Summary
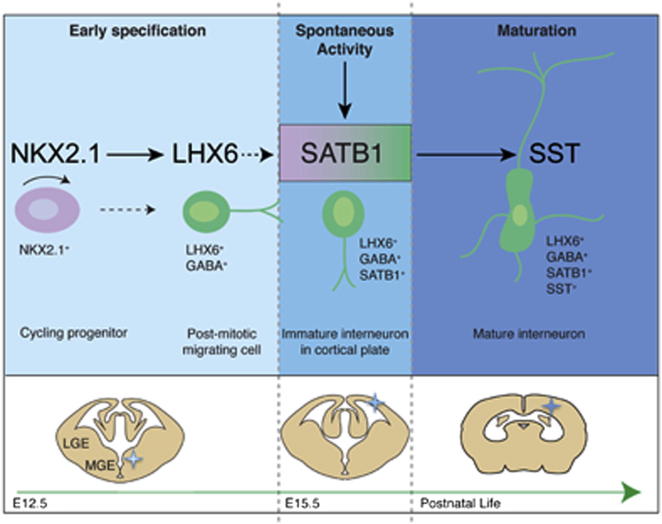
The generation of cortical interneuron subtypes is controlled by genetic programs that are activated in the ventral forebrain and unfold during the prolonged period of inhibitory neuron development. The LIM-homeodomain protein LHX6 is critical for the development of all cortical interneurons originating in the medial ganglionic eminence, but the molecular mechanisms that operate downstream of LHX6 to control the terminal differentiation of somatostatin- and parvalbumin-expressing interneurons within the cortex remain unknown. Here, we provide evidence that the nuclear matrix and genome organizer protein SATB1 is induced by neuronal activity and functions downstream of Lhx6 to control the transition of tangentially migrating immature interneurons into the terminally differentiated Somatostatin (SST)-expressing subtype. Our experiments provide a molecular framework for understanding the genetic and epigenetic mechanisms by which specified but immature cortical interneurons acquire the subtype-defining molecular and morphophysiological characteristics that allow them to integrate and function within cortical circuits.
Graphical Abstract
Highlights
► Satb1 is specifically expressed in MGE-derived cortical interneurons ► Satb1 is required for the maturation of MGE-derived cortical interneurons in vivo ► Satb1 promotes the maturation of SST+ cortical interneurons downstream of Lhx6 ► Neuronal activity induces expression of Satb1 in immature cortical interneurons
Cortical interneurons play a crucial role in the function of cerebral circuits, and abnormal development of interneurons results in severe neurodevelopmental disorders. Pachnis and colleagues demonstrate that the genome organizer SATB1 can be activated by neuronal activity and functions downstream of the transcription factor LHX6 to promote the differentiation of somatostatin-expressing interneurons. Mice deficient in SATB1 fail to differentiate this critical group of interneurons. These findings identify a key regulator of neural circuit formation in the mammalian cortex.
Introduction
Motor planning and execution, sensory representation, and cognitive activity depend on the integrated function of cortical circuits that consist of glutamatergic projection neurons and GABAergic local circuit interneurons. In rodents, cortical interneurons constitute approximately 20% of all cortical neurons and are extremely diverse (Markram et al., 2004). They are generated during embryogenesis in two transient subpallial structures, the medial and caudal ganglionic eminence (MGE and CGE), and reach the cortex by tangential migration (Wonders and Anderson, 2006; Welagen and Anderson, 2011). Although the different groups of interneurons are endowed with subtype-specifying genetic programs at their sites of origin in the ventral forebrain, during tangential migration they are morphologically indistinguishable and lack many of the properties of functionally mature inhibitory neurons (Batista-Brito and Fishell, 2009; Cossart, 2011). Termination of tangential migration and entry into the cortical plate heralds the onset of terminal differentiation and maturation, which culminates in the generation of distinct interneuron subtypes identified by characteristic morphological, electrophysiological and molecular properties (Butt et al., 2005). For example, the neuropeptide somatostatin (SST) and the calcium-binding protein parvalbumin (PV) mark nonoverlapping groups that originate in the MGE, while the vasoactive intestinal peptide (VIP) is expressed specifically by CGE-derived interneurons (Kawaguchi and Kubota, 1997; Markram et al., 2004; Rudy et al., 2011).
We and others have previously demonstrated that the LIM homeodomain (HD) transcription factor LHX6 plays a pivotal role in the generation of MGE-derived cortical interneurons. Lhx6 is induced early in all MGE-derived interneuron precursors and is required in a dose-dependent manner for the differentiation of the SST+ and PV+ groups of interneurons and the assembly of functional inhibitory circuits (Liodis et al., 2007; Zhao et al., 2008; Neves et al., 2012). Although Lhx6 controls the development of all MGE-derived interneurons, the unfolding of Lhx6-dependent differentiation and maturation programs follows subtype-specific spatiotemporal patterns. This is underscored by the differential spatiotemporal expression profile of subtype markers, such as PV and SST. Both Pv mRNA and protein are expressed only at relatively late stages of interneuron development in the postnatal cortex (del Río et al., 1994; Taniguchi et al., 2011). In contrast, Sst mRNA is expressed during embryogenesis by tangenitally migrating immature interneurons, although the SST peptide is detected exclusively in postnatal animals (Taniguchi et al., 2011; Neves et al., 2012). These observations suggest that region- and stage-specific extrinsic factors are likely to interact with intrinsic genetic determinants to regulate the different phases of cortical interneuron development in the mammalian forebrain. The identity of such extrinsic signals and the intrinsic factors that regulate interneuron differentiation downstream of LHX6 remain largely unknown.
In this study, we identify the genome organizer Special AT-rich DNA Binding Protein 1 (SATB1) as a key regulator of cortical interneuron development. We provide evidence that Satb1 is specifically activated in MGE-derived interneurons within the cortical plate in an activity-dependent manner. By manipulating its expression in the MGE, we demonstrate that SATB1 inhibits the tangential migration and controls the differentiation of SST+ interneurons downstream of LHX6. In addition, genetic ablation of Satb1 leads to differentiation deficits primarily of the SST+ group of cortical interneurons in vivo. Together, our studies identify SATB1 as a critical regulator of interneuron maturation and terminal differentiation in the mammalian cortex.
Results
Satb1 Is Expressed in MGE-Derived Cortical Interneurons following Entry into the Cortical Plate
To identify genes implicated in the development of cortical inhibitory circuits, we carried out a microarray-based comparative profiling of gene expression of dorsal forebrain from embryonic day (E) 15.5 wild-type and Lhx6-null mice (Liodis et al., 2007) (see Experimental Procedures). Our screen identified genes already known to be expressed in cortical interneurons in an Lhx6-dependent manner (such as Kcnc1, also referred to as Kv3.1, Npas1, Npy, Sst, Sox6) (Liodis et al., 2007; Zhao et al., 2008; Batista-Brito et al., 2009) and others that have been implicated in the differentiation of diverse cell types, but their role in the development of cortical interneurons is currently unknown. Among the genes of the latter group was Satb1, which encodes a HD nuclear matrix protein that controls T cell differentiation and cancer metastasis via chromatin organization and epigenomic modifications (Alvarez et al., 2000; Cai et al., 2003, 2006; Han et al., 2008). The complete set of genes affected by deletion of Lhx6 in the dorsal forebrain is shown in Table S1.
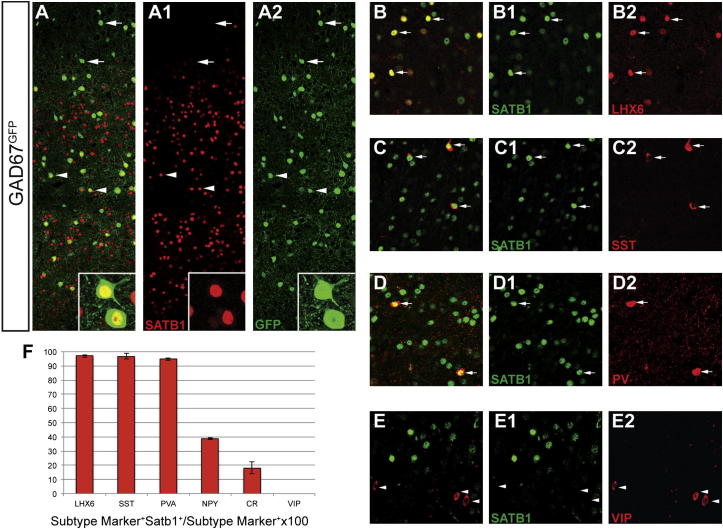
To determine whether Satb1 is expressed in cortical interneurons, we analyzed Gad67GFP mice, which express green fluorescent protein (GFP) in all GABA+ neurons of the cortex (Tanaka et al., 2003). Double immunostaining of brain sections from postnatal day (P) 30 Gad67GFP heterozygous mice for SATB1 and GFP showed that Satb1 was expressed in a large subset of cortical interneurons (Figures 1A–1A2). Virtually all LHX6+ (97.1% ± 1.1%), SST+ (96.4% ± 2.1%), and PV+ (94.8% ± 1.1%) interneurons were also positive for SATB1 (Figures 1B–1D2 and 1F). In addition, SATB1 was detected in 38.7% ± 0.6% of NPY+ and 18.1% ± 4.1% of CR+ interneurons (Figure 1F). The fractions of CR+ and NPY+ interneurons expressing Satb1 are comparable to those coexpressing Lhx6 (Fogarty et al., 2007), highlighting further the overlap between the LHX6+ and SATB1+ subsets of cortical interneurons. Finally, CGE-derived VIP+ interneurons were negative for SATB1 (Figures 1E–1E2 and 1F). We conclude that Satb1 is specifically expressed by MGE-derived cortical interneurons.
Figure 1.
Satb1 Is Specifically Expressed by MGE-Derived Cortical Interneurons
(A) Double immunostaining of brain sections from GAD67GFP heterozygous mice for SATB1 (A1) and GFP (A2). (A) shows a merge of images in A1 and A2. Arrows indicate GFP+SATB1+ and arrowheads indicate GFP+SATB1+ neurons. Insets in (A)–(A2) show GFP+SATB1+ cells.
(B–E) Double immunostaining of cortical sections from GAD67GFP animals for SATB1 (B1, C1, D1, and E1) and LHX6 (B2), SST (C2), PV (D2), and VIP (E2). Images in (B)–(E) are merges of (B1) and (B2), (C1) and (C2), (D1) and (D2), and (E1) and (E2), respectively.
(F) Quantification of the percentage of LHX6+, SST+, PV+, NPY+, CR+, and VIP+ interneurons coexpressing SATB1 (n = 3 animals per each interneuron marker analysis).
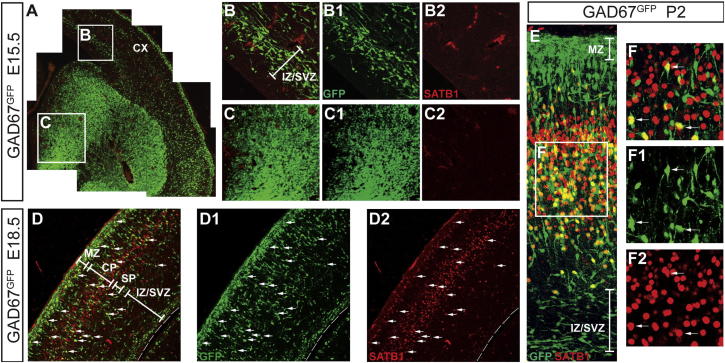
Figure 2.
