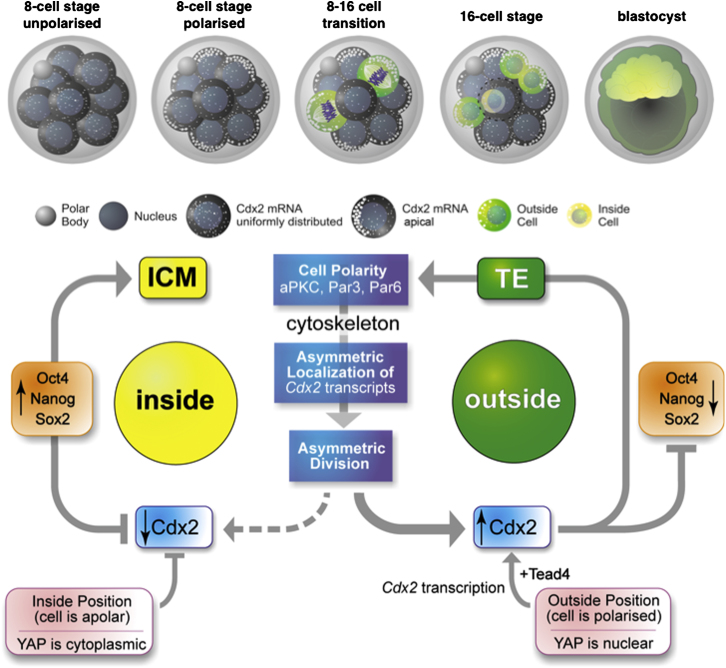
Summary
A longstanding question in mammalian development is whether the divisions that segregate pluripotent progenitor cells for the future embryo from cells that differentiate into extraembryonic structures are asymmetric in cell-fate instructions. The transcription factor Cdx2 plays a key role in the first cell-fate decision. Here, using live-embryo imaging, we show that localization of Cdx2 transcripts becomes asymmetric during development, preceding cell lineage segregation. Cdx2 transcripts preferentially localize apically at the late eight-cell stage and become inherited asymmetrically during divisions that set apart pluripotent and differentiating cells. Asymmetric localization depends on a cis element within the coding region of Cdx2 and requires cell polarization as well as intact microtubule and actin cytoskeletons. Failure to enrich Cdx2 transcripts apically results in a significant decrease in the number of pluripotent cells. We discuss how the asymmetric localization and segregation of Cdx2 transcripts could contribute to multiple mechanisms that establish different cell fates in the mouse embryo.
Graphical Abstract
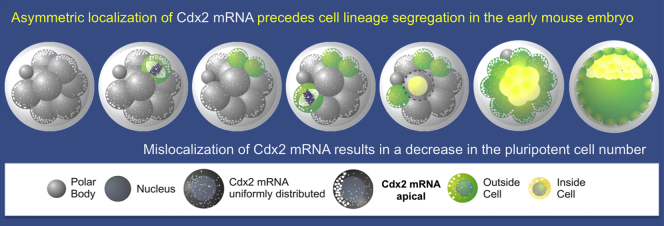
Highlights
► Cdx2 mRNA localizes apically upon embryo compaction at the eight-cell stage ► Cdx2 mRNA is inherited asymmetrically during asymmetric divisions ► Localization requires cell polarization and intact cytoskeletal components ► Mislocalization of Cdx2 mRNA decreases the number of pluripotent cells
A longstanding question in mouse development is whether divisions that set apart pluripotent and differentiating cells are asymmetric in cell-fate instructions. Cdx2 is a key factor in this first fate decision. Zernicka-Goetz and colleagues use live embryo imaging to show that localization and inheritance of Cdx2 transcripts become asymmetric as pluripotent and differentiating lineages segregate. They demonstrate that asymmetric transcript localization requires cell polarization as well as intact microtubule and actin cytoskeletons, and that mislocalization of Cdx2 transcripts significantly decreases the number of pluripotent cells.
Introduction
Asymmetric localization of specific transcripts is a common posttranscriptional mechanism for regulating gene activity in various model systems (Holt and Bullock, 2009; St Johnston, 2005, Meignin and Davis, 2010). Such asymmetric localization and then segregation of messenger RNA (mRNA) in cell division are often important for cell-fate determination (Li et al., 1997; Melton, 1987; Neuman-Silberberg and Schüpbach, 1993). However, whether any asymmetric localization and segregation of transcripts occur in early mammalian embryos is currently unknown.
Segregation of the first two cell lineages in the mouse embryo is initiated at the eight- to 16-cell-stage transition when blastomeres undertake divisions to generate inside cells that will form pluripotent inner cell mass (ICM) and outside cells that will form trophectoderm (TE) (Johnson and Ziomek, 1981; Bruce and Zernicka-Goetz, 2010). The ICM gives rise to cells of the future body, and the TE gives rise to an extraembryonic tissue with an essential role in patterning the embryo and building the placenta. Divisions that generate ICM and TE progenitor cells were cautiously termed “differentiative” (Johnson and Ziomek, 1981) because it was unknown whether such divisions are asymmetric in transmitting cell-fate instructions or whether inside and outside cells follow different fates only because of the differential positions assumed by the cells. In contrast, divisions that generate only TE progenitor cells were termed “conservative.”
