Summary
Growing evidence suggests that aggregation-prone proteins are both harmful and functional for a cell. How do cellular systems balance the detrimental and beneficial effect of protein aggregation? We reveal that aggregation-prone proteins are subject to differential transcriptional, translational, and degradation control compared to nonaggregation-prone proteins, which leads to their decreased synthesis, low abundance, and high turnover. Genetic modulators that enhance the aggregation phenotype are enriched in genes that influence expression homeostasis. Moreover, genes encoding aggregation-prone proteins are more likely to be harmful when overexpressed. The trends are evolutionarily conserved and suggest a strategy whereby cellular mechanisms specifically modulate the availability of aggregation-prone proteins to (1) keep concentrations below the critical ones required for aggregation and (2) shift the equilibrium between the monomeric and oligomeric/aggregate form, as explained by Le Chatelier’s principle. This strategy may prevent formation of undesirable aggregates and keep functional assemblies/aggregates under control.
Graphical Abstract
Highlights
► mRNA encoding aggregation-prone proteins is complex, suggesting greater translational regulation ► Aggregation-prone proteins are present in low abundance and for short periods of time ► Tight control is evolutionarily conserved and provides robustness against aggregation ► Aggregation-prone proteins are subject to tight regulation
Although deposits of protein aggregates are the hallmark of various neurodegenerative diseases, recent studies have demonstrated that protein aggregation is exploited in different physiological processes. These observations raise the question of how cells balance the detrimental and beneficial effects of aggregation. Gsponer and Babu reveal that aggregation-prone structured and disordered proteins are subject to differential regulation compared to non-aggregation-prone proteins. Their results provide a unifying framework for understanding the control of functional and nonfunctional protein aggregation in cells.
Introduction
The process of protein aggregation has been linked to several human pathologies, such as Alzheimer’s and Parkinson’s disease (Chiti and Dobson, 2006). While the potentially harmful effects of protein aggregation have been well established by several studies, it is less often emphasized that protein aggregation can also have beneficial effects to cellular systems. A number of recent studies have shown that several human physiological processes depend on protein aggregation or even fibril formation (Fowler et al., 2007; Reijns et al., 2008; Salazar et al., 2010). Remarkably, the dynamic formation of a variety of cellular bodies, such as stress granules and processing bodies, has been shown to depend on protein aggregation (Balagopal and Parker, 2009). For instance, assembly of stress granules is mediated by aggregation of a glutamine-rich domain in the RNA-binding proteins TIA-1 (Gilks et al., 2004) and Pum (Salazar et al., 2010). Similarly, glutamine/asparagine (Q/N)-rich segments have been shown to be essential for the formation of processing bodies. Although it is unlikely that all aggregates formed in these cellular bodies have a fibrillar character, it is certain that the aggregation propensity of proteins has been exploited to mediate the formation of these assemblies (Fiumara et al., 2010; Salazar et al., 2010). Nonetheless, recent studies have shown that certain protein interactions (for example, hdm2-arf) indeed involve formation of amyloid-like structures (Sivakolundu et al., 2008) and that several peptide and protein hormones are stored in an amyloid-like conformation within cells (Maji et al., 2009).
The observations that extant genomes contain a significant proportion of proteins with the potential to form aggregates and that stretches of aggregation-prone regions are evolutionarily conserved (see Extended Results; Figure S1) suggest that, though potentially harmful, such regions might be structurally and functionally important (Goldschmidt et al., 2010; Linding et al., 2004; Monsellier et al., 2008). For instance, they may be part of the essential hydrophobic core of globular proteins (Linding et al., 2004) or may form patches that mediate protein interactions (Masino et al., 2011; Pechmann et al., 2009). Taken together, these considerations raise the following fundamental questions: (1) how do cells minimize the likelihood of spontaneous aggregation of proteins containing aggregation-prone regions? (2) How are functional aggregates kept under control? The fact that protein aggregation can have harmful effects suggests that “nonfunctional” aggregation should be avoided and “functional” aggregation has to be highly regulated. Indeed, for individual cases of functional aggregates, control mechanisms that regulate the aggregation process have been identified (Fowler et al., 2007). However, very little is known about the regulation of the majority of proteins that are known to form aggregates in a cell or that contain evolutionarily conserved aggregation-prone segments. We hypothesized that cellular systems could have evolved regulatory mechanisms to keep protein aggregation under control by ensuring that the levels of these proteins are low and that they are turned over rapidly. In this work, we present evidence that supports this hypothesis, define a framework for protein aggregation regulation, and discuss its implications.
Extended Results.
Extant Genomes Contain Aggregation-Prone Proteins and Stretches of Aggregation-Prone Amino Acids Are Evolutionarily Conserved
We investigated how many proteins in S. cerevisiae. S. pombe, D. melanogaster and H. sapiens contain at least one aggregation prone stretch of at least seven consecutive residues that TANGO assigns a high score. Interestingly, we found that between 30% and 39% of all proteins in these organisms contain at least one aggregation prone stretch (Table S4A). Even more importantly, an analysis of the conservation of aggregation prone and non-aggregation prone residues in nine yeast strains revealed that the aggregation promoting residues in S. cerevisiae are more often conserved in the other strains than the non-aggregation prone residues (Figure S1). This result, together with recent published work (David et al., 2010; Demontis and Perrimon, 2010; Goldschmidt et al., 2010; Linding et al., 2004; Monsellier et al., 2008; Tartaglia et al., 2005), suggests that (i) aggregation prone residues have been conserved for functional reasons that may not directly be related to aggregation and/or (ii) organisms may exploit controlled aggregation for biological function. Certain conserved stretches of aggregation prone hydrophobic residues may be found in many prokaryotic and eukaryotic proteins because they are necessary to stabilize the hydrophobic core of their globular folds (Lim and Sauer, 1991; Linding et al., 2004; Mendel et al., 1992; Niwa et al., 2009). In addition, it has been established that hydrophobic patches mediate many high-affinity protein-protein interactions (Pechmann et al., 2009; Sivakolundu et al., 2008). Recently, it has also been shown that interface regions of protein complexes are more aggregation prone than other surfaces (Pechmann et al., 2009).
The Requirement to Classify Proteins into Structured and Unstructured Proteins and Validity of the Predicted (Non-) Aggregation-Prone Proteins
Requirement for the Classification into the Structured and Unstructured Group of Proteins
We previously showed that highly unstructured proteins are, in general, differently regulated from highly structured proteins (Gsponer et al., 2008). To investigate the effect of the presence of aggregation prone residues on the availability of proteins, we therefore had to analyze proteins in the unstructured (U) and the structured (S) group separately. For the analysis presented in the main text, we did not consider proteins in the moderately unstructured (M group) in order to (i) ensure that we are unambiguously dealing with highly unstructured and structured proteins and (ii) to identify the effect of the presence of aggregation prone stretches on the availability of proteins in each group. However, all the trends we observed between aggregation prone and non-aggregation prone proteins are valid for the whole proteome of S. cerevisiae not only proteins in the S and U group (see below, Table S3F).
Structured Class
The likelihood of a protein to form β sheet aggregates can vary significantly depending on the amino acid sequence composition of a polypeptide chain (Bemporad et al., 2006; López de la Paz and Serrano, 2004; Pastor et al., 2005; Pawar et al., 2005). A key determinant of the aggregation likelihood is the hydrophobicity of the polypeptide chain (Chiti et al., 2003; Fernandez-Escamilla et al., 2004; Gsponer and Vendruscolo, 2006). In folded proteins, hydrophobic residues are generally buried in the core of structured protein domains (Lim and Sauer, 1991; Mendel et al., 1992). Therefore, protein misfolding or partial (un)folding of a structured domain, which can be triggered by changes in the cellular environment, is often required for structured protein to assemble non-specifically with other molecules or aggregate (Chow et al., 2004). It has also been established that aggregation after unfolding/misfolding is initiated by stretches of exposed highly hydrophobic, aggregation prone residues (Chiti and Dobson, 2006; Münch and Bertolotti, 2010). Hence, highly hydrophobic, aggregation prone stretches facilitate aggregation of structured proteins, but thermodynamic stability and the fording/unfolding mechanisms will affect the real “aggregation potential” of these hydrophobic residues in a given protein. However, assessing thermodynamic stability and folding mechanism experimentally at the proteome scale is not yet reliably feasible nor is such data available on a genomic scale.
Unstructured Class
Proteins that lack a unique three-dimensional structure, so-called intrinsically unstructured or disordered proteins (IUPs), have far fewer hydrophobic residues than structured ones and often have a high net charge (Dunker et al., 2001). These sequence properties generally reduce the risk of undesired aggregation of IUPs under physiological condition (Monsellier et al., 2008). However, some IUPs contain long repetitive sequence elements like Q/N-enriched stretches that have a high aggregation propensity and such stretches of the polypeptide chain can mediate the formation of beta-sheet aggregates (Altschuler et al., 1997; Chen et al., 2001; DePace et al., 1998). Although some of these Q/N-enriched segments may be flanked with prolines or charged residues that can reduce the likelihood of aggregation, it is established that long Q/N-enriched segments are essential for aggregation. Thus it is clear that for such proteins, there is no requirement to unfold in order to aggregate since the protein is unstructured already. However, the Q/N enriched stretches could also mediate interactions with other proteins that are not of the β-aggregate type. Indeed, recently such regions have also been shown to form coiled-coils (Fiumara et al., 2010). Such interactions may affect the real “aggregation potential” of the Q/N stretches by preventing their exposure. There is no available large-scale experimental data that allows assessing the direct involvement of Q/N enriched stretches in protein interactions.
Given (i) the major differences between the kinds of requirements to form aggregates involving structured proteins and unstructured proteins and (ii) structured proteins are generally regulated differently from unstructured proteins (see Gsponer et al., 2008), it is therefore important to investigate these two groups of proteins independently.
Validity of Predicted (Non-) Aggregation-Prone Proteins in the Two Structural Classes
Structured Class
How well does TANGO predict aggregation prone proteins? TANGO was benchmarked against 175 peptides of over 20 proteins and was able to predict the sequences experimentally observed to contribute to the aggregation of these proteins (Fernandez-Escamilla et al., 2004). In addition, we tested the performance of TANGO in identifying proteins in S. cerevisiae that form punctate foci upon nutrient starvation (Narayanaswamy et al., 2009). These reversible protein assemblies have been observed for both tagged and untagged proteins by using multiple independent techniques. As can be seen in Table S4B, proteins that form foci have a significantly higher TANGO score than proteins that do not form foci. This was also noted by Narayanaswamy et al. (2009). In addition, we observed that the former group harbors aggregation prone stretches more often than the latter. This is also the case when we used AGGRESCAN instead of TANGO (Table S4B).
Unstructured Class
In order to test whether K or E enriched domains are a common feature of intrinsically unstructured proteins that remain soluble even under harsh conditions and do not aggregate, we analyzed a set of intrinsically unstructured proteins that have recently been identified in the heat-treated, soluble extract from mouse fibroblast cells (Galea et al., 2009). While intrinsically unstructured proteins in mouse (disorder predicted with Disopred2) have on average 19 lysines or glutamic acids in any continuous segment of 80 residues, the heat-treated and soluble subgroup has on average 23 lysines or glutamic acids in any continuous segment of 80 residues (p < 10−16; t test).
mRNA Encoding Aggregation-Prone Proteins Have Lower Translation Efficiency
We also analyzed recently published ribosome-profiling data to investigate translation efficiency of aggregation prone and non-aggregation prone proteins. However, many of the short reads obtained from the foot printing experiment will be discarded if they map to repeat regions. These are the kind of regions that many unstructured proteins contain (e.g., regions coding for the polyK/E and polyQ/N segments), thus potentially making the data less reliable for testing a general principle. Therefore, we used ribosome-profiling data from Ingolia et al. only to compare the translational efficiency of structured aggregation and non-aggregation prone proteins (Figure S3).
Partial Least Square Regression Analysis
In order to obtain an estimate of the relative contribution of transcript abundance, transcription rate and protein-half-life to protein abundance of aggregation prone and non-aggregation prone proteins, we carried out a partial least square regression analysis (PLSR, see Extended Experimental Procedures). The PLSR analysis revealed that protein abundance mainly depended on transcript levels for both the groups of aggregation prone (SA and UA) and non-aggregation prone (SNA and UNA) proteins; it should be noted that transcript abundance primarily depends on transcription rate (see Table S2D). However, the contribution of protein half-life to predict protein abundance was twice as high for the group of aggregation prone proteins compared to the non-aggregation prone proteins. Indeed, removing transcript abundance from the PLSR analysis revealed that the prediction of protein abundance for both groups of proteins was affected. However, removal of protein half-life data did not affect the prediction of protein abundance of the non-aggregation prone proteins but did significantly affect the prediction of abundance of aggregation prone proteins (see Table S2E). These results indicate that protein half-life has a significant impact on the abundance of aggregation-pone proteins.
Reported Differences Are Not Primarily a Consequence of Cellular Localization of the Proteins
The different groups of proteins analyzed in this study are predominantly found in differing cellular compartments according to their gene ontology (GO) annotation (see Table S3A); highly aggregation prone proteins have an enriched occurrence in the ER (structured proteins) and nucleus (unstructured proteins), whereas proteins with low aggregation likelihood are predominantly found in the cytosol (structured proteins) and the nucleolus (unstructured proteins). However, proteins that are mainly found in the cytosol and the ER respectively do not show a difference in abundance (xcytosol: 2730; xER: 3100, p = 0.1) or half-live (xcytosol: 42.0; xER: 40.5, p = 0.9). Interestingly, proteins located in the nucleolus are present in higher amounts than those found in the rest of the nucleus (xnucleolus: 4490; xnucleus: 2270, p = 3x10−9), but their half-lives (xnucleolus: 46; xnucleus:40, p = 0.03) do not differ significantly. Overall, these tests confirm that the observed differences in the regulation of aggregation prone and non-aggregation prone proteins are unlikely to be determined only by intracellular localization of the proteins.
Comparison with Previous Results on Overexpression Toxicity
Recently, Vavouri et al. (Vavouri et al., 2009) identified properties that are linked to dosage sensitive genes. They reported a lack of significant relationship between dosage sensitivity and aggregation load. This result appears to be in contrast to our findings, but Vavouri et al. used only (i) the pure TANGO score of the entire protein and (ii) the number of aggregation prone residues (defined by TANGO as > 5%) to investigate the relationship between aggregation propensity and dosage sensitivity. The authors also did not consider stretches of aggregation prone residues or do not explicitly consider poly-Q/N stretches as aggregation prone proteins.
In our study, we used a strict criterion of the presence or absence of aggregation prone hydrophobic stretches (i.e., presence of at least 7 consecutive aggregation prone residues with a score > 50% reported by TANGO) to distinguish aggregation prone and non-aggregation prone proteins (see also Extended Experimental Procedures, above and Extended Discussion). Since continuous stretches of aggregation prone amino acids have been shown to be the driving force behind protein aggregation (Chiti and Dobson, 2006), we believe that the total number of aggregation prone residues in a protein is a less relevant criteria compared to the presence of aggregation prone stretches of amino-acids. In addition to using TANGO that identifies hydrophobic stretches, which contribute to aggregation, we also identified the aggregation prone unstructured group of proteins that employ poly-Q/N rich stretches for aggregation. Thus, we believe that classification of aggregation prone proteins is more extensive in the current study, thereby allowing us to more reliably identify the relationship between protein aggregation and overexpression toxicity.
Figure S1.
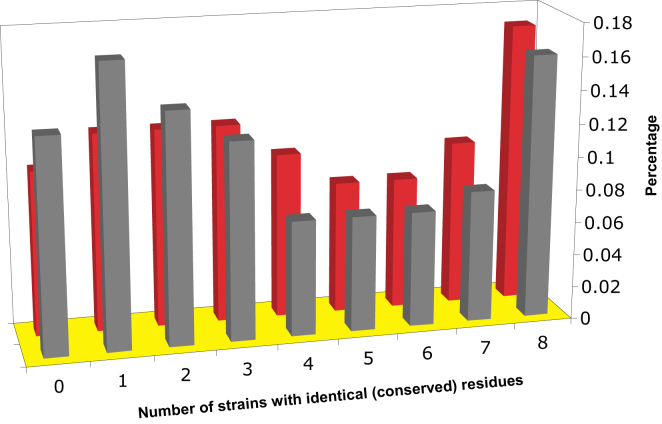
Conservation of Aggregation-Prone and Nonaggregation-Prone Residues in Nine Different Fungi, Related to Figure 1
The program TANGO was used to determine aggregation prone residues in the nine strains. The aggregation profile of S. cerevisiae was taken as a reference. The alignments of the orthologous sequences for the nine yeast strains were obtained from Tuller and Ruppin (Tuller et al., 2009). The x-axis shows the fractions of residues that are present in all eight (category 8), only a fraction (categories 7 to 1) or none of the other fungal species (category 0). The y-axis shows the % of residues in aggregation prone (red bars) or non-aggregation-prone (grey bars) segments, respectively. The distributions of conservation of aggregation prone and non-aggregation prone residues are significantly different with aggregation prone residues more conserved than non-aggregation prone residues (p = 1x10−6; Wilcoxon test).