Satb1 Is Expressed in Cortical Interneurons that Have Terminated Tangential Migration
(A) Dorsolateral quadrant of a coronal forebrain section of an E15.5 GAD67GFP mouse embryo double immunostained for SATB1 (red) and GFP (green).
(B and C) (B)–(B2) and (C)–(C2) show in higher magnification the boxes B and C, respectively, from (A). (B) and (C) are overlays of images in (B1) and (B2) and (C1) and (C2), respectively. At this embryonic stage, Satb1 is not expressed in the MGE or in tangentially migrating interneurons within the cortex.
(D) Cortical section from an E18.5 GAD67GFP mouse embryo double immunostained for GFP (green) and SATB1 (red). (D) is an overlay of images shown in (D1) (GFP) and (D2) (SATB1). At this stage, Satb1 is expressed in interneurons present within the cortical plate.
(E and F) Double immunostaining of cortical sections from a GAD67GFP P2 animal double immunostained for SATB1 (red) and GFP (green). (F) shows a single optical section from the box F in (E) and is an overlay of (F1) and (F2). Arrows indicate GFP+ neurons expressing Satb1. CP, cortical plate; CTX, cortex; IZ, intermediate zone; MGE, medial ganglionic eminence; MZ, marginal zone, SP, subplate; VZ, ventricular zone.
To determine the developmental profile of Satb1 expression in cortical interneurons, brain sections from E12.5-P2 Gad67GFP mice were double immunostained for SATB1 and GFP. SATB1+ interneurons were not detected in the dorsal forebrain of E12.5 embryos and were infrequent in the cortex of E15.5 embryos (Figure 2A; data not shown). At these stages, Satb1 was not expressed in the MGE or in tangentially migrating interneurons (Figures 2A–2C2). In contrast, a large number of GFP+SATB1+ interneurons was detected within the CP and MZ of E18.5 Gad67GFP embryos (Figures 2D–2D2). At later stages (P2), SATB1+ interneurons were absent from the MZ and IZ/SVZ but were abundant within the cortex (Figures 2E–2F2). Finally, and in agreement with previous reports (Huang et al., 2011; Balamotis et al., 2012), Satb1 was also expressed by pyramidal neurons (Figures 1A–1A2 and 2E–2F2). By demonstrating that during cortical development Satb1 is expressed by an increasing number of MGE-derived GABAergic neurons, our results suggest that SATB1 is implicated in the terminal differentiation and maturation of cortical interneurons.
Ectopic Expression of SATB1 in the MGE Induces a Gene Expression Profile Characteristic of Mature Cortical Interneurons
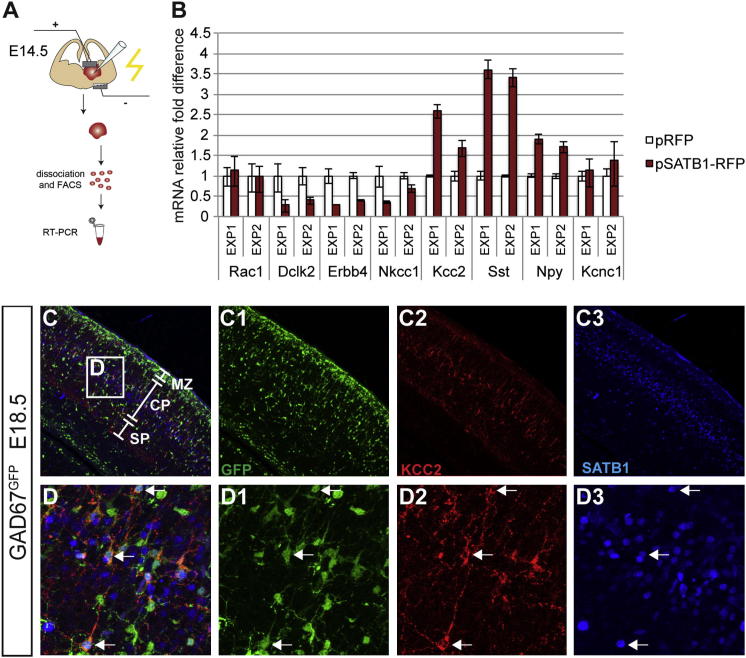
To determine whether SATB1 promotes the maturation of cortical interneuron precursors, we conducted a series of misexpression experiments in the MGE of E14.5 embryos and analyzed changes in expression of selected marker genes by qPCR (Figures 3A and 3B and Extended Experimental Procedures). As expected (Figure 2), SATB1 was undetectable in control MGE, but electroporation of a pCAGGs-based SATB1-encoding expression vector (pSATB1-RFP) led to high-level expression of nuclear-localized SATB1 protein (Figures S1B–S1C2). Twenty-four hours after electroporation, the level of mRNAs encoding RAC1, a small Rho GTPase with multiple roles in the development of inhibitory neurons in the forebrain (Chen et al., 2007) and KCNC1, a pottasium channel that marks PV+ interneurons (Chow et al., 1999), remained unchanged, while transcritps for PV and CR were undetectable in both control and Satb1-expressing MGEs (Figure 3B and data not shown). In contrast, transcripts encoding DCLK2, which is transiently detected in tangentially migrating interneuron precursors (our observations and Tuy et al., 2008), were downregulated (Figure 3B). In addition, ERBB4, a tyrosine kinase receptor widely expressed by tangentially migrating interneurons (Flames et al., 2004) but restricted postnatally mainly to PV+ interneurons (Fazzari et al., 2010; Neddens and Buonanno, 2010), and NKCC1, a Na-K-Cl cotransporter expressed in immature GABAergic interneurons (Yamada et al., 2004; Inada et al., 2011), were also downregulated (Figure 3B). Conversely, mRNAs for KCC2, a K+/Cl− exchanger expressed in mature interneurons and the overlapping subtype markers SST and NPY, were significantly upregulated (Figure 3B). The upregulation of Kcc2 by SATB1 in MGE is likely to reflect an in vivo regulatory scenario since all KCC2+ interneurons in the cortex of E18.5 GAD67GFP embryos coexpressed SATB1 (Figures 3C–3D3). The SATB1-induced downregulation of genes associated with tangentially migrating immature cortical interneurons (Dclk2, ErbB4, and Nkcc1) and the concomitant upregulation of Kcc2 and the subtype markers SST and NPY in MGE precursors support the idea that SATB1 drives the maturation of the SST+ group of cortical interneurons.
Figure 3.
Overexpression of SATB1 in MGE Precursors Induces a Gene Expression Profile Associated with Mature Cortical Interneurons
(A) Schematic presentation of the experimental strategy.
(B) Quantification (fold change) of transcripts in two independent experiments for the indicated genes following electroporation of control (pRFP; white bars) and Satb1-expressing (pSATB1-RFP; red bars) vectors.
(C and D) Triple immunostaining of a cortical section from GAD67GFP E18.5 embryo for GFP (green), KCC2 (red), and SATB1 (blue). (D) Magnification of the box in (C). (C1)–(C3) and (D1)–(D3) show the single colors of (C) and (D), respectively. Note that all GFP+SATB1+ interneurons coexpress KCC2.
See also Figures S1 and S3.
Extended Experimental Procedures.
Animals
Two independently generated ES cell clones with the Satb1EUCOMM were transmitted through the germline to establish Satb1EUCOMM heterozygous mice. Targeting of the Satb1 locus was validated by Southern blot and PCR analysis of genomic DNA from ES cells and heterozygous animals (data not shown). Immunostaining of brain sections from Satb1EUCOMM/EUCOMM mice with a SATB1-specific antibody confirmed deletion of Satb1 in homozygous mice (Figure S7). Similar to the phenotype previously reported for an independently generated null mutant strain (Alvarez et al., 2000), Satb1EUCOMM homozygous mice were born but failed to thrive and died within the first three weeks.
Gene Expression Profiling of Dorsal Forebrain from Lhx6-Null Mutant Mice
Dorsal forebrain from E15.5 embryos resulting from intercrossing Lhx6+/− mice was dissected and stored in RNAlater (QIAGEN). Total RNA was isolated using RNeasy (QIAGEN) according to the manufacturer’s protocol. Microarray experiments and bioinformatics analysis was performed by Oryzon (Oryzon Genomics, Parc Cient’fic de Barcelona, Spain). RNA samples were labeled with specific fluorochromes and hybridized to cRNA controls on the DNA microarrays. The microarray images were acquired using an Agilent scanner and the raw data were processed with proprietary Oryzon software. Bioinformatics analysis was applied to the results using the proprietary Oryzon tool Polyphemous™, which normalizes the raw data, conducts an analysis of the copies, defines the list of genes that undergo a significant change in the level of gene expression, and defines the ontology and the metabolic or regulatory pathways that are involved. From that point, data mining was used to carry out our biological analysis.
Quantification of MGE Transcripts
For quantification of transcripts following electroporation with pSatb1-RFP, the area of MGE expressing the highest levels of RFP was dissected 24 hr after transfection, and RFP+ cells were isolated by flowcytometry (FACS ARIAII, Becton Dickinson; typically a yield of 10000-20000 cells). Total RNA was purified (RNeasy microKit, QIAGEN) and subjected to reverse transcription (High-Capacity cDNA Reverse Transcription Kit, Applied Biosystems) and quantitative PCR according to manufacturer’s instructions (Taqman Gene Expression Assays, Applied Biosystems). Each independent experiment represents a triplicate of qPCRs.
Expression Constructs
Full length cDNAs for Satb1 (IMAGE clone ID: 4164993), Kcc2 (IMAGE clone ID: 6838880) and Lhx6 (Grigoriou et al., 1998) were cloned into modified pCAGGs-based expression vectors. The pCAGGS-IRES-GFP was a gift from James Briscoe (NIMR) and pCAGGS-IRES-RFP was a gift from Francois Guillemot (NIMR). Expression was confirmed by Western blot analysis or immunostaining of transfected P19 embryonal carcinoma cells or brain slices (Figure S3 and data not shown). Satb1-specific shRNA expression pGFP-V-RS vectors were obtained from Origene (TG501973) and their efficiency to downregulate expression of Satb1 was tested by Western blot analysis in P19 cells transfected with pSatb1-RFP (Figure S1D).
Electroporations, MGE Cell Culture. and Transplantation
Ex vivo electroporation of MGE in embryonic brain slices was conducted as described previously (Stühmer et al., 2002). Twelve hours after electroporation, MGE regions with the strongest RFP or GFP signal were dissociated as previously described (Du et al., 2008). The resulting cell suspension was either cocultured with feeder cells prepared from neonatal cortex (P0-P1) (Xu et al., 2004) or grafted into neonatal (P0-P1) cortex as described (Xu et al., 2010). Transplanted animals were transcardially perfused at P30, and dissected brains were processed for immunohistochemistry. In utero electroporations were carried out as previously described (Nguyen et al., 2006).
P19 Cells and Western Blotting
Mouse embryonal carcinoma P19 cells were grown in DMEM supplemented with 10% FCS, 2 mM glutamin and 1% penicillin/streptomycin (all from GIBCO) and transfected with lipofectamin according to the manufacturer’s protocol (Lipofectamin® 2000 reagent, Invitrogen). Cells were then mechanically homogenized in RIPA buffer supplemented with protease and phosphatase inhibitors (all from SIGMA) and total protein extractions were obtained. The proteins were separated on 4%–12% gradient gels using the XCell Surelock Mini-Cell and NuPAGE® MES SDS Running Buffer (Invitrogen) and transferred to nitrocellulose membranes (Amersham) using the XCell II Blot module and NuPAGE® Transfer Buffer (Invitrogen). Membranes were blocked with Blotto A (Santa-Cruz) and incubated O/N with primary antibodies for Satb1 (goat polyclonal anti-Satb1, Santa-Cruz; 1:1000) and actin (Sigma; 1:1000) diluted in the blocking buffer. The next day after washing in TBST, membranes were incubated for 1 hr at RT with HRP-conjugated secondary antibodies (Santa-Cruz; 1:2000). The immunoreactive bands were observed with ECL according to manufacturer’s instructions (Pierce® ECL Western Blotting Substrate, ThermoScientific).
