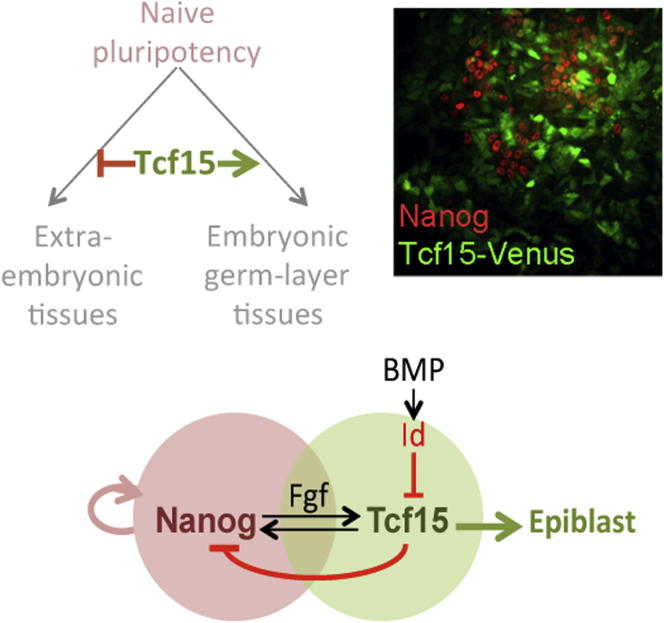
Summary
The events that prime pluripotent cells for differentiation are not well understood. Inhibitor of DNA binding/differentiation (Id) proteins, which are inhibitors of basic helix-loop-helix (bHLH) transcription factor activity, contribute to pluripotency by blocking sequential transitions toward differentiation. Using yeast-two-hybrid screens, we have identified Id-regulated transcription factors that are expressed in embryonic stem cells (ESCs). One of these, Tcf15, is also expressed in the embryonic day 4.5 embryo and is specifically associated with a novel subpopulation of primed ESCs. An Id-resistant form of Tcf15 rapidly downregulates Nanog and accelerates somatic lineage commitment. We propose that because Tcf15 can be held in an inactive state through Id activity, it may prime pluripotent cells for entry to somatic lineages upon downregulation of Id. We also find that Tcf15 expression is dependent on fibroblast growth factor (FGF) signaling, providing an explanation for how FGF can prime for differentiation without driving cells out of the pluripotent state.
Graphical Abstract
Highlights
► Tcf15 marks a subpopulation of pluripotent cells primed for somatic lineages ► Tcf15 expression is regulated by FGF signaling ► Tcf15 activity is repressed by Id proteins ► Tcf15 represses Nanog and drives differentiation once released from Id inhibition
Little is known about the transcription factors that oppose the pluripotency network to drive cells toward differentiation. Lowell and colleagues show that the basic helix-loop-helix (bHLH) transcription factor Tcf15 is expressed in the late preimplantation embryo and in a differentiation-primed subpopulation of embryonic stem cells and that an activated form of Tcf15 drives embryonic stem cells into a differentiation-primed state. These findings suggest a role for Tcf15 in manipulating and monitoring the earliest transition from pluripotency toward lineage commitment.
Introduction
Considerable progress has been made in establishing the factors that maintain pluripotency (Chambers and Smith, 2004). In contrast, little is known about the transcription factors that guide the transition from pluripotency to somatic lineage commitment. Pluripotent cells are maintained by a network of pluripotency factors that include Oct4, Sox2, Nanog, Klf4, and Esrrb. In the early blastocyst, fibroblast growth factor (FGF) 4 drives a subpopulation of cells toward a primitive endoderm fate (Nichols et al., 2009; Yamanaka et al., 2010). Cells that escape FGF action and retain high levels of Nanog go on to become restricted to an epiblast fate by around embryonic day 4.25 (E4.25) (Nichols and Smith, 2009; Yamanaka et al., 2010). Experiments using embryonic stem cells (ESCs) show that FGF signaling is required not only for primitive endoderm differentiation but also for competence to differentiate into somatic cell types (Kunath et al., 2007). FGF is necessary but not sufficient to drive lineage commitment: further progression to overt differentiation is restrained by the combination of leukemia inhibitory factor (LIF) and bone morphogenetic protein (BMP) signaling, both of which restrict cells from progressing to a postimplantation epiblast-like state (Ying et al., 2003).
The transcription factors that act downstream of FGF in order to drive epiblast cells toward this differentiation-primed state are not known. A clue to their identity comes from the finding that inhibitor of DNA binding/differentiation (Id) proteins are able to block the transition of ESCs to epiblast stem cells (EpiSC) (Zhang et al., 2010). Id proteins classically function through the inhibition of active basic helix-loop-helix (bHLH) transcription factors. We thus hypothesized that epiblast priming is driven by specific bHLH factors that are expressed in pluripotent cells but held in an inactive state through the action of Id proteins. As soon as Id proteins are downregulated, the bHLH activity of these “primed” cells would be released from inhibition, allowing epiblast maturation to proceed.
In other cell types, Id proteins act through either direct binding and inhibition of bHLH transcription factors or indirect inhibition of bHLH transcription factor function through binding and sequestration of their essential heterodimerization partners E proteins (including E47 and E12) (Norton, 2000). Thus, we set out to identify the targets of Id inhibition by determining the direct binding partners of both Id and E proteins in ESCs. To achieve this, we performed a series of yeast two-hybrid (Y2H) screens for binding partners of Id1, E47, and E12 within a library generated from the messenger RNA (mRNA) of pluripotent mouse ESCs. This revealed three Id-regulated bHLH factors that are expressed in ESCs, of which one, Tcf15, is also expressed in the inner cell mass of the E4.5 embryo.
Despite a known function in controlling somite development (Burgess et al., 1996), a role for Tcf15 at this earlier development stage has been unknown. Here, we demonstrate a distinct wave of Tcf15 expression in the late preimplantation embryo in vivo and a transient spike of expression during the early stages of ESC differentiation in vitro. We show that an Id-resistant form of Tcf15 rapidly downregulates Nanog and accelerates the transition of ESCs through the epiblast state while suppressing primitive endoderm differentiation.
Efforts to understand the balance between pluripotency and lineage commitment have been hampered by the lack of a marker that can be used to monitor exit from the pluripotent state toward somatic lineages. Tcf15 acts as a marker of this transition state: it is rapidly upregulated as ESCs transit from a naive to a primed state, and is associated with a subpopulation of epiblast-primed Oct4+ Nanog/Klf4-low cells. Transcription of Tcf15 is driven by FGF signaling, whereas its activity is suppressed by Id proteins, which are direct targets of BMP signaling (Nakashima et al., 2001; Ying et al., 2003; Wilson-Rawls et al., 2004); this helps explain how these extrinsic signals allow pluripotent cells to become primed for, but restrained from, somatic differentiation.
Results
Identification of Id Protein Targets in ESCs through Y2H Screening of an ES-Cell cDNA Library
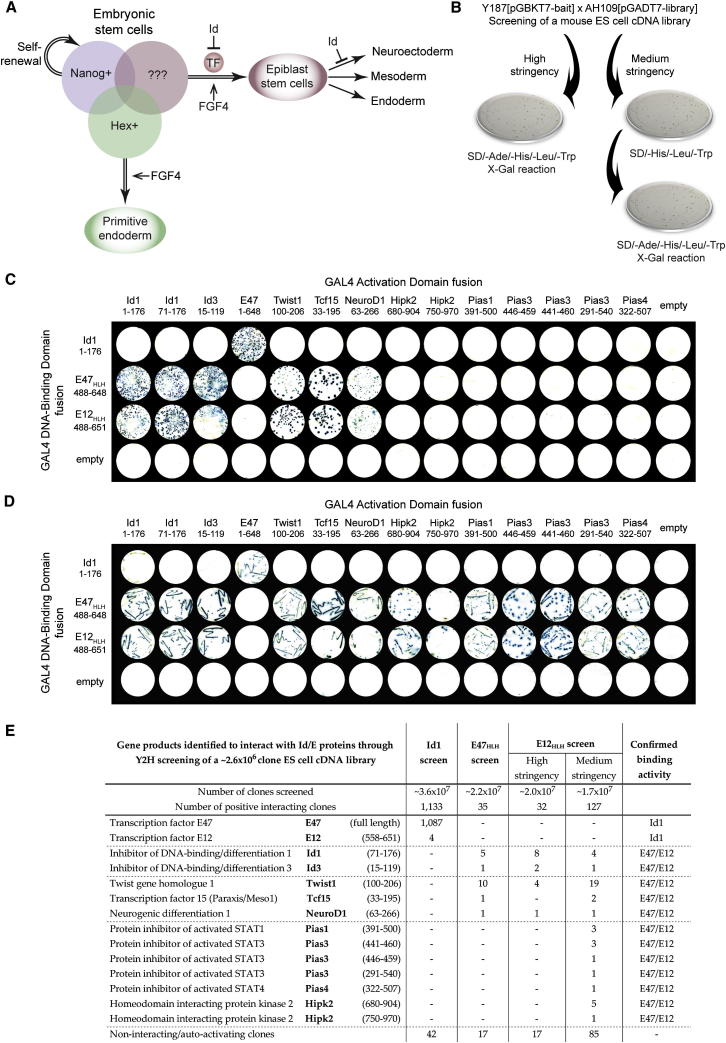
Id1 is expressed in ESCs and can block the transition of ESCs to differentiation-primed epiblast (Ying et al., 2003; Pollard et al., 2006; Zhang et al., 2010). However, the transcription factor targets of Id, and thus the mechanisms by which Id controls these early transitions, remain unknown (Figure 1A). We used a Y2H screening strategy to identify the direct binding partners of Id and E proteins in ESCs. We constructed a random primed complimentary DNA (cDNA) library of ∼2.6 × 106 independent clones from the mRNA extracted from a homogenous monolayer culture of self-renewing ESCs (maintained in media containing LIF and fetal calf serum [FCS]). Y2H screens of this library were performed for Id1, E47, and E12 baits using the highly stringent Matchmaker (Clontech) system in which positive interactions are dependent on the activation of three independent reporter genes (Figure 1B).