Results
Identification of Aggregation and Nonaggregation Prone Proteins
Protein aggregates that have been linked to human disease and those found in several functional complexes are primarily beta-sheet aggregates (Chiti and Dobson, 2006; Fowler et al., 2007; Maji et al., 2009). Though the morphologies (e.g., fibrillar or amorphous) of aggregates may differ, their formation depends on the propensity to form beta-sheets (Rousseau et al., 2006a). We therefore aimed to identify proteins that are likely to form beta-sheet aggregates, irrespective of the morphology of the resulting aggregate. Increased beta-sheet aggregation potential of proteins is associated with the presence of aggregation-prone elements, such as hydrophobic and Q/N-rich stretches (Krobitsch and Lindquist, 2000; Michelitsch and Weissman, 2000). The former are predominantly found buried within folded domains and may need to be exposed to form aggregates, while the latter are often part of unstructured segments and do not have the requirement to unfold to form aggregates (Chen et al., 2001; Linding et al., 2004). Therefore, we first distinguished the proteins in S. cerevisiae that are highly structured (S) or highly unstructured (U) (Gsponer et al., 2008) in order to identify the aggregation-prone proteins in this proteome (see Extended Experimental Procedures).
We then identified aggregation-prone, structured proteins by detecting stretches of hydrophobic amino acids using the TANGO algorithm (Fernandez-Escamilla et al., 2004). As proteins with low aggregation likelihood, we identified those highly structured proteins that lack any stretch of consecutive hydrophobic residues. In the highly unstructured proteins, we detected Q/N-rich and lysine/glutamate (K/E)-rich regions using the algorithm described by Michelitsch and Weissman (2000), as their presence in unstructured proteins has been associated with increased and decreased aggregation likelihood, respectively (Lawrence et al., 2007; Santner et al., 2012) (see Extended Results).
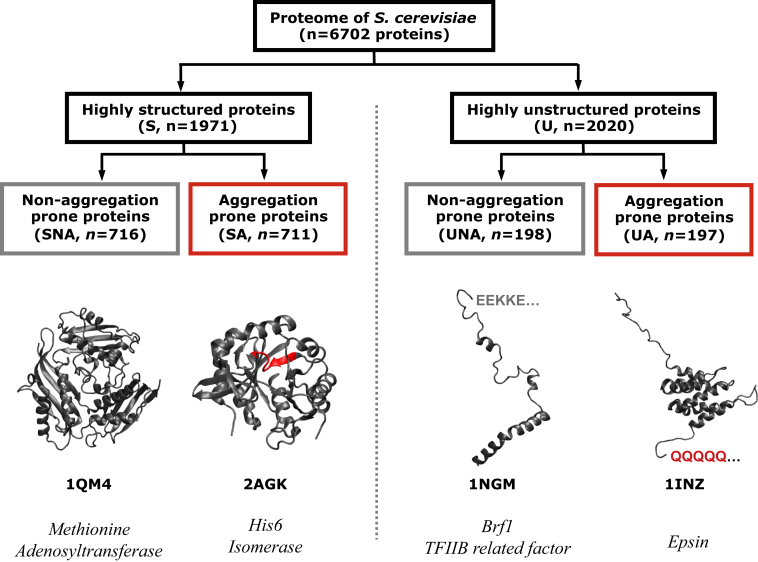
In this manner, we divided the S. cerevisiae proteome into four groups (Figure 1) and investigated whether the aggregation-prone proteins are regulated differently from the nonaggregation-prone proteins by integrating this structural information with different genome-scale data sets that describe most of the regulatory steps that influence protein synthesis or degradation (Tables 1 and S1). Additional sequence and structure features, such as the thermodynamic stability of a protein, its folding/unfolding pathway, and involvement in physical interactions with other proteins in a cell, will affect the manifestation of the aggregation-prone elements described above and, ultimately, the likelihood of the protein to aggregate in vivo. While the importance of these features has been investigated using individual proteins (Masino et al., 2011; Münch and Bertolotti, 2010), they are not trivial to assess on a genomic scale.
Figure 1.
Identification of Structured and Unstructured Nonaggregation-Prone Proteins
The S. cerevisiae proteome was grouped into four categories: highly structured protein without aggregation prone elements (SNA), highly structured proteins with aggregation prone elements (SA), highly unstructured proteins with nonaggregating K/E-rich stretches (UNA), and highly unstructured proteins with aggregation prone Q/N-rich stretches (UA). PDB codes are provided for the structures as the four-letter code.
Table 1.
Compendium of Data Sets Used in Our Study
| Type of Information and Citation [PubMed ID] | Description of the Method Used to Obtain the Data |
|---|---|
| Histone modifications | Database of published ChIP-microarray experiments that gives the relative enrichment of each histone modification at selected promoter regions or ORFs. |
| O’conner and Wyrick [17485428] | |
| Transcriptional rate | Transcriptional rates for yeast grown in YPD were calculated by the authors based on the transcript abundances and mRNA half-lives. These were in turn determined by obtaining and comparing transcript levels of the wild-type and the temperature-sensitive RNA polymerase rpb1-1 mutant strains using an Affymetrix microarray. For mouse cells, transcription rate was computed from experimentally obtained transcript steady state levels and turnover rates through next generation sequencing of mRNA. |
| Holstege et al. [9845373] and Schwanhausser et al. [21593866] | |
| Transcript abundance | Transcript abundances for the yeast and human cells were determined using high-density oligonucleotide arrays. For mouse cells, next generation sequencing was used to quantify transcript levels. |
| Holstege et al. [9845373], Vogel et al. [20739923], Lackner et al. [17434133], and Schwanhausser et al. [21593866] | |
| Transcript half-life | Transcript half-lives were determined by measuring transcript levels over several minutes after inhibiting transcription. This was estimated using (1) the temperature-sensitive RNA polymerase rpb1-1 mutant S. cerevisiae strain, (2) 300 μg/ml 1,10-phenanthroline to block transcription in S. pombe, and (3) actinomycin D for human cell lines. For mouse cells, mRNA was labeled using 4-thiouridine. mRNA abundance was monitored over time using next generation sequencing to obtain turnover rates of transcripts. |
| Wang et al. [11972065], Lackner et al. [17434133], Yang et al. [12902380], and Schwanhausser et al. [21593866] | |
| RBP-bound transcripts | To identify RNAs associated with RNA-binding proteins (RBPs), (TAP)-tagged proteins were affinity purified from whole-cell extracts of cultures grown to mid-log phase in rich medium. RNA was extracted from the extracts, reverse transcribed, and then hybridized to DNA microarrays. |
| Hogan et al. [18959479] | |
| Transcript 5′ UTR length | To map transcribed regions of the yeast genome, polyadenylate [poly(A)] RNA was isolated from yeast cells grown in rich media and used to generate double-stranded complementary DNA (cDNA) by reverse transcription. The double-stranded cDNA was fragmented and subjected to high-throughput Illumina sequencing, in which 35 base pairs of sequence were determined from one end of each fragment and mapped back onto the genome. |
| Nagalakshmi et al. [18451266] and Vogel et al. [20739923] | |
| Transcript secondary structure and G-quadruplexes | Parallel analysis of RNA structure: To identify RNA secondary structure location, in vitro-folded RNAs were first treated with different structure-specific enzymes, fragmented, and then determined by deep sequencing. G-quadruplexes were computationally identified using G4 DNA motif pattern and the loop length threshold approach. |
| Kertesz et al. [20811459] and Capra et al. [20676380] | |
| Translational efficiency/codon bias | The tRNA adaptation index (tAi) is determined by calculating a weight for each of the sense codons, derived from the copy number of all tRNA types that recognize it (including wobble interactions). For a given coding sequence, the tAi value is the geometric mean of the weights of all its sense codons. The tAi of a coding sequence ranges theoretically from 0 to 1 (0.2–0.7 for S. cerevisiae genome), with high values corresponding to high levels of translational efficiency. Ingolia et al. performed a ribosome foot-printing experiment to enrich for protected parts of the mRNA and subsequently performed a next generation sequencing experiment to obtain nucleotide resolution ribosome occupancy data in yeast. |
| Man and Pilpel [17277776] and Ingloia et al. [19213877] | |
| Translational rate | To obtain a profile of ribosome association for the yeast transcriptome, which is an indicator for translational rate, the authors fractionated polysomes using velocity sedimentation. Following this, a quantitative microarray analysis of several fractions across the gradient was used to estimate the translational status of each mRNA. Translation rates for mouse proteins were computed from experimentally obtained steady state levels and turnover rates of proteins using SILAC and mass spectrometry. |
| Arava et al. [12660367], Lackner et al. [17434133], and Schwanhausser et al. [21593866] | |
| Protein abundance | Estimates of the endogenous protein expression levels during log-phase were obtained by tagging every protein with TAP-tag and/or green fluorescent protein and measuring the intensity for S. cerevisiae and S. pombe. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to obtain the abundance of proteins in the medulloblastoma Daoy cell line for human and NIH 3T3 cells for mouse. |
| Ghaemmaghami et al. [14562106], Newman et al. [16699522], Matsuyama et al. [16823372], Vogel et al. [20739923], and Schwanhausser et al. [21593866] | |
| Protein half-life | Protein half-lives were determined by first inhibiting protein synthesis via the addition of cyclohexamide and by monitoring the abundance of each TAP-tagged protein in the yeast genome as a function of time. For the mouse cells, SILAC labeling of proteins followed by LC-MS/MS over time was employed to obtain protein half-life. |
| Belle et al. [16916930] and Schwanhausser et al. [21593866] | |
| Overexpression phenotypes | To examine gene overexpression effects, an ordered array of 5,280 yeast strains was constructed, each conditionally overexpressing a unique yeast gene, covering 85% of all yeast genes. To catalog the spectrum of genes that affect cellular fitness when overexpressed, the array was transferred to a medium containing galactose, and each strain was examined for defects in colony formation. |
| Sopko et al. [16455487] | |
| Genetic screen for aggregation | For S. cerevisiae, a gene deletion set (YGDS) of 4,850 viable mutant haploid strains was used to identify genes that enhance toxicity of a mutant huntingtin fragment or alpha-synuclein. In C. elegans, genome-wide RNA interference was used to identify genes that, when suppressed, resulted in the premature appearance of protein aggregates. |
| Willingham et al. [14657499] and Nollen et al. [15084750] |
See also Table S1.
Transcripts Encoding Aggregation-Prone Proteins Are Present in Low Levels due to Slower Transcription Rate
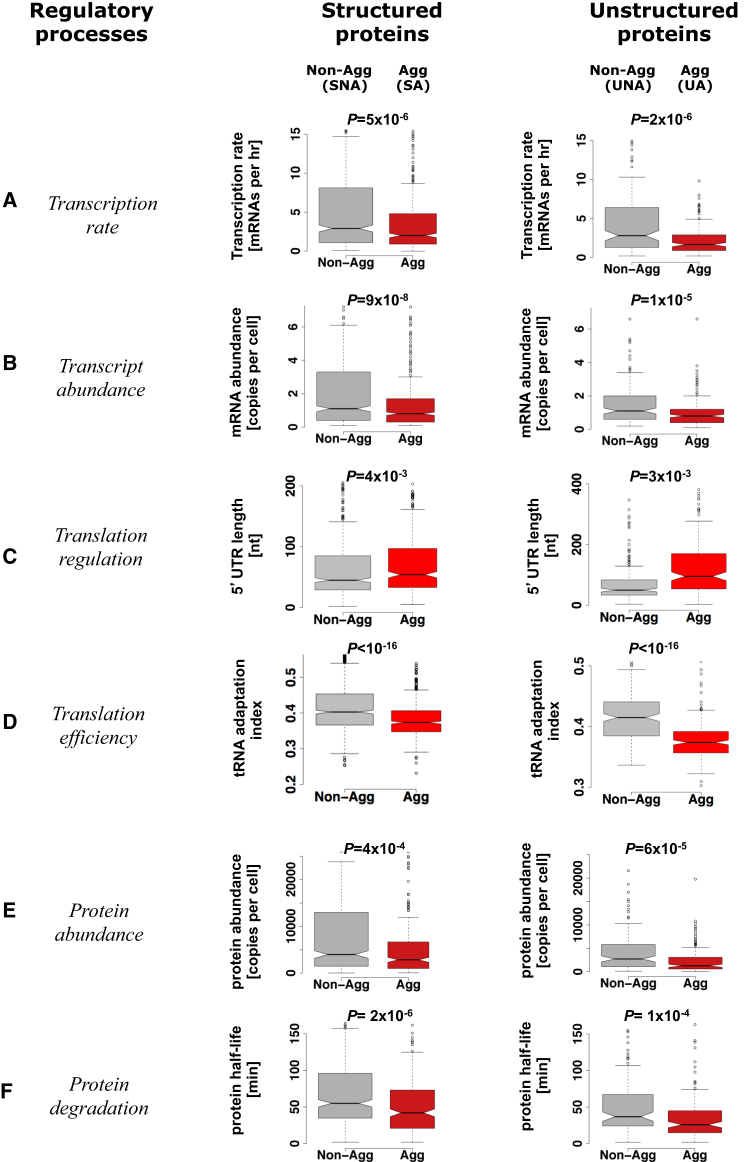
A comparison of transcript levels revealed that messenger RNAs (mRNA) encoding aggregation-prone proteins are expressed at lower levels than transcripts encoding nonaggregation-prone proteins (Figure 2B), which is consistent with recent reports (de Groot and Ventura, 2010; Tartaglia et al., 2007, 2009). This difference in transcript levels appears to primarily result from a differential rate of transcription (Figure 2A), because we did not observe a statistically significant difference in transcript half-lives between the groups (Table S2A). An analysis of histone modification data did not reveal a consistent difference in the promoter or the open reading frame (ORF) between genes that encode aggregation-prone or nonaggregation-prone proteins, suggesting that the differential rate of transcription cannot be explained due to global differences in histone modification alone. However, we do observe statistically significant differences for a few histone modification marks that are associated with transcription within the ORF region between the two groups of unstructured proteins (Table S2C).
Figure 2.
Box-Plot of the Distribution of Values for Various Regulatory and Cellular Properties
This is shown for the different groups of proteins that are nonaggregation-prone (structured SNA, unstructured UNA, gray boxes) and aggregation-prone (structured SA, unstructured UA, red boxes) in S. cerevisiae. Box-plot identifies the middle 50% of the data, the median, and the extreme points.
(A–F) (A) Transcription rate, (B) transcript abundance, (C) translational regulation, (D) translational efficiency, (E) protein abundance, and (F) protein degradation. p values were computed using the Wilcoxon test.
See also Tables 1 and S2, and Figures S2 and S3.
Aggregation-Prone Proteins Are Present in Low Abundance and for a Short Time
A comparison of protein levels showed that the intracellular concentration of proteins that are likely to form aggregates is significantly lower than that of nonaggregation-prone proteins (Figure 2E). Reduced protein abundance could be the result of low transcript abundance, increased protein turnover, or decreased protein synthesis, due to tight translational regulation. An analysis of differences in protein half-lives showed that aggregation-prone proteins have a shorter half-life than the nonaggregation-prone proteins, suggesting that a rapid turnover of such proteins could contribute to their limited availability in a cell (Figure 2F). Reduced protein synthesis due to translational regulation may be mediated by (1) protein-mRNA interaction, (2) complex 5′ untranslated regions (UTRs), (3) decreased ribosomal density on transcripts, and (4) restricted choice of codon usage. We therefore systematically investigated each of these regulatory steps.