Immunostaining
For immunostaining on brain sections from mice P15 or older, animals were transcardially perfused with 4% PFA and brains were postfixed overnight (O/N). Vibratome (70 μm) or cryostat (14 μm) sections were permeabilized with PBT (0.5% Triton X-100 in PBS) for 1 hr at room temperature (RT), washed in PBT, blocked in 10% FCS in PBT (2 h; RT), and incubated with primary antibodies diluted in blocking solution at 4°C, O/N. After 3 washes with PBT, sections were incubated with secondary antibodies diluted in blocking solution at RT for 2 hr, washed in PBT, and mounted using Vectashield (Vector) medium. For immunostaining on embryonic sections, dissected brains were fixed in 4% PFA in PBS at 4°C, O/N. Cryostat sections (12 μm) were permeabilized in 0.1% Triton X-100 in PBS (PBT) for 5 min, and processed as above with the exception of the blocking buffer which was made up of 1% BSA (Sigma) and 0.1% glycine (BDH) in PBT. For immunostaining of primary cultures, cells were fixed with 4% PFA in PBS at RT for 10 min, and processed as in cryostat sections. The following antibodies were used: rabbit polyclonal anti-CR (Chemicon; 1:1000), rabbit polyclonal anti-cFOS (Calbiochem; 1:10.000), mouse monoclonal anti-GAD67 (Chemicon; 1:1000), rabbit polyclonal anti-GFP (Invitrogen; 1:1000), rat monoclonal anti-GFP (Invitrogen; 1:1000), rabbit polyclonal anti-KCC2 (Millipore; 1:2000), rabbit polyclonal anti-LHX6 (1:250–1000) (Lavdas et al., 1999), rabbit polyclonal anti-nNOS (Invitrogen; 1:200), rabbit polyclonal anti-NPY (Biogenesis), mouse monoclonal anti-PV (Chemicon; 1:1000), rabbit polyclonal anti-RFP (Abcam; 1:500), goat polyclonal anti-SATB1 (Santa-Cruz; 1:100), rabbit polyclonal anti-SATB1 (gift from V. Tarabykin; 1:500), rabbit polyclonal anti-SATB2 (gift from V. Tarabykin; 1:500), rat monoclonal anti-SST (Millipore; 1:1000) and rabbit polyclonal anti-VIP (Immunostar). Secondary antibodies used were as follows: Alexa Fluor 488-conjugated donkey anti-mouse, anti-rat, anti-goat and anti-rabbit and Alexa Fluor 568-conjugated donkey anti-mouse, anti-rat, anti-goat and anti-rabbit (all from Invitrogen; all 1:500).
In Situ Hybridization Histochemistry
In situ hybridization histochemistry (ISHH) was carried out essentially as described (Schaeren-Wiemers and Gerfin-Moser, 1993). Riboprobes used were specific for: Cux2 (gift from F. Guillemot); Dclk2 (IMAGE 6822681); Er81 (gift from S. Arber); Kcnc1 (gift from N. Kessaris); Lhx6 (Grigoriou et al., 1998), and Sst (gift from N. Kessaris).
Quantification
Images of in situ hybridized sections were acquired with a digital color camera (ProgRes C14) attached to an Axioplan 2 microscope (Zeiss) and processed with Openlab software. The number of GAD67+, LHX6+, SST+ and CRH+ cells was determined in the primary motor (M1, bregma 0.38 mm), somatosensory (S1BF, bregma 0.38 mm and bregma −1.94 mm) and primary visual (V1, bregma −2.92 mm) cortex of 2 week-old Satb1−/− (n = 3) and Satb1+/+ (n = 3) animals. For each region we counted cells present in 675.66 μm–wide columns on 14 μm thick sections which corresponded to equivalent bregma positions (Paxinos and Franklin, 2001). Fluorescent images spanning the whole cortical length were captured with a Leica TCS SP5 MP confocal microscope and processed with Adobe Photoshop CS4. We counted cells from 775 μm–wide columns on 14 μm thick sections which corresponded to equivalent bregma positions (Paxinos and Franklin, 2001). For the SST subset analysis (CR, NPY, nNOS) and VIP, from each of the three animals analyzed we counted cell numbers from four consecutive sections of the somatosensory cortex (between bregma positions 0.38 mm and −1.94 mm). For both the in situ hybridization and immunofluoresence countings, data are given as means ± standard deviation of the mean (STDEV). Statistical significance between data sets was determined by using the Student’s t test function (two-tailed distribution, two-sample equal variance) in Excel (Microsoft) and they were considered significant when p < 0.05 (∗), p < 0.01 (∗∗) and p < 0.001 (∗∗∗). Colocalization of SATB1 and other markers for cortical interneuron subtypes was evaluated in three adult wild-type mice as described previously (Liodis et al., 2007). The percentage of YFP+ cells expressing Satb1 was evaluated in the primary motor, primary sensory, barrel and visual cortex of P15 Nkx2.1CRE;Rosa26EYFP control (Lhx6+/+) and mutant (Lhx6−/−) mice (3 for each genotype). In each region we counted the number of YFP+ cells present in 1000-μm-wide columns (16 μm sections; 300 μm apart). The number of RFP+ cells coexpressing SST or PV was evaluated in the whole pallium of mice grafted with control or Satb1-overexpressing MGE-derived cells (n = 4 for each condition). Analysis of primary cultures was conducted on a Axioplan 2 microscope (Zeiss) or in fluorescent images captured by confocal laser microscopy (Bio-Rad/Radiance 2100 or Leica SP5II). More than 500 cells were analyzed per condition. In all experiments statistical significance was established using the Student’s homoscedastic t test with two-tailed distibution where ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Figure S1.
Manipulation of Satb1 Expression in the MGE, Related to Figures 3, 4, and 6
(A) Schematic representation of MGE electroporation and culture of forebrain slices from E14.5 mouse embryos.
(B–C2) Sections of slices in which the MGE was transfected with pRFP (B–B2) or pSATB1-RFP (C–C2) and immunostained 24 hr later for RFP (B1, C1) or SATB1 (B2, C2). (B) and (C) are overlays of (B1) and (B2) and (C1) and (C2), respectively. SATB1 is detected only in sections from slices electroporated with pSATB1-RFP vector.
(D) Western blot analysis of protein extracts from P19 embryonal carcinoma cells cotransfected with pSATB1-RFP and constructs expressing SATB1- or scrambled (control) shRNAs. Note the efficient downregulation of SATB1 upon cotransfection of Satb1-specific shRNAs.
SATB1 Promotes the Maturation of SST+ Interneurons
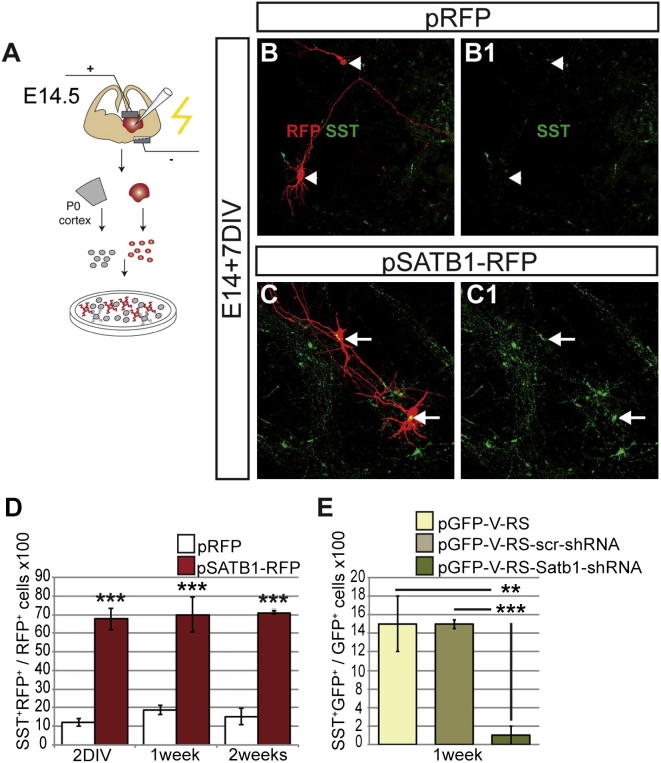
To examine whether misexpression of Satb1 in immature interneuron precursors leads to SST production, a feature of mature interneurons of the postnatal cortex, dissociated E14.5 MGEs transfected with pSATB1-RFP were plated onto feeder layers of neonatal mouse cortex and analyzed by immunostaining after 2, 7, and 14 days in vitro (DIV; Figure 4A). Expression of SATB1 increased dramatically the percentage of MGE cells that produced SST at all stages analyzed (Figures 4B–4D). Interestingly, the maximal increase in the percentage of RFP+SST+ cells was evident by 2DIV (Figure 4D), suggesting that upregulation of Sst mRNA (by 24 hr; Figure 3B) and SST protein in this assay represent immediate early effects of Satb1 misexpression in interneuron precursors. SATB1 also led to a significant increase in the fraction of transfected cells that coexpress NPY, a neuropeptide expressed by a subset of SST+ interneurons (Fogarty et al., 2007) (at 2DIV, pRFP: 7.2% ± 2.1% versus pSATB1-RFP: 29.3% ± 1.4%, p < 0.01, n = 3 independent electroporations). In contrast to the upregulation of SST and NPY, no PV+ cells were detected during the 2 week period we examined (data not shown). Taken together, our results suggest that SATB1 is sufficient to induce in immature MGE precursors a molecular phenotype that is normally encountered in mature SST+ cortical interneurons.
Figure 4.
SATB1 Promotes the Expression of SST in Immature Interneuron Precursors
(A) Schematic presentation of the experimental strategy.
(B and C) Images of cultured MGE cells transfected with control (pRFP; B) or Satb1-expressing (pSATB1-RFP; C) vectors, immunostained for RFP (red) and SST (green). Images in (B1) and (C1) show only the green channel of the images shown in (B) and (C), respectively.
(D) Percentage of Sst-expressing MGE cells electroporated with control (white bars) and Satb1-expressing (red bars) vectors and cultured for 2, 7, and 14 days on P0 cortical feeder layers.
(E) Percentage of Sst-expressing MGE cells transfected with pGFP-V-RS, pGFP-V-RS-scrShRNA, or pGFP-V-RS-SATB1ShRNA.
n = 3 independent electroporations for each experimental condition, ∗∗p < 0.01, ∗∗∗p < 0.001 for each test (Student’s homoscedastic t test with two-tailed distribution). See also Figures S1 and S2.
To examine the requirement of Satb1 activity for SST induction, we adopted a short-hairpin (sh)RNA approach to knock down endogenous Satb1 in interneuron precursors (Figure S1D). pGFP-V-RS (control), pGFP-V-RS-scrShRNA (scrambled small hairpin RNA [ShRNA] control) and pGFP-V-RS-Satb1ShRNA constructs were transfected into E14.5 MGEs, which were subsequently dissociated and cultured for 7 days on cortical feeder layers (Figure 4A). Immunostaining for GFP (to identify transfected cells) and SST showed that expression of the Satb1 ShRNA resulted in a dramatic decrease in the percentage of SST+ cells relative to controls (Figure 4E). These experiments argue that Satb1 activity is required in our coculture system for the differentiation of MGE precursors into SST+ interneurons.