Figure 1.
A Y2H Screen to Identify Transcription Factors Regulated by Id1 in ES Cells
(A) FGF signaling primes pluripotent cells for differentiation. Id1 inhibits subsequent transitions: first to epiblast and then to neuroepithelium. We hypothesize that unknown Id-regulated transcription factors are expressed in epiblast-primed cells.
(B) A Y2H library screen was performed by mating the Y187 yeast strain carrying the pGBKT7-bait vector (Id1, E47HLH, or E12HLH) with the AH109 yeast strain carrying the pGADT7-library vector (mouse ESC cDNA library). For high-stringency selection, positive interactors were determined by growth of mated yeast on SD/-Ade/-His/-Leu/-Trp media and positive X-Gal reaction (indicating activation of ADE2, HIS1 and LacZ reporter genes). For medium-stringency selection, positive interactions were determined by initial growth of mated yeast on SD/-His/-Leu/-Trp media (indicating activation of HIS1 reporter gene), with subsequent growth on SD/-Ade/-His/-Leu/-Trp media and positive X-Gal reaction (indicating activation of ADE2, HIS1 and LacZ reporter genes).
(C) High-stringency Y2H analysis of interactions among Id1, E47HLH, and E12HLH, and genes identified through library screens; positive interactors are determined by growth on SD/-Ade/-His/-Leu/-Trp media and positive X-Gal reaction.
(D) Medium-stringency Y2H analysis of interactions among Id1, E47HLH, and E12HLH, and genes identified through library screens; positive interactions are determined by initial growth on SD/-His/-Leu/-Trp media with subsequent growth on SD/-Ade/-His/-Leu/-Trp media and positive X-Gal reaction.
(E) The genes identified as pGADT7 vector inserts that interact with Id1, E47HLH, or E12HLH at the protein level within yeast are shown along with the number of times clones were identified in each screen, and whether each gene was confirmed as a true binding partner of Id1 or E47/E12 by subsequent analysis. The total numbers of noninteracting and autoactivating inserts identified are also shown.
Identification of E47/E12 as Binding Partners of Id1
We first screened for Id1 binding partners within our ES-cell cDNA library. In total, ∼3.6 × 107 clones were screened. Of 1,133 positively interacting clones, the vast majority (1,091) were found to correspond to coding sequences of the E2A gene products E47 and E12. All putative Id1-interacting partners were tested through a series of controlled Y2H reactions. E47 and E12 exhibited strong and specific interactions with Id1, whereas all other candidates were ruled out owing to nonspecific positive interactions or inability to bind Id1 (Figures 1C and 1E). We conclude that the only true binding partners identified are E47/E12. Id1 is therefore likely to function in ESCs through E protein sequestration, with downstream bHLH targets constituting direct binding partners of E47/E12.
Identification of bHLH Factors Tcf15, Twist1, and NeuroD1 as Binding Partners of E47/E12
We next screened for binding partners of E47 and E12; having determined that the full-length proteins are toxic in yeast, screens were performed using the bHLH domains of these proteins (E47488-648 and E12488-651). Of ∼2.2 × 107 and ∼2.0 × 107 clones tested, 35 and 32 positive interacting clones were detected as putative binding partners of E47 and E12, respectively. Upon further testing, five factors were confirmed as true interacting partners; the remainder were excluded owing to nonspecific interactions or inability to bind E47/E12 (Figures 1C and 1E). Of the five true interacting partners, two correspond to Id1 and Id3, confirming the results of the Id1 screen; the remaining three correspond to bHLH transcription factors Tcf15, Twist1, and NeuroD1. These bHLH factors have recognized functions in other cell types at later developmental stages and are known targets of Id protein inhibition (Naya et al., 1995; Wilson-Rawls et al., 2004; Connerney et al., 2006) but have no previously identified functions in ESCs or in the preimplantation embryo.
To ensure we had not missed any important binding partners, we also performed an E12488-651 library screen using a reduced-stringency method that is designed to favor the detection of lower-affinity interactions (Figure 1B). Of ∼1.7 × 107 clones screened, 127 positive interacting clones were detected. In addition to Id1, Id3, Tcf15, Twist1, and NeuroD1, we also confirmed protein inhibitor of activated STAT factors Pias1, Pias2, and Pias4, and homeodomain-interacting protein kinase Hipk2, as true but weaker binding partners of E47 and E12 (Figures 1D and 1E). Pias1 has already been reported as an E12 binding partner (Kawai-Kowase et al., 2005), further validating our screening strategy. However, no additional transcription factors were identified from this lower-strategy screen. Our Y2H studies therefore led to the identification of three bHLH transcription factors as candidates for mediating the effect of Id in pluripotent cells: Tcf15, Twist1, and NeuroD1.
Tcf15 Is Expressed in Pluripotent Populations
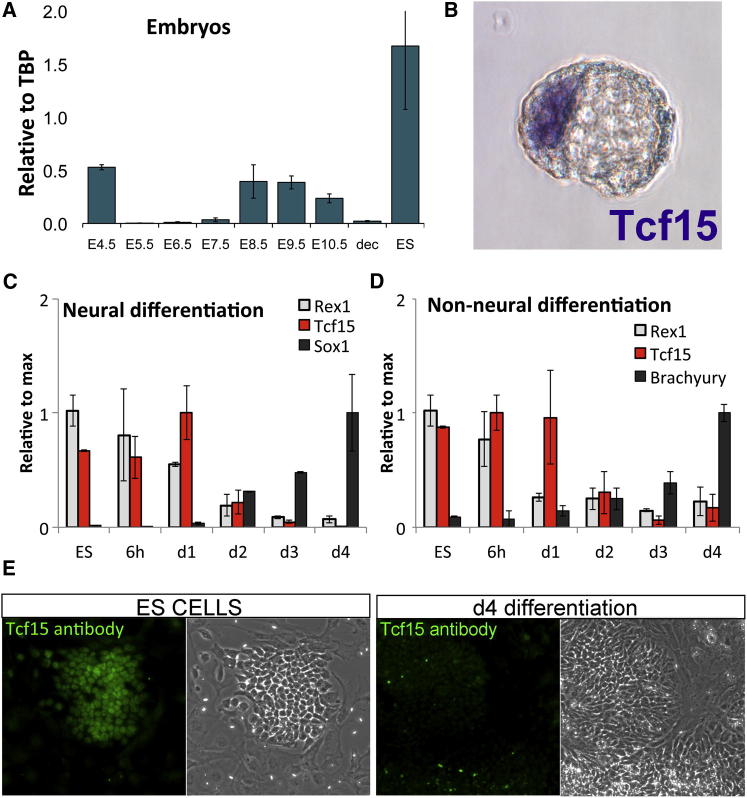
We next checked expression of the three candidates in preimplantation embryos. NeuroD1 expression was not detected in the E4.5 blastocyst, and although we did detect Twist1 expression, it was at significantly lower levels than in ESCs. In contrast, Tcf15 expression was readily detected in the E4.5 blastocyst at levels comparable to those found in ESCs (Figure 2A). Expression dramatically declines postimplantation before rising again at E8.5, in keeping with the previously reported expression of Tcf15 in the developing somites (Burgess et al., 1995) (Figure 2A). Expression of Tcf15 prior to E7.5 has not been previously reported in the literature, although the early wave of Tcf15 expression in the blastocyst is apparent in the Unigene EST expression database. In situ hybridization confirmed that Tcf15 is expressed in the E4.5 blastocyst and revealed that it is specifically detected in the inner cell mass (Figure 2B).
Figure 2.
Tcf15 Is Expressed in Pluripotent Cells
(A) qPCR analysis of Tcf15 expression in embryos at different developmental ages. dec, decidual tissue; ES, embryonic stem cells.
(B) In situ hybridization of an E4.5 mouse embryo with a Tcf15 probe.
(C) qPCR analysis of ESCs undergoing differentiation into the neural lineage in serum-free media.
(D) qPCR analysis of ESCs undergoing differentiation into a mixture of predominantly nonneural lineages in the presence of FCS and the absence of LIF.
(E) Antibody staining for Tcf15 in ESCs cultures containing a mixture of ESCs and spontaneously differentiating cells or after 4 days differentiation in serum-free media.
All data are represented as mean ±SD.
Upon directed differentiation of ESCs to either neural or nonneural lineages, high Tcf15 expression persists after the marker of naive pluripotency Rex1 is downregulated (Figures 2C and 2D). Tcf15 is downregulated at around the time that cells commit to differentiation, as measured by the upregulation of overt neural marker Sox1 (Wood and Episkopou, 1999) (Figure 2C). Tcf15 protein is readily detectable within the nucleus of ESCs but not in spontaneously differentiating cells within the same culture or in cells that have been cultured for 4 days under differentiation conditions (Figure 2E).