RNA-Binding Proteins Preferentially Bind mRNA of Aggregation-Prone Proteins
An investigation of protein-mRNA interaction data for 45 RNA-binding proteins in yeast revealed that several RNA-binding proteins show a significant enrichment for interactions with transcripts that encode aggregation-prone proteins (Table S2B). Interestingly, the translation initiation repressor protein KHD1p binds to more than 65% (PUA: 2 × 10−9; Fisher’s test) of all transcripts encoding aggregation-prone, Q/N-rich, unstructured proteins, but only 19% of the nonaggregation-prone, unstructured proteins. In addition, KHD1p shows enrichment for binding transcripts that encode aggregation-prone, structured proteins (PSA: 2 × 10−4; Fisher’s exact test). These observations suggest that translational regulation via protein-mRNA interaction is an important factor that might influence the availability of some aggregation-prone proteins.
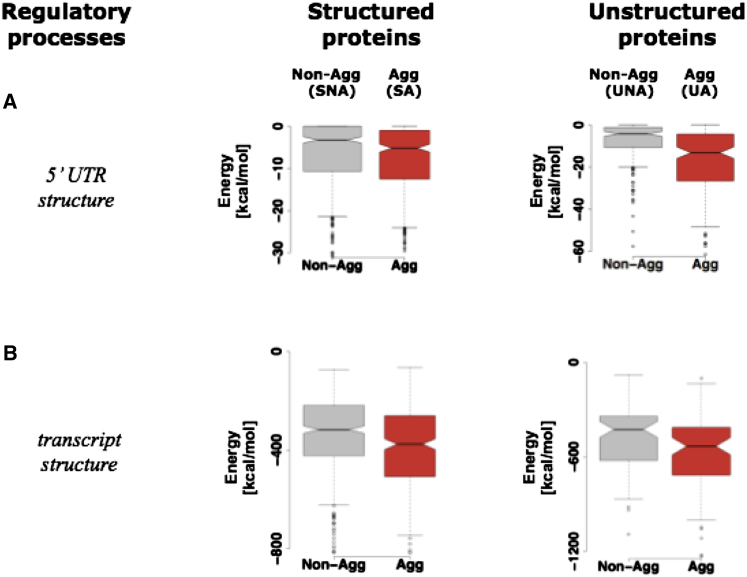
mRNA of Aggregation-Prone Proteins Have Complex 5′UTR and RNA Structure
An analysis of the 5′UTR regions of transcripts showed that mRNA encoding aggregation-prone proteins tend to have much larger 5′UTR sequences (Figure 2C). The longer 5′UTR sequences form energetically more favorable secondary structures when compared to mRNA encoding non-aggregation-prone proteins (Figure S2A). Consistent with these finding, an analysis of the secondary structure profile of transcripts of S. cerevisiae that was recently measured using a high-throughput experimental approach (Table 1) revealed more secondary structure in the transcripts of aggregation-prone proteins when compared to nonaggregation-prone proteins (Figure S2B). Moreover, transcripts of aggregation-prone, unstructured proteins contain G-quadruplex-forming sequences more often than nonaggregation-prone proteins (PSA-SNA: 0.1; PUA-UNA: 1 × 10−4; Fisher’s exact test). Such folded structures might contribute to the observed reduced protein levels by regulating translational initiation (Kudla et al., 2009; Kumari et al., 2007). These findings suggest that translation initiation is likely to be differently regulated for mRNA encoding aggregation-prone proteins.
Figure S2.
5′UTR and Transcript Structure and Aggregation Propensity of the Encoded Proteins, Related to Figure 2
Box-plot of the distribution of the energies of 5′UTR structures (which indicate the stability of the structure) for the different groups of proteins that are non-aggregation prone (structured SNA, unstructured UNA, gray boxes) and aggregation prone (structured SA, unstructured UA, red boxes) in S. cerevisiae using the predicted structure (A) and experimentally estimated structure on the whole mRNA from Kertesz et al. (Kertesz et al., 2010) (B). Box-plot identifies the middle 50% of the data, the median, and the extreme points. The entire set of data points is divided into quartiles and the inter-quartile range (IQR) is calculated as the difference between × 0.75 and × 0.25. The range of the 25% of the data points above ( × 0.75) and below ( × 0.25) the median ( × 0.50) is displayed as a filled box. The horizontal line and the notch represent the median and confidence intervals, respectively. Data points greater or less than 1.5·IQR represent outliers and are shown as dots. The horizontal line that is connected by dashed lines above and below the filled box (whiskers) represents the largest and the smallest non-outlier data points, respectively.
(A): PSA-SNA: 3x10−9; PUA-UNA: 7x10−9; Wilcoxon test.
(B): PSA-SNA: 4x10−5; PUA-UNA: 2x10−2; Wilcoxon test.
mRNA Encoding Aggregation-Prone Proteins Have Lower Translation Efficiency
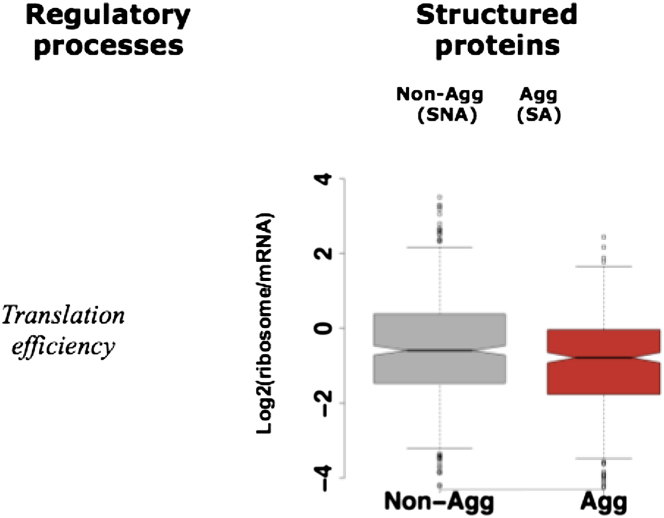
In order to investigate the role of codon bias, we compared the transfer RNA (tRNA) adaptation index (tAi) of genes encoding aggregation-prone and nonaggregation-prone proteins. The tAi is based on the copy number of each of the tRNAs in a given genome and can be used to establish translational selection as well as to score objectively translational efficiency (dos Reis et al., 2004; Gingold and Pilpel, 2011). We find that transcripts encoding aggregation-prone proteins have a significantly lower tAi compared to those encoding nonaggregation-prone proteins (Figure 2D). It has been noted recently that translationally optimal codons are associated with buried residues in proteins, irrespective of their expression level, possibly to minimize protein misfolding (Drummond and Wilke, 2008; Lee et al., 2010). We also analyzed recently published ribosome-profiling data to further investigate this difference in translational efficiency. Indeed, aggregation-prone, structured proteins are less efficiently translated than nonaggregation-prone ones (Figure S3; Extended Results). An analysis of the polysome profiling data revealed that mRNA encoding aggregation-prone, unstructured proteins have a lower density of ribosomes per transcript (Table S2A). These observations collectively suggest that transcripts that encode aggregation-prone proteins are globally less efficiently translated when compared to those encoding nonaggregation-prone proteins.
Figure S3.
Translation Efficiency and Aggregation Propensity of the Encoded Proteins, Related to Figure 2
Box-plot of the translation efficiency derived from ribosome profiling for the structured proteins that are non-aggregation prone (SNA) and aggregation prone (SA) in S. cerevisiae. Unstructured proteins were not analyzed because of the repetitive sequences in the Q/N and K/E-enriched groups. Plotted are the ratios of ribosome footprints to mRNA fragments. The difference in translational efficiency between the SNA and SA groups is statistically significant (PSA-SNA: 3x10−4). We thank Nicholas T. Ingolia for providing the ribosome profiling data. See Extended Results.
Synthesis and Degradation of Aggregation-Prone Proteins Is Tightly Regulated
While transcript and protein abundance may be related quantities, they are not strictly correlated, and recent genome-scale studies show extensive evidence for posttranscriptional regulation (Vogel, 2011). Thus it is important to study them independently and identify the influence of the different steps that affect protein abundance. Therefore, we performed a comprehensive statistical analysis of the major contributors in the gene expression process that affect protein abundance for the (non-) aggregation-prone proteins through a detailed partial least square regression (PLSR) analysis (see Extended Experimental Procedures and Extended Results; Tables S2D and S2E). The results of the PLSR calculations and the reported findings consistently suggest that (1) the cellular regulation of the aggregation-prone proteins is different compared to the nonaggregation-prone proteins and (2) a combination of reduced transcript abundance, rapid protein turnover, and translational regulation contributes to the low availability of aggregation-prone proteins.
The Observed Trends Are Evolutionary Conserved
We then assessed whether the tight regulation of aggregation-prone proteins is likely to be an evolutionarily conserved mechanism. To this end, we analyzed several published data sets (Table 1) for Schizosaccharomyces pombe and Homo sapiens and found similar trends to those observed for S. cerevisiae for the available data (Table 2). We also analyzed a recently published data set for Mus musculus and found that aggregation-prone and nonaggregation-prone proteins are regulated significantly differently at the protein level (Tables 2 and S3G). Overall these results suggest that the tight regulation of aggregation-prone proteins may be an evolutionarily conserved strategy.
Table 2.
Comparison of Aggregation-Prone and Non-Aggregation-Prone Proteins in S. pombe, M. musculus, and H. sapiens
| Cellular Quantity | SNA | SA | ||
|---|---|---|---|---|
| S. pombe | ||||
| n | n | |||
| Transcript abundance [signal intensity] | 2318 ± 264 | 676 | 1,867 ± 193 (3 × 10−5) | 488 |
| Transcript half-life [% with long half-life] | 77% | 246 | 47% (3 × 10−11) | 189 |
| Translational efficiency [tAi] | 0.40 ± 0.00 | 633 | 0.38 ± 0.00 (3 × 10−11) | 451 |
| Protein abundance [signal intensity] | 0.50 ± 0.09 | 621 | 0.17 ± 0.04 (<10−16) | 289 |
| H. sapiens | ||||
| Transcript abundance [arbitrary unit] | 2,530 ± 1,005 | 107 | 345 ± 330 (3 × 10−7) | 89 |
| Translation regulation [5′UTR length in nt] | 85 ± 15 | 107 | 113 ± 30 (4 × 10−2) | 89 |
| Protein abundance [molecules/cell] | 1,776 ± 1,100 | 107 | 505 ± 247 (3 × 10−5) | 89 |
| M. musculus | ||||
| Transcriptional rate [mRNAs/cell∗h] | 2.0 ± 0.2 | 505 | 1.6 ± 0.2 (0.3) | 359 |
| Transcript abundance [copies/cell] | 21 ± 1 | 497 | 20 ± 2 (0.9) | 357 |
| Transcript half-life [h] | 11 ± 1 | 542 | 11 ± 1 (0.02) | 392 |
| Translational rate [proteins/mRNA∗h] | 63 ± 6 | 486 | 39 ± 5 (3 × 10−8) | 346 |
| Protein abundance [proteins/cell] | 36 k ± 7 k | 570 | 13 k ± 5 k (6 × 10−9) | 408 |
| Protein half-life [h] | 79 ± 7 | 570 | 56 ± 6 (5 × 10−7) | 408 |
Median values and their confidence intervals (C.I. , where IQR is the inter-quartile range and n the group sample size) are reported for highly structured proteins without aggregation-prone elements (SNA) and highly structured proteins with aggregation-prone elements (SA). There are only a low number of Q/N-enriched proteins in the data set for S. pombe and few proteins with Q/N-enriched domains in the data set available for H. sapiens. Therefore, no statistically significant analysis on the U group was possible. Results for the U group in M. musculus can be seen in Table S3G. “n” is the number of data points. Statistical significance was calculated with the Wilcoxon test and Fischer’s exact test. Statistically significant differences are highlighted in bold. See also Table S3.
Control Calculations for Alternative Explanations and Confounding Factors
We observed that the trends were not a result of differences in protein length and intrinsic disorder in the respective groups (Table S3A). Elimination of membrane proteins in the structured group, which have stretches of hydrophobic amino acids, does not affect the observed differences (Table S3B). While we observed that the four groups of proteins defined here are enriched to occur in different subcellular compartments (Table S3A), the observed differences are not primarily a consequence of their location within a cell (see Extended Results). As unstructured proteins are more tightly regulated than structured proteins due to their involvement in regulatory and signaling roles (Babu et al., 2011; Gsponer et al., 2008), we investigated only the subset of unstructured proteins that were not associated with regulatory or signaling roles and found similar trends (Table S3C).
Although TANGO has been benchmarked on very different aggregating and nonaggregating peptides, our selection of aggregation-prone, structured proteins may contain an unknown bias toward those with low abundance. Therefore, we used an additional predictor, PASTA (Trovato et al., 2007), to identify aggregation-prone, structured proteins and found consistent trends (Table S3D). Similarly, we selected a recently identified list of prionogenic proteins (Alberti et al., 2009) in S. cerevisiae for the aggregation-prone, unstructured proteins and found that they have the expected low abundances and short half-lives (Table S3E). The four groups of proteins that we selected allowed for a clear distinction between aggregation-prone and nonaggregation-prone proteins as well as the necessary distinction between structured and unstructured proteins (see above). However, the four groups cover only part of the S. cerevisiae proteome, and a more continuous classification with respect to aggregation likelihood is required to assess the regulation of all yeast proteins. To address this, we used the algorithm AGGRESCAN (Conchillo-Solé et al., 2007) that identifies hydrophobic and nonhydrophobic aggregation-prone segments in structured and unstructured proteins. Analysis using the alternative classification scheme confirmed all the trends that we identified (Table S3F). The differences reported here are consistently determined to be statistically significant by two independent statistical tests: the Wilcoxon rank-sum test and the Kolmogorov–Smirnov test (Table S3H).
Modulators Enhancing Aggregation Phenotype Are Enriched in Genes Influencing Expression Homeostasis
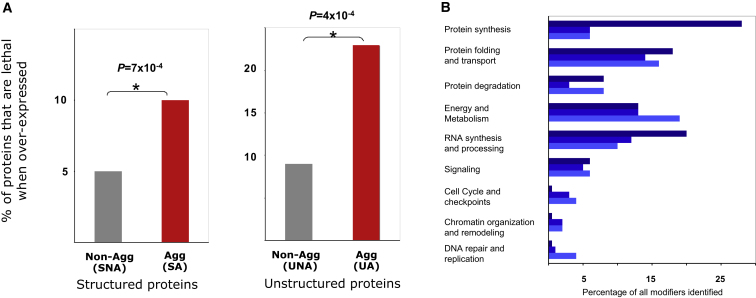
The reported observations lead to the following predictions: (1) If the regulation at multiple stages is crucial for minimizing protein aggregation in a cell, then overexpression of the aggregation-prone proteins should be more often detrimental than overexpression of nonaggregation-prone proteins. Indeed, an investigation of the overexpression phenotype data (Sopko et al., 2006) revealed that aggregation-prone proteins are twice as often lethal when overexpressed compared to nonaggregation-prone proteins (Figure 3A; Extended Results). (2) In addition to genes that influence protein folding, multiple genetic loci that participate in gene expression homeostasis, such as RNA-binding proteins, should modulate protein aggregation in vivo. To test this hypothesis, we investigated published screens in yeast (Willingham et al., 2003) and C. elegans (Nollen et al., 2004) that systematically identified genetic backgrounds in which the phenotype due to the expression of an aggregation-prone protein is enhanced. We found, in both organisms, that alterations in genetic background that enhance the aggregation phenotype were enriched for multiple genes that directly or indirectly influence transcript and protein availability (Figure 3B). Thus our findings support the emerging view that, in addition to mutations in the proteins themselves, mutations which affect their expression level or the genes that influence the expression level of an aggregation-prone protein may contribute to the disease phenotype involving protein aggregation (Powers et al., 2009).
Figure 3.
Overexpression Toxicity and Genetic Modulators of Protein Aggregation
(A) Overexpression toxicity phenotype. P values obtained from Fisher’s test.
(B) Distribution of the functional categories of the genes, which when deleted or downregulated result in enhanced lethality upon expression of an aggregation-prone protein (expression of huntingtin fragment in C. elegans [dark blue], α-synuclein [blue], and huntingtin [light blue] in S. cerevisiae). The genes were grouped according to the GO annotation of the proteins they encode. The GO annotations influencing transcript and protein availability are terms in the figure other than DNA repair and replication, cell cycle and checkpoints, signaling, energy, and metabolism. All these terms influencing transcript and protein availability are significantly enriched (p < 4 × 10−2) for the C. elegans data set, except for chromatin organization and remodeling, for which the number of genes was small. For the yeast data sets, only the term protein synthesis was enriched significantly (p < 10−3). However, p values for this data set have to be interpreted with care, as the number of identified genes in the screen is small.
