxy radical that enhances superoxide anion formation, which can have an impact on the extent of DNA damage induced by HQ.
We found that BT is the only benzene metabolite that induces scission in supercoiled DNA in the absence of •NO. The induction of DNA damage by BT is partially inhibited by adding phenol, HQ or CAT. BT is a strong reducing agent which readily participates in co-oxidation and auto-oxidation, generating a reactive oxygen species (O2•−, H2O2, •OH and semiquinone) (44). The mechanisms by which BT induces DNA damage are apparently distinct from those involving HQ or CAT.
Our results described herein, suggest that excess •NO generated by the metabolism of benzene in vivo may have an important role in DNA damage induced by benzene, likely via peroxynitrite formation. This effect may be responsible for bone marrow cell toxicity. Furthermore, peroxynitrite can also convert benzene metabolites to toxic nitrated derivatives (30). In general, nitro substitution is known to alter the mutagenic and carcinogenic activity of parent hydrocarbons (45,46).
Acknowledgments
The authors would like to thank Brian Pitman for his assistance with the statistical analyses. This work was supported by the National Cancer Institute grants CA70972 and CA17613.
Abbreviations
- BZ
benzene
- BQ
1,4-benzoquinone
- BT
1,2,4-trihydroxybenzene
- carboxy-PTIO
1H-imidazol-1-yloxy,2-(4-carboxyphenyl)-4,5-dihydro-4,4,5,5-tetramethyl-3-oxide potassium salt
- CAT
1,2-dihydroxybenzene
- DEA-NO
sodium 1-(N,N-diethylamino)diazen-1-ium-1,2-diolate
- DTPA
diethylenetriamine-penta-acetic acid
- DHR
dihydrorhodamine
- HQ
1,4-dihydroxy-benzene
- •NO
nitric oxide
- iNOS
inducible nitric oxide synthase
- O2•−
superoxide anion radical
- SSB
single-strand DNA breaks
- SOD
superoxide dismutase
- ONOO−
peroxynitrite
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- trans,trans-MA
trans,trans-muconaldehyde
References
- 1.Snyder R. Benzene and leukemia. Crit Rev Toxicol. 2002;32:155–210. doi: 10.1080/20024091064219. [DOI] [PubMed] [Google Scholar]
- 2.Schnatter AR, Rosamilia K, Wojcik NC. Review of the literature on benzene exposure and leukemia subtypes. Chem Biol Interact. 2005;153–154:9–21. doi: 10.1016/j.cbi.2005.03.039. [DOI] [PubMed] [Google Scholar]
- 3.Huff J. Chemicals studied and evaluated in long-term carcinogenesis bioassays by both the Ramazzini Foundation and the National Toxicology Program: in tribute to Cesare Maltoni and David Rall. Ann NY Acad Sci. 2002;982:208–230. doi: 10.1111/j.1749-6632.2002.tb04935.x. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann D, Hoffmann I, El-Bayoumy K. The less harmful cigarette: a controversial issue. A tribute to Ernest L. Wynder. Chem Res Toxicol. 2001;14:767–790. doi: 10.1021/tx000260u. [DOI] [PubMed] [Google Scholar]
- 5.Thomas X, Chelghoum Y. Cigarette smoking and acute leukemia (review) Leuk Lymphoma. 2004;45:1103–1109. doi: 10.1080/10428190310001638904. [DOI] [PubMed] [Google Scholar]
- 6.Korte JE, Hertz-Picciotto I, Schulz MR, Ball LM, Duell EJ. The contribution of benzene to smoking-induced leukemia. Environ Health Perspect. 2000;108:333–339. doi: 10.1289/ehp.00108333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brownson R, Novotny TE, Perry CP. Cigarette smoking and adult leukemia. A meta-analysis. Arch Intern Med. 1993;153:469–474. [PubMed] [Google Scholar]
- 8.Valentine JL, Lee SS, Seaton MJ, Asgharian B, Farris G, Corton JC, Gonzalez FJ, Medinsky MA. Reduction of benzene metabolites and toxicity in mice that lack CYP2E1 expression. Toxicol Appl Pharmacol. 1996;141:205–213. doi: 10.1006/taap.1996.0277. [DOI] [PubMed] [Google Scholar]
- 9.Rana SV, Verma Y. Biochemical toxicity of benzene (review) J Environ Biol. 2005;26:157–168. [PubMed] [Google Scholar]
- 10.Snyder R. Xenobiotic metabolism and the mechanism(s) of benzene toxicity (review) Drug Metab Rev. 2004;36:531–547. doi: 10.1081/dmr-200033445. [DOI] [PubMed] [Google Scholar]