Tcf15 Marks a Population of Primed ESCs
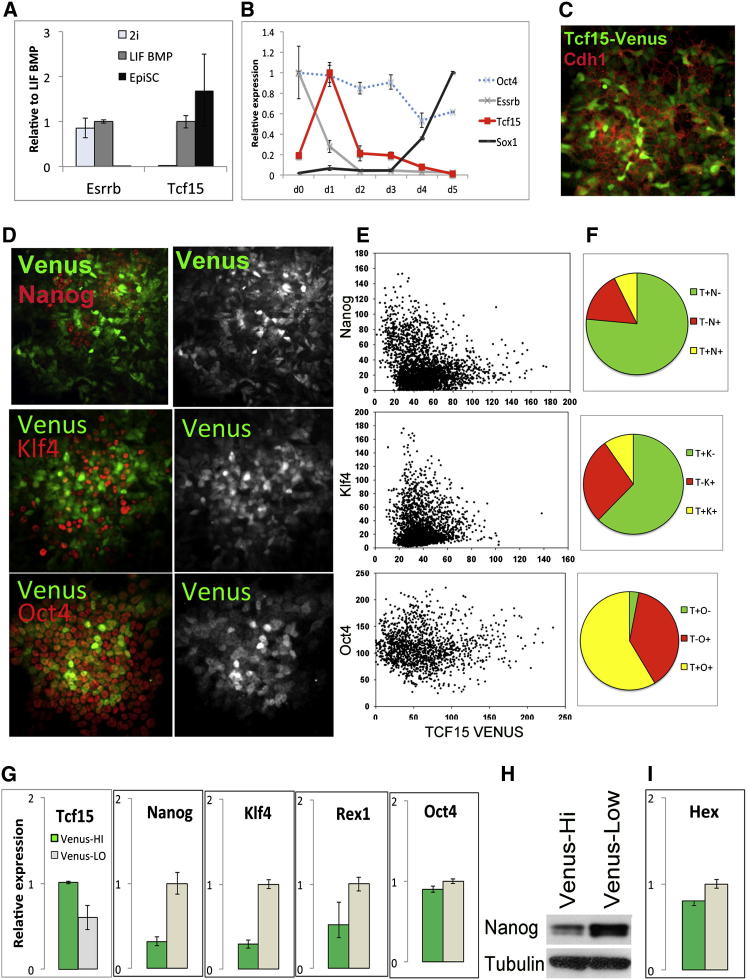
Pluripotent cells can exist in a “naive” state, possibly representing cells from the early blastocyst or in a “differentiation primed state” that may correspond to cells within the mature blastocyst or the early postimplantation epiblast (Nichols and Smith, 2009). Single-cell quantitative PCR (qPCR) analysis has revealed that Tcf15 is upregulated as ESCs are derived from E3.5 blastocysts and that Tcf15 is predominantly associated with the Gata6-negative subpopulation during the derivation process (Tang et al., 2010). This suggests that Tcf15 may become specifically upregulated in cells that are maturing into epiblast-primed ESCs but not into primitive endoderm. Pharmacological inhibitors of Erk and GSK3b can be used to maintain ESCs in a naive state (Ying et al., 2008), whereas culture in FGF2 plus activin drives them into differentiation-primed EpiSC (Brons et al., 2007; Tesar et al., 2007). Microarray analysis of the transition of naive ESCs to EpiSC indicates that Tcf15 expression increases during the early transition period (Hayashi et al., 2011). Similarly, we find a strong increase in Tcf15 expression as naive ESCs move into a primed ES-cell state (Figure 3A) and an early transient pulse of Tcf15 expression as naive ESCs move toward neural differentiation (Figure 3B).
Figure 3.
Tcf15 Marks a Subpopulation of Differentiation-Primed ES Cells
(A) qPCR analysis of puro-selected Oct4GIP cells after at least 1 week of culture in 2i, LIF + BMP, or EpiSC conditions.
(B) qPCR analysis of ESCs undergoing neural differentiation starting from 2i conditions.
(C) Antibody staining for E-cadherin (Cdh1) in a population of Tcf15-Venus reporter ESCs in LIF + FCS.
(D) Tcf15-gtxIRES-Venus reporter cells stained for Nanog, Klf4, or Oct4.
(E) Quantitative immunofluorescence analysis of Tcf15-gtxIRES-Venus reporter cells stained for Nanog, Klf4, or Oct4.
(F) Proportion of cells positive for Tcf15-Venus only (green) or Nanog/Klf4/Oct4 (red) or both.
(G) qPCR analysis of subpopulations of ESCs gated for PECAM+ and sorted on the basis of Tcf15-Venus expression.
(H) Western blot for Nanog in subpopulations of ESCs gated for PECAM+ and sorted on the basis of Tcf15-Venus expression.
(I) qPCR analysis of subpopulations of ESCs gated for PECAM+ and sorted on the basis of Tcf15-Venus expression.
All data are represented as mean ±SD. See also Figure S1.
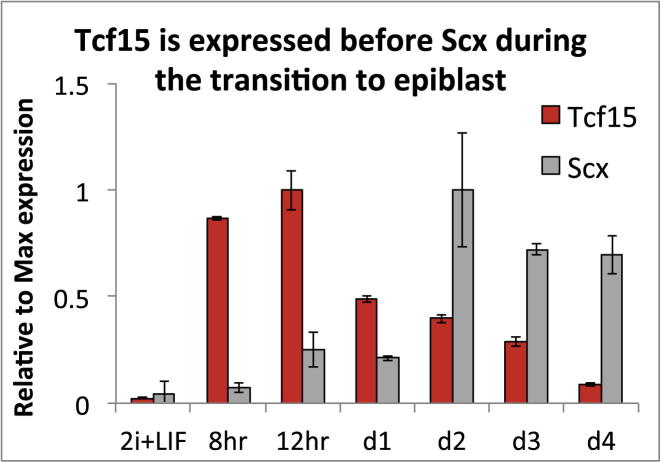
Tcf15 shares an almost identical DNA binding domain with the closely related bHLH transcription factor Scleraxis (Scx) (Burgess et al., 1995). Although Scx did not emerge from our screen (Figure 1), we considered the possibility that it could act as an early prodifferentiation factor in ESCs. We found, however, that Scx is not consistently detectable by qPCR in ESCs maintained in LIF + FCS (data not shown) although it does become upregulated in EpiSC (Figure S1), in keeping with its reported expression in the postimplantation epiblast (Brown et al., 1999). Because Scx is not expressed in ESCs, we eliminated it as a candidate determinant of early primed pluripotent state.
Figure S1.
Tcf15 Is Expressed Earlier than Scleraxis during the Transition to Epiblast, Related to Figure 3
qPCR analysis for Tcf15 (red) and Scx (gray) during the transition from naive pluripotency (2i+LIF) toward epiblast (FGF+Activin). All data are represented as mean ±SD.
ESCs cultured in LIF + FCS are heterogeneous mixtures of primed cells and naive cells (Nichols and Smith, 2009), with naive cells marked by Klf4 and Nanog. In order to find out whether Tcf15 is associated with the primed state in these mixed cultures, we made use of a sensitive reporting strategy for Tcf15 based on translational amplification of a fluorescent protein. The 216D1 Tcf15 reporter cell line was generated by gene trapping: a gtx-IRES-Venus cassette was incorporated into the 3′ UTR of Tcf15 (Tanaka et al., 2008). Venus shows a heterogeneous distribution within ESC cultures (Figure 3C) and becomes undetectable in the majority of cells after differentiation, in keeping with the downregulation of Tcf15 upon differentiation (Figures 2C and 2D).
We plated Tcf15-Venus reporter cells at clonal density and allowed colonies to form over 5 days. Many colonies were morphologically undifferentiated yet were mosaic for Venus expression, supporting the idea that Tcf15 may be associated with a dynamic subpopulation of ESCs (Figure 3D).
Both the Venus-high subpopulation and the Venus-low subpopulations within these colonies of ESCs express Oct4, whereas Nanog and Klf4 were predominantly expressed only within the Venus-low subpopulation (Figure 3D). We used quantitative immunofluorescence in order to further examine the relationship between Tcf15-Venus and Nanog, Klf4, or Oct4 in ESCs (Figures 3E and 3F). This confirmed that less than 15% of Venus-high cells express Nanog or Klf4 whereas over 95% express Oct4 (Figure 3F). This indicates that Tcf15 marks an undifferentiated but differentiation-primed state.
We confirmed that Venus correlates with Tcf15 mRNA in subpopulations of Venus-high and Venus-low ESCs sorted by flow cytometry (Figure 3G). In keeping with our quantitative immunofluorescence analysis, we found transcripts for naive pluripotency markers to be enriched in the Venus-low subpopulation of ESCs (Figure 3G). Nanog protein was also enriched in the Venus-low subpopulation (Figure 3H). These data suggest that Tcf15 marks an undifferentiated but differentiation-primed state.
Hex has been reported to mark a subpopulation of ESCs that are primed for primitive endoderm (Canham et al., 2010); however, microarray analysis of Hex-high and Hex-low subpopulations indicates that Tcf15 is not specifically associated with the Hex-high population (Canham et al., 2010). Furthermore, we find that Hex is not associated with the Tcf15-high population (Figure 3I). Taken together, these data suggest that Tcf15 is associated with a differentiation-primed state that differs from the previously described primitive endoderm-primed subpopulation.
Tcf15 Accelerates Differentiation
We next tested whether Tcf15 has prodifferentiation activity in ESCs. We engineered a form of Tcf15 that is tethered by a flexible linker to its heterodimerization partner E47 (Figure S2A). This strategy renders Tcf15 more resistant to inhibition by Id (Neuhold and Wold, 1993) and also avoids disrupting the balance of endogenous bHLH factors in the cell. This construct was placed under the control of a tetracycline (tet)-responsive promoter and integrated into a cell line that constitutively expresses a reverse tetracycline-controlled transactivator (rtTA) from the Rosa26 (R26) locus (Figure S2B). This allowed Tcf15 to be artificially maintained under differentiation conditions at levels similar to those in ESCs (Figure S3C), with expression being lost within 24 hr of removing doxycycline (dox) (Figure S3D). Using this system, we find that Tcf15 accelerates neural differentiation: Tuj1+ neurites were abundant by the third day in dox-treated but not control cultures (Figure 4A). Tcf15-E47 is effective at accelerating differentiation even if expressed only during the first 24 hr of differentiation (Figures 4A and 4B). This suggests that Tcf15 activity may be a limiting factor at an early stage of neural induction.