Discussion
We have provided a study that analyses the control of aggregation-prone proteins at multiple levels of gene expression regulation in evolutionarily diverse organisms within a single framework. Previous studies on individual data sets have indicated that aggregation-prone proteins may be regulated differently than non-aggregation-prone ones, but our findings reveal that S. cerevisiae keeps aggregation-prone proteins at low abundance by combining several strategies at nearly every regulatory level: from the initiation of transcription up to degradation of proteins. The differential regulation of aggregation-prone and nonaggregation-prone proteins seems to be evolutionarily conserved, as several of the trends are also found in S. pombe, H. sapiens, and M. musculus. This conservation of a differential regulation of aggregation-prone proteins in multiple organisms may underline the significance and generality of our findings. Considering the growing evidence for the importance of aggregation in various physiological processes, the conserved differential control of aggregation-prone proteins may be part of a general regulatory framework that not only minimizes unwanted/potentially harmful aggregation, but also keeps functional aggregation in check (see below).
The results presented here were possible only because of a conceptual framework, in which we distinguish between structured and unstructured aggregation-prone proteins. The trends we report would be missed by grouping them together. The fact that we find similar regulatory differences between aggregation-prone and nonaggregation-prone structured and unstructured proteins is, nevertheless, quite intriguing, particularly considering that hydrophobic stretches in structured proteins and Q/N-enriched stretches in unstructured proteins have different functions and are likely to have potentially different pathological consequences. These observations emphasize the importance of future studies to investigate the detailed molecular mechanisms that underlie the regulation of aggregation-prone proteins. In this direction, our analyses provide interesting pointers for regulatory mechanisms that may be of particular interest. For instance, we find that the 5′UTR structure of transcripts encoding aggregation-prone proteins is more complex and longer than that of nonaggregation-prone ones and that specific RNA-binding proteins target a large fraction of the transcripts that encode aggregation-prone proteins. It is likely that the fine-tuning of the different regulatory steps that keep protein concentrations low is different for individual aggregation-prone proteins or that even additional, specific control mechanisms are in place (Fowler et al., 2007). We wish to note that, while the general outcome from this study is biologically meaningful, as we observe consistent differences across many tests, the reader should be aware that, for specific individual comparisons, a significant p value does not always mean that the difference will be biologically significant.
Below, we discuss how these findings fit in with the current understanding of avoidance of protein aggregation and regulating functional aggregates.
Evolution by Negative Design Minimizes Nonfunctional Protein Aggregation
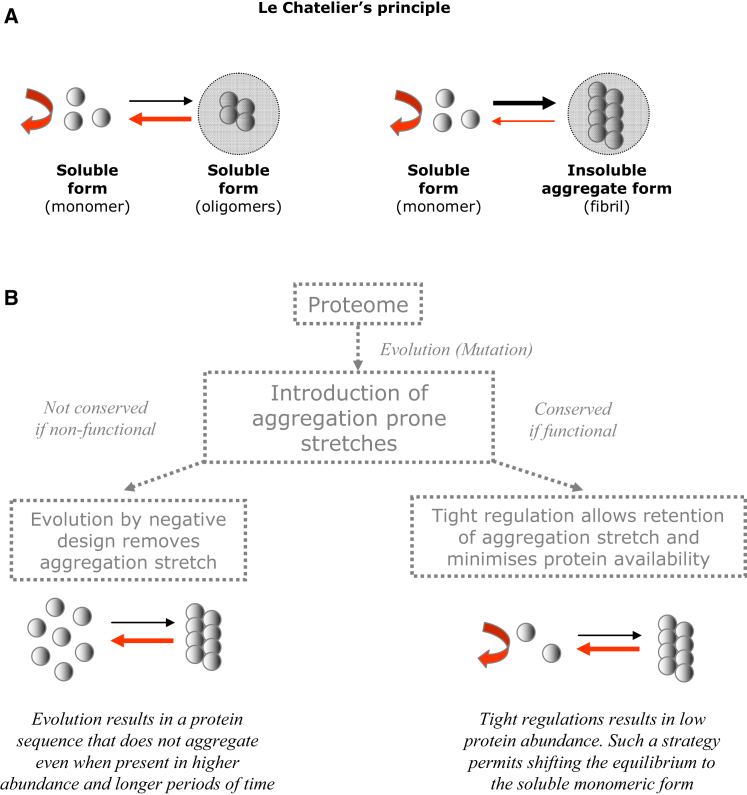
During the course of evolution, when an aggregation promoting mutation is introduced in a gene, two main scenarios can be envisioned: either the mutated protein with increased aggregation likelihood provides a fitness advantage or it does not. If the mutated protein does not provide a fitness advantage, but forms nonfunctional or toxic aggregates, individuals harboring such sequences are likely to be eliminated from the population, resulting in selection for sequences that are less likely to form aggregates. Reports that support this outcome have been accumulated in recent years (Geiler-Samerotte et al., 2011; Morell et al., 2011). Accordingly, sequence motifs that are highly aggregation prone are significantly underrepresented (Broome and Hecht, 2000; Patki et al., 2006). These findings have been interpreted as strong indicators for avoidance of aggregation as a major evolutionary driving force in the design of protein sequences (Rousseau et al., 2006b) (Figure 4; Extended Discussion).
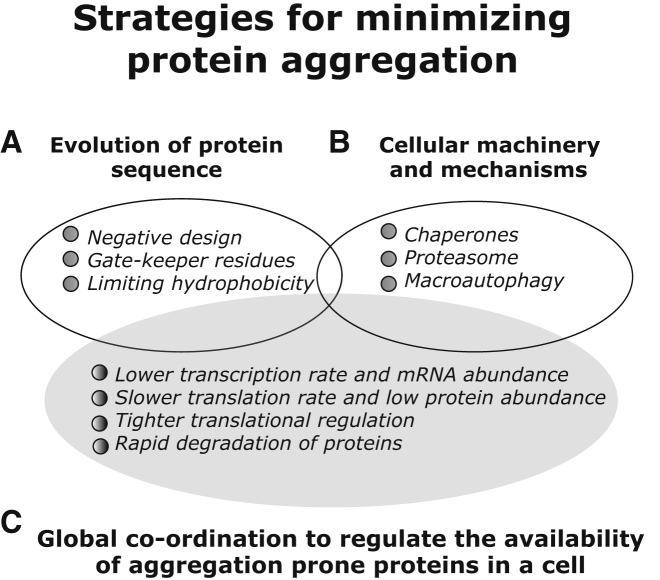
Figure 4.
Evolutionary and Cellular Strategies to Deal with Proteins that Are Likely to Aggregate
Two major solutions to explain how cells are robust to the presence of aggregation-prone proteins have been proposed.
(A) The sequences of such proteins have been subjected to rigorous selection such that they do not easily form aggregates (i.e., avoidance of aggregation is a major evolutionary driving force in the design of protein sequences).
(B) Cellular systems have evolved machineries and mechanisms to either avoid or efficiently clear aggregates.
(C) The results from our integrated analysis provide insights into a third possible solution to this problem. Cellular systems have evolved regulatory strategies that control the availability of aggregation-prone proteins and their encoding transcripts such that they are present for short periods, low quantities, and in precise amounts in a cell. This strategy may ensure that the abundance of aggregation-prone proteins is below the critical concentration.
Extended Discussion.
Aggregation Is Not Always Harmful: Functional Aggregates Exist in the Cell
Human pathologies caused by the failure of proteins to adopt or remain in their functional conformational state are called protein conformational diseases (Chiti and Dobson, 2006; Dobson, 2006; Selkoe, 2003). The biggest group of these diseases is associated with the conversion of soluble proteins into amorphous aggregates and ultimately highly organized amyloid fibrillar aggregates. These aggregates could be found as extracellular or intracellular deposits in different human tissues and have been linked to human pathologies such as neurodegenerative diseases or systemic amyloidosis (Caughey and Lansbury, 2003; Chiti and Dobson, 2006).
While the harmful effects of aggregation are generally accepted, recent findings indicate that a variety of organisms take advantage of the particular properties (e.g., yield strength, protease and detergent resistance) of amyloid-like protein aggregates, i.e., they use them as “functional” aggregates (Badtke et al., 2009; Decker et al., 2007; Fiumara et al., 2010; Fowler et al., 2006; Fowler et al., 2007; Franks and Lykke-Andersen, 2008; Herter et al., 2005; Kaiser et al., 2008; Kentsis et al., 2002; Kopito, 2000; Lelouard et al., 2002; Maji et al., 2009; Olzscha et al., 2011; Ozgur et al., 2010; Reijns et al., 2008; Salazar et al., 2010; Sivakolundu et al., 2008; Spector, 2006). Bacteria such as Escherichia coli or Steptomyces coelicolor use fibrils formed from different proteins to mediate binding to host proteins and enable hyphae growth into the air, respectively. In yeast, a variety of proteins, so-called prions, have been found that exist in both a soluble state and an insoluble aggregated form. The latter form is self-perpetuating, infectious, and can serve as a non-chromosomal genetic element (Tessier and Lindquist, 2009; Uptain and Lindquist, 2002; Wiltzius et al., 2009). In addition to globular domains, prions contain intrinsically unstructured segments with Q/N-enriched stretches, which mediate aggregation and thereby inactivate the protein. Importantly, the resulting phenotypes, though not generally beneficial, can be advantageous to the organism under specific, often challenging conditions such as stress (Alberti et al., 2009; Jarosz et al., 2010). The diversity of the molecular functions different yeast prions carry out (Sup35; translation termination, Ure2p; nitrogen catabolism, Swi1 and Cyc8; transcriptional regulation) indicates that the switch between a soluble and an insoluble aggregated protein form has been exploited by yeast in order to possess an additional level of control on several physiological processes, which can be advantageous when environmental conditions are challenging (Du et al., 2008; Halfmann et al., 2010; Halfmann and Lindquist, 2010; Jarosz et al., 2010; Patel et al., 2009; Uptain and Lindquist, 2002).
Despite the fact that protein β sheet aggregation and amyloid formation in humans is predominantly referred in the context of disease (Chiti and Dobson, 2006, 2009; Conway et al., 2001; Goedert, 2001), it is important to note that an increasing number of human physiological processes have been identified to depend on protein aggregation or even fibril formation (Badtke et al., 2009; Decker et al., 2007; Fiumara et al., 2010; Fowler et al., 2006; Fowler et al., 2007; Franks and Lykke-Andersen, 2008; Herter et al., 2005; Kaiser et al., 2008; Kentsis et al., 2002; Kopito, 2000; Lelouard et al., 2002; Maji et al., 2009; Olzscha et al., 2011; Ozgur et al., 2010; Reijns et al., 2008; Salazar et al., 2010; Sivakolundu et al., 2008; Spector, 2006). For instance, aggregation and fibril formation of the protein Pmel17 has been shown to be important in the biosynthesis of melanin. Remarkably, the dynamic formation of a variety of cellular bodies has been shown to depend on protein aggregation (Balagopal and Parker, 2009). These cellular bodies include stress granules and processing bodies. Stress granules, for instance, are dynamically assembled in cells subjected to environmental stress. It has been also shown that their assembly is mediated by prion-like aggregation of a glutamine rich domain in the RNA-binding proteins TIA-1 (Gilks et al., 2004) and Pum (Salazar et al., 2010). Similarly, Q/N-enriched segments have been shown to be pivotal to the formation of processing bodies (Reijns et al., 2008). Though it is not known whether the aggregates formed in these cellular bodies have a fibrillar character, it is unambiguous that the aggregation propensity of the Q/N-enriched segments has been exploited to mediate the formation of different, physiologically relevant assemblies (Fiumara et al., 2010; Jahn and Radford, 2005, 2008; Kopito, 2000; Linding et al., 2004; Rousseau et al., 2006a; Salazar et al., 2010; Turoverov et al., 2010).
The fact that the aggregation process can have cytotoxic effects and that fibrils can seed or even cross-seed further aggregation and thereby promote toxicity clearly implies that (i) “non-functional” aggregation should be avoided and (ii) “functional” aggregation has to be highly regulated. Indeed, for individual cases of functional amyloid, control mechanisms could be identified that regulate the aggregation process via binding proteins or post-translational modifications (Chamot-Rooke et al., 2011; Fowler et al., 2007). However, so far we know very little about the regulation of many proteins that are known to form β sheet aggregates in a cell or proteins that contain aggregation prone segments that are conserved for functional reasons. As eukaryotic cells have evolved a variety of machineries that assist protein in folding or clear the cell from protein aggregates (see also below) (Balch et al., 2008; Powers et al., 2009), we hypothesized that they could also have evolved regulatory mechanisms at the level of protein synthesis and degradation in order to keep aggregation under control, i.e., by ensuring that the levels of these proteins are low and that they are turned over rapidly. Such specific regulation of concentration and turnover kinetics of aggregation prone proteins would contribute significantly to prevent “non-functional” aggregation and keep “functional” aggregation/assemblies under control.
Avoiding Unwanted Aggregation and Keeping Functional Aggregation Under Control
The fact that more than 40 pathological conditions in humans have been linked with the deposition of insoluble fibrillar protein aggregates has prompted the hypothesis that living systems have evolved strategies to avoid protein aggregation (i.e., evolved by negative design) (Dobson, 1999; Monsellier and Chiti, 2007). Indeed, research on the occurrence of aggregation prone amino acid revealed that amino-acid stretches of more than 3 consecutive hydrophobic residues are present but significantly underrepresented (Patki et al., 2006; Schwartz et al., 2001). Moreover, the aggregation prone pattern of alternating polar and non-polar residues is the least represented among all possible patterns of polar and non-polar residues in proteins (Broome and Hecht, 2000). This under-representation of aggregation prone hydrophobic sequence motifs has been interpreted as a strong indicator for the existence of a negative selection pressure against stretches that would promote protein aggregation. Further support for evolutionary pressures against stretches that promote aggregation comes from the finding that, if present, the stretches are often flanked by amino acids that prevent aggregation or reduce its likelihood - so called “gate-keepers” or “β-brakers” (Dehay and Bertolotti, 2006; Richardson and Richardson, 2002; Rousseau et al., 2006b). Moreover, the aggregation likelihood of proteins decreases significantly when they are folded into a stable structure (Kelly, 1998) and the existence of a selection pressure for sequences to fold into a stable 3D structure is well established (Xia and Levitt, 2004). In fact selection against mutations that result in misfolding induced defects is now well accepted as a driving force for sequence evolution (Bershtein et al., 2012; Couñago et al., 2006; Drummond and Wilke, 2008; Geiler-Samerotte et al., 2011; Peña et al., 2010). However, many proteins need assistance for their folding in vivo (Hartl et al., 2011; Kopito, 2000). Therefore, cellular systems have evolved numerous machineries that influence the folding of proteins, which not only help proteins to fold (via chaperones, for instance) but also assist in trafficking, compartmental localization, and degradation. Hence, this network of protein machineries, which has recently been named “proteostasis” network, ensures that proteins can fulfil their function in a cell and do not misfold or aggregate (Balch et al., 2008; Roth and Balch, 2011). If and when they do aggregate, the proteostasis network ensures that potentially dangerous aggregates can be removed via mechanisms involving the proteasome or through autophagy (García-Arencibia et al., 2010; Korolchuk et al., 2010; Rubinsztein, 2006; Williams et al., 2006). See Figure 4.
Despite an evolutionary pressure against aggregation prone stretches and the evolution of cellular machineries that prevent and clear protein aggregation, recent results indicate that aggregation prone stretches are still abundant in eukaryotic proteomes for functional reasons and that fibrillar aggregates mediated by structured and unstructured proteins can even play physiological roles (Badtke et al., 2009; Fowler et al., 2007; Goldschmidt et al., 2010; Maji et al., 2009). An interesting example represents proteins that have stretches enriched for glutamine or even polyQ tracts. Proteins with expanded polyQ stretches have been associated with at least nine neurodegenerative disorders, the most well known being Huntington’s disease (von Mikecz, 2009). Although only extended polyQ stretches are pathogenic, biophysical data shows that both expanded and non-expanded polyQ stretches aggregate (Kar et al., 2011; Klein et al., 2007). Despite this, polyQ regions and particularly stretches enriched for glutamine are abundant in eukaryotic proteomes (Faux et al., 2005; Michelitsch and Weissman, 2000). Why is this the case? PolyQ tracts have been shown to play a role in transcription activation (Faux et al., 2005; Gerber et al., 1994), and the propensity to aggregate or form coiled-coil structures (Fiumara et al., 2010) has been exploited to mediate the assembly of cellular bodies such as P-bodies or stress granules. Hence, the ability of glutamine-enriched stretches to aggregate is likely to facilitate the assembly of large protein assemblies, but at the same time this comes along with the potential for the formation of cytotoxic non-functional aggregates. These observations raise the question how cells balance the benefits and risks involving aggregation prone proteins?