A number of transcription factors that are important for distinguishing the ICM and TE lineages become differentially expressed between inside and outside cells, which are precursors of these first two cell lineages. Of these, Cdx2 and Eomes have increased expression in outside cells (Russ et al., 2000; Strumpf et al., 2005), and Oct4, Sox2, and Nanog have progressively increased expression in inside cells (Avilion et al., 2003; Chambers et al., 2003; Mitsui et al., 2003; Palmieri et al., 1994). Cdx2 is critical for initiating this cell-fate diversification. Preventing Cdx2 expression leads to an increased expression of pluripotency genes in outside cells and improper development of the TE (Strumpf et al., 2005; Jedrusik et al., 2008). One established means of regulating the differential expression of Cdx2 between inside and outside cells is to exert transcriptional control in response to differential cell position when the ICM and TE precursors segregate from each other. This is achieved through nuclear localization of Yes-associated protein (YAP) in outside cells to permit Tead4-mediated transcription of Cdx2 (Yagi et al., 2007; Nishioka et al., 2008). The retention of YAP in the cytoplasm of inside cells, in part through the activity of the Hippo pathway, restrains transcription of Cdx2 in inside cells (Nishioka et al., 2009). As a result of differential expression of Cdx2, factors required for pluripotency are downregulated in outside but not inside cells (Niwa et al., 2005). The effect of cell position upon the regulation of Cdx2 expression is therefore well established.
The mechanisms behind the first differential expression of Cdx2 among blastomeres at the eight-cell stage are less well understood (Jedrusik et al., 2008; Ralston and Rossant, 2008). However, this differential expression is important because it significantly biases cell fate: cells with higher levels of CDX2 contribute preferentially to the TE, and those with lower levels contribute preferentially to the ICM (Jedrusik et al., 2008). The orientation of cell divisions that govern whether cells are allocated to the TE or ICM is influenced by cell polarization (Plusa et al., 2005). However, the effect of cell polarity on CDX2 remains largely unknown.
Here, we provide evidence for temporal and spatial localization and segregation of Cdx2 transcripts that precede the segregation of the ICM and TE lineages. Using a sensitive assay to follow transcripts in living mouse embryos, we found that Cdx2 mRNA becomes preferentially localized apically at the late eight-cell stage and inherited asymmetrically at the eight- to 16-cell transition. We define the localization sequence within the Cdx2 mRNA and show that Cdx2 mRNA localization depends on apical polarization and the microtubule and actin cytoskeletons. Preventing Cdx2 mRNA localization leads to the upregulation of CDX2 protein in inside cells, which consequently decreases the number of pluripotent cells. Our results indicate that “differentiative” divisions in the mouse embryo contribute to the asymmetry of cell-fate instruction: they prevent inside cells from inheriting Cdx2 transcripts, thereby permitting the development of pluripotency while adding support to the differentiation program of outside cells.
Results
Spatial and Temporal Localization of Cdx2 Transcripts
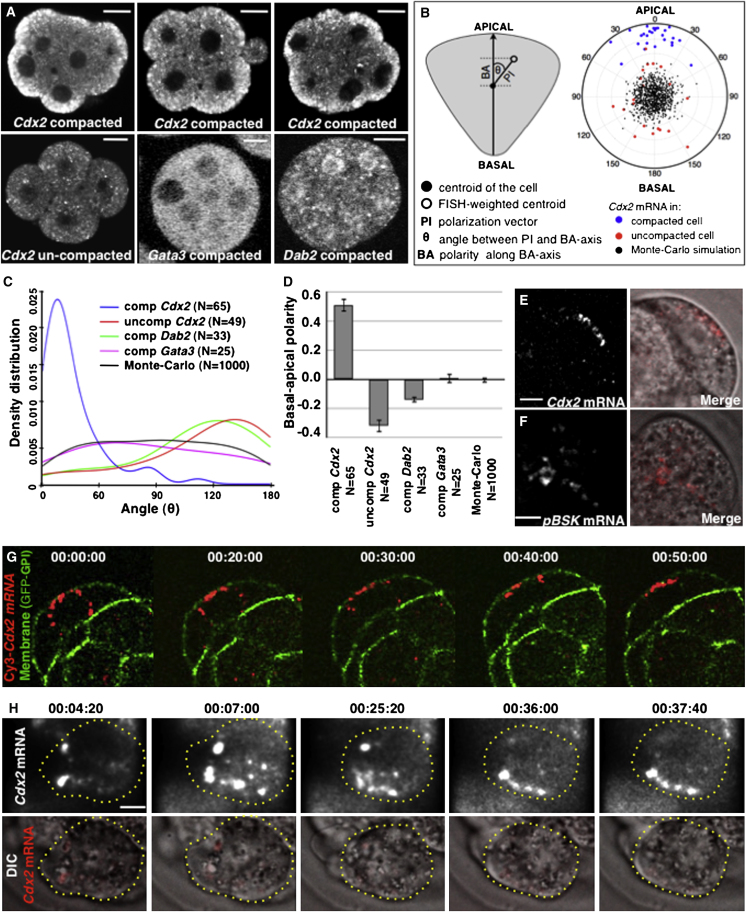