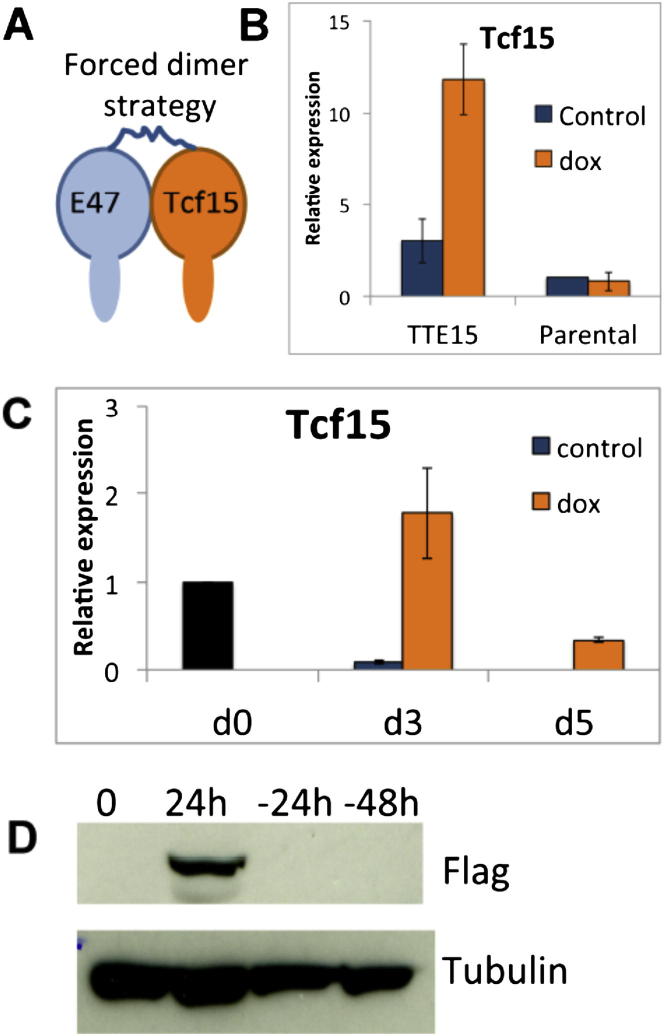
Figure S2.
Validation of Dox-Inducible Cell Lines, Related to Figure 4
(A) Forced dimer strategy: Tcf15 is tethered to E47 through a flexible linker.
(B) qPCR analysis for Tcf15 in dox-inducible Tcf15-E47 ESCs or parental control ESCs: 48h treatment with dox in LIF+FCS conditions.
(C) qPCR analysis for Tcf15 in dox-inducible Tcf15-E47 cells during neural differentiation.
(D) Western blot to detect flag-tagged Tcf15 in dox-inducible Tcf15-E47 ESCs after 24h induction with dox or 24 or 48 hr after removal of dox.
All data are represented as mean +/− standard deviation.
Figure S3.
Tcf15 Accelerates Non-neural Differentiation, Related to Figure 5
qPCR analysis of dox- inducible Tcf15-E47 ESCs undergoing differentiation in FCS in the absence of LIF. All data are represented as mean +/− standard deviation.
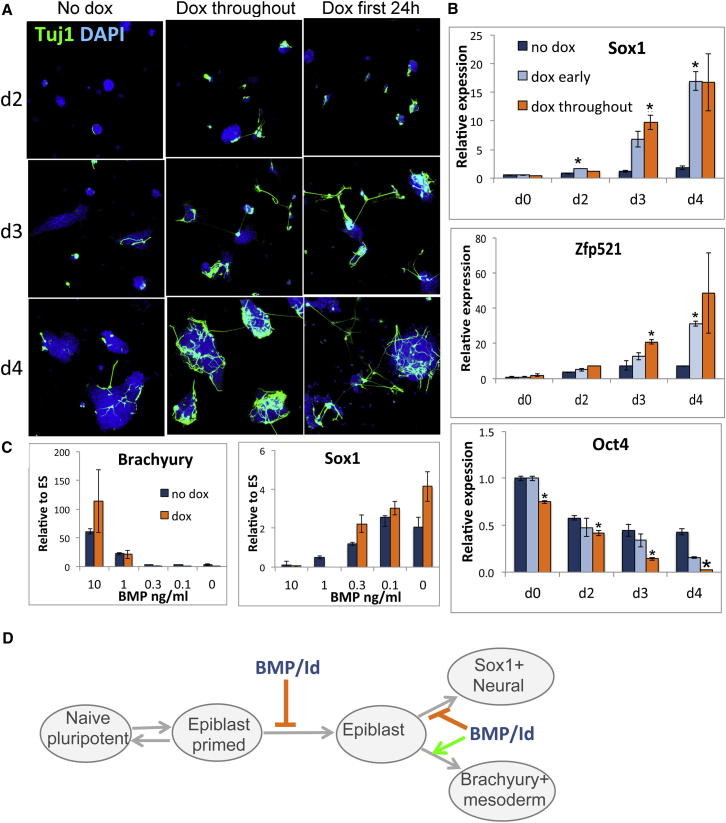
Figure 4.
Tcf15 Accelerates Differentiation
(A) Tuj1 staining to detect neurons during neural differentiation of dox-inducible Tcf15-E47 ESCs.
(B) qPCR analysis of dox-inducible Tcf15-E47 ESCs undergoing neural differentiation.
(C) qPCR analysis of dox-inducible Tcf15-E47 ESCs after 4 days in N2B27 in the presence of various doses of BMP4.
(D) BMP/Id suppresses the transition of ESCs to EpiSC and then the transition of EpiSC toward neural progenitors.
All data are represented as mean ±SD. See also Figure S2.
The ability of Tcf15 to accelerate differentiation was not restricted to the neural lineage. LIF withdrawal in the presence of FCS causes ESCs to differentiate into a mixture of different nonneural cell types; under this protocol, differentiation was also accelerated (Figure S3) without promoting neural differentiation (data not shown). Furthermore, somatic lineage choice did not appear to be biased significantly by Tcf15 as measured by the dose-dependent ability of BMP to suppress neural differentiation (marked by Sox1) and instead promote nonneural differentiation (marked by Brachyury) (Figure 4C). Tcf15 therefore does not appear to mediate the ability of BMP/Id to suppress neural induction (Ying et al., 2003) but may mediate the earlier role of Id to inhibit the transition of ESCs to differentiation-primed epiblast (Zhang et al., 2010) (see these two transitions illustrated in Figure 4D).
Tcf15 Drives the Transition to Epiblast and Suppresses Primitive Endoderm Differentiation
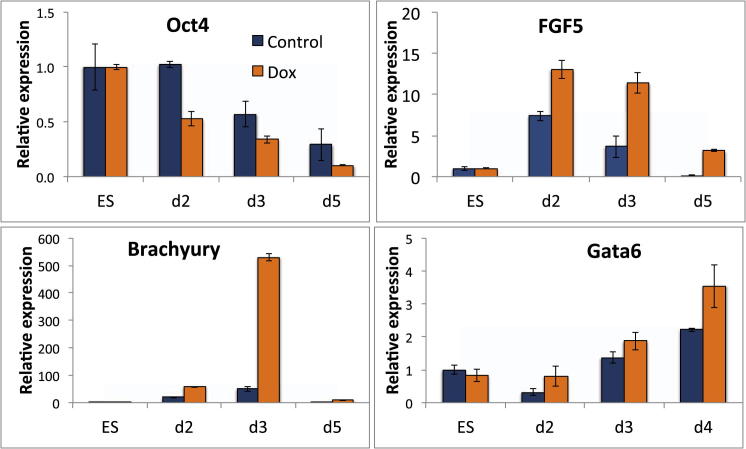
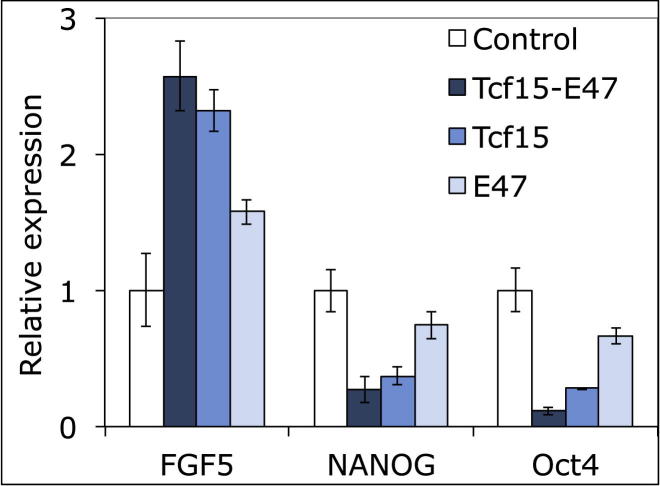
ESCs cultured in LIF + FCS or LIF + BMP are restrained from progressing to overt differentiation but free to explore naive and primed states of pluripotency (Nichols and Smith, 2009). We used these conditions to ask whether Tcf15-E47 can drive the transition to a primed state. Under these conditions, Tcf15-E47 is able to downregulate markers of naive ESCs and upregulate the epiblast marker Fgf5 (Figure 5A). A 24 hr pulse of dox drives irreversible commitment to differentiation in a significant proportion of cells even in the continued presence of LIF + BMP (Figure 4B). These cells were morphologically similar to cells from which LIF has been withdrawn. This suggests that Tcf15-E47 may drive the loss of LIF responsiveness, which is a characteristic feature of the transition toward postimplantation EpiSC (Brons et al., 2007; Tesar et al., 2007). We confirmed that Tcf15 alone, but not E47 alone, recapitulates the ability of Tcf15-E47 to favor differentiation (Figure S4), in keeping with reports that E47 cannot act as a transcription factor in its own right in most cell types, with the single exception of B cells. (Benezra, 1994; Shen and Kadesch, 1995; Sloan et al., 1996; Markus and Benezra, 1999).
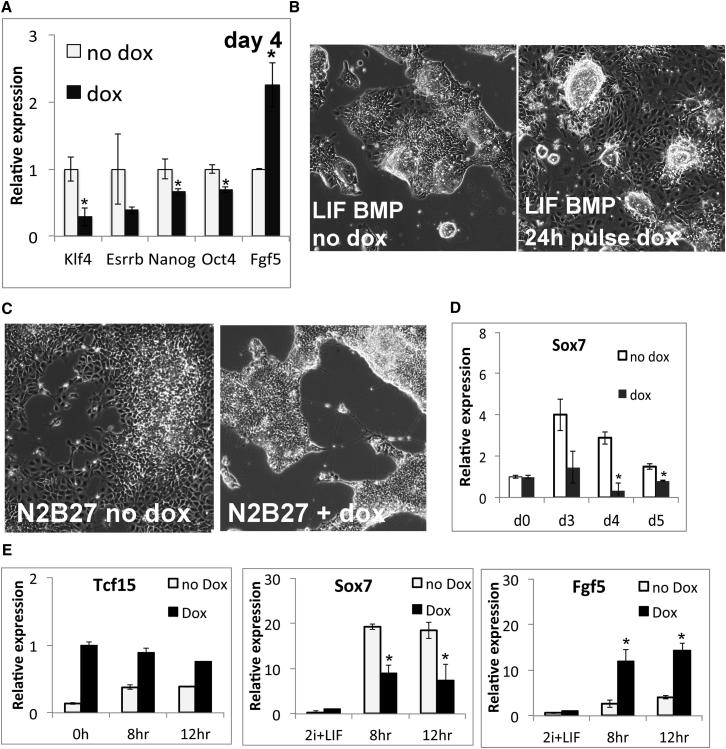
Figure 5.