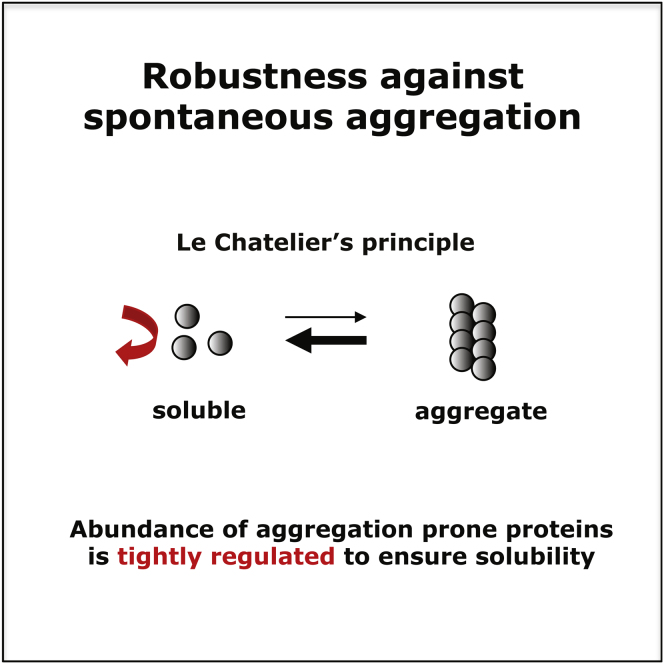
Tight Regulation of Aggregation-Prone Proteins Suggests that Cells Balance the Risk and Benefits of Aggregation-Prone Proteins Using Le Chatelier’s Principle
Ordered protein aggregation such as fibril formation occurs via a nucleation dependent polymerization (Harper and Lansbury, 1997). The characteristics of a nucleation-dependent polymerization are that (i) no aggregation occurs at a protein concentration below the critical concentration, which is determined by the protein sequence and the solution conditions and (ii) at concentrations that exceed the critical one, there is a lag time (lag phase) before an oligomeric nucleus of critical size is formed and polymerization occurs. We propose that the observed tight regulation of aggregation prone proteins at the transcriptional, translational, and degradation level permits to prevent undesirable aggregate formation and keeps functional aggregates under control. How is this achieved? By keeping the effective intracellular concentration of an aggregation prone protein generally below its critical concentration, the cell may prevent unwanted formation of aggregation nuclei of critical size and polymerization. When protein aggregation is required, local (super-) saturation, which is an increase of the effective local concentration above the critical one, can be mediated by specific regulatory proteins that “scaffold” aggregation prone proteins in one location, or by the confinement of the proteins within cellular compartments (Decker et al., 2007; Fowler et al., 2006; Ganusova et al., 2006; Harper and Lansbury, 1997; van Ham et al., 2010). Release of the aggregation-prone proteins from the confinement before a nucleus of critical size has formed, i.e., before the end of the lag time, will resolve the high molecular weight oligomeric aggregates by Le Chatelier’s principle (Figure 5 and Figure S4A). This principle states that if any change (e.g., altered concentration due to rapid turnover) is imposed on a system that is in equilibrium (e.g., soluble versus oligomeric or fibrillar) then the system tends to adjust to a new equilibrium counteracting the change. Even in the case where the release occurs after the lag time has passed, the formation of fibrillar structures may still be reversible, because it has been shown that monomers can recycle in and out of the fibrillar aggregate in a concentration dependent manner or by active mechanisms (Bieschke et al., 2009; Carulla et al., 2005; Duennwald et al., 2012; Shorter and Lindquist, 2004) (Figure S4). If aggregation, nevertheless, gets out of control, the proteostasis network will further ensure, under physiological conditions, that the potentially dangerous protein aggregates or polymers get degraded.
The cellular strategies to minimize protein aggregation can be summarized as follows: During evolution, if mutations result in increased aggregation-likelihood of a protein and do not increase cellular fitness, cells harboring such mutated sequences are likely to be eliminated from the population (evolution by negative design). If the introduced mutations do increase cellular fitness, the mutated sequence is likely to be conserved and a tight regulation of the availability of such sequence will prevent undesired aggregation and keep functional aggregation under control. The latter is likely to operate together with the proteostasis network to minimize the likelihood of protein aggregation by ensuring protein solubility and folding homeostasis (Figure S4B).
Proteostasis-Chaperone Network Minimizes Undesirable Protein Aggregation
If the mutated protein provides a fitness advantage, individuals that are able to prevent nonfunctional or undesirable aggregation of the mutated protein will be selected for in a population. Evidence for this outcome has also been presented in the literature. Despite the underrepresentation of sequence motifs that are highly aggregation-prone, extant genomes still contain a significant proportion of proteins that have aggregation-prone stretches (Linding et al., 2004; Monsellier et al., 2008) (see Figure S1; Table S4A). This suggests that cells minimize the harmful effects of aggregation-prone proteins. Accordingly, a significant body of work has shown that a substantial part of any organism’s proteome (the proteostasis network) is dedicated to minimizing nonfunctional aggregation by ensuring protein folding, solubility, and removal of aggregates by specific cellular mechanisms (De Baets et al., 2011; Gidalevitz et al., 2010; Glover and Lindquist, 1998; Hishiya and Takayama, 2008; Powers et al., 2009; Roth and Balch, 2011) (Figure 4; Extended Discussion).
Tight Regulation as a Means to Control Functional and Toxic Aggregates by Le Chatelier’s Principle
While the proteostasis-chaperone network may ensure folding of aggregation-prone proteins, too much buffering may be harmful for a cell. For instance, increased chaperone activity can minimize aggregation (Kitamura et al., 2006), but too high levels of chaperones may lead to tumorigenesis (Dai et al., 2007). These considerations, together with the fact that protein aggregation can be functional, raises the fundamental question: how do cells balance the benefits and risks of protein aggregation?
We suggest that the observed regulation of aggregation-prone proteins facilitates prevention of the formation of undesirable aggregates and may keep functional assemblies/aggregates under control, as explained by Le Chatelier’s principle (Figure 5). This principle states that if any change (e.g., altered concentration due to rapid turnover) is imposed on a system that is in equilibrium (e.g., soluble versus oligomeric or fibrillar), then the system tends to adjust to a new equilibrium, counteracting the change. Since (1) aggregate formation is a nucleation-dependent process that relies on the amount (i.e., critical concentration) of aggregation-prone proteins in the cell (Harper and Lansbury, 1997; Serio et al., 2000) and (2) individual subunits can recycle in and out of fibrillar or amorphous aggregates in a concentration-dependent manner or by active mechanisms (Carulla et al., 2005; Colby et al., 2006; Kim et al., 2002; Stenoien et al., 2002), control of the abundance of aggregation-prone proteins may (1) ensure that levels are lower than the critical concentration and (2) permit shifting of the dynamic equilibrium between the soluble monomeric form and the aggregate form, as explained by Le Chatelier’s principle (Figures 5 and S4; Extended Discussion).
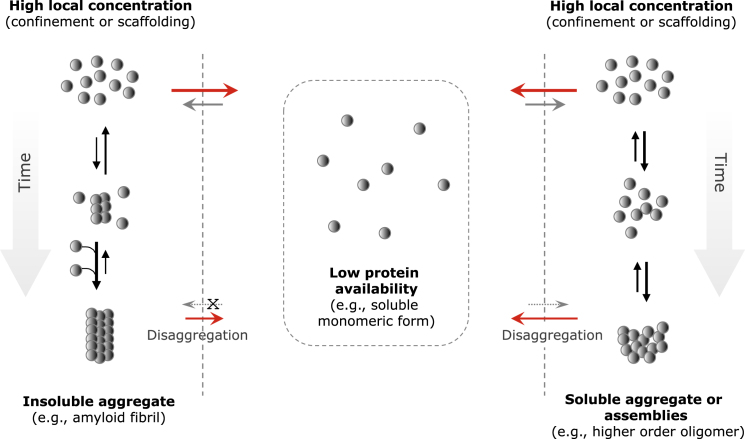
Figure 5.
Proposed Model for the Avoidance of Undesirable Aggregation and the Control of Functional Aggregation as Explained by Le Chatelier’s Principle
The differential regulation of aggregation-prone protein results in low intracellular abundance of these proteins. We suggest that it is below the critical concentration, thereby preventing undesired aggregation (center panel). Local increase in monomer concentration above the critical one (i.e., supersaturation) via confinement or a scaffolding protein can facilitate formation of aggregates. This local supersaturation would start the aggregation process and, depending on the time span of supersaturation, allow the formation of oligomers or even fibrils (left and right panels, respectively). Release from the confinement/scaffold (gray dashed vertical lines) before an aggregation nucleus has formed will cause dissociation of all soluble oligomeric assemblies/aggregates (bottom right). As a result, the equilibrium will shift significantly toward the soluble monomeric form (red horizontal arrow). Even in the case where insoluble aggregates (e.g., fibrillar structures) have been formed (bottom left), their formation may be partially reversed (small red arrow) by the low abundance of the soluble monomeric form achieved by tight regulation, because it has been shown that monomers can recycle in and out of the fibrillar aggregate in a concentration-dependent manner or disaggregate by active mechanisms.
Figure S4.
Le Chatelier’s Principle and Evolutionary Strategies for Minimizing Protein Aggregation, Related to Figure 4
(A) Tight regulation of aggregation prone proteins may keep functional and non-functional aggregates under control. Intracellular concentrations below the critical one, as a result of reduced transcript synthesis, tight translational control and rapid protein turnover (curved bold red arrow), prevent undesired aggregation. Local super-saturation by confinement (indicated by the gray circles) can initiate aggregation. Release of the aggregation-prone proteins from the confinement before a nucleus of critical size has formed, i.e., before the end of the lag time, will resolve all oligomeric aggregates by the principle of mass action (left). As a result of Le Chatelier’s principle, the equilibrium will shift significantly toward the monomers outside of the confinement (thick red straight arrow). Even in the case where the release occurs after the lag time has passed, the formation of highly stable fibrillar structures may still be at least partially reversible (thin red straight arrow), because it has been shown that monomers can recycle in and out of the fibrillar aggregate in a concentration dependent manner or by active mechanisms such as the disaggresome (Duennwald et al., 2012; Murray et al., 2010) (right).
(B) Cellular strategies to minimize protein aggregation. During evolution, if the introduced mutations results in increased aggregation-likelihood of a protein and do not increase cellular fitness, cells harboring such mutated sequences are likely to be eliminated from the population and the sequence thereby removed (evolution by negative design (Dobson, 1999)). If the introduced mutations do increase cellular fitness, the mutated sequence is likely to be conserved and a tight regulation of the availability of such sequence will prevent undesired aggregation and keep functional aggregation under control. The latter is likely to operate in addition to the proteostasis network that minimizes the likelihood of protein aggregation by ensuring protein-folding homeostasis.
The critical concentration above which fibrillar aggregation takes place is largely determined by the amino-acid composition and the environment of the protein. These properties are evolvable as long as protein function is not compromised during sequence changes. However, if sequence evolution is constrained due to functional reasons, evolution of a tight control over the availability of aggregation-prone proteins can keep their effective intracellular concentration below their critical concentration and minimize the chances of aggregation, even when the chaperone-system or clearance mechanisms fail due to stress or functional overload (i.e., upon failure of the chaperone proteostasis network) (Bence et al., 2001; Satyal et al., 2000). Hence, proteins that are kept at low concentrations can “accept” mutations that provide functional advantages, but equally increase the aggregation propensity. In other words, proteins that are kept at low concentrations for functional purposes can, by evolutionary drift, end up with a higher aggregation propensity without incurring significant negative selection. This concept is consistent with the observation of Drummond et al. (2005) that highly abundant proteins evolve slowly. Thus the tight regulation of aggregation-prone proteins offers a distinct solution to the problem of minimizing protein aggregation as compared to evolution by negative design and the proteostasis network. Supporting this hypothesis, it has been shown that (1) an iron response element (IRE) in the 5′UTR of α-synuclein and the amyloid precursor protein (APP) ensures a tight translational control, which when disrupted results in increased protein abundance and leads to protein aggregation (Avramovich-Tirosh et al., 2008; Friedlich et al., 2007; Rogers et al., 2002), and (2) increased expression of full-length TIA-1 that contains a Q/N-enriched region induces stress granule formation, and overexpression of the Q/N-rich region of TIA-1 alone forms cytoplasmic microaggregates that sequester endogenous TIA-1 (Gilks et al., 2004).
Importantly, if protein aggregation, higher-order oligomerization, or even amyloid-like fibrillation is required for functional reasons, local (super) saturation, which is an increase in the effective local concentration above the critical one, can be mediated. This can be achieved, for example, by specific regulatory proteins that “scaffold” aggregation-prone proteins in a particular location or by confinement of the protein within cellular compartments (Brangwynne et al., 2009; Decker et al., 2007; Fowler et al., 2006; Harper and Lansbury, 1997; Hu et al., 2009; Li et al., 2012; van Ham et al., 2010). Release of the aggregation-prone proteins from the confinement will reverse the aggregation process and likely resolve soluble aggregates and partially insoluble aggregates, as explained by Le Chatelier’s principle. This may be further enhanced by active mechanisms, such as the disaggresome (Bieschke et al., 2009) (Figure 5; Extended Discussion).
Support for this concept is provided by exciting recent reports. (1) Intrinsically disordered low complexity regions (LCR) of several RNA-binding proteins have been shown to enable the formation of reversible granule-like macromolecular assemblies by promoting amyloid-like interactions. Though the regulation of the formation of the highly dynamic aggregates is not yet understood, it has been proposed that RNA may act as a scaffold, allowing the LCRs to reach high local concentrations that are necessary for aggregation to occur (Han et al., 2012; Kato et al., 2012). (2) Under normal conditions, the yeast protein Lsb2p is expressed at low levels, but under stress conditions, its expression increases. Importantly, as it is a scaffold for Sup35p, the increase in Lsb2p concentration promotes local accumulation of soluble Sup35 and its conversion into the fibrillar state (Chernova et al., 2011). Taken together, these recent findings suggest that a phenomenon similar to Le Chatelier’s principle is exploited for functional and reversible assembly/aggregation. While it is clear that Le Chatelier’s principle is only valid for closed thermodynamic systems and cells are not “closed” in the strict sense, it is certain that changes in local concentration of proteins in a cell can affect the equilibrium between monomeric and multimeric states of proteins.
Implications and Significance of the Proposed Strategy to Regulate Aggregation-Prone Proteins
The observation that the availability of aggregation-prone proteins is likely to be tuned by several factors at multiple levels during transcription, translation, and degradation complements and underscores the emerging view that the pathophysiology of aggregation-related diseases is multifactorial in nature. In addition, the process of aging is also likely to alter the mechanisms that help maintain protein homeostasis and thereby increases the risk for aggregation-related diseases (Gidalevitz et al., 2010; Powers et al., 2009; Roth and Balch, 2011). In addition to this aspect, our findings have important implications:
-
(1)
Mutations that disrupt the tight control of aggregation-prone proteins, thereby altering the dynamic equilibrium between the soluble monomeric and aggregated states, are likely to be a common mechanism that may underlie several expression level–dependent disease phenotypes that involve protein aggregation. Cross-seeding by other aggregation-prone proteins may also affect such equilibrium (Sandefur and Schnell, 2011). Indeed, it has been shown that huntingtin aggregates cross-seed TIA-1 and is likely to repress the physiological function of TIA-1 due to loss of function caused by sequestration of the functional protein into aggregates (Furukawa et al., 2009).
-
(2)
Our findings suggest potential candidate loci (for example, 5′ and 3′UTR regions of aggregation-prone proteins, RNA-binding proteins, E3 ubiquitin ligases, etc.), which are likely to manifest as low-frequency mutant alleles with partial penetrance, that should be prioritized for further detailed investigation in whole-genome association studies aimed at identifying causal mutations of aggregation diseases. Indeed, a recent work has identified a single nucleotide mutation in the 5′UTR of puratrophin-1 to be associated with autosomal-dominant cerebellar ataxia, a group of heterogeneous neurodegenerative disorders (Ishikawa et al., 2005).