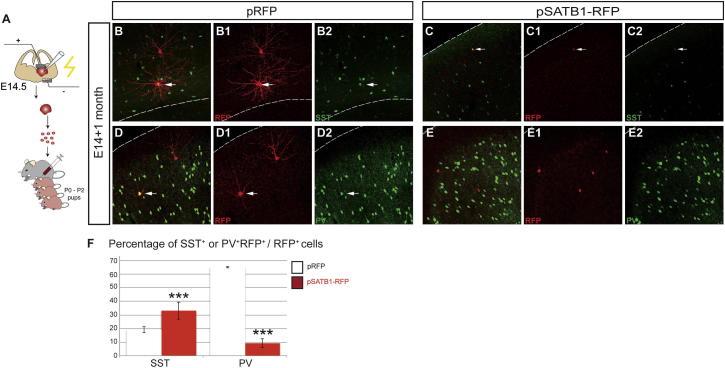
To explore the effect of Satb1 on the differentiation of interneurons in vivo, E14.5 MGEs transfected with control or pSATB1-RFP vectors were mechanically dissociated and the resulting cell suspension was grafted into the cortex of P0 mice (Figure S2A) (Xu et al., 2010). One month later, the transplanted cells were identified in the cortex by RFP expression and their subtype identity was analyzed. Consistent with previous reports (Miyoshi et al., 2007), the majority of MGE cells transfected with the control vector dispersed throughout the cortex and differentiated predominantly into PV+ and (to a lesser extent) SST+ interneurons (65% and 20%, respectively; Figures S2B–S2B2, S2D–S2D2, and S2F). In contrast, fewer pSATB1-RFP-expressing MGE cells were identified within the cortex, suggesting that premature overexpression of Satb1 in MGE precursors inhibits their migration and dispersion (see below). Interestingly though, we observed a significant increase in the representation of SST+ cells (33.1% ± 0.4%) and a corresponding decrease of PV+ cells (9.27% ± 3.3%) within the population of grafted MGE cells that expressed Satb1 within the cortical layers (Figures S2C–S2C2, S2E–S2E2, and S2F). Although this analysis is confounded by the apparent effect of SATB1 overexpression on the migration of grafted MGE cells, it reinforces the idea that SATB1 promotes the maturation of interneurons into the SST+ subtype. Together, these experiments demonstrate that overexpression of SATB1 in MGE precursors induces a neuropeptide phenotype that characterizes mature cortical interneurons of the SST+ group.
Figure S2.
SATB1 Promotes the Differentiation of MGE Cells into SST+ Interneurons In Vivo, Additional Results Related to Figure 4
(A) Schematic representation of the experimental strategy for transfection and transplantation of MGE cells to the cortex of P0 mice. B–E2, immunostaining for RFP (B1, C1, D1, and E1) and SST (B2 and C2) or PV (D2 and E2) of cortical sections from P30 animals grafted at P0 with E14.5 MGE cells transfected with pRFP (B–B2 and D–D2) or pSATB1-RFP (C–C2 and E–E2). Arrows point to double-positive cells. (F) Quantification of the percentage of SST+ and PV+ interneurons within the population of RFP-expressing grafted cells transfected with control (white bar) or SATB1-expressing (red bar) vector. n = 4 mice for each experimental condition, ∗∗∗p < 0.001 (Student’s homoscedastic t test with two-tailed distribution).
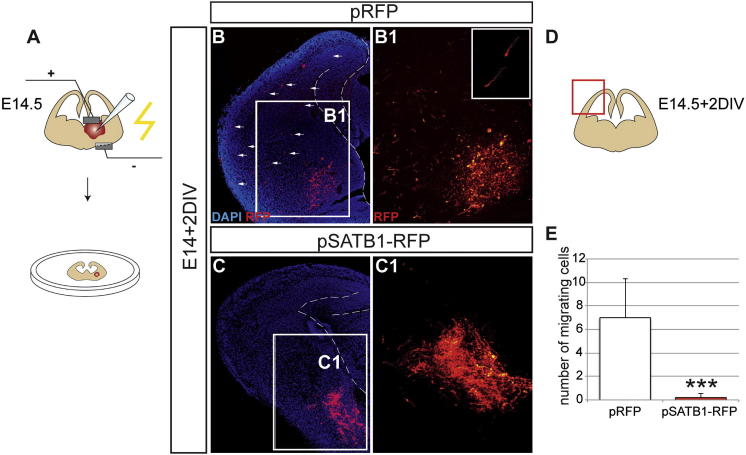
Overexpression of SATB1 Blocks Tangential Migration of Interneuron Precursors
Maturation of interneurons is associated with termination of tangential migration (Bortone and Polleux, 2009). To examine whether SATB1 is sufficient to promote the sessile state of interneuron precursors, pSATB1-RFP was electroporated into the MGE and its effect on the tangential migration of interneuron precursors was analyzed by culturing forebrain slices for 48 hr (Figure S3A). As expected, MGE cells expressing a control vector showed robust tangential migration toward the cortex (Figures S3B and S3B1). Moreover, the majority of these cells were characterized by the typical morphology of tangentially migrating interneurons, highlighted by the presence of a long leading process (arrow in inset of Figure S3B1; Bellion et al., 2005). In all cases, MGE cells expressing Satb1 failed to migrate tangentially and were localized exclusively at the site of electroporation in the MGE (Figures S3C–S3E). We conclude that SATB1 inhibits the tangential migration and promotes the sessile state of cortical interneurons.
Figure S3.
Expression of Satb1 in the MGE Inhibits the Tangential Migration of Interneurons, Additional Results Related to Figure 3
(A) Schematic presentation of forebrain slices from E14.5 mouse embryos electroporated in the MGE and cultured for 2 days.
(B) Image of a slice which was electroporated with the control vector pRFP (transfected cells shown in red), cultured for two days and counterstained with DAPI (blue). B1 shows an enlargement of the box B1 in (B). Inset in (B1) shows images of tangentially migrating interneurons with a prominent leading process.
(C) Image of a slice electroporated with the pSatb1-RFP vector and cultured in parallel to the control slice shown in (B). Note the absence of tangentially migrating SATB1+ interneurons. C1, shows an enlargement of the box C1 in (C).
(D and E) Schematic of a cultured slice 48 hr after electroporation. The effect of SATB1 overexpression on tangential migration of interneurons was quantified by counting RFP+ cells within the indicated boxed area in the pallium. Data obtained are shown as a histogram in (E); n = 12 slices per condition, ∗∗∗p < 0.001 (Student’s homoscedastic t test with two-tailed distribution).
Satb1 Activity Is Required In Vivo for the Differentiation of SST+ Cortical Interneurons
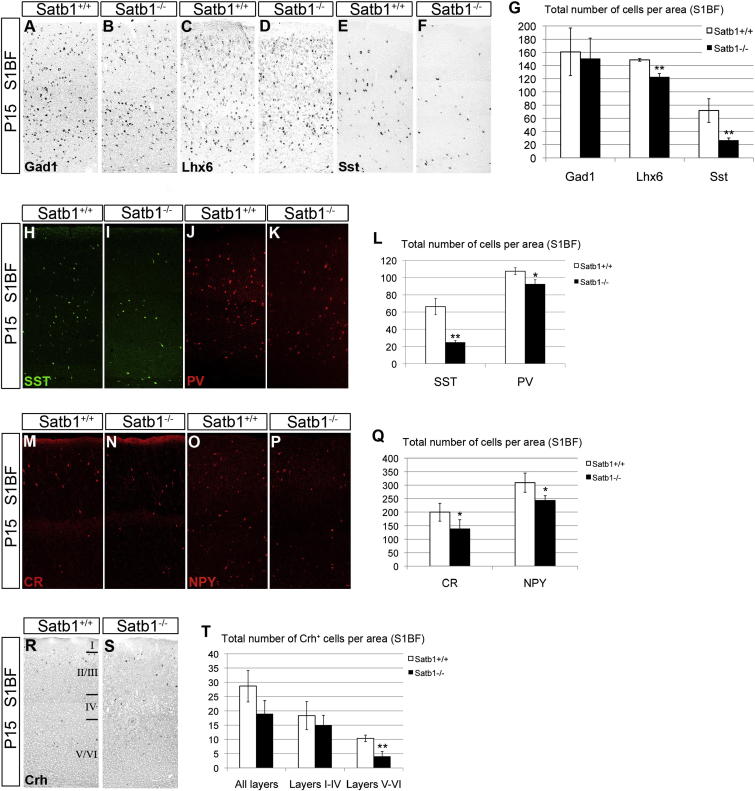
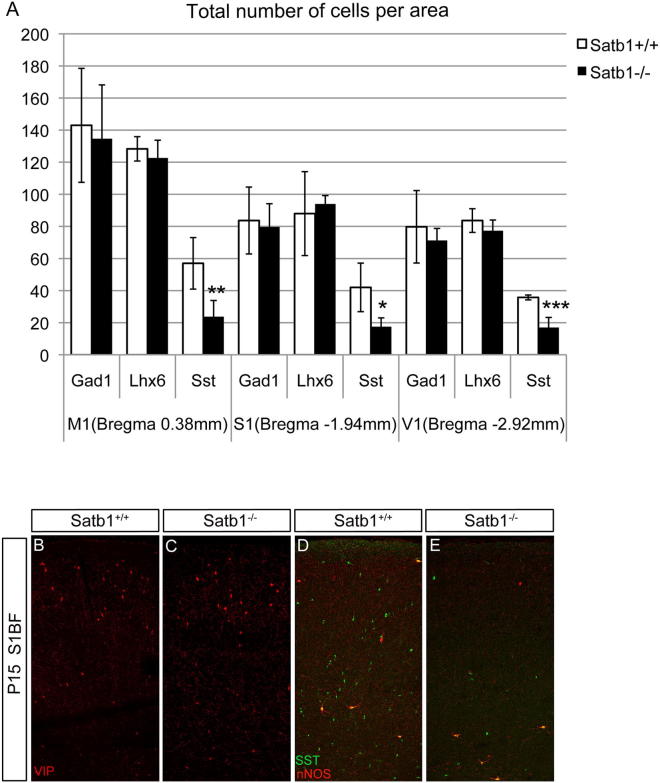
To examine whether SATB1 is required for the development of cortical interneurons in vivo, we analyzed mice homozygous for a knockout allele of Satb1 (Satb1tm1a(EUCOMM)Hmgu; called hereafter Satb1EUCOMM) (for details on the generation and analysis of the phenotype of Satb1EUCOMM mice see Experimental Procedures and Supplemental Information). No reduction in the total number of cortical interneurons (as assessed by in situ hybridization histochemistry for Gad1) was observed in different cortical areas of P15 Satb1EUCOMM homozygous mice (Figures 5A, 5B, 5G, and S4A). In addition, the number of LHX6+ interneurons in the motor, somatosensory (bregma −1.94 mm) and visual cortex of Satb1 mutants was normal, although a modest (∼18%) reduction of these cells was observed in the somatosensory cortex (S1BF; bregma 0.38 mm) of Satb1-deficient animals (Figures 5C, 5D, 5G, and S4A). In contrast, we observed a dramatic reduction in the number of neurons expressing Sst mRNA (∼63%) and SST protein (∼62%) in all cortical areas of P15 Satb1 mutants (Figures 5E–5I, 5L, and S4A). In addition, we observed a small decrease (∼14%) in the number of PV+ interneurons in all cortical areas of Satb1 mutants we examined (Figures 5J–5L). Finally, no significant difference was observed in the number or distribution of CGE-derived VIP-expressing interneurons (Figures S4B and S4C) between control (99.7 ± 20.6) and Satb1-deficient animals (87.7 ± 9.3, n = 3 animals per genotype). These results argue that Satb1 activity is required in vivo primarily for the development of the SST+ group of cortical interneurons.