Tcf15 Drives the Transition to Primed Epiblast and Suppresses Primitive Endoderm Differentiation
(A) qPCR analysis of dox-inducible Tcf15-E47 ESCs after 4 days in N2B27 + LIF + BMP4.
(B) Dox-inducible Tcf15-E47 ESCs after 4 days in N2B27 + LIF + BMP4. Second panel shows cells that received a 24 hr pulse of dox during the first day.
(C) Dox-inducible Tcf15-E47 ESCs after 4 days in N2B27. Treatment with dox eliminates spontaneous differentiation into primitive-endoderm-like cells.
(D) qPCR analysis of dox-inducible Tcf15-E47 cells during differentiation in N2B27.
(E) qPCR analysis of dox-inducible Tcf15-E47 cells after transfer to EpiSC conditions.
All data are represented as mean ±SD. See also Figures S3 and S4.
Figure S4.
Tcf15 but Not E47 Can Recapitulate the Pro-differentiation Activity of Tcf15-E47, Related to Figure 5
qPCR analysis of wild-type ESCs following transient transfection with plasmids encoding Tcf15-E47, Tcf15 monomer, E47 monomer, or empty vector control. Cells were cultured for four days in LIF+BMP in the absence of selection before qPCR analysis. All data are represented as mean +/− standard deviation.
We next asked whether Tcf15-E47 primes ESCs for differentiation into extraembryonic endoderm as well as into epiblast-derived somatic cell types. Upon removal of LIF and BMP, a subpopulation of ESCs spontaneously differentiate into primitive endoderm as determined both by their characteristic morphology and upregulation of the extraembryonic endoderm marker Sox7: both indicators of primitive endoderm differentiation were almost completely eliminated upon induction of Tcf15-E47 (Figures 5C and 5D). We confirmed that Tcf15-E47 accelerates upregulation of epiblast marker FGF5 and suppresses expression of primitive endoderm marker Sox7 when ESCs are placed in N2B27 + basic FGF + activin, conditions that favor conversion of ESCs into EpiSC (Tesar et al., 2007).
We conclude that Tcf15 primes pluripotent cells for the transition to mature differentiation-primed epiblast but not for differentiation into extraembryonic endoderm. Consistent with this idea, Id1 gene expression is high in the primitive endoderm (Canham et al., 2010), suggesting that Tcf15 activity is usually suppressed in this cell type.
Tcf15 Expression Is Dependent on FGF Signaling
Tcf15 is able to prime cells for differentiation by driving the transition to a mature epiblast-like state; because Tcf15 is an Id-regulated factor, this explains why Id is able to suppress the conversion of ESCs into EpiSC (Zhang et al., 2010). We show above that Tcf15 is expressed specifically in cells that are primed for differentiation. We next investigated the mechanism by which Tcf15 expression becomes associated with this primed state.
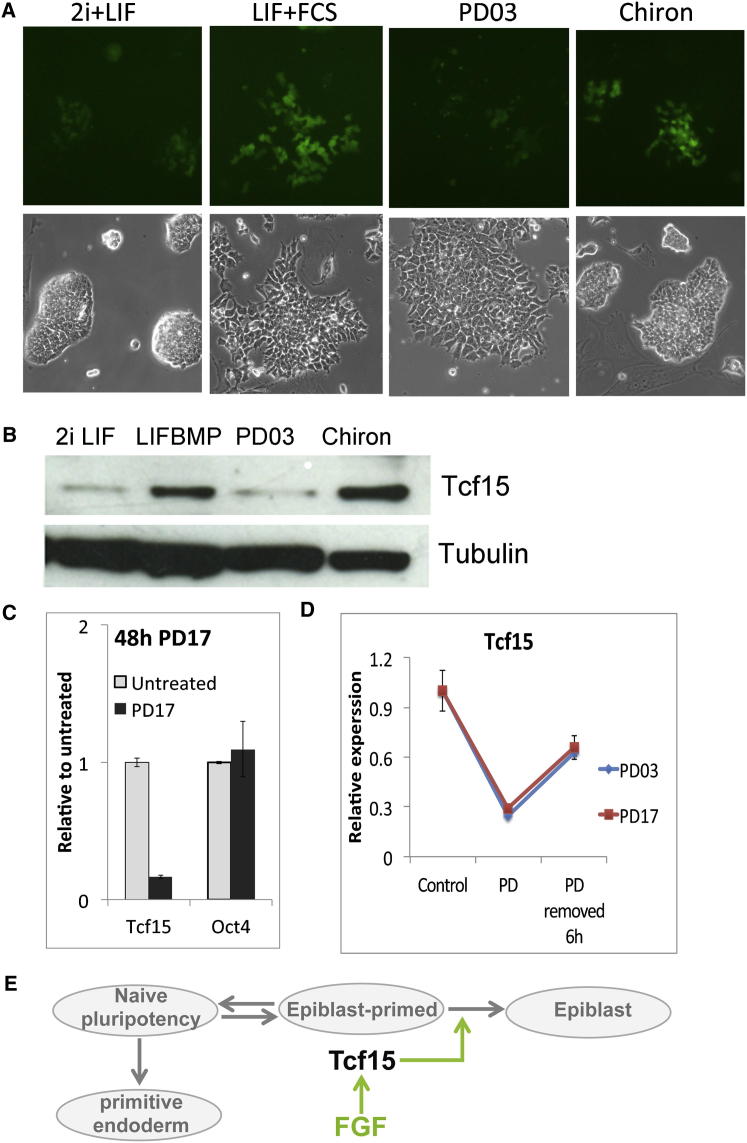
We used Tcf15-Venus reporter cells to investigate which of the components of 2i culture is responsible for suppressing Tcf15 expression. We first confirmed that Venus expression is undetectable in 2i conditions and expressed in a subset of cells in LIF + FCS (Figure 6A). Transfer to basal media (N2B27) or basal media with Chiron alone did not have any obvious impact on Venus expression, but PD0325901 reduced Venus to almost undetectable levels (Figure 6A). We conclude that Erk signaling is required for maintaining Tcf15 expression in ESCs. We confirmed that endogenous Tcf15 protein was also lost when cells were cultured in the presence of PD0325901 but maintained when cells were cultured in the presence of Chiron (Figure 6B). Fibroblast growth factor receptor (FGFR) inhibitor PD17 was also able to suppress Tcf15 expression (Figure 6C). The response of Tcf15 to FGF is rapid: Tcf15 is strongly upregulated within 6 hr of removing FGFR or Erk inhibitors. We conclude that Tcf15 expression is dependent on autoinductive FGF signaling and that it responds rapidly to increases in FGF activity (Figure 6D). FGF could therefore prime cells for differentiation by upregulating the prodifferentiation factor Tcf15 (Figure 6E), whereas BMP/Id may restrain progression to overt differentiation from this primed state by inhibiting Tcf15 activity.
Figure 6.
Tcf15 Is Regulated by FGF Signaling
(A) Tcf15-Venus cells cultured for 6 days in N2B27 under the stated conditions.
(B) Western blot analysis of E14tg2a cells cultured for 6 days in N2B27 under the stated conditions.
(C) qPCR analysis of E14tg2a cells treated for 48 hr with the FGFR inhibitor PD17.
(D) qPCR analysis of E14tg2a cells in N2B27 + LIF + BMP treated for 48 hr with the FGFR inhibitor PD17 or the ERK inhibitor PD0325901 or 6 hr after washout of inhibitors.
(E) FGF upregulates Tcf15 expression and drives the transition to epiblast.
All data are represented as mean ±SD.