- 11.Ross D. Functions and distribution of NQO1 in human bone marrow potential clues to benzene toxicity (review) Chem Biol Interact. 2005;153–154:137–146. doi: 10.1016/j.cbi.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Ross D. The role of metabolism and specific metabolites in benzene-induced toxicity: evidence and issues. J Toxicol Environ Health. 2000;61A:357–372. doi: 10.1080/00984100050166361. [DOI] [PubMed] [Google Scholar]
- 13.Subrahmanyam VV, Ross D, Eastmond DA, Smith MT. Potential role of free radicals in benzene-induced myelotoxicity and leukemia. Free Radic Biol Med. 1991;11:495–515. doi: 10.1016/0891-5849(91)90063-9. [DOI] [PubMed] [Google Scholar]
- 14.Sabourin PJ, Muggenburg BA, Couch RC, Lefler D, Lucier G, Birnbaum LS, Henderson RF. Metabolism of [14C]benzene by cynomolgus monkeys and chimpanzees. Toxicol Appl Pharmacol. 1992;14:277–284. doi: 10.1016/0041-008x(92)90078-7. [DOI] [PubMed] [Google Scholar]
- 15.National Toxicology Program (NTP) bioassay of phenol for possible carcinogenicity. NCI/NTP; Bethesda, MD: 1986. [Google Scholar]
- 16.Koachana P, Subrahmanyam VV, Meyer KB, Zhang L, Smith MT. Benzene and its phenolic metabolites produce oxidative DNA damage in HL60 cells in vitro and in bone marrow in vivo. Cancer Res. 1993;53:1023–1026. [PubMed] [Google Scholar]
- 17.Rao NR, Snyder R. Oxidative modifications produced in HL-60 cells on exposure to benzene metabolites. J Appl Toxicol. 1995;15:403–409. doi: 10.1002/jat.2550150511. [DOI] [PubMed] [Google Scholar]
- 18.Horita M, Wang DH, Tsutsui K, Sano K, Masuoka N, Kira S. Involvement of oxidative stress in hydroquinone-induced cytotoxicity in catalase-deficient Escherichia coli mutants. Free Rad Res. 2005;39:1035–1041. doi: 10.1080/10715760500232008. [DOI] [PubMed] [Google Scholar]
- 19.Wan J, Badham HJ, Winn L. The role of c-MYB in benzene-initiated toxicity (review) Chem Biol Interact. 2005;153–154:171–178. doi: 10.1016/j.cbi.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 20.Fabiani R, De Bartolomeo A, Morozzi G. Involvement of oxygen-free radicals in the serum-mediated increase of benzo-quinone genotoxicity. Environ Mol Mutagen. 2005;46:156–163. doi: 10.1002/em.20146. [DOI] [PubMed] [Google Scholar]
- 21.Shen Y, Shen HM, Shi CY, Ong CN. Benzene metabolites enhance reactive oxygen species generation in HL60 human leukemia cells. Hum Exp Toxicol. 1996;15:422–427. doi: 10.1177/096032719601500510. [DOI] [PubMed] [Google Scholar]
- 22.Laskin DL, Heck DE, Punjabi CJ, Laskin JD. Nitric oxide as mediator of benzene-induced hematosuppression and toxicity. J Toxicol Environ Health. 2000;61:413–417. doi: 10.1080/00984100050166433. [DOI] [PubMed] [Google Scholar]
- 23.Vestergaard S, Loft S, Möller P. Role of inducible nitrogen oxide synthase in benzene-induced oxidative DNA damage in the bone marrow of mice. Free Rad Biol Med. 2002;32:481–484. doi: 10.1016/s0891-5849(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 24.White KA, Marletta MA. Nitric oxide synthase is cytochrome P-450 type hemoprotein. Biochemistry. 1992;31:6627–6631. doi: 10.1021/bi00144a001. [DOI] [PubMed] [Google Scholar]
- 25.Beckman JS, Koppenol WH. Nitric oxide, superoxide, and peroxynitrite: The good, the bad and the ugly. Am J Physiol. 1996;271:C1424–C1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 26.Tamir S, Tannenbaum SR. The role of nitric oxide (NO) in the carcinogenic process (review) Biochim Biophys Acta. 1996;1288:F31–F36. doi: 10.1016/0304-419x(96)00021-2. [DOI] [PubMed] [Google Scholar]
- 27.Ducrocq C, Blanchard B, Pignatelli B, Ohshima H. Peroxynitrite: an endogenous oxidizing and nitrating agent (review) Cell Mol Life Sci. 1999;55:1068–1077. doi: 10.1007/s000180050357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radi R, Beckman JS, Bush KM, Freeman BA. Peroxynitrite-induced membrane lipid peroxidation: the cytotoxic potential of superoxide and nitric oxide. Arch Biochem Biophys. 1991;288:481–487. doi: 10.1016/0003-9861(91)90224-7. [DOI] [PubMed] [Google Scholar]
- 29.Ohshima H, Bartsch H. Chronic infection and inflammatory processes as cancer risk factor; possible role of nitric oxide in carcinogenesis. Mutat Res. 1994;305:253–264. doi: 10.1016/0027-5107(94)90245-3. [DOI] [PubMed] [Google Scholar]