Figure 5.
Satb1 Deletion Results in Impaired Differentiation of SST+ Cortical Interneurons
(A–F) In situ hybridization histochemistry of P15 cortical sections from wild-type (A, C, and E) and Satb1-deficient (B, D, and F) mice with riboprobes specific for Gad1 (A and B), Lhx6 (C and D), and Sst (E and F).
(G) Quantification of Gad1-, Lhx6-, and Sst-expressing cells in the somatosensory cortex (S1BF, bregma 0.38 mm) of wild-type (white bars) and Satb1-deficient mice (black bars).
(H–K) Immunofluorescence of P15 cortical sections from wild-type (H and J) and Satb1-deficient (I and K) mice with antibodies specific for SST (H and I) and PV (J and K).
(L) Quantification of SST+ and PV+ cells in the somatosensory cortex (S1BF, bregma 0.38 mm) of wild-type (white bars) and Satb1-deficient mice (black bars).
(M–P) immunofluorescence of P15 cortical sections from wild-type (M and O) and Satb1-deficient (N and P) mice with antibodies specific for CR (M and N) or NPY (O and P).
(Q) Percentages of CR+ and NPY+ cells in wild-type (white bars) and Satb1-deficient cortex (black bars).
(R and S) In situ hybridization histochemistry of P15 cortical sections from wild-type (R) and Satb1 mutant (S) mice with a Crh-specific riboprobe.
(T) Quantification of CRH+ neurons in the layers of the somatosensory cortex (S1BF, bregma 0.38 mm) of wild-type (white bars) and Satb1-deficient mice (black bars).
n = 3 animals per each interneuron marker analysis, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 for each test (Student’s homoscedastic t test with two-tailed distibution). See also Figures S4, S5, and S7.
Figure S4.
Analysis of Interneuron Markers in P15 Wild-Type and Satb1-Deficient Mice, Additional Results Related to Figure 5
(A) Quantification of GAD1+, LHX6+ and SST+ interneurons in the motor (M1, bregma 0.38 mm), somatosensory (S1, bregma −1.94 mm) and visual (V1, bregma −2.92 mm) cortex of wild-type (white bars) and Satb1-deficient (black bars) mice. The number of Sst mRNA-expressing interneurons is significantly reduced in all areas examined.
(B–E) Immunofluoresence on P15 cortical sections from wild-type (Satb1+/+; B and D) and Satb1-deficient (Satb1−/−; C and E) mice with antibodies specific for VIP (B and C) and nNOS and SST (D and E). n = 3 animals per genotype, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student’s homoscedastic t test with two-tailed distribution).
To examine whether deletion of Satb1 affects selectively expression of Sst, we analyzed additional markers, which are expressed by subsets of SST+ interneurons. Among them are the neuropeptides corticotropin releasing hormone (CRH) and NPY and the proteins CR and nNOS (Wonders and Anderson, 2006; Kubota et al., 2011). We observed a significant decrease in the number of CR+ and NPY+ interneurons in the S1BF area of Satb1 mutants (Figures 5M–5Q). We also observed a consistent trend toward reduced numbers of CRH+ interneurons when all cortical layers of S1BF were analyzed (Figures 5R–5T). However, a clear reduction of CRH+ interneurons was documented in the bottom (V–VI) cortical layers (Figure 5T), in which interneurons coexpressing CRH and SST are located (Kubota et al., 2011). Finally, no difference was observed in the number of type I nNOS+ projection interneurons (Tomioka et al., 2005; Perrenoud et al., 2012) in the cortex of control (22.6 ± 0.6 per surface area) versus Satb1-deficient mice (19.1 ± 3.2 per surface area, n = 3 animals per genotype) (Figures S4D and S4E). These findings indicate that the effect of Satb1 deletion is not restricted to the expression of Sst arguing that the gene controls multiple aspects of the maturation program of SST+, interneurons.
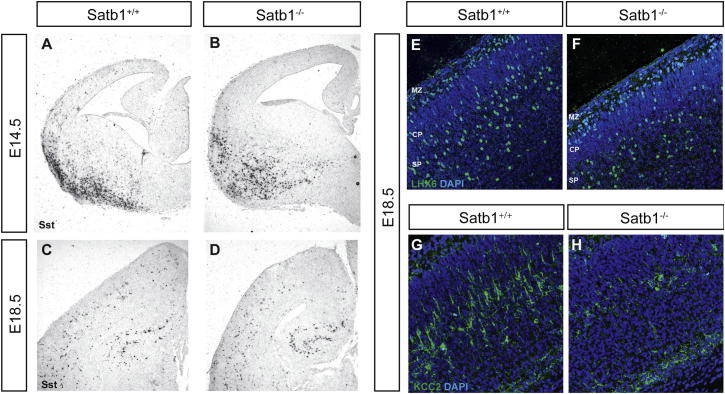
Interestingly, despite the dramatic reduction of both Sst mRNA and protein in the cortex of Satb1EUCOMM mice, the number and distribution of tangentially migrating Sst(mRNA)+ interneurons during embryogenesis were indistinguishable between control and mutants (Figures S5A–S5D). In addition, no difference in the expression of Lhx6 was observed in tangentially migrating interneurons or in the cortex of E18.5 wild-type and Satb1 mutant embryos (Figures S5E and S5F; data not shown). Since expression of Lhx6 and Sst mRNA are early markers of MGE-derived interneuron precursors (Liodis et al., 2007; Neves et al., 2012), these experiments suggest that Satb1 activity is dispensable for the early specification of the SST group of interneurons and is required for their maturation within the pallium.
Figure S5.
SATB1 Is Not Required for the Specification or Tangential Migration of Cortical Interneurons, Additional Results Related to Figure 5
(A–D) In situ hybridization histochemistry on forebrain brain sections from E14.5 (A and B) or cortical sections from E18,5 (C and D) wild-type (A and C) or Satb1−/− (B and D) mouse embryos using an Sst-specific riboprobe.
(E and F) Immunostaining of cortical sections from E18.5 wild-type (E and G) and Satb1-deficient (F and H) mice, immunostained for LHX6 (E and F) or KCC2 (G and H) and counterstained with DAPI. Although there is no difference in the number of LHX6+ cells at this stage between the two genotypes, note the dramatic reduction of KCC2-specific signal in Satb1-deficient relative to control embryos.
To further explore this hypothesis, we analyzed the expression of KCC2, a nonspecific marker of maturation of multiple groups of interneurons and pyramidal neurons, in the cortex of control and Satb1 mutant embryos. At E18.5, we observed a dramatic decrease in the expression of KCC2 in mutant cortex (Figures S5G and S5H). Since at this stage the majority of KCC2+ cells are interneurons (Figure 3), these experiments provide further evidence that Satb1 activity is required in vivo for the maturation of cortical interneurons. The expression of Satb1 in principal cortical neurons raises the possibility that the cortical interneuron defects observed in Satb1 mutants are secondary to deficits in cortical lamination. However, this is unlikely as we failed to detect any changes in cortical lamination in the cortex of Satb1EUCOMM homozygous mice using in situ hybridization for the layer-specific markers ER81 and CUX2 (data not shown; see also Liodis et al., 2007). Taken together, our experiments indicate that Satb1 activity is required in vivo for the maturation of the SST+ group of MGE-derived cortical interneurons.
Lhx6-Dependent Differentiation of SST+ Cortical Interneurons Requires Satb1 Activity
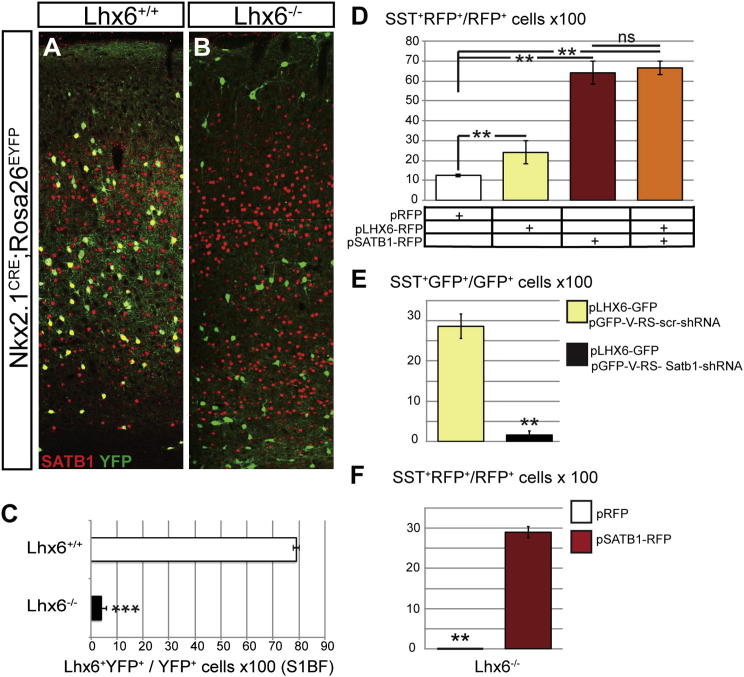
Satb1 was identified in our genome-wide microarray screen as a gene downregulated in Lhx6-deficient mouse brain, suggesting that its expression depends on Lhx6 activity. To investigate the epistatic relationship between Satb1 and Lhx6, we analyzed the levels of SATB1 in the cortex of Lhx6 mutant mice (Liodis et al., 2007) that were also transgenic for the Nkx2.1CRE;Rosa26EYFP reporter. Consistent with our gene expression profiling, we found that, in the brain of P15 Lhx6−/− mice, the proportion of YFP+ cells expressing Satb1 was dramatically reduced relative to that of control mice (Figures 6A–6C). These experiments indicate that Lhx6 activity is necessary for Satb1 expression in MGE-derived cortical interneurons.
Figure 6.
SATB1 is an Effector of Lhx6 in Cortical Interneuron Differentiation
(A and B) Double immunostaining of cortical sections from Nkx2.1Cre;Rosa2EYFP transgenic mice homozygous for the wild-type (A) or null (B) Lhx6 allele with antibodies for GFP (green) and SATB1 (red).
(C) Percentage of SATB1+ cells within the YFP+ cell population of Lhx6+/+ and Lhx6−/− mice. Note the dramatic reduction of Satb1-expressing cortical interneurons in Lhx6-deficient animals.
(D) Percentage of SST+ wild-type MGE cells electroporated with control (pRFP; white bar), Lhx6-expressing (pLHX6-RFP; yellow bar), Satb1-expressing (pSATB1-RFP; red bar), or Lhx6- and Satb1-expressing vectors (orange bar) and cultured for 7 days on P0 cortical feeder layers.
(E) Percentage of SST+ wild-type MGE cells transfected with pLhx6-GFP alone or pLhx6-GFP and pGFP-V-RS-Satb1ShRNA vectors.
(F) Percentage of SST+ Lhx6-deficient MGE cells transfected with pRFP or pSatb1-RFP. The percentage of Lhx6-deficient MGE cells that express SST increases dramatically upon expression of SATB1.