Tcf15 Downregulates the Pluripotency Determinant Nanog and Upregulates the Epiblast Determinant Otx2
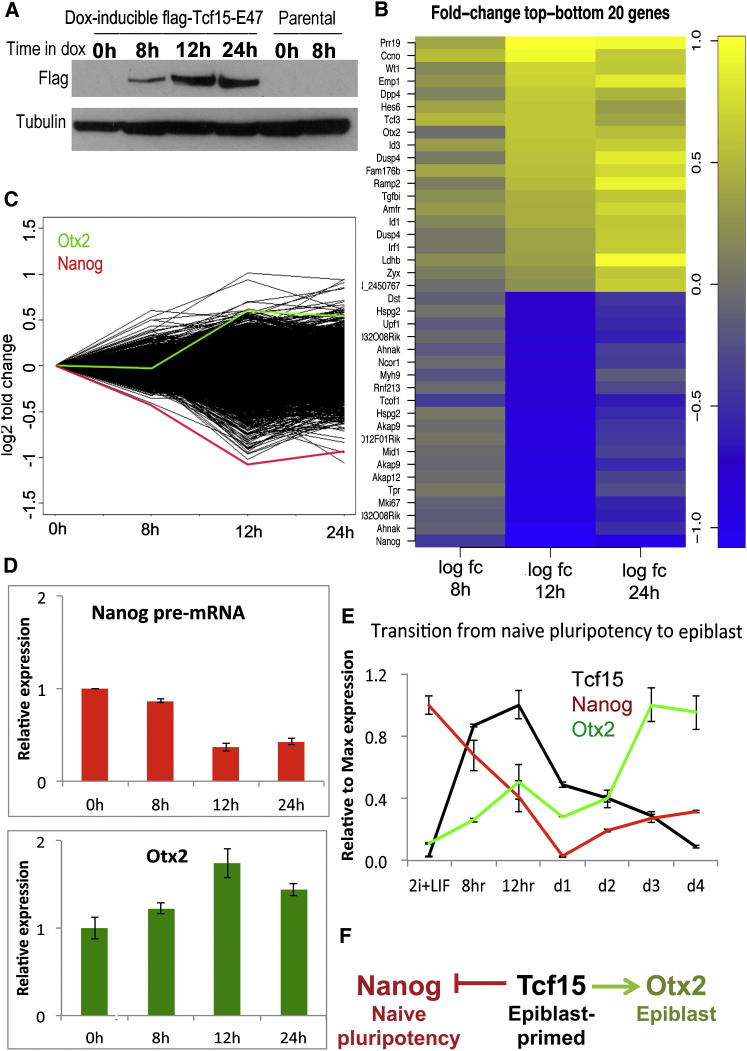
We next set out to ask how Tcf15 drives pluripotent cells away from naive pluripotency and toward epiblast. We carried out a time-course microarray analysis aimed at capturing the earliest changes in gene expression in response to Tcf15. Using our dox-inducible Tcf15-E47 ESC lines, we found that Tcf15-E47 protein becomes weakly detectable at around 8 hr after addition of dox and reaches robust levels after 12 hr (Figure 7A). A total of only 61 genes changed significantly over these two early time points. Of these, the most strongly downregulated gene at both time points is Nanog (Figures 7B–7D). This further validates our hypothesis that Tcf15 activation represents the first step toward differentiation and suggests a mechanism by which it drives cells away from a naive pluripotent state. Furthermore, the epiblast-determinant Otx2 (Acampora et al., 2012) was one of the top-ten most significantly upregulated genes at early time points (Figures 7B–7D), explaining how Tcf15 specifically favors epiblast rather than primitive endoderm differentiation after loss of Nanog. We confirmed that a burst of Tcf15 expression accompanies the loss of Nanog and precedes the upregulation of Otx2 during the transition of ESCs to EpiSC (Figure 7E).
Figure 7.
Tcf15 Downregulates Nanog and Upregulates Otx2
(A) Western blot analysis of flag-Tcf15-E47 or parental cells in ESCs at various time points after addition of dox.
(B) Genes that respond most strongly to Tcf15-E47 after 12 hr dox treatment. Note that Tcf3 refers to the E2A gene and detects the E47 transgene in this experiment.
(C) Fold change in all genes within the microarray probe set. Nanog is highlighted in red and Otx2 is highlighted in green.
(D) qPCR validation showing downregulation of Nanog pre-mRNA and Otx2 mRNA in response to dox-mediated induction of Tcf15-E47 in ESCs.
(E) qPCR analysis showing the kinetics of Tcf15, Nanog, and Otx2 expression during the transition toward EpiSC.
(F) Model. We propose that Tcf15 suppresses Nanog and activates Otx2 to drive pluripotent cells away from naive pluripotency and toward epiblast.
All data are represented as mean ±SD.
These data support a model in which Tcf15 drives ESCs out of a naive pluripotent state by suppressing expression of Nanog and toward a mature epiblast state by upregulating Otx2 (Figure 7F).
Discussion
Pluripotent cells navigate a route toward lineage commitment through a complex network of positive and negative cues. The negative cues that restrain cells in a pluripotent state include transcription factors Nanog and Klf4 and signaling pathways activated by BMP and LIF. In contrast, little is known about the positive cues that drive progression toward lineage commitment. The transition from naive epiblast to mature differentiation-primed epiblast spans the peri-implantation period and is therefore technically challenging to study in vivo. Conversion of ESCs to EpiSC in culture recapitulates the progression from naive preimplantation epiblast to epithelialized egg cylinder in vivo (Rossant and Tam, 2009) and provides a useful tool for interrogating this process. This system has identified Id1 as a key negative regulator of the transition of ESCs to EpiSC (Zhang et al., 2010) implicating unknown bHLH transcription factors in priming pluripotent cells for differentiation.
We have used a Y2H screening strategy to identify prodifferentiation factors that are likely to be regulated by Id1 in ESCs. Searching within an ESC cDNA library, the only proteins that we found to directly bind to Id1 were the E proteins E12 and E47. Although it does remains possible that there may be direct binding partners of Id1 in ESCs that were not picked up in our screen, we conclude that Id1 is likely to be acting at least in part through sequestration of these two E proteins. Regulators of ESC differentiation are therefore likely to include direct binding partners of these E proteins. We have identified one such factor, Tcf15, as a bHLH transcription factor that is expressed in the late preimplantation embryo and within a differentiation-primed subpopulation of ESCs, and which can accelerate differentiation when rendered immune to Id inhibition.
What relevance might our findings have for the development of the epiblast in vivo? Tcf15 is best known as a regulator of somite development (Burgess et al., 1996), and there are some intriguing parallels between somitogenesis and epiblast formation. In somites, Tcf15 is not required for cell fate specification; rather, Tcf15 is necessary and sufficient for remodeling of mesenchyme into an N-cadherin+ polarized epithelial structure (Burgess et al., 1996; Linker et al., 2005; Takahashi et al., 2005). Epithelialization in the somites is a consequence of N-cadherin-mediated cell-cell adhesion (Duband et al., 1987). Transition of the inner cell mass of the blastocyst undergoes a similar remodeling process as it matures into postimplantation epiblast, including the upregulation of N-cadherin (Hatta et al., 1987; Bao et al., 2009) and formation of a polarized N-cadherin E-cadherin double-positive epithelium. We speculate that Tcf15 may therefore play a similar role in both scenarios to help trigger the morphological remodeling events that characterize these changes in cell identity.
Tcf15 mutant embryos survive to birth, but die shortly afterward as a consequence of muscular-skeletal defects due to the failure in somite morphogenesis earlier in development. We speculate that by analogy with its role in the somites and based on our own ESC data, Tcf15 may perhaps also help to coordinate epithelialization of the epiblast. The consequence of removing this earlier function of Tcf15 could be to delay epiblast formation and consequently to disrupt the kinetics of subsequent lineage allocation. However, the early embryo is highly regulative, and such phenotypes may therefore not be readily apparent when examining mutant embryos at late stages. Indeed, a phenotype may only manifest itself as a lowering of the robustness of the early developmental process, which may only become apparent under suboptimal conditions. Furthermore, any putative Tcf15 mutant phenotype may be masked by compensatory premature upregulation of the closely related transcription factor Scx, which shares an almost identical DNA binding domain with Tcf15 (Burgess et al., 1995). Scx has an established role in later stages of epiblast maturation: from around E6.5 the epiblast of Scx mutants fails to develop further and does not properly epithelialize. Intriguingly, the Scx mutant phenotype begins to manifest itself shortly after Tcf15 is downregulated after implantation (Brown et al., 1999), supporting the idea that these two transcription factors act sequentially during development and may exhibit some functional redundancy. However, Scx is not expressed in ESCs and therefore, unlike Tcf15, does not mark an early epiblast-primed pluripotent state. It will be of interest in future studies to explore the relevance of our ES-based studies for early embryonic development.
Even in the continued presence of LIF, a subset of ESCs downregulate markers of naive pluripotency. This subset of ESCs is consequently primed toward differentiation (Chambers et al., 2007; Toyooka et al., 2008; Kalmar et al., 2009). Recent work shows that some of these Nanog-low cells, marked by the endodermal transcription factor Hex, are primed for primitive endoderm, but a third population expresses neither Hex nor naive pluripotency markers (Canham et al., 2010). It has been speculated (Canham et al., 2010; Lanner and Rossant, 2010) that this third population is primed for differentiation into mature epiblast and therefore ultimately for differentiation into somatic cell types, but a marker of this early primed state has proved elusive. We find that Tcf15-Venus marks a Klf4-low Nanog-low subpopulation but is not specifically associated with Hex. Taken together with the ability of Tcf15 to drive epiblast priming, this indicates that Tcf15 marks the epiblast-primed subpopulation of ESCs and identifies cells taking the first step toward embryonic rather than extraembryonic lineages. In support of this idea, our genome-wide analysis reveals Nanog to be the gene most rapidly downregulated in response to Tcf15 and the epiblast-determinant Otx2 to be one of the genes most rapidly upregulated in response to Tcf15. This further validates our hypothesis that Tcf15 activation represents the first step toward differentiation of ESCs, and suggests a mechanism by which it acts.
How does FGF prime cells for differentiation without pushing them out of the pluripotent state? Tcf15 is a prodifferentiation factor whose expression is driven by the priming factor FGF but whose activity is likely to be restrained by the antidifferentiation factor BMP4 through upregulation of Id. Although other targets of FGF and Id are likely to cooperate with Tcf15 to prime pluripotent cells for differentiation, our work provides one clear example of how this priming mechanism operates. FGF/Erk activity is required for somatic commitment from ESCs (Kunath et al., 2007). However, it is not clear how much Erk activity there is in late preimplantation or peri-implantation epiblast: certainly activity is not as high as in extraembryonic tissues at these stages (Corson et al., 2003). Any ERK signaling within the epiblast is therefore likely to be relatively transient. It is interesting in this respect that Tcf15 expression is also transient in the early embryo: it is readily detectable at E4.5 but returns to background levels by E5.5. In contrast, the widely used epiblast marker T-Brachyury (Lanner and Rossant, 2010) is expressed later than Tcf15 and persists as cells exit the epiblast and commit to mesodermal lineages. Tcf15 is a transcription factor that specifically marks a transient FGF-primed intermediate on the route from pluripotency toward somatic differentiation of ESCs.
In summary, we have used a Y2H screen to identify a marker, Tcf15, of a transient cell state that stands midway between naive pluripotency and somatic lineage commitment. We propose that Tcf15 contributes toward steering pluripotent cells in the direction of somatic lineages and that it will serve as a valuable tool for monitoring and interrogating early developmental transitions.