-
(3)
The observations provide a framework for identifying intracellular pathways that regulate levels of specific aggregation-prone proteins (e.g., the presence of IRE in 5′UTR of α-synuclein and APP). Therefore, it could provide avenues and enumerate strategies for tailoring drugs that modulate expression of specific aggregation-prone proteins rather than develop generic drugs that may disrupt both harmful and functional aggregates indiscriminately.
Finally, the reported strategy to control protein aggregates provides robustness to cellular systems by minimizing the potentially harmful effects of aggregation-prone proteins and, at the same time, permits their vital contribution to the functioning of a cell.
Experimental Procedures
See Extended Experimental Procedures for more details.
Extended Experimental Procedures.
Data Sets
The complete proteome sequences of S. cerevisiae, S. pombe, D. melanogaster, M. musculus and H. sapiens were obtained from the NCBI and ENSEMBL websites. Information on mRNA abundance, transcriptional rate, transcript half-life, histone modifications, transcripts bound to RNA binding proteins, 5′UTR length, RNA secondary structure, and codon bias for S. cerevisiae was obtained from Holstege et al. (Holstege et al., 1998), Wang et al. (Wang et al., 2002), ChromatinDB (O’Connor and Wyrick, 2007), Hogan et al. (Hogan et al., 2008), Nagalakshmi et al. (Nagalakshmi et al., 2008), Kertesz et al. (Kertesz et al., 2010), and Man and Pilpel (Man and Pilpel, 2007), respectively. RNAfold (Hofacker and Stadler, 2006) was used to calculate the free energy structure of 5′UTR RNA. Data on protein abundance, protein half-life, polysome profiling, ribosome profiling, and overexpression phenotype for S. cerevisiae was obtained from Ghaemmaghami et al. (Ghaemmaghami et al., 2003), Belle et al. (Belle et al., 2006), Arava et al. (Arava et al., 2003), Ingolia et al. (Ingolia et al., 2009), and Sopko et al. (Sopko et al., 2006), respectively. Data on mRNA half-life, codon bias and protein abundance for S. pombe was obtained from Lackner et al. (Lackner et al., 2007), Man and Pilpel (Man and Pilpel, 2007) and Matsuyama et al. (Matsuyama et al., 2006), respectively. Data on mRNA abundance, 5′UTR length and protein abundance for H. sapiens was obtained from Vogel et al. (Vogel et al., 2010). Data on mouse transcriptional rate, transcript abundance, transcript half-life, translational rate, protein abundance and protein half-life was obtained from Schwanhausser et al. (Schwanhäusser et al., 2011). Information on genetic backgrounds that enhance the lethality due to the expression of an aggregation prone protein in S. cerevisiae and C. elegans was obtained from Willingham et al. (Willingham et al., 2003) and Nollen et al. (Nollen et al., 2004), respectively (see Table 1).
Identification of Highly Structured and Unstructured Groups of Proteins
The prediction of intrinsic disorder was carried out using Disopred (Ward et al., 2004), which is a support vector machine based prediction method. For every protein, we filtered out coiled-coil regions and transmembrane segments using the program pfilt (http://bioinf.cs.ucl.ac.uk/downloads/pfilt). We then calculated the fraction of the sequence that was predicted to be unstructured. Depending on this fraction, we classified each protein of the S. cerevisiae proteome into one of the three classes: Highly structured, S (1971 sequences; 0 – 10% of the sequence is unstructured), moderately unstructured, M (2711 sequences; 10% – 30%) and highly unstructured, U (2020 sequences; 30% - 100%). Using the same criteria, we obtained three groups for S. pombe (S = 1576 sequences, M = 2005 sequences, and U = 1409 sequences). To ensure equally sized groups in H. sapiens, the criteria was adjusted to: Highly structured, S (8619 sequences; 0 – 15% of the sequence is unstructured), moderately unstructured, M (8511 sequences; 15% – 35%) and highly unstructured, U (7934 sequences; 35% - 100%). The numbers for M. musculus were S (5938 sequences; 0 – 15% of the sequence is unstructured), M (5616 sequences; 15% – 35%) and U (4754 sequences; 35% - 100%). For the analysis presented in the main text, we did not consider proteins in the M group in order to (i) ensure that we are unambiguously dealing with highly unstructured and structured proteins and (ii) identify the effect of the presence of aggregation prone stretches on the cellular availability of proteins in each group.
Identification of Aggregation-Prone and Non-Aggregation-Prone Proteins in Each Structural Class
We identified aggregation prone stretches in the proteins of the S group by using the TANGO algorithm (Fernandez-Escamilla et al., 2004) and aggregation prone stretches in the proteins of the U group by using the algorithm described by Michelitsch and Weissman (Michelitsch and Weissman, 2000). See Extended Results for rationalization of the requirement to group proteins into the structured and unstructured proteins classes.
Structured (S) Group of Proteins:
TANGO was used to define aggregation prone and non-aggregation prone structured proteins. TANGO is based on physico-chemical principles of secondary structure formation extended by the principle that the core regions of an aggregate are fully buried. TANGO returns a score that represents the tendency for each amino acid to be part of a beta-sheet aggregate; it ranges from 0% for no aggregation to 100% for strong aggregation tendency. Extensive comparison with experimental data revealed that a peptide with a stretch of more than 5 consecutive residues with a TANGO score of more than 50% have an 85% chance on average to aggregate in vitro (Fernandez-Escamilla et al., 2004). Based on these observations and in order to get equally sized groups of aggregation prone and non-aggregation prone proteins, we used a criterion that is more stringent and classified a structured protein as aggregation prone if it contained at least one stretch of 7 consecutive residues that TANGO identifies as aggregation prone (i.e., stretches of 7 residues with a score > 50% each). Proteins were considered as non-aggregation prone if they did not have a single residue that TANGO identified to have an aggregation tendency (i.e., no residue with a score > 50%).
As a control, we also identified aggregation prone structured proteins with PASTA (Trovato et al., 2007). PASTA is based on the working principle that the same interactions that stabilize β sheet structures in globular proteins are responsible for protein aggregation. Hence, sequence-specific interaction energies between pairs of protein fragments have been derived from a statistical analysis of the native folds of globular proteins. These energies are then used to assign aggregation scores to protein sequences. Here, we considered structured proteins as aggregation prone if they had at least one segment that had an aggregation score of E ≤ −15kcal/mol and non-aggregation prone if they had no segment that had an aggregation score E < −8kcal/mol.
Unstructured (U) Group of Proteins:
For the unstructured group (U), we used another strategy to identify aggregation prone proteins. TANGO is an excellent predictor of aggregation nucleating segments in proteins, in particular if they contain hydrophobic residues. Intrinsically unstructured proteins are, however, often depleted of hydrophobic residues (Dunker et al., 2001). For instance, TANGO does not identify Sup35, a well-characterized S. cerevisiae prion protein, as aggregation prone. Sup35 is largely unstructured and its prion domain is enriched in glutamines and asparagines, not hydrophobic residues.
In order to unambiguously identify aggregation prone proteins in the U group, we used the algorithm described by Michelitsch and Weissman (Michelitsch and Weissman, 2000) that detects stretches of amino-acids enriched for Q/N. Although some of these Q/N-enriched segments may be flanked with proline or charged residues that can reduce the likelihood of aggregation, it is established that long Q/N-enriched segments have a high propensity to beta-sheet aggregate and form self-propagating amyloid fibrils (Alberti et al., 2009; Fiumara et al., 2010; Kar et al., 2011; Klein et al., 2007; Lakhani et al., 2010). Indeed, new aggregation prone prions have been identified based on screens for Q/N-enriched proteins and subsequently experimentally validated in the labs of Lindquist and Weissman (Alberti et al., 2009; Michelitsch and Weissman, 2000). Following the procedure of Michelitsch and Weismann (Michelitsch and Weissman, 2000), we classified an unstructured protein as aggregation prone if it contained at least 25 glutamines and/or asparagines in any continuous segment of 80 residues.
As unstructured proteins with low aggregation likelihood, we identified those proteins that contained K or E-enriched segments in the U group (at least 30 lysine or glutamic acid in 80 residues). Apart from being the most enriched amino acids in unstructured proteins (Dunker et al., 2001), large numbers of charged residues such as K and E have been shown to prevent aggregation even under conditions that normally cause proteins to aggregate (e.g., heating or chemical denaturation) (Lawrence et al., 2007; Vendruscolo and Dobson, 2007). We did not consider proteins with enrichment in R and D in a stretch of 80 residues as the number of proteins harboring such stretches and with relevant experimental measurements available were too low to make meaningful statistical comparisons. A cut-off of 25 Q and/or N and 30 K and/or E has been chosen for the aggregation prone and non-aggregation prone group, respectively, because at these cut-offs, the two groups contain proteins of similar average length and are similar in the number of members. Importantly, the quantity most relevant to this paper, protein abundance, is always lower for proteins enriched in Q/N compared to proteins enriched in K/E, independent of the cut-off used. As a control, we also selected as aggregation prone unstructured proteins a recently identified list of prionogenic proteins (Alberti et al., 2009).
Identification of Aggregation-Prone and Nonaggregation-Prone Proteins Using AGGRESCAN
While the analysis presented in the main text was based on subgroups that cover only part of the proteome of S. cerevisiae, we aimed, as a control, at confirming the observed differences in regulation between proteins with different likelihoods of aggregation for all members of the proteome of S. cerevisiae. Therefore we used the algorithm AGGRESCAN (Conchillo-Solé et al., 2007) to identify highly aggregation prone (XA), moderately aggregation prone (XM) and non-aggregation prone (XNA) proteins in the highly structured, S moderately unstructured, M, and highly unstructured, U, groups of S. cerevisiae (where X is S, M or U). We chose AGGRESCAN because it identifies at least one aggregation prone residue in all proteins of S. cerevisiae and, therefore, allows for a classification on a spectrum of aggregation likelihood. AGGRESCAN calculates the average amino-acid aggregation propensity value (a4v) over sliding windows of 5, 7, 9 or 11 residues, depending on the sequence length. In order to identify highly aggregation prone proteins (XA), we selected those proteins that had at least one segment of 9 consecutive residues with a4v score higher than 0.5. Since AGGRESCAN reports aggregation prone residues in all proteins of S. cerevisiae, we classified proteins as non-aggregation prone (XNA), if they had no aggregation prone stretches, defined as 4 or less consecutive residues that have a4v score greater than 0.5. The remaining proteins (those with 5-8 consecutive residues with a4v score higher than 0.5) in each structural group (S, M or U) were classified as moderately aggregation prone (XM).
Identification of Evolutionarily Conserved Residues in Aggregation-Prone Stretches
In order to calculate the conservation of aggregation prone residues in nine fungi strains, we analyzed the presence of aggregation prone residues in a set of 1372 orthologous proteins from the following fungal species: A. nidulans, C. albicans, C. glabrata, D. hansenii, K. lactis, S. bayanus, S. cerevisiae, S. pombe and Y. lipolytica. The set of aligned orthologs was kindly provided by Eytan Ruppin and Tamir Tuller (Tuller et al., 2009). Aggregation prone and non-aggregation prone residues in the 1372 proteins of all strains were determined by using TANGO. Taking the aggregation profile of S. cerevisiae as reference, we determined the percentage of the aggregation prone and non-aggregation prone residues that are present in the other strains (Figure S1).
Estimation of Statistical Significance
All analyses to estimate statistical significance (Wilcoxon rank-sum, Kolmogorov-Smirnov and Fisher’s exact test) were carried out using the R statistical analysis package. Wilcoxon rank-sum and Kolmogorov-Smirnov tests are non-parametric tests that evaluate whether two samples of observations come from the same distribution or not. Importantly, both do not require assumptions about the form of the distribution of the values. As most data sets used in this analysis are not normally distributed, we determined the significance of the difference between the aggregation prone and non-aggregation prone subgroups by using Wilcoxon’s test. Consistently, we find similar results by using the Kolmogorov-Smirnov test (Table S3H). In order to evaluate the significance of the enrichment for certain properties that don’t have distributions but only have percentages, we used Fisher’s exact test.
Partial Least Square Regression Analysis
In order to obtain an estimate of the relative contribution of transcript abundance, transcription rate and protein-half-life to protein abundance of aggregation prone and non-aggregation prone proteins, we carried out a partial least square regression analysis (PLSR). For the PLSR analysis, several linear observable variables (e.g., transcript abundance, protein translational rate, protein half-life, etc.) are first scaled to zero mean and unit variance. Subsequent PLSR analysis finds latent variables of a linear model that describes the predicted variable (protein abundance) in terms of the observable variables. The proportion of the predicted variable’s variance, R2, explained by each latent variable, the significance of R2 and the fractional contribution of each observable variable to the latent variable were determined (Tables S2D and S2E).
Identification of Highly Structured and Unstructured Group of Proteins
The proteome of S. cerevisiae was divided into proteins that are highly structured and those that are highly unstructured. The prediction of intrinsic disorder was carried out using Disopred2 (Ward et al., 2004). We then calculated the fraction of the sequence that was predicted to be unstructured. Depending on this fraction, we classified each protein as highly structured (S) (1,971 proteins with 0%–10% of all residues unstructured), highly unstructured (U) (2,020 proteins with 30%–100% unstructured residues), and not fitting within each group (2,711 proteins).
Identification of Aggregation-Prone and Nonaggregation-Prone Proteins in Each Structural Class
To identify aggregation-prone proteins among the highly structured and highly unstructured proteins, we used TANGO (Fernandez-Escamilla et al., 2004) for the structured proteins and the algorithm described by Michelitsch and Weissman (2000) for the unstructured proteins. We divided the group of highly structured proteins into those that are highly aggregation prone (SAG; 711 proteins with more than seven consecutive residues identified as aggregation-prone by TANGO) and those with very low aggregation likelihood (SNA; 716 proteins with no residues identified as aggregation-prone by TANGO; for details, see Extended Experimental Procedures). We identified aggregation-prone stretches in the highly unstructured proteins by searching for segments that contain large Q- or N-enriched segments. We used the algorithm described by Michelitsch and Weissman (2000) and identified 197 aggregation-prone, unstructured proteins (UAG) that contained 25 glutamines or asparagines in a segment of 80 residues. As unstructured proteins with low aggregation likelihood (UNA), we identified 198 proteins that contain 30 lysines or glutamic acids in a segment of 80 residues (see Extended Experimental Procedures). Large numbers of charged amino acids, such as K and E, have been shown to prevent aggregation, even under conditions that normally cause proteins to aggregate, such as heating or chemical denaturation (Lawrence et al., 2007).
Data Set and Statistical Analysis
The complete proteome sequences of S. cerevisiae, S. pombe, human, and mouse were obtained from the National Center for Biotechnology Information (NCBI). Information on mRNA abundance, transcriptional rate, transcript half-life, transcripts bound to RNA-binding proteins, protein abundance, protein half-life, translational rate, overexpression phenotype, and data on genetic background that enhanced protein aggregation were all obtained from published literature (see Table 1). All statistical analyses to estimate significance (Wilcoxon, Kolmogorov–Smirnov, and Fisher’s exact test) and the partial least square regression (PLSR) analysis were carried out using the R statistical analysis package.
Acknowledgments
This work was supported by the Medical Research Council (U105185859). We thank A. Bertolotti, E. Levy, J. Scott-Brown, M. Buljan, M. Garcia-Alai, M. Goedert, R. van der Lee, S. Michnick, S. Radford, S. Teichmann, and S. Sarkar for providing helpful comments. We apologize for, because of lack of space, not citing the work that describes the datasets and other references. M.M.B. acknowledges Darwin College, Schlumberger Ltd, Trinity College, HFSP (RGY0073/2010), ERASysBio+ (GRAPPLE; BBSRC), and the EMBO YI Programme for support. J.G. is funded by PrioNet Canada and the University of British Columbia.
Published: November 15, 2012
Footnotes
Supplemental Information includes Extended Results, and Extended Discussion, Extended Experimental Procedures, four figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2012.09.036.
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Jörg Gsponer, Email: gsponer@chibi.ubc.ca.