In (C), n = 3 animals; in (D)–(F), n = 3 independent electroporations for each experimental condition, ∗∗p < 0.01, ∗∗∗p < 0.001 for each test (Student’s homoscedastic t test with two-tailed distibution). See also Figures S1 and S6.
A common feature of Lhx6 (Liodis et al., 2007; Zhao et al., 2008) and Satb1 (this study) mutant mice is the differentiation deficit of MGE-derived cortical interneurons. To further examine whether LHX6 and SATB1 function in a common cascade, we overexpressed combinations of control (RFP-only), pSATB1-RFP and pLHX6-RFP expression vectors in the MGE of E14.5 mouse embryos and immunostained for SST following 7 days coculture with P0 cortical feeder layers (Figure 4A). Expression of pLHX6-RFP alone resulted in an ∼2-fold increase in the percentage of RFP+ cells coexpressing SST, while expression of pSATB1-RFP led to an ∼6-fold increase in the percentage of SST+RFP+ interneurons (Figure 6D). Interestingly, cotransfection of SATB1- and LHX6-encoding vectors did not result in a further increase in the percentage of SST+RFP+ cells beyond the levels achieved by expression of SATB1 alone (Figure 6D). These findings are consistent with our phenotypic analysis of Satb1 mutants and support the idea that LHX6 and SATB1 are components of a common developmental cascade.
The Satb1-dependent upregulation of SST in dissociated MGE cultures (Figure 4) allowed us to test whether Satb1 activity mediates the effect of LHX6 on SST induction. For this, LHX6 was overexpressed in E14.5 MGEs along with control (GFP-scrShRNA) or Satb1-specific (GFP-Satb1ShRNA) ShRNA constructs, and SST expression was analyzed following dissociation and culture of transfected cells on cortical feeder layers for 7 days. Coelectroporation of pGFP-V-RS-Satb1ShRNA and pLHX6-GFP vectors resulted in a dramatic reduction in the fraction of SST+ cells relative to the pLHX6-GFP and pGFP-V-RS-scrShRNA combination (Figure 6E), suggesting that Satb1 is required for the Lhx6-mediated upregulation of SST and differentiation of MGE progenitors.
To test whether SATB1 is sufficient to rescue SST expression in Lhx6-deficient interneuron precursors, pSATB1-RFP was introduced into Lhx6-deficient MGEs that were subsequently cultured for 7 days. Relative to control vectors, expression of SATB1 increased dramatically the percentage of MGE cells that were positive for SST (Figure 6F), indicating that SATB1 is sufficient to drive expression of Sst downstream of LHX6. Nevertheless, the efficiency of SST induction in mutant MGE cells was considerably smaller relative to that observed in their wild-type counterparts (Figure 6D), arguing that additional factors missing from Lhx6 mutant MGE cells are implicated in the control of SST expression.
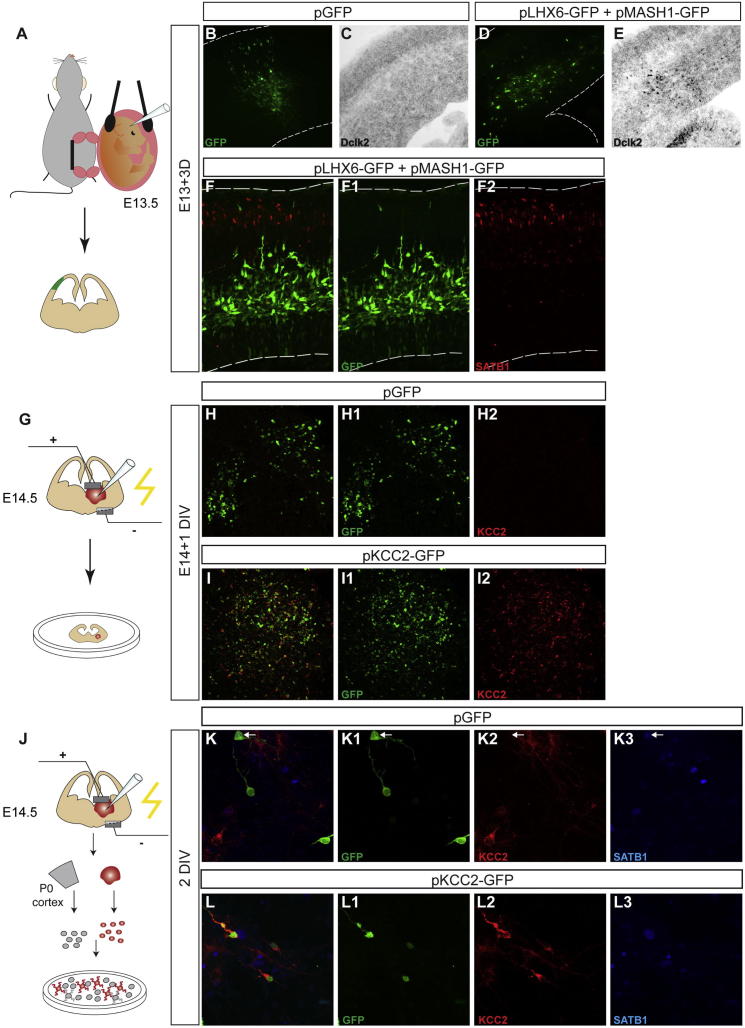
LHX6 Is Not Sufficient to Drive Expression of Satb1
During development, Satb1 is induced in MGE-derived cortical interneurons several days after the expression of Lhx6 (Figure 2), suggesting that LHX6, even though necessary (Figure 6), is not sufficient to induce expression of Satb1. Consistent with this idea, overexpression of LHX6 in MGE precursors (by electroporating pLHX6-GFP) did not upregulate Satb1 (data not shown). Moreover, in utero coelectroporation of pLHX6-GFP and a MASH1-expressing plasmid into cortical projection neuron progenitors, which is sufficient to upregulate the early interneuron marker DCLK2, failed to induce expression of Satb1 (Figures S6A–S6F2). To further explore the mechanisms that regulate expression of Satb1, we tested the idea that KCC2, which is coexpressed in mature cortical interneurons with SATB1, is an upstream regulator of Satb1. However, electroporation of the Kcc2-encoding pKcc2-GFP vector (Figures S6G–S6I2) into MGE precursors did not result in induction of SATB1 (Figures S6J–S6L3 and data not shown).
Figure S6.
LHX6 and KCC2 Are Not Sufficient to Induce Satb1 Expression, Additional Results Related to Figure 6
(A) schematic representation of in utero electroporation of pallial progenitors.
(B–F2) Sections from E16.5 embryos which have been in utero electroporated at E13.5 with either pGFP (B-C) or pLHX6-GFP and pMASH1-GFP (D and E). Sections shown in (B) and (D) were immunostained for GFP, while their corresponding serial sections (C) and (E) were in situ hybridized with a Dclk2-specific riboprobe. Coexpression of LHX6 and MASH1 in cortical progenitors upregulates Dclk2. Sections shown in (F1) and (F2) have been immunostained for GFP and SATB1, respectively. (F) shows an overlay of images in (F1) and (F2). Lines in (B)–(F2) mark the edge of the tissue.
(G–I) (G) Schematic representation of MGE electroporation and culture of forebrain slices from E14.5 embryos. H-I2, sections of slices in which MGE was transfected with GFP (H–H2) or pKCC2-GFP (I–I2) and immunostained 24hrs later for GFP (green, H1, I1) or KCC2 (red, H2, I2). (H) and (I) are overlays of (H1) and (H2) and (I1) and (I2), respectively.
(J–L) (J) Schematic representation of the experimental procedure. (K)–(L3) Images of cultured MGE cells transfected with pGFP (K) or KCC2-expressing (pKCC2-GFP; L) vectors, immunostained for GFP (K1 and L1), KCC2 (K2 and L2), and SATB1 (K3 and L3). (K) and (L) are overlays of (K1), (K2), (K3), and (L1), (L2), (L3), respectively. Arrow in (K) points to a GFP+ MGE cell expressing SATB1 but negative for KCC2.
Activity-Mediated Induction of Satb1
Spontaneous neuronal activity, a feature of the developing CNS, influences fundamental processes of brain development, such as cell migration, neuronal maturation, and synaptogenesis (Moody and Bosma, 2005). In the mouse neocortex, spontaneous activity is observed from around E16.5 and peaks at early postnatal stages (Corlew et al., 2004), raising the possibility that neuronal excitability controls the upregulation of SATB1 observed at perinatal stages. To explore this possibility, we established cultures of dissociated dorsal telencephalon from E14.5 mouse embryos and analyzed the expression of Satb1 in GABAergic interneurons at various intervals after plating (Figure 7A). At 24 hr, only a small fraction of interneurons were positive for SATB1 but the percentage of SATB1+GAD67+ cells increased considerably at subsequent stages (Figures 7B–7E and 7N), reflecting the developmental profile of SATB1 in endogenous cortical interneurons. Interestingly, treatment of 24 hr cultures with KCl (40 mM), which is capable of depolarizing immature neurons, led to a significant upregulation of c-FOS and SATB1 within 24 hr (Figures 7F–7K and 7O). KCl-induced depolarization can trigger Ca2+ influx through voltage-gated calcium channels, a subtype of which (L-type) is known to be expressed in cortical interneurons and have been implicated in several activity-dependent developmental processes in the brain (Jiang and Swann, 2005). Consistent with this, KCl treatment of cortical cultures in the presence of the Ca2+ channel blocker nifedipine (10 μM) abrogated the KCl-mediated induction of SATB1 (Figures 7K, 7L, and 7O).
Figure 7.
Neuronal Activity Induces Expression of Satb1 in Cortical Interneurons
(A) Schematic representation of experimental strategy. E14.5 cortices were dissected, dissociated, and cultured for the indicated times.
(B–E) Images of cortical cultures immunostained for GAD67 (green) and SATB1 (red) and counterstained with DAPI (blue) at different time points. Note the increase of the number of SATB1+ neurons during the 5 day culture period.
(F–I) Immunostaining of 48 hr control (F and H) and KCL-treated (G and I) cortical cultures with antibodies for cFOS (F and G) and SATB1 (H and I).
(J–M) Immunostaining of 48 hr control (J), KCl- (K), KCl+Nifedipine- (L), or Bicuculine-treated (M) cortical cultures with antibodies for SATB1 (red) and GAD67 (green). Arrowheads in (J)–(M) indicate double-positive GAD67+SATB1+ interneurons, while arrows indicate interneurons negative for SATB1.
(N–P) Quantification of the percentage of GAD67+ cells that coexpress SATB1 under the indicated culture conditions. n = 3 independent experiments for (N) and (P) and n = 4 for (O), ∗p < 0.05, ∗∗∗p < 0.001 for each test (Student’s homoscedastic t test with two-tailed distibution).
In immature cortical interneurons, GABA has an excitatory effect due to a reverse chloride gradient and several studies suggest that this neurotransmitter influences neuronal migration, maturation, and integration into circuits (Ben-Ari, 2002; Ben-Ari and Spitzer, 2010). To explore whether GABA is part of the activity stimulus that upregulates Satb1 in our cortical cultures, we treated 24 hr cortical cultures with two independent GABA(A) receptor (GABAAR) antagonists, bicuculine (10 μM) and picrotoxin (10 μM). Both resulted in a relatively small but statistically significant reduction in the percentage of SATB1+GAD67+ neurons at 48 hr (Figures 7M and 7P). Taken together, these studies suggest that neuronal excitability and Ca2+ activity are implicated in the upregulation of Satb1 in cortical interneurons and that this upregulation is partially mediated by GABA receptors.