Experimental Procedures
Y2H Analysis
The DNA sequences corresponding to mouse Id1 (1–176) and the HLH regions of E47 (E47HLH: 488–648) and E12 (E12HLH: 488–651) were cloned into the Y2H vectors pGBKT7 and pGADT7 (Clontech). Y2H analysis was performed using the Matchmaker 3 Y2H system (Clontech), with protocols based on the manufacturer’s instructions. pGBKT7 and pGADT7 vectors were transformed into the yeast strains Y187 and AH109, respectively, through a polyethylene glycol/single-stranded DNA/lithium acetate procedure. Interactions between bait and target proteins were tested by mating the Y187[pGBKT7-bait] strain with the AH109[pGADT7-target] strain; single colonies from each strain were resuspended in 0.5 ml 2xYPDA and incubated at 30°C, 50 rpm for 24 hr. Cultures were then diluted 1 in 10 using 0.5xYPDA, and 100 μl was plated onto SD/−Leu/−Trp (double dropout) to select for mated colonies, and onto SD/−Ade/−His/−Leu/−Trp (quadruple dropout) to select for mated colonies with the activation of ADE and HIS reporter genes. Plates were incubated at 30°C for 5 days. Colonies were lifted onto filters (Whatman No. 5, 70 mm), lysed by two cycles of freeze-thaw at −80°C, and β-galactosidase assays were performed by incubation in 100 mM sodium phosphate (pH 7.0), 10 mM KCl, 1 mM MgSO4, 0.3% β-mercaptoethanol, 1 mg/ml X-Gal (Promega) at 30°C for 5 hr. All filter lift assays shown are representative of at least three replicates performed using independent yeast transformants.
ES Cell cDNA Y2H Library Construction
A mouse ESC cDNA library was generated using the Matchmaker library construction and screening kit (Clontech). Total RNA was extracted from a homogenous monolayer culture of self-renewing mouse ESCs (E14Tg2α strain) using an Absolutely RNA miniprep kit (Stratagene), and purified for poly(A)+ RNA using an Oligotex mRNA mini kit (QIAGEN). cDNA was generated by first strand synthesis using Moloney murine leukemia virus reverse transcriptase with a random primer that inserts a 3′ CDS III sequence; a 5′ SMART III sequence was then inserted and amplification performed by long-distance PCR using Advantage 3 DNA polymerase (Clontech). cDNA was recombined into the pGADT7 vector through cotransformation of the AH109 yeast strain with cDNA and linearized vector using a high-efficiency polyethylene glycol/single-stranded DNA/lithium acetate procedure; transformants were selected on SD/-Leu plates. The resulting ∼2.6 × 106 colonies were pooled in 1l of YPDA media supplemented with 10% glycerol, and frozen in 1 ml aliquots.
Y2H Library Screening
To perform a library screen, a Y187[pGBKT7-bait] strain was mixed with an AH109[pGADT7-library] aliquot in 50 ml 2xYPDA and incubated at 30°C, 50 rpm for 24 hr. For high-stringency screening, cultures were plated on SD/−Ade/−His/−Leu/−Trp and incubated at 30°C for 7 days; colonies were subsequently grown three times on SD/−Ade/−His/−Leu/−Trp. For medium-stringency screens, cultures were initially plated on SD/-His/-Leu/-Trp and incubated at 30°C for 7 days; colonies were then grown three times on SD/−Ade/−His/−Leu/−Trp. The cDNA identities of positively interacting pGADT7 vectors were determined through single-colony PCR with gel purification and DNA sequencing. Vectors were extracted from positive colonies using Zymoprep yeast plasmid miniprep kit II (Zymo Research), with pGADT7 vectors selected and amplified through ampicillin resistance in bacteria. Interactions were then confirmed through the transformation of fresh AH109 yeast with sequence-verified pGADT7 vectors, and with mating and β-galactosidase assays performed as described above.
Mouse ES Cell Culture and Differentiation
ESCs were maintained as previously described (Smith, 1991). Neural differentiation was carried out as previously described (Pollard et al., 2006).
Pharmacological Inhibitors
PD0325901 (used at 1 μM) and PD173074 (used at 100 ng/ml) were obtained from Axon Medchem.
Western Blot Analysis
Blots were probed using conjugated anti-FLAG M2 peroxidase antibody (1:1,000; A8592, Sigma), and monoclonal mouse anti-β-tubulin primary antibody (1:4,000; T5293, Sigma) with monoclonal anti-mouse horseradish peroxidase secondary antibody (1:3,000; Promega).
Immunofluorescence and FACS
Primary antibodies were obtained from the following sources Oct4 (Santa Cruz: sc-5279); green florescent protein (Molecular Probes); Nestin (DSHB); Sox2 (Chemicon); Neuronal beta-III tubulin (Covance); Nanog (gift of I. Chambers); Tcf15 (Aviva).
Fluorescence-activated cell sorting (FACS) analysis was performed using a Becton Dickinson FACS Calibur flow cytometer. FACS was carried out on a Becton Dickinson FACS Aria flow cytometer. For sorting of Tcf15-Venus cells, the top 30% of Venus+ cells and the bottom 30% of Venus-low cells were sorted from within a platelet endothelial cell adhesion molecule (PECAM)+ gated population, excluding differentiated cells.
Quantification of Nuclear Immunostaining
In order to quantify the fluorescence signal in each individual cell, we generated an automated pipeline for image analysis, described in full in Extended Experimental Procedures.
Extended Experimental Procedures.
Quantification of Nuclear Immunostaining
In order to quantify the fluorescence signal in each individual cell, we generated an automated pipeline for image analysis. Briefly, RGB pictures were registered and preprocessed using a plugin in ImageJ (http://rsbweb.nih.gov/ij/) developed by Dr. Guillaume Blin. The preprocessing step consisted in a background subtraction in each channel as well as a gamma correction of the blue channel to reveal low intensity nuclei. Then, to detect single cell nuclei, the blue channel (dapi) was segmented using a previously published algorithm (Li et al., 2007) with the following parameters values: sigma = 0.15, minimum nucleus size = 350 pixels and fusion threshold = 1. This algorithm provides a picture within which each nucleus is labeled with a unique color in the image. Using a homemade java application that we developed with eclipse (http://www.eclipse.org), the signal in the red and green channels of the preprocessed RGB picture was measured in the superimposed area of each nucleus to calculate the average intensity. Finally, the red and green average intensities were plotted using R.
In order to quantify neurite density after neural differentiation five randomly selected fields (for each condition: treatment and day of differentiation) stained for Tuj1 were pictured. Each field was scored depending on the number of neurites present from 0 to 9, where 0 means no neurites can be observed and 9 neurites cover almost the entire field; the scores for each condition were averaged.
Microarray Analysis
TTE15 and parental AW2 cells were plated in 6well plates (IWAKI) one day before stimulation. Cells were stimulated by 1 μg/mL doxycycline for the indicated time and lysed for RNA. Sample preparation and data analysis is described in Supplemental Information.
RNA was prepared using Absolutely RNA Purification Kits (Agilent). 100 ng of RNA were reverse transcribed into ds cDNA and transcribed/amplified into biotin labeled cRNA using an Illumina TotalPrep RNA Amplification Kit (AMIL1791, Ambion). Labeled RNA was submitted to the WTCRF MRC Human Genetics Unit (University of Edinburgh) for further processing. cRNA quality was checked using a Agilent 2100 Bioanalyser and hybridization performed on an MouseWG-6 v2 BeadChip (Illumina). Raw data was processed in R using the beadarray (Dunning et al., 2007) and limma (Wettenhall and Smyth, 2004) packages from the Bioconductor suite (Gentleman et al., 2004). Briefly, we removed low-quality probes from the input data. The data was subsequently quantile-normalized and log2-transformed before assessing differential expression with the limma algorithms. We considered genes differentially expressed which had an FDR-adjusted p-value of at most 0.05 and a fold change of 1.5 or more for at least one time point in comparison to the 0 hr baseline. Microarray data is available as supplemental information.
Primers Used for qPCR
Esrrb CGATTCATGAAATGCCTCAA, CCTCCTCGAACTCGGTCA
Fgf5 AAAACCTGGTGCACCCTAGA, CATCACATTCCCGAATTAAGC
Gata6 GGTCTCTACAGCAAGATGAATGG, TGGCACAGGACAGTCCAAG
Hex CTACACGCACGCCCTACTC, CAGAGGTCGCTGGAGGAA
Klf4 CGGGAAGGGAGAAGACACT, GAGTTCCTCACGCCAACG
Nanog CCTCCAGCAGATGCAAGAA, GCTTGCACTTCATCCTTTGG
Oct4 GTTGGAGAAGGTGGAACCAA, CTCCTTCTGCAGGGCTTTC
Nanog pre mRNA N26 GGTGATACGTTGGCCTTCTAGT, TTCTCAAATACACACAAGAGCCTTA
Nanog pre mRNA N28 AGCCCAGTACTCAGGCTTGT, AGCATCACAACACGCACCT
Nanog pre mRNA N29 GCCAGCAGATGGCATAATTT, TGATGGCAATGCTGAGGTTA
Nanog pre mRNA N32 GATTCTATTCACCCAGCACCA, CCTTCTGAGTGGAGGTTTATCC
Nanog pre mRNA N33 CCATCTCAGCTACTGGAGCA, ATTAGAACCGTGACCGCATC
Otx2 GACCCGGTACCCAGACATC, GCTCTTCGATTCTTAAACCATACC
Rex1 CAGCTCCTGCACACAGAAGA, ACTGATCCGCAAACACCTG
Scx ACACCCAGCCCAAACAGAT, TCTGTCACGGTCTTTGCTCA
Sox1 GTGACATCTGCCCCCATC, GAGGCCAGTCTGGTGTCAG
Sox7 GCGGAGCTCAGCAAGATG, GGGTCTCTTCTGGGACAGTG
T Brachyury CAGCCCACCTACTGGCTCTA, GAGCCTGGGGTGATGGTA
TBP GGGGAGCTGTGATGTGAAGT, CCAGGAAATAATTCTGGCTCA
Tcf15 GTGTAAGGACCGGAGGACAA, GATGGCTAGATGGGTCCTTG
Tcf15 transgene GCTCCATCTGCACCTTCTG, GGCTACACCCCTCACTTTCA
Zfp521 AAGCAAGCGAAACCGAGAT, TTCTGGCCTCTTCTTGCAGT
In Situ Hybridization
Whole-mount in situ hybridization on MF1 × MF1 mouse blastocysts was performed according to the protocol of Rosen and Beddington (1993). Digoxigenin-labeled anti-Tcf15 riboprobes were synthesized from Tcf15 cDNA clones (Burgess et al., 1995).