M. Madan Babu, Email: madanm@mrc-lmb.cam.ac.uk.
Supplemental Information
References
- Alberti S., Halfmann R., King O., Kapila A., Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramovich-Tirosh Y., Amit T., Bar-Am O., Weinreb O., Youdim M.B. Physiological and pathological aspects of Abeta in iron homeostasis via 5’UTR in the APP mRNA and the therapeutic use of iron-chelators. BMC Neurosci. 2008;9(Suppl 2):S2. doi: 10.1186/1471-2202-9-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M.M., van der Lee R., de Groot N.S., Gsponer J. Intrinsically disordered proteins: regulation and disease. Curr. Opin. Struct. Biol. 2011;21:432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- Balagopal V., Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence N.F., Sampat R.M., Kopito R.R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Bieschke J., Cohen E., Murray A., Dillin A., Kelly J.W. A kinetic assessment of the C. elegans amyloid disaggregation activity enables uncoupling of disassembly and proteolysis. Protein Sci. 2009;18:2231–2241. doi: 10.1002/pro.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Broome B.M., Hecht M.H. Nature disfavors sequences of alternating polar and non-polar amino acids: implications for amyloidogenesis. J. Mol. Biol. 2000;296:961–968. doi: 10.1006/jmbi.2000.3514. [DOI] [PubMed] [Google Scholar]
- Carulla N., Caddy G.L., Hall D.R., Zurdo J., Gairí M., Feliz M., Giralt E., Robinson C.V., Dobson C.M. Molecular recycling within amyloid fibrils. Nature. 2005;436:554–558. doi: 10.1038/nature03986. [DOI] [PubMed] [Google Scholar]
- Chen S., Berthelier V., Yang W., Wetzel R. Polyglutamine aggregation behavior in vitro supports a recruitment mechanism of cytotoxicity. J. Mol. Biol. 2001;311:173–182. doi: 10.1006/jmbi.2001.4850. [DOI] [PubMed] [Google Scholar]
- Chernova T.A., Romanyuk A.V., Karpova T.S., Shanks J.R., Ali M., Moffatt N., Howie R.L., O’Dell A., McNally J.G., Liebman S.W. Prion induction by the short-lived, stress-induced protein Lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol. Cell. 2011;43:242–252. doi: 10.1016/j.molcel.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Colby D.W., Cassady J.P., Lin G.C., Ingram V.M., Wittrup K.D. Stochastic kinetics of intracellular huntingtin aggregate formation. Nat. Chem. Biol. 2006;2:319–323. doi: 10.1038/nchembio792. [DOI] [PubMed] [Google Scholar]
- Conchillo-Solé O., de Groot N.S., Avilés F.X., Vendrell J., Daura X., Ventura S. AGGRESCAN: a server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinformatics. 2007;8:65. doi: 10.1186/1471-2105-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C., Whitesell L., Rogers A.B., Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Baets G., Reumers J., Delgado Blanco J., Dopazo J., Schymkowitz J., Rousseau F. An evolutionary trade-off between protein turnover rate and protein aggregation favors a higher aggregation propensity in fast degrading proteins. PLoS Comput. Biol. 2011;7:e1002090. doi: 10.1371/journal.pcbi.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot N.S., Ventura S. Protein aggregation profile of the bacterial cytosol. PLoS ONE. 2010;5:e9383. doi: 10.1371/journal.pone.0009383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., Teixeira D., Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M., Savva R., Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D.A., Wilke C.O. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D.A., Bloom J.D., Adami C., Wilke C.O., Arnold F.H. Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. USA. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Escamilla A.M., Rousseau F., Schymkowitz J., Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- Fiumara F., Fioriti L., Kandel E.R., Hendrickson W.A. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell. 2010;143:1121–1135. doi: 10.1016/j.cell.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D.M., Koulov A.V., Alory-Jost C., Marks M.S., Balch W.E., Kelly J.W. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D.M., Koulov A.V., Balch W.E., Kelly J.W. Functional amyloid—from bacteria to humans. Trends Biochem. Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Friedlich A.L., Tanzi R.E., Rogers J.T. The 5′-untranslated region of Parkinson’s disease alpha-synuclein messengerRNA contains a predicted iron responsive element. Mol. Psychiatry. 2007;12:222–223. doi: 10.1038/sj.mp.4001937. [DOI] [PubMed] [Google Scholar]
- Furukawa Y., Kaneko K., Matsumoto G., Kurosawa M., Nukina N. Cross-seeding fibrillation of Q/N-rich proteins offers new pathomechanism of polyglutamine diseases. J. Neurosci. 2009;29:5153–5162. doi: 10.1523/JNEUROSCI.0783-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte K.A., Dion M.F., Budnik B.A., Wang S.M., Hartl D.L., Drummond D.A. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc. Natl. Acad. Sci. USA. 2011;108:680–685. doi: 10.1073/pnas.1017570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T., Kikis E.A., Morimoto R.I. A cellular perspective on conformational disease: the role of genetic background and proteostasis networks. Curr. Opin. Struct. Biol. 2010;20:23–32. doi: 10.1016/j.sbi.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H., Pilpel Y. Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 2011;7:481. doi: 10.1038/msb.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J.R., Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Goldschmidt L., Teng P.K., Riek R., Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. USA. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsponer J., Futschik M.E., Teichmann S.A., Babu M.M. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., McKnight S.L. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Harper J.D., Lansbury P.T., Jr. Models of amyloid seeding in Alzheimer’s disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu. Rev. Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- Hishiya A., Takayama S. Molecular chaperones as regulators of cell death. Oncogene. 2008;27:6489–6506. doi: 10.1038/onc.2008.314. [DOI] [PubMed] [Google Scholar]
- Hu X., Crick S.L., Bu G., Frieden C., Pappu R.V., Lee J.M. Amyloid seeds formed by cellular uptake, concentration, and aggregation of the amyloid-beta peptide. Proc. Natl. Acad. Sci. USA. 2009;106:20324–20329. doi: 10.1073/pnas.0911281106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K., Toru S., Tsunemi T., Li M., Kobayashi K., Yokota T., Amino T., Owada K., Fujigasaki H., Sakamoto M. An autosomal dominant cerebellar ataxia linked to chromosome 16q22.1 is associated with a single-nucleotide substitution in the 5′ untranslated region of the gene encoding a protein with spectrin repeat and Rho guanine-nucleotide exchange-factor domains. Am. J. Hum. Genet. 2005;77:280–296. doi: 10.1086/432518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Nollen E.A., Kitagawa K., Bindokas V.P., Morimoto R.I. Polyglutamine protein aggregates are dynamic. Nat. Cell Biol. 2002;4:826–831. doi: 10.1038/ncb863. [DOI] [PubMed] [Google Scholar]
- Kitamura A., Kubota H., Pack C.G., Matsumoto G., Hirayama S., Takahashi Y., Kimura H., Kinjo M., Morimoto R.I., Nagata K. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat. Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- Krobitsch S., Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G., Murray A.W., Tollervey D., Plotkin J.B. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S., Bugaut A., Huppert J.L., Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.S., Phillips K.J., Liu D.R. Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 2007;129:10110–10112. doi: 10.1021/ja071641y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Zhou T., Tartaglia G.G., Vendruscolo M., Wilke C.O. Translationally optimal codons associate with aggregation-prone sites in proteins. Proteomics. 2010;10:4163–4171. doi: 10.1002/pmic.201000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Banjade S., Cheng H.C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J.V., King D.S., Banani S.F. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R., Schymkowitz J., Rousseau F., Diella F., Serrano L. A comparative study of the relationship between protein structure and beta-aggregation in globular and intrinsically disordered proteins. J. Mol. Biol. 2004;342:345–353. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- Maji S.K., Perrin M.H., Sawaya M.R., Jessberger S., Vadodaria K., Rissman R.A., Singru P.S., Nilsson K.P., Simon R., Schubert D. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masino L., Nicastro G., Calder L., Vendruscolo M., Pastore A. Functional interactions as a survival strategy against abnormal aggregation. FASEB J. 2011;25:45–54. doi: 10.1096/fj.10-161208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitsch M.D., Weissman J.S. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. USA. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsellier E., Ramazzotti M., Taddei N., Chiti F. Aggregation propensity of the human proteome. PLoS Comput. Biol. 2008;4:e1000199. doi: 10.1371/journal.pcbi.1000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell M., de Groot N.S., Vendrell J., Avilés F.X., Ventura S. Linking amyloid protein aggregation and yeast survival. Mol. Biosyst. 2011;7:1121–1128. doi: 10.1039/c0mb00297f. [DOI] [PubMed] [Google Scholar]
- Münch C., Bertolotti A. Exposure of hydrophobic surfaces initiates aggregation of diverse ALS-causing superoxide dismutase-1 mutants. J. Mol. Biol. 2010;399:512–525. doi: 10.1016/j.jmb.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen E.A., Garcia S.M., van Haaften G., Kim S., Chavez A., Morimoto R.I., Plasterk R.H. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. USA. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patki A.U., Hausrath A.C., Cordes M.H. High polar content of long buried blocks of sequence in protein domains suggests selection against amyloidogenic non-polar sequences. J. Mol. Biol. 2006;362:800–809. doi: 10.1016/j.jmb.2006.07.055. [DOI] [PubMed] [Google Scholar]
- Pechmann S., Levy E.D., Tartaglia G.G., Vendruscolo M. Physicochemical principles that regulate the competition between functional and dysfunctional association of proteins. Proc. Natl. Acad. Sci. USA. 2009;106:10159–10164. doi: 10.1073/pnas.0812414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers E.T., Morimoto R.I., Dillin A., Kelly J.W., Balch W.E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Reijns M.A., Alexander R.D., Spiller M.P., Beggs J.D. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J.T., Randall J.D., Cahill C.M., Eder P.S., Huang X., Gunshin H., Leiter L., McPhee J., Sarang S.S., Utsuki T. An iron-responsive element type II in the 5′-untranslated region of the Alzheimer’s amyloid precursor protein transcript. J. Biol. Chem. 2002;277:45518–45528. doi: 10.1074/jbc.M207435200. [DOI] [PubMed] [Google Scholar]
- Roth D.M., Balch W.E. Modeling general proteostasis: proteome balance in health and disease. Curr. Opin. Cell Biol. 2011;23:126–134. doi: 10.1016/j.ceb.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F., Schymkowitz J., Serrano L. Protein aggregation and amyloidosis: confusion of the kinds? Curr. Opin. Struct. Biol. 2006;16:118–126. doi: 10.1016/j.sbi.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Rousseau F., Serrano L., Schymkowitz J.W. How evolutionary pressure against protein aggregation shaped chaperone specificity. J. Mol. Biol. 2006;355:1037–1047. doi: 10.1016/j.jmb.2005.11.035. [DOI] [PubMed] [Google Scholar]
- Salazar A.M., Silverman E.J., Menon K.P., Zinn K. Regulation of synaptic Pumilio function by an aggregation-prone domain. J. Neurosci. 2010;30:515–522. doi: 10.1523/JNEUROSCI.2523-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandefur C.I., Schnell S. A model of threshold behavior reveals rescue mechanisms of bystander proteins in conformational diseases. Biophys. J. 2011;100:1864–1873. doi: 10.1016/j.bpj.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A.A., Croy C.H., Vasanwala F.H., Uversky V.N., Van Y.Y., Dunker A.K. Sweeping away protein aggregation with entropic bristles: intrinsically disordered protein fusions enhance soluble expression. Biochemistry. 2012;51:7250–7262. doi: 10.1021/bi300653m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyal S.H., Schmidt E., Kitagawa K., Sondheimer N., Lindquist S., Kramer J.M., Morimoto R.I. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio T.R., Cashikar A.G., Kowal A.S., Sawicki G.J., Moslehi J.J., Serpell L., Arnsdorf M.F., Lindquist S.L. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- Sivakolundu S.G., Nourse A., Moshiach S., Bothner B., Ashley C., Satumba J., Lahti J., Kriwacki R.W. Intrinsically unstructured domains of Arf and Hdm2 form bimolecular oligomeric structures in vitro and in vivo. J. Mol. Biol. 2008;384:240–254. doi: 10.1016/j.jmb.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Huang D., Preston N., Chua G., Papp B., Kafadar K., Snyder M., Oliver S.G., Cyert M., Hughes T.R. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Stenoien D.L., Mielke M., Mancini M.A. Intranuclear ataxin1 inclusions contain both fast- and slow-exchanging components. Nat. Cell Biol. 2002;4:806–810. doi: 10.1038/ncb859. [DOI] [PubMed] [Google Scholar]
- Tartaglia G.G., Pechmann S., Dobson C.M., Vendruscolo M. Life on the edge: a link between gene expression levels and aggregation rates of human proteins. Trends Biochem. Sci. 2007;32:204–206. doi: 10.1016/j.tibs.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Tartaglia G.G., Pechmann S., Dobson C.M., Vendruscolo M. A relationship between mRNA expression levels and protein solubility in E. coli. J. Mol. Biol. 2009;388:381–389. doi: 10.1016/j.jmb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Trovato A., Seno F., Tosatto S.C. The PASTA server for protein aggregation prediction. Protein Eng. Des. Sel. 2007;20:521–523. doi: 10.1093/protein/gzm042. [DOI] [PubMed] [Google Scholar]
- van Ham T.J., Holmberg M.A., van der Goot A.T., Teuling E., Garcia-Arencibia M., Kim H.E., Du D., Thijssen K.L., Wiersma M., Burggraaff R. Identification of MOAG-4/SERF as a regulator of age-related proteotoxicity. Cell. 2010;142:601–612. doi: 10.1016/j.cell.2010.07.020. [DOI] [PubMed] [Google Scholar]
- Vogel C. Translation’s coming of age. Mol. Syst. Biol. 2011;7:498. doi: 10.1038/msb.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J.J., Sodhi J.S., McGuffin L.J., Buxton B.F., Jones D.T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Willingham S., Outeiro T.F., DeVit M.J., Lindquist S.L., Muchowski P.J. Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha-synuclein. Science. 2003;302:1769–1772. doi: 10.1126/science.1090389. [DOI] [PubMed] [Google Scholar]
Supplemental References
- Altschuler, E.L., Hud, N.V., Mazrimas, J.A., and Rupp, B. (1997). Random coil conformation for extended polyglutamine stretches in aqueous soluble monomeric peptides. J. Pept. Res. 50, 73–75. [DOI] [PubMed]
- Arava, Y., Wang, Y., Storey, J.D., Liu, C.L., Brown, P.O., and Herschlag, D. (2003). Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100, 3889–3894. [DOI] [PMC free article] [PubMed]
- Badtke, M.P., Hammer, N.D., and Chapman, M.R. (2009). Functional amyloids signal their arrival. Sci. Signal. 2, pe43. [DOI] [PMC free article] [PubMed]
- Balch, W.E., Morimoto, R.I., Dillin, A., and Kelly, J.W. (2008). Adapting proteostasis for disease intervention. Science 319, 916–919. [DOI] [PubMed]
- Belle, A., Tanay, A., Bitincka, L., Shamir, R., and O’Shea, E.K. (2006). Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. USA 103, 13004–13009. [DOI] [PMC free article] [PubMed]
- Bemporad, F., Calloni, G., Campioni, S., Plakoutsi, G., Taddei, N., and Chiti, F. (2006). Sequence and structural determinants of amyloid fibril formation. Acc. Chem. Res. 39, 620–627. [DOI] [PubMed]