Discussion
Development of cortical interneurons in mammals is a prolonged process that is initiated during embryogenesis in the ventral telencephalon and completed in juvenile animals with the functional integration of highly divergent GABAergic neurons into cortical circuits (Batista-Brito et al., 2009). The astonishing diversity of cortical interneurons is ultimately determined by genetic programs activated in the ventral forebrain but the full spectrum of molecular, morphological and physiological properties of mature interneurons becomes apparent only at relatively late stages of interneuron development and coincides with the onset of neuronal activity (Butt et al., 2005; Miyoshi et al., 2010; Cossart, 2011). Considerable recent progress has identified genetic cascades that control the specification of cortical interneurons, but the molecular mechanisms that underlie the maturation of different subtypes remain largely unknown (Batista-Brito et al., 2009). Here, we demonstrate that the chromatin organizer and transcription factor SATB1 has a pivotal role in the maturation and terminal differentiation of a major group of MGE-derived cortical interneurons and provide evidence that spontaneous neuronal activity is a critical regulator of Satb1 expression in the pallium. Our combined loss- and gain-of-function studies demonstrate that SATB1 coordinately controls the expression of subtype-specific and interneuron-wide genes in a manner that characterizes mature interneurons of the Sst group. Based on these findings, we argue that SATB1 links subtype-specific genetic programs that are activated in the ventral forebrain and control the specification of SST+ interneurons with molecular cascades that promote the terminal differentiation and maturation of all classes of inhibitory neurons within the pallium.
Consistent with the observation that Satb1 is specifically expressed in interneurons within the cortical plate, a Satb1-null mutation in mice resulted in a dramatic downregulation of Sst mRNA and protein in cortical interneurons of postnatal animals but had no effect on Sst expression in their tangentially migrating precursors. These findings argue for a two-stage regulation of Sst expression: an early SATB1-independent phase, which denotes subtype commitment of SST+ interneurons, and a subsequent SATB1-dependent phase, which reflects the maturation of SST-specified MGE precursors into functional interneurons. This biphasic mode of regulation of Sst is consistent with the complex regulatory landscape of the locus (Burbach, 2002) and suggests that distinct stage-specific transcriptional regulators control the expression of Sst during the course of cortical interneuron development. A recent report has shown that in mouse brain SATB1 occupies an upstream regulatory region of Sst (Balamotis et al., 2012), suggesting direct transcriptional regulation of this locus by SATB1. Although this finding provides a pertinent explanation for our current observations, the detailed mechanisms by which SATB1 controls the coordinate expression of interneuron-specific genes requires further investigation. During T cell development and in breast cancer cells, it is thought that target loci of SATB1 are tethered via specialized genomic sequences to a common regulatory framework that recruits chromatin-modifying factors and allows the simultaneous response to transcriptional complexes (Alvarez et al., 2000; Cai et al., 2003; Han et al., 2008). These studies and our current findings suggest that SATB1 functions as a global genome organizer to coordinately control the expression of multiple loci that promote maturation and integration of SST+ interneurons into functional cortical circuits.
SST is detectable exclusively in cortical interneurons of postnatal animals suggesting that in mammals productive expression of Sst is associated with the later SATB1-dependent phase of expression. This is supported by our demonstration that overexpression of SATB1 in MGE precursors is sufficient to induce expression of both Sst mRNA and protein. These findings argue that the absence of SST from the MGE and tangentially migrating interneurons is not due to the nonspecific inhibition of Sst mRNA translation in these cells and raise the intriguing possibility that, in addition to its transcriptional role, SATB1 is implicated in the posttranscriptional regulation of Sst expression in cortical interneurons. Although the mechanisms of such regulation are currently unclear, it has been previously reported that nNOS1 and PSD95 are posttranscriptionally regulated in pyramidal neurons in an activity-dependent manner by the product of the fragile X syndrome-associated FMR1 locus (Todd et al., 2003; Kwan et al., 2012). It appears therefore that in the mammalian cortex posttranscriptional gene regulation is a feature of both excitatory and inhibitory neurons and that SATB1 could be a key mediator of activity-dependent posttranscriptional regulation in cortical interneurons.
Although both major groups of MGE-derived interneurons express Satb1, our analysis of Satb1-deficient mice revealed a relatively modest deficit of PV+ inhibitory neurons. Such deficit could reflect a secondary effect of the developmental abnormalities of SST+ interneurons observed in these mutants, an idea which is supported by the emerging view that abnormal maturation of specific subsets of GABAergic interneurons in the cortex often lead to generalized defects of other subpopulations of interneurons across the entire neuronal network (Cossart, 2011). The additional dendritic and synaptic transmission deficits of principal cortical neurons observed in Satb1 mutants (Balamotis et al., 2012) could also contribute to the PV+ interneuron deficit. Alternatively, the reduced number of PV+ interneurons in Satb1-deficient animals could result from a cell autonomous requirement of this factor for the differentiation of a subset of PV+ interneurons. Subtype-specific deletion of Satb1 in vivo should distinguish between these possibilities.
Previous studies from our and other groups have demonstrated that the LIM HD transcription factor LHX6 is an upstream regulator of divergent molecular cascades that control the development of MGE-derived cortical interneurons (Liodis et al., 2007; Zhao et al., 2008). Downstream of LHX6, the transcriptional regulator SOX6 controls predominantly the differentiation of PV+ interneurons (Azim et al., 2009; Batista-Brito et al., 2009). Here, we provide evidence that expression of Satb1 and the consequent effect on the development of SST+ interneurons depend on Lhx6 activity. The considerable delay of Satb1 activation relative to Lhx6 in vivo and our overexpression studies indicate that LHX6 is not sufficient to drive expression of Satb1 and suggest that additional stage-specific regulators may be involved. The demonstration that neuronal excitability is sufficient to upregulate Satb1 expression in cultured interneurons provides a framework for understanding the mechanisms by which LHX6 controls later stages of interneuron development. We hypothesize that LHX6 is required cell autonomously in cortical interneurons for the advent of a gene expression profile that allows these cells to develop and/or respond to the emerging activity of the developing brain. Previous studies have indicated that ambient GABA plays a key role in the development of early activity of the brain (Ben-Ari et al., 2007), raising the possibility that GABA-mediated activity controls the expression of Satb1 in an LHX6-dependent manner in cortical interneurons. In support of this hypothesis, we have previously reported dramatic changes in the release of GABA by inhibitory neurons in the hippocampus of Lhx6-null mutant mice (Neves et al., 2012). Moreover, our current pharmacological experiments suggest a requirement for GABAAR for the activation of Satb1 in cortical interneuron cultures. Nevertheless, the observed inhibition of Satb1 expression in these experiments was only partial, suggesting that additional mechanisms may be implicated in the induction of this gene. In summary, our experiments argue that SATB1 constitutes an activity-dependent molecular switch downstream of Lhx6, which controls the maturation and terminal differentiation of cortical interneurons. These findings have important implications for understanding the molecular control of cortical interneuron development and inhibitory network assembly.
Experimental Procedures
Animals
Embryonic stem cells (ESCs) heterozygous for the Satb1EUCOMM allele were obtained from the EUCOMM consortium (http://www.knockoutmouse.org/about/eucomm). The oligonucleotide primers used for genotyping were R10 (5′-CAG GCC ACA TTG TCC TAA CT-3′), R11 (5′-GAA TAG GAA CTT CGG TTC CG-3′), and F9 (5′-TGC TCA TGT GGA ATG TCG AG-3′). The generation of the following alleles has been described previously: Nkx2.1Cre (Kessaris et al., 2006), Gad67GFP (Tanaka et al., 2003), Rosa26YFP (Srinivas et al., 2001), and Lhx6− (Liodis et al., 2007). For timed pregnancies, the day of vaginal plug was considered E0.5. All experiments were carried out under a United Kingdom Home Office Project License and were approved by the Institutional Ethics Committee.
Embryonic Cortical Cultures
Cortices from E14.5 mouse (Parkes) embryos were mechanically dissociated in the presence of DNase (100 ng/ml, Sigma-Aldrich), plated on poly-D-lysine (10 μg/ml) and laminin-coated (5 μg/ml) 8-well chamber slides (Labtek, VWR) (1 × 105 cells per well), and cultured in Neurobasal medium (1XB-27, 0.5% glucose, 1 × glutamine, 1 × PenStrep in Neurobasal medium, all from Invitrogen) for 1–5 days. All drugs (nifedipine and picrotoxin from Sigma and bicuculline methochloride from Tocris) were stored at −20°C as 1,000× stocks and diluted to the final concentration just prior to application.
Additional methods and any associated references are available in the Extended Experimental Procedures.
Figure S7.

SATB1 Is Not Detected in the Cortex of Satb1−/− Mice, Additional Results Related to Figure 5
Immunostaining of cortical sections from P2 control (Satb1+/+, A) and Satb1-null mutant (Satb1−/−, B) animals immunostained with SATB1-specific antisera.
Acknowledgments
We thank Troy Margrie, Francois Guillemot, Holger Apitz, and members of the Pachnis laboratory for useful discussions and insightful comments on the manuscript. J. Briscoe, F. Guillemot, N. Kessaris, and V. Tarabykin provided reagents, and we are grateful to them. We also thank Francesca Subirada and Olga Durany of Oryzon Genomics for their contributions to the microarray data analysis. We are grateful to Lydia Teboul, who provided critical advice on culturing conditions for the Satb1-targeted C57Bl/6 ESC lines, and Mauro Tollaini, who carried out the blastocyst injections of ESCs. We also thank members of the NIMR Biological Services for expert animal husbandry and Hayley Wood from NIMR Photographic Services for generating the schematics used in this manuscript. This work was funded by the Medical Research Council (UK; Grant-in-Aid no. A2555RP50) and an FP6 EU grant (INTERDEVO Consortium).
Published: November 8, 2012
Footnotes
Supplemental Information includes Extended Experimental Procedures, seven figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2012.10.003.