Dox-Inducible Tcf15-E47 Cell Line
The DNA sequence corresponding to N-terminally FLAG-tagged mouse Tcf15 tethered at its C terminus to the N terminus of mouse E47 through a 13-amino-acid flexible linker of sequence TGSTGSKTGSTGS was generated by overlapping extension PCR. This sequence was placed under a tet-responsive promoter and transfected into the E14tg2A_AW2 cell line, which contains the coding sequence for the rtTA integrated into the Rosa 26 and expressed from the R26 promoter. The E14tg2A_AW2 cell line will be described in more detail elsewhere (A.W., S.L., and A.J.H.S., unpublished data).
Microarray Analysis
TTE15 and parental AW2 cells were plated in six-well plates (IWAKI) 1 day before stimulation. Cells were stimulated by 1 μg/ml doxycycline for the indicated time and lysed for RNA. Sample preparation and data analysis are described in Extended Experimental Procedures.
Acknowledgments
We thank Val Wilson for embryo dissection, Mattias Malaguti and Florian Halbritter for microarray analysis, Nicola Festuccia for providing plasmids, and Pablo Navarro for providing primers and for help with experiments. We are grateful to Ian Chambers, Nicola Festuccia, Josh Brickman, Tilo Kunath, Mattias Malaguti, Paul Nistor, and Val Wilson for helpful discussions and comments on the manuscript. This work was supported by the Welcome Trust (WT082232AIA) and the BBSRC (BB/I006680/1 to S.L., G15381 to A.J.H.S.). O.R.D. was funded by a postdoctoral research fellowship from the Royal Commission for the Exhibition of 1851. S.L. is a Wellcome Trust Research Career Development Fellow.
Published: February 7, 2013
Footnotes
Supplemental Information includes Extended Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.01.017.
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Accession Numbers
The GEO accession number for the microarray data reported in this paper is GSE42539.
Supplemental Information
References
- Acampora D., Di Giovannantonio L.G., Simeone A. Otx2 is an intrinsic determinant of the embryonic stem cell state and is required for transition to a stable epiblast stem cell condition. Development. 2012;140:43–55. doi: 10.1242/dev.085290. [DOI] [PubMed] [Google Scholar]
- Bao S., Tang F., Li X., Hayashi K., Gillich A., Lao K., Surani M.A. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R. An intermolecular disulfide bond stabilizes E2A homodimers and is required for DNA binding at physiological temperatures. Cell. 1994;79:1057–1067. doi: 10.1016/0092-8674(94)90036-1. [DOI] [PubMed] [Google Scholar]
- Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A., Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Brown D., Wagner D., Li X., Richardson J.A., Olson E.N. Dual role of the basic helix-loop-helix transcription factor scleraxis in mesoderm formation and chondrogenesis during mouse embryogenesis. Development. 1999;126:4317–4329. doi: 10.1242/dev.126.19.4317. [DOI] [PubMed] [Google Scholar]
- Burgess R., Cserjesi P., Ligon K.L., Olson E.N. Paraxis: a basic helix-loop-helix protein expressed in paraxial mesoderm and developing somites. Dev. Biol. 1995;168:296–306. doi: 10.1006/dbio.1995.1081. [DOI] [PubMed] [Google Scholar]
- Burgess R., Rawls A., Brown D., Bradley A., Olson E.N. Requirement of the paraxis gene for somite formation and musculoskeletal patterning. Nature. 1996;384:570–573. doi: 10.1038/384570a0. [DOI] [PubMed] [Google Scholar]
- Canham M.A., Sharov A.A., Ko M.S.H., Brickman J.M. Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol. 2010;8:e1000379. doi: 10.1371/journal.pbio.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Connerney J., Andreeva V., Leshem Y., Muentener C., Mercado M.A., Spicer D.B. Twist1 dimer selection regulates cranial suture patterning and fusion. Dev. Dyn. 2006;235:1345–1357. doi: 10.1002/dvdy.20717. [DOI] [PubMed] [Google Scholar]
- Corson L.B., Yamanaka Y., Lai K.-M.V., Rossant J. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–4537. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
- Duband J.L., Dufour S., Hatta K., Takeichi M., Edelman G.M., Thiery J.P. Adhesion molecules during somitogenesis in the avian embryo. J. Cell Biol. 1987;104:1361–1374. doi: 10.1083/jcb.104.5.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta K., Takagi S., Fujisawa H., Takeichi M. Spatial and temporal expression pattern of N-cadherin cell adhesion molecules correlated with morphogenetic processes of chicken embryos. Dev. Biol. 1987;120:215–227. doi: 10.1016/0012-1606(87)90119-9. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Kalmar T., Lim C., Hayward P., Muñoz-Descalzo S., Nichols J., Garcia-Ojalvo J., Martinez Arias A. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai-Kowase K., Kumar M.S., Hoofnagle M.H., Yoshida T., Owens G.K. PIAS1 activates the expression of smooth muscle cell differentiation marker genes by interacting with serum response factor and class I basic helix-loop-helix proteins. Mol. Cell. Biol. 2005;25:8009–8023. doi: 10.1128/MCB.25.18.8009-8023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T., Saba-El-Leil M.K., Almousailleakh M., Wray J., Meloche S., Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- Lanner F., Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- Linker C., Lesbros C., Gros J., Burrus L.W., Rawls A., Marcelle C. beta-Catenin-dependent Wnt signalling controls the epithelial organisation of somites through the activation of paraxis. Development. 2005;132:3895–3905. doi: 10.1242/dev.01961. [DOI] [PubMed] [Google Scholar]
- Markus M., Benezra R. Two isoforms of protein disulfide isomerase alter the dimerization status of E2A proteins by a redox mechanism. J. Biol. Chem. 1999;274:1040–1049. doi: 10.1074/jbc.274.2.1040. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Takizawa T., Ochiai W., Yanagisawa M., Hisatsune T., Nakafuku M., Miyazono K., Kishimoto T., Kageyama R., Taga T. BMP2-mediated alteration in the developmental pathway of fetal mouse brain cells from neurogenesis to astrocytogenesis. Proc. Natl. Acad. Sci. USA. 2001;98:5868–5873. doi: 10.1073/pnas.101109698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya F.J.F., Stellrecht C.M.C., Tsai M.J.M. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- Neuhold L.A., Wold B. HLH forced dimers: tethering MyoD to E47 generates a dominant positive myogenic factor insulated from negative regulation by Id. Cell. 1993;74:1033–1042. doi: 10.1016/0092-8674(93)90725-6. [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Nichols J., Silva J., Roode M., Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton J.D. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J. Cell Sci. 2000;113:3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- Pollard S.M., Benchoua A., Lowell S. Neural stem cells, neurons, and glia. Methods Enzymol. 2006;418:151–169. doi: 10.1016/S0076-6879(06)18010-6. [DOI] [PubMed] [Google Scholar]
- Rosen B., Beddington R.S. Whole-mount in situ hybridization in the mouse embryo: gene expression in three dimensions. Trends Genet. 1993;9:162–167. doi: 10.1016/0168-9525(93)90162-b. [DOI] [PubMed] [Google Scholar]
- Rossant J., Tam P.P.L. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- Shen C.P., Kadesch T. B-cell-specific DNA binding by an E47 homodimer. Mol. Cell. Biol. 1995;15:4518–4524. doi: 10.1128/mcb.15.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan S.R., Shen C.P., McCarrick-Walmsley R., Kadesch T. Phosphorylation of E47 as a potential determinant of B-cell-specific activity. Mol. Cell. Biol. 1996;16:6900–6908. doi: 10.1128/mcb.16.12.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.G. Culture and differentiation of embryonic stem cells. J. Tissue Cult. Methods. 1991;13:89–94. [Google Scholar]
- Takahashi Y., Sato Y., Suetsugu R., Nakaya Y. Mesenchymal-to-epithelial transition during somitic segmentation: a novel approach to studying the roles of Rho family GTPases in morphogenesis. Cells Tissues Organs. 2005;179:36–42. doi: 10.1159/000084507. [DOI] [PubMed] [Google Scholar]
- Tanaka T.S., Davey R.E., Lan Q., Zandstra P.W., Stanford W.L. Development of a gene-trap vector with a highly sensitive fluorescent protein reporter system for expression profiling. Genesis. 2008;46:347–356. doi: 10.1002/dvg.20404. [DOI] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Bao S., Lee C., Nordman E., Wang X., Lao K., Surani M.A. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Wilson-Rawls J., Rhee J.M., Rawls A. Paraxis is a basic helix-loop-helix protein that positively regulates transcription through binding to specific E-box elements. J. Biol. Chem. 2004;279:37685–37692. doi: 10.1074/jbc.M401319200. [DOI] [PubMed] [Google Scholar]
- Wood H.B., Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev. 1999;86:197–201. doi: 10.1016/s0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y., Lanner F., Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Nichols J., Chambers I., Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Li L., Huang C., Shen C., Tan F., Xia C., Liu P., Rossant J., Jing N. Distinct functions of BMP4 during different stages of mouse ES cell neural commitment. Development. 2010;137:2095–2105. doi: 10.1242/dev.049494. [DOI] [PubMed] [Google Scholar]
Supplemental References
- Dunning M.J., Smith M.L., Ritchie M.E., Tavaré S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Liu T., Tarokh A., Nie J., Guo L., Mara A., Holley S., Wong S.T.C. 3D cell nuclei segmentation based on gradient flow tracking. BMC Cell Biol. 2007;8:40. doi: 10.1186/1471-2121-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall J.M., Smyth G.K. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.