- Bershtein, S., Mu, W., and Shakhnovich, E.I. (2012). Soluble oligomerization provides a beneficial fitness effect on destabilizing mutations. Proc. Natl. Acad. Sci. USA 109, 4857–4862. [DOI] [PMC free article] [PubMed]
- Caughey, B., and Lansbury, P.T. (2003). Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 26, 267–298. [DOI] [PubMed]
- Chamot-Rooke, J., Mikaty, G., Malosse, C., Soyer, M., Dumont, A., Gault, J., Imhaus, A.F., Martin, P., Trellet, M., Clary, G., et al. (2011). Posttranslational modification of pili upon cell contact triggers N. meningitidis dissemination. Science 331, 778–782. [DOI] [PubMed]
- Chiti, F., and Dobson, C.M. (2009). Amyloid formation by globular proteins under native conditions. Nat. Chem. Biol. 5, 15–22. [DOI] [PubMed]
- Chiti, F., Stefani, M., Taddei, N., Ramponi, G., and Dobson, C.M. (2003). Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature 424, 805–808. [DOI] [PubMed]
- Chow, M.K., Lomas, D.A., and Bottomley, S.P. (2004). Promiscuous beta-strand interactions and the conformational diseases. Curr. Med. Chem. 11, 491–499. [DOI] [PubMed]
- Conway, K.A., Rochet, J.C., Bieganski, R.M., and Lansbury, P.T., Jr. (2001). Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science 294, 1346–1349. [DOI] [PubMed]
- Couñago, R., Chen, S., and Shamoo, Y. (2006). In vivo molecular evolution reveals biophysical origins of organismal fitness. Mol. Cell 22, 441–449. [DOI] [PubMed]
- David, D.C., Ollikainen, N., Trinidad, J.C., Cary, M.P., Burlingame, A.L., and Kenyon, C. (2010). Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 8, e1000450. [DOI] [PMC free article] [PubMed]
- Dehay, B., and Bertolotti, A. (2006). Critical role of the proline-rich region in Huntingtin for aggregation and cytotoxicity in yeast. J. Biol. Chem. 281, 35608–35615. [DOI] [PubMed]
- Demontis, F., and Perrimon, N. (2010). FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143, 813–825. [DOI] [PMC free article] [PubMed]
- DePace, A.H., Santoso, A., Hillner, P., and Weissman, J.S. (1998). A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 93, 1241–1252. [DOI] [PubMed]
- Dobson, C.M. (1999). Protein misfolding, evolution and disease. Trends Biochem. Sci. 24, 329–332. [DOI] [PubMed]
- Dobson, C.M. (2006). Protein aggregation and its consequences for human disease. Protein Pept. Lett. 13, 219–227. [DOI] [PubMed]
- Du, Z., Park, K.W., Yu, H., Fan, Q., and Li, L. (2008). Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 40, 460–465. [DOI] [PMC free article] [PubMed]
- Duennwald, M.L., Echeverria, A., and Shorter, J. (2012). Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol. 10, e1001346. [DOI] [PMC free article] [PubMed]
- Dunker, A.K., Lawson, J.D., Brown, C.J., Williams, R.M., Romero, P., Oh, J.S., Oldfield, C.J., Campen, A.M., Ratliff, C.M., Hipps, K.W., et al. (2001). Intrinsically disordered protein. J. Mol. Graph. Model. 19, 26–59. [DOI] [PubMed]
- Faux, N.G., Bottomley, S.P., Lesk, A.M., Irving, J.A., Morrison, J.R., de la Banda, M.G., and Whisstock, J.C. (2005). Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res. 15, 537–551. [DOI] [PMC free article] [PubMed]
- Franks, T.M., and Lykke-Andersen, J. (2008). The control of mRNA decapping and P-body formation. Mol. Cell 32, 605–615. [DOI] [PMC free article] [PubMed]
- Galea, C.A., High, A.A., Obenauer, J.C., Mishra, A., Park, C.G., Punta, M., Schlessinger, A., Ma, J., Rost, B., Slaughter, C.A., and Kriwacki, R.W. (2009). Large-scale analysis of thermostable, mammalian proteins provides insights into the intrinsically disordered proteome. J. Proteome Res. 8, 211–226. [DOI] [PMC free article] [PubMed]
- Ganusova, E.E., Ozolins, L.N., Bhagat, S., Newnam, G.P., Wegrzyn, R.D., Sherman, M.Y., and Chernoff, Y.O. (2006). Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol. Cell. Biol. 26, 617–629. [DOI] [PMC free article] [PubMed]
- García-Arencibia, M., Hochfeld, W.E., Toh, P.P., and Rubinsztein, D.C. (2010). Autophagy, a guardian against neurodegeneration. Semin. Cell Dev. Biol. 21, 691–698. [DOI] [PMC free article] [PubMed]
- Gerber, H.P., Seipel, K., Georgiev, O., Höfferer, M., Hug, M., Rusconi, S., and Schaffner, W. (1994). Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science 263, 808–811. [DOI] [PubMed]
- Ghaemmaghami, S., Huh, W.K., Bower, K., Howson, R.W., Belle, A., Dephoure, N., O’Shea, E.K., and Weissman, J.S. (2003). Global analysis of protein expression in yeast. Nature 425, 737–741. [DOI] [PubMed]
- Goedert, M. (2001). Alpha-synuclein and neurodegenerative diseases. Nat. Rev. Neurosci. 2, 492–501. [DOI] [PubMed]
- Gsponer, J., and Vendruscolo, M. (2006). Theoretical approaches to protein aggregation. Protein Pept. Lett. 13, 287–293. [DOI] [PubMed]
- Halfmann, R., and Lindquist, S. (2010). Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science 330, 629–632. [DOI] [PubMed]
- Halfmann, R., Alberti, S., and Lindquist, S. (2010). Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 20, 125–133. [DOI] [PMC free article] [PubMed]
- Hartl, F.U., Bracher, A., and Hayer-Hartl, M. (2011). Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332. [DOI] [PubMed]
- Herter, S., Osterloh, P., Hilf, N., Rechtsteiner, G., Höhfeld, J., Rammensee, H.G., and Schild, H. (2005). Dendritic cell aggresome-like-induced structure formation and delayed antigen presentation coincide in influenza virus-infected dendritic cells. J. Immunol. 175, 891–898. [DOI] [PubMed]
- Hofacker, I.L., and Stadler, P.F. (2006). Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics 22, 1172–1176. [DOI] [PubMed]
- Hogan, D.J., Riordan, D.P., Gerber, A.P., Herschlag, D., and Brown, P.O. (2008). Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6, e255. [DOI] [PMC free article] [PubMed]
- Holstege, F.C., Jennings, E.G., Wyrick, J.J., Lee, T.I., Hengartner, C.J., Green, M.R., Golub, T.R., Lander, E.S., and Young, R.A. (1998). Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728. [DOI] [PubMed]
- Ingolia, N.T., Ghaemmaghami, S., Newman, J.R., and Weissman, J.S. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. [DOI] [PMC free article] [PubMed]
- Jahn, T.R., and Radford, S.E. (2005). The Yin and Yang of protein folding. FEBS J. 272, 5962–5970. [DOI] [PMC free article] [PubMed]
- Jahn, T.R., and Radford, S.E. (2008). Folding versus aggregation: polypeptide conformations on competing pathways. Arch. Biochem. Biophys. 469, 100–117. [DOI] [PMC free article] [PubMed]
- Jarosz, D.F., Taipale, M., and Lindquist, S. (2010). Protein homeostasis and the phenotypic manifestation of genetic diversity: principles and mechanisms. Annu. Rev. Genet. 44, 189–216. [DOI] [PubMed]
- Kaiser, T.E., Intine, R.V., and Dundr, M. (2008). De novo formation of a subnuclear body. Science 322, 1713–1717. [DOI] [PubMed]
- Kar, K., Jayaraman, M., Sahoo, B., Kodali, R., and Wetzel, R. (2011). Critical nucleus size for disease-related polyglutamine aggregation is repeat-length dependent. Nat. Struct. Mol. Biol. 18, 328–336. [DOI] [PMC free article] [PubMed]
- Kelly, J.W. (1998). The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr. Opin. Struct. Biol. 8, 101–106. [DOI] [PubMed]
- Kentsis, A., Gordon, R.E., and Borden, K.L. (2002). Control of biochemical reactions through supramolecular RING domain self-assembly. Proc. Natl. Acad. Sci. USA 99, 15404–15409. [DOI] [PMC free article] [PubMed]
- Kertesz, M., Wan, Y., Mazor, E., Rinn, J.L., Nutter, R.C., Chang, H.Y., and Segal, E. (2010). Genome-wide measurement of RNA secondary structure in yeast. Nature 467, 103–107. [DOI] [PMC free article] [PubMed]
- Klein, F.A., Pastore, A., Masino, L., Zeder-Lutz, G., Nierengarten, H., Oulad-Abdelghani, M., Altschuh, D., Mandel, J.L., and Trottier, Y. (2007). Pathogenic and non-pathogenic polyglutamine tracts have similar structural properties: towards a length-dependent toxicity gradient. J. Mol. Biol. 371, 235–244. [DOI] [PubMed]
- Kopito, R.R. (2000). Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530. [DOI] [PubMed]
- Korolchuk, V.I., Menzies, F.M., and Rubinsztein, D.C. (2010). Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 584, 1393–1398. [DOI] [PubMed]
- Lackner, D.H., Beilharz, T.H., Marguerat, S., Mata, J., Watt, S., Schubert, F., Preiss, T., and Bähler, J. (2007). A network of multiple regulatory layers shapes gene expression in fission yeast. Mol. Cell 26, 145–155. [DOI] [PMC free article] [PubMed]
- Lakhani, V.V., Ding, F., and Dokholyan, N.V. (2010). Polyglutamine induced misfolding of huntingtin exon1 is modulated by the flanking sequences. PLoS Comput. Biol. 6, e1000772. [DOI] [PMC free article] [PubMed]
- Lelouard, H., Gatti, E., Cappello, F., Gresser, O., Camosseto, V., and Pierre, P. (2002). Transient aggregation of ubiquitinated proteins during dendritic cell maturation. Nature 417, 177–182. [DOI] [PubMed]
- Lim, W.A., and Sauer, R.T. (1991). The role of internal packing interactions in determining the structure and stability of a protein. J. Mol. Biol. 219, 359–376. [DOI] [PubMed]
- López de la Paz, M., and Serrano, L. (2004). Sequence determinants of amyloid fibril formation. Proc. Natl. Acad. Sci. USA 101, 87–92. [DOI] [PMC free article] [PubMed]
- Man, O., and Pilpel, Y. (2007). Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species. Nat. Genet. 39, 415–421. [DOI] [PubMed]
- Matsuyama, A., Arai, R., Yashiroda, Y., Shirai, A., Kamata, A., Sekido, S., Kobayashi, Y., Hashimoto, A., Hamamoto, M., Hiraoka, Y., et al. (2006). ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 24, 841–847. [DOI] [PubMed]
- Mendel, D., Ellman, J.A., Chang, Z., Veenstra, D.L., Kollman, P.A., and Schultz, P.G. (1992). Probing protein stability with unnatural amino acids. Science 256, 1798–1802. [DOI] [PubMed]
- Monsellier, E., and Chiti, F. (2007). Prevention of amyloid-like aggregation as a driving force of protein evolution. EMBO Rep. 8, 737–742. [DOI] [PMC free article] [PubMed]
- Murray, A.N., Solomon, J.P., Wang, Y.J., Balch, W.E., and Kelly, J.W. (2010). Discovery and characterization of a mammalian amyloid disaggregation activity. Protein Sci. 19, 836–846. [DOI] [PMC free article] [PubMed]
- Nagalakshmi, U., Wang, Z., Waern, K., Shou, C., Raha, D., Gerstein, M., and Snyder, M. (2008). The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349. [DOI] [PMC free article] [PubMed]
- Narayanaswamy, R., Levy, M., Tsechansky, M., Stovall, G.M., O’Connell, J.D., Mirrielees, J., Ellington, A.D., and Marcotte, E.M. (2009). Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. USA 106, 10147–10152. [DOI] [PMC free article] [PubMed]
- Niwa, T., Ying, B.W., Saito, K., Jin, W., Takada, S., Ueda, T., and Taguchi, H. (2009). Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc. Natl. Acad. Sci. USA 106, 4201–4206. [DOI] [PMC free article] [PubMed]
- O’Connor, T.R., and Wyrick, J.J. (2007). ChromatinDB: a database of genome-wide histone modification patterns for Saccharomyces cerevisiae. Bioinformatics 23, 1828–1830. [DOI] [PubMed]
- Olzscha, H., Schermann, S.M., Woerner, A.C., Pinkert, S., Hecht, M.H., Tartaglia, G.G., Vendruscolo, M., Hayer-Hartl, M., Hartl, F.U., and Vabulas, R.M. (2011). Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144, 67–78. [DOI] [PubMed]
- Ozgur, S., Chekulaeva, M., and Stoecklin, G. (2010). Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol. Cell. Biol. 30, 4308–4323. [DOI] [PMC free article] [PubMed]
- Pastor, M.T., Esteras-Chopo, A., and López de la Paz, M. (2005). Design of model systems for amyloid formation: lessons for prediction and inhibition. Curr. Opin. Struct. Biol. 15, 57–63. [DOI] [PubMed]
- Patel, B.K., Gavin-Smyth, J., and Liebman, S.W. (2009). The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat. Cell Biol. 11, 344–349. [DOI] [PMC free article] [PubMed]
- Pawar, A.P., Dubay, K.F., Zurdo, J., Chiti, F., Vendruscolo, M., and Dobson, C.M. (2005). Prediction of “aggregation-prone” and “aggregation-susceptible” regions in proteins associated with neurodegenerative diseases. J. Mol. Biol. 350, 379–392. [DOI] [PubMed]
- Peña, M.I., Davlieva, M., Bennett, M.R., Olson, J.S., and Shamoo, Y. (2010). Evolutionary fates within a microbial population highlight an essential role for protein folding during natural selection. Mol. Syst. Biol. 6, 387. [DOI] [PMC free article] [PubMed]
- Richardson, J.S., and Richardson, D.C. (2002). Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. USA 99, 2754–2759. [DOI] [PMC free article] [PubMed]
- Rubinsztein, D.C. (2006). The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786. [DOI] [PubMed]
- Schwanhäusser, B., Busse, D., Li, N., Dittmar, G., Schuchhardt, J., Wolf, J., Chen, W., and Selbach, M. (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed]
- Schwartz, R., Istrail, S., and King, J. (2001). Frequencies of amino acid strings in globular protein sequences indicate suppression of blocks of consecutive hydrophobic residues. Protein Sci. 10, 1023–1031. [DOI] [PMC free article] [PubMed]
- Selkoe, D.J. (2003). Folding proteins in fatal ways. Nature 426, 900–904. [DOI] [PubMed]
- Shorter, J., and Lindquist, S. (2004). Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304, 1793–1797. [DOI] [PubMed]
- Spector, D.L. (2006). SnapShot: Cellular bodies. Cell 127, 1071. [DOI] [PubMed]
- Tartaglia, G.G., Pellarin, R., Cavalli, A., and Caflisch, A. (2005). Organism complexity anti-correlates with proteomic beta-aggregation propensity. Protein Sci. 14, 2735–2740. [DOI] [PMC free article] [PubMed]
- Tessier, P.M., and Lindquist, S. (2009). Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+]. Nat. Struct. Mol. Biol. 16, 598–605. [DOI] [PMC free article] [PubMed]
- Tuller, T., Kupiec, M., and Ruppin, E. (2009). Co-evolutionary networks of genes and cellular processes across fungal species. Genome Biol. 10, R48. [DOI] [PMC free article] [PubMed]
- Turoverov, K.K., Kuznetsova, I.M., and Uversky, V.N. (2010). The protein kingdom extended: ordered and intrinsically disordered proteins, their folding, supramolecular complex formation, and aggregation. Prog. Biophys. Mol. Biol. 102, 73–84. [DOI] [PMC free article] [PubMed]
- Uptain, S.M., and Lindquist, S. (2002). Prions as protein-based genetic elements. Annu. Rev. Microbiol. 56, 703–741. [DOI] [PubMed]
- Vavouri, T., Semple, J.I., Garcia-Verdugo, R., and Lehner, B. (2009). Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell 138, 198–208. [DOI] [PubMed]
- Vendruscolo, M., and Dobson, C.M. (2007). Chemical biology: More charges against aggregation. Nature 449, 555. [DOI] [PubMed]
- Vogel, C., Abreu, Rde.S., Ko, D., Le, S.Y., Shapiro, B.A., Burns, S.C., Sandhu, D., Boutz, D.R., Marcotte, E.M., and Penalva, L.O. (2010). Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol. Syst. Biol. 6, 400. [DOI] [PMC free article] [PubMed]
- von Mikecz, A. (2009). PolyQ fibrillation in the cell nucleus: who’s bad? Trends Cell Biol. 19, 685–691. [DOI] [PubMed]
- Wang, Y., Liu, C.L., Storey, J.D., Tibshirani, R.J., Herschlag, D., and Brown, P.O. (2002). Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA 99, 5860–5865. [DOI] [PMC free article] [PubMed]
- Williams, A., Jahreiss, L., Sarkar, S., Saiki, S., Menzies, F.M., Ravikumar, B., and Rubinsztein, D.C. (2006). Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr. Top. Dev. Biol. 76, 89–101. [DOI] [PubMed]
- Wiltzius, J.J., Landau, M., Nelson, R., Sawaya, M.R., Apostol, M.I., Goldschmidt, L., Soriaga, A.B., Cascio, D., Rajashankar, K., and Eisenberg, D. (2009). Molecular mechanisms for protein-encoded inheritance. Nat. Struct. Mol. Biol. 16, 973–978. [DOI] [PMC free article] [PubMed]
- Xia, Y., and Levitt, M. (2004). Simulating protein evolution in sequence and structure space. Curr. Opin. Struct. Biol. 14, 202–207. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.