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information
References
- Alvarez J.D., Yasui D.H., Niida H., Joh T., Loh D.Y., Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521–535. [PMC free article] [PubMed] [Google Scholar]
- Azim E., Jabaudon D., Fame R.M., Macklis J.D. SOX6 controls dorsal progenitor identity and interneuron diversity during neocortical development. Nat. Neurosci. 2009;12:1238–1247. doi: 10.1038/nn.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamotis M.A., Tamberg N., Woo Y.J., Li J., Davy B., Kohwi-Shigematsu T., Kohwi Y. Satb1 ablation alters temporal expression of immediate early genes and reduces dendritic spine density during postnatal brain development. Mol. Cell. Biol. 2012;32:333–347. doi: 10.1128/MCB.05917-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Brito R., Fishell G. The developmental integration of cortical interneurons into a functional network. Curr. Top. Dev. Biol. 2009;87:81–118. doi: 10.1016/S0070-2153(09)01203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Brito R., Rossignol E., Hjerling-Leffler J., Denaxa M., Wegner M., Lefebvre V., Pachnis V., Fishell G. The cell-intrinsic requirement of Sox6 for cortical interneuron development. Neuron. 2009;63:466–481. doi: 10.1016/j.neuron.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellion A., Baudoin J.P., Alvarez C., Bornens M., Métin C. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J. Neurosci. 2005;25:5691–5699. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y. Excitatory actions of gaba during development: the nature of the nurture. Nat. Rev. Neurosci. 2002;3:728–739. doi: 10.1038/nrn920. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y., Spitzer N.C. Phenotypic checkpoints regulate neuronal development. Trends Neurosci. 2010;33:485–492. doi: 10.1016/j.tins.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y., Gaiarsa J.L., Tyzio R., Khazipov R. GABA: a pioneer transmitter that excites immature neurons and generates primitive oscillations. Physiol. Rev. 2007;87:1215–1284. doi: 10.1152/physrev.00017.2006. [DOI] [PubMed] [Google Scholar]
- Bortone D., Polleux F. KCC2 expression promotes the termination of cortical interneuron migration in a voltage-sensitive calcium-dependent manner. Neuron. 2009;62:53–71. doi: 10.1016/j.neuron.2009.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbach J.P. Regulation of gene promoters of hypothalamic peptides. Front. Neuroendocrinol. 2002;23:342–369. doi: 10.1016/s0091-3022(02)00005-5. [DOI] [PubMed] [Google Scholar]
- Butt S.J., Fuccillo M., Nery S., Noctor S., Kriegstein A., Corbin J.G., Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Cai S., Han H.J., Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- Cai S., Lee C.C., Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Chen L., Liao G., Waclaw R.R., Burns K.A., Linquist D., Campbell K., Zheng Y., Kuan C.Y. Rac1 controls the formation of midline commissures and the competency of tangential migration in ventral telencephalic neurons. J. Neurosci. 2007;27:3884–3893. doi: 10.1523/JNEUROSCI.3509-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow A., Erisir A., Farb C., Nadal M.S., Ozaita A., Lau D., Welker E., Rudy B. K(+) channel expression distinguishes subpopulations of parvalbumin- and somatostatin-containing neocortical interneurons. J. Neurosci. 1999;19:9332–9345. doi: 10.1523/JNEUROSCI.19-21-09332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corlew R., Bosma M.M., Moody W.J. Spontaneous, synchronous electrical activity in neonatal mouse cortical neurones. J. Physiol. 2004;560:377–390. doi: 10.1113/jphysiol.2004.071621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart R. The maturation of cortical interneuron diversity: how multiple developmental journeys shape the emergence of proper network function. Curr. Opin. Neurobiol. 2011;21:160–168. doi: 10.1016/j.conb.2010.10.003. [DOI] [PubMed] [Google Scholar]
- del Río J.A., de Lecea L., Ferrer I., Soriano E. The development of parvalbumin-immunoreactivity in the neocortex of the mouse. Brain Res. Dev. Brain Res. 1994;81:247–259. doi: 10.1016/0165-3806(94)90311-5. [DOI] [PubMed] [Google Scholar]
- Fazzari P., Paternain A.V., Valiente M., Pla R., Luján R., Lloyd K., Lerma J., Marín O., Rico B. Control of cortical GABA circuitry development by Nrg1 and ErbB4 signalling. Nature. 2010;464:1376–1380. doi: 10.1038/nature08928. [DOI] [PubMed] [Google Scholar]
- Flames N., Long J.E., Garratt A.N., Fischer T.M., Gassmann M., Birchmeier C., Lai C., Rubenstein J.L., Marín O. Short- and long-range attraction of cortical GABAergic interneurons by neuregulin-1. Neuron. 2004;44:251–261. doi: 10.1016/j.neuron.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Fogarty M., Grist M., Gelman D., Marín O., Pachnis V., Kessaris N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J. Neurosci. 2007;27:10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H.J., Russo J., Kohwi Y., Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- Huang Y., Zhang L., Song N.N., Hu Z.L., Chen J.Y., Ding Y.Q. Distribution of Satb1 in the central nervous system of adult mice. Neurosci. Res. 2011;71:12–21. doi: 10.1016/j.neures.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Inada H., Watanabe M., Uchida T., Ishibashi H., Wake H., Nemoto T., Yanagawa Y., Fukuda A., Nabekura J. GABA regulates the multidirectional tangential migration of GABAergic interneurons in living neonatal mice. PLoS ONE. 2011;6:e27048. doi: 10.1371/journal.pone.0027048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Swann J.W. A role for L-type calcium channels in the maturation of parvalbumin-containing hippocampal interneurons. Neuroscience. 2005;135:839–850. doi: 10.1016/j.neuroscience.2005.06.073. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y., Kubota Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb. Cortex. 1997;7:476–486. doi: 10.1093/cercor/7.6.476. [DOI] [PubMed] [Google Scholar]
- Kessaris N., Fogarty M., Iannarelli P., Grist M., Wegner M., Richardson W.D. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat. Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y., Shigematsu N., Karube F., Sekigawa A., Kato S., Yamaguchi N., Hirai Y., Morishima M., Kawaguchi Y. Selective coexpression of multiple chemical markers defines discrete populations of neocortical GABAergic neurons. Cereb. Cortex. 2011;21:1803–1817. doi: 10.1093/cercor/bhq252. [DOI] [PubMed] [Google Scholar]
- Kwan K.Y., Lam M.M., Johnson M.B., Dube U., Shim S., Rašin M.R., Sousa A.M., Fertuzinhos S., Chen J.G., Arellano J.I. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell. 2012;149:899–911. doi: 10.1016/j.cell.2012.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liodis P., Denaxa M., Grigoriou M., Akufo-Addo C., Yanagawa Y., Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J. Neurosci. 2007;27:3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H., Toledo-Rodriguez M., Wang Y., Gupta A., Silberberg G., Wu C., Monyer H. Interneurons of the neocortical inhibitory system. Nat. Rev. Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- Miyoshi G., Butt S.J., Takebayashi H., Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J. Neurosci. 2007;27:7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G., Hjerling-Leffler J., Karayannis T., Sousa V.H., Butt S.J., Battiste J., Johnson J.E., Machold R.P., Fishell G. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J. Neurosci. 2010;30:1582–1594. doi: 10.1523/JNEUROSCI.4515-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody W.J., Bosma M.M. Ion channel development, spontaneous activity, and activity-dependent development in nerve and muscle cells. Physiol. Rev. 2005;85:883–941. doi: 10.1152/physrev.00017.2004. [DOI] [PubMed] [Google Scholar]
- Neddens J., Buonanno A. Selective populations of hippocampal interneurons express ErbB4 and their number and distribution is altered in ErbB4 knockout mice. Hippocampus. 2010;20:724–744. doi: 10.1002/hipo.20675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves G., Shah M.M., Liodis P., Achimastou A., Denaxa M., Roalfe G., Sesay A., Walker M.C., Pachnis V. The LIM homeodomain protein Lhx6 regulates maturation of interneurons and network excitability in the mammalian cortex. Cereb. Cortex. 2012 doi: 10.1093/cercor/bhs159. Published online June 17, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrenoud Q., Geoffroy H., Gauthier B., Rancillac A., Alfonsi F., Kessaris N., Rossier J., Vitalis T., Gallopin T. Characterization of type I and type II nNOS-expressing interneurons in the barrel cortex of mouse. Frontiers Neural Circuits. 2012;6:36. doi: 10.3389/fncir.2012.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy B., Fishell G., Lee S., Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev. Neurobiol. 2011;71:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka D., Nakaya Y., Yanagawa Y., Obata K., Murakami F. Multimodal tangential migration of neocortical GABAergic neurons independent of GPI-anchored proteins. Development. 2003;130:5803–5813. doi: 10.1242/dev.00825. [DOI] [PubMed] [Google Scholar]
- Taniguchi H., He M., Wu P., Kim S., Paik R., Sugino K., Kvitsiani D., Fu Y., Lu J., Lin Y. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd P.K., Mack K.J., Malter J.S. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc. Natl. Acad. Sci. USA. 2003;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka R., Okamoto K., Furuta T., Fujiyama F., Iwasato T., Yanagawa Y., Obata K., Kaneko T., Tamamaki N. Demonstration of long-range GABAergic connections distributed throughout the mouse neocortex. Eur. J. Neurosci. 2005;21:1587–1600. doi: 10.1111/j.1460-9568.2005.03989.x. [DOI] [PubMed] [Google Scholar]
- Tuy F.P., Saillour Y., Kappeler C., Chelly J., Francis F. Alternative transcripts of Dclk1 and Dclk2 and their expression in doublecortin knockout mice. Dev. Neurosci. 2008;30:171–186. doi: 10.1159/000109861. [DOI] [PubMed] [Google Scholar]
- Welagen J., Anderson S. Origins of neocortical interneurons in mice. Dev. Neurobiol. 2011;71:10–17. doi: 10.1002/dneu.20857. [DOI] [PubMed] [Google Scholar]
- Wonders C.P., Anderson S.A. The origin and specification of cortical interneurons. Nat. Rev. Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Xu Q., Guo L., Moore H., Waclaw R.R., Campbell K., Anderson S.A. Sonic hedgehog signaling confers ventral telencephalic progenitors with distinct cortical interneuron fates. Neuron. 2010;65:328–340. doi: 10.1016/j.neuron.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada J., Okabe A., Toyoda H., Kilb W., Luhmann H.J., Fukuda A. Cl- uptake promoting depolarizing GABA actions in immature rat neocortical neurones is mediated by NKCC1. J. Physiol. 2004;557:829–841. doi: 10.1113/jphysiol.2004.062471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Flandin P., Long J.E., Cuesta M.D., Westphal H., Rubenstein J.L. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J. Comp. Neurol. 2008;510:79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- Alvarez, J.D., Yasui, D.H., Niida, H., Joh, T., Loh, D.Y., and Kohwi-Shigematsu, T. (2000). The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 14, 521–535. [PMC free article] [PubMed]
- Du, T., Xu, Q., Ocbina, P.J., and Anderson, S.A. (2008). NKX2.1 specifies cortical interneuron fate by activating Lhx6. Development 135, 1559–1567. [DOI] [PubMed]
- Grigoriou, M., Tucker, A.S., Sharpe, P.T., and Pachnis, V. (1998). Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development 125, 2063–2074. [DOI] [PubMed]
- Lavdas, A.A., Grigoriou, M., Pachnis, V., and Parnavelas, J.G. (1999). The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J. Neurosci. 19, 7881–7888. [DOI] [PMC free article] [PubMed]
- Liodis, P., Denaxa, M., Grigoriou, M., Akufo-Addo, C., Yanagawa, Y., and Pachnis, V. (2007). Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J. Neurosci. 27, 3078–3089. [DOI] [PMC free article] [PubMed]
- Nguyen, L., Besson, A., Heng, J.I., Schuurmans, C., Teboul, L., Parras, C., Philpott, A., Roberts, J.M., and Guillemot, F. (2006). p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 20, 1511–1524. [DOI] [PMC free article] [PubMed]
- Paxinos, G.F., and Franklin, K.B.J. (2001). The Mouse Brain in Stereotaxic Coordinates, Second Edition (New York: Academic Press).
- Schaeren-Wiemers, N., and Gerfin-Moser, A. (1993). A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry 100, 431–440. [DOI] [PubMed]
- Stühmer, T., Puelles, L., Ekker, M., and Rubenstein, J.L. (2002). Expression from a Dlx gene enhancer marks adult mouse cortical GABAergic neurons. Cereb. Cortex 12, 75–85. [DOI] [PubMed]
- Xu, Q., Cobos, I., De La Cruz, E., Rubenstein, J.L., and Anderson, S.A. (2004). Origins of cortical interneuron subtypes. J. Neurosci. 24, 2612–2622. [DOI] [PMC free article] [PubMed]
- Xu, Q., Guo, L., Moore, H., Waclaw, R.R., Campbell, K., and Anderson, S.A. (2010). Sonic hedgehog signaling confers ventral telencephalic progenitors with distinct cortical interneuron fates. Neuron 65, 328–340. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.