- 30.Chen K-M, El-Bayoumy K, Cunningham J, Aliaga C, Li H, Melikian AA. Detection of nitrated benzene metabolites in bone marrow of B6C3F1 mice treated with benzene. Chem Res Toxicol. 2004;17:370–377. doi: 10.1021/tx030039s. [DOI] [PubMed] [Google Scholar]
- 31.Chen K-M, El-Bayoumy K, Hosey J, Cunningham J, Aliaga C, Li H, Melikian AA. Benzene increase protein-bound 3-nitrotyrosine in bone marrow of B6C3F1 mice. Chem Biol Interact. 2005;156:81–91. doi: 10.1016/j.cbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Pryor WA, Cueto R, Jin X, Koppenol WH, Ngu-Schwemlein M, Squadrito GL, Uppu PL, Uppu RM. A practical method for preparing peroxynitrite solutions of low ionic strength and free of hydrogen peroxide. Free Rad Biol Med. 1995;18:75–83. doi: 10.1016/0891-5849(94)00105-s. [DOI] [PubMed] [Google Scholar]
- 33.Golding BT, Kennedy G, Watson WP. Simple synthesis of isomers of muconaldehydes, D-tetrahydrophtalaldehyde and related compounds. J Am Chem Soc. 1952;74:3014–3018. [Google Scholar]
- 34.Yoshie Y, Ohshima H. Nitric oxide synergistically enhances DNA strand breakage induced by polyhydroxyaromatic compounds, but inhibits that induced by Fenton reaction. Arch Biochem Biophys. 1997;342:13–21. doi: 10.1006/abbi.1997.0100. [DOI] [PubMed] [Google Scholar]
- 35.Epe B, Hegler J. Oxidative DNA damage: Endonuclease fingerprinting. Methods Enzymol. 1994;234:122–131. doi: 10.1016/0076-6879(94)34083-8. [DOI] [PubMed] [Google Scholar]
- 36.Kooy NW, Royall JA, Ischiropoulos H, Beckman JS. Peroxynitrite-mediated oxidation of dihydrorhodamine 123. Free Rad Biol Med. 1994;16:149–156. doi: 10.1016/0891-5849(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 37.Dunnett CW. A multiple comparisons procedure for comparing several treatments with a control. J Am Stat Assoc. 1955;50:1096–1121. [Google Scholar]
- 38.WinNonlin Professional, version 3.1, Pharsight Corporation, Mountain View, CA.
- 39.Kennedy LJ, Moor K, Jr, Caulfield JL, Tannenbaum SR, Dedon PC. Quantitation of 8-oxoguanine and strand breaks produced by four oxidizing agents. Chem Res Toxicol. 1997;10:386–392. doi: 10.1021/tx960102w. [DOI] [PubMed] [Google Scholar]
- 40.Yoshie Y, Ohshima H. Synergistic induction of DNA strands breakage by catechol-estrogen and nitric oxide: implications for hormonal carcinogenesis. Free Rad Biol Med. 1998;24:341–348. doi: 10.1016/s0891-5849(97)00269-4. [DOI] [PubMed] [Google Scholar]
- 41.Marrazzini A, Chelotti L, Barrai I, Loprieno N, Barale R. In vivo genotoxic interaction among three phenolic benzene metabolites. Mutat Res. 1994;341:29–46. doi: 10.1016/0165-1218(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, Eastmond DA. Synergistic increase in chromosomal breakage within the euchromatin induced by an interaction of benzene metabolites phenol and hydroquinone in mice. Carcinogenesis. 1995;16:1963–1969. doi: 10.1093/carcin/16.8.1963. [DOI] [PubMed] [Google Scholar]
- 43.Miles AM, Bohle DS, Glassbrenner PA, Hansert B, Wink DA, Grisham MB. Modulation of superoxide-dependent oxidation and hydroxylation reactions by nitric oxide. J Biol Chem. 1996;271:40–47. doi: 10.1074/jbc.271.1.40. [DOI] [PubMed] [Google Scholar]
- 44.Li ASH, Bandy B, Tsang SS, Davison AJ. DNA breakage induced by 1,2,4-benzenetriol: relative contribution of oxygen-derived active species and transition metal ion. Free Rad Biol Med. 2001;30:943–956. doi: 10.1016/s0891-5849(01)00478-6. [DOI] [PubMed] [Google Scholar]
- 45.El-Bayoumy K. Environmental carcinogens that may be involved in human breast cancer etiology. Chem Res Toxicol. 1992;5:585–590. doi: 10.1021/tx00029a001. [DOI] [PubMed] [Google Scholar]
- 46.Amin S, El-Bayoumy K. Tumorigenicity of polycyclic aromatic hydrocarbons. On the possible contribution of PAHs and their nitro-derivatives to the development of human breast cancer. In: Luch A, editor. The Carcinogenic Effects of Polycyclic Aromatic Hydrocarbons. Imperial College Press; London: 2005. pp. 315–351. [Google Scholar]