CDX2 is a key transcription factor that constitutes an essential switch in the first cell-fate decision. It becomes expressed differentially upon the differentiative divisions at the eight- to 16-cell transition. Cdx2 expression in outside cells is critical for TE specification and has to be suppressed in inside cells to allow development of pluripotency. In order to determine the mechanism that regulates this differential Cdx2 expression, we first examined in detail the spatial and temporal distribution of Cdx2 transcripts by fluorescent in situ hybridization (FISH). We found that before embryo compaction occurs, Cdx2 mRNA distributes evenly within the cells (n = 56), but in contrast, upon compaction at the late eight-cell stage, Cdx2 mRNA becomes localized asymmetrically along the apical-basal axis (n = 144; Figure 1A). Apical localization of Cdx2 mRNA was evident in 81% (117/144) of cells in compacted embryos (a significantly greater proportion than in uncompacted embryos in which none of the cells showed apical localization of Cdx2 mRNA; n = 0/56, χ2 test, p < 0.001). The remaining 19% of cells (27/144) showed either no localization (8%) or a distribution that was ambiguous (11%; Figure S1A). In order to quantitate the spatial distribution of Cdx2 transcripts, we used a computational approach to measure the polarization of the FISH signal (Park et al., 2012). In this method, apical distribution is characterized by small values of the angle between the polarization index (PI) vector and the apical-basal axis (θ), and by relatively high positive values of the basal-apical polarization (BA) vector (see Extended Experimental Procedures). The analyses yielded mean values for θ of 126° ± 7° /SEM/ and 23° ± 3°, and the BA values of −0.32 ± 0.05 /SEM/ and 0.51 ± 0.04 for uncompacted (n = 49) and compacted (n = 65) embryos, respectively (t test, p < 0.001; Figures 1B–1D), indicating that Cdx2 transcripts localize toward the apical region upon compaction. Apical localization was not observed for transcripts of two other transcription factors examined at this stage, Dab2 (n = 33, θ = 121° ± 8° and BA = −0.14 ± 0.02) and Gata3 (n = 25, θ = 89° ± 11° and BA = 0.01 ± 0.03), which distributed evenly (Figures 1A and 1C). Thus, Cdx2 transcripts acquire a polarized distribution preceding ICM and TE lineage segregation.
Figure 1.
Exogenous Cdx2 Transcripts Recapitulate Endogenous Cdx2 Transcript Localization
(A) Representative images of endogenous Cdx2 (n = 56 uncompacted and n = 144 compacted), Dab2 (n = 17 compacted), and Gata3 (n = 14 compacted) mRNAs detected by FISH in eight-cell embryos. Scale bar, 10 μm.
(B) Quantification of the distribution of Cdx2 transcripts. Polar coordinates for individual cells are expressed as a single point defined by vector PI and angle θ on a polar graph. The PI vector is defined by the center of gravity of the cell and the center of gravity of the FISH signal. This vector shows an angular displacement, θ (the angular coordinate), from the apical-basal (polar) axis. The radial coordinate (PI) can be alternatively expressed along the apical-basal (polar) axis as the value BA. Apical localization of Cdx2 mRNA in compacted blastomeres (n = 27) is represented by the clustering of blue dots around 0°; red dots represent Cdx2 mRNA in individual cells before compaction (n = 20), and black dots represent a Monte Carlo simulation of random localization.
(C) Density distributions of values for angles θ for indicated transcripts (measured as described in B for individual cells).
(D) Representation of the distribution as the average value (±SEM) of the polar coordinate (BA) for the indicated transcripts. This has a positive value when directed apically and is negative when basal.
(E and F) Localization of exogenous labeled RNAs for full-length Cdx2 mRNA (E; n = 52) and control mRNA (F; n = 12) injected into eight-cell embryo cells and imaged 1.5–2 hr after injection. Scale bar, 10 μm.
(G) Stills from a representative movie of a compacted eight-cell blastomere injected with labeled Cdx2 mRNA. Time points are presented in hr:min:s format. Scale bar, 20 μm.
(H) Dynamics of Cdx2 mRNA localization after cell division of the eight-cell blastomere. A cell outline is presented with a dashed line. Scale bar, 5 μm.
Figure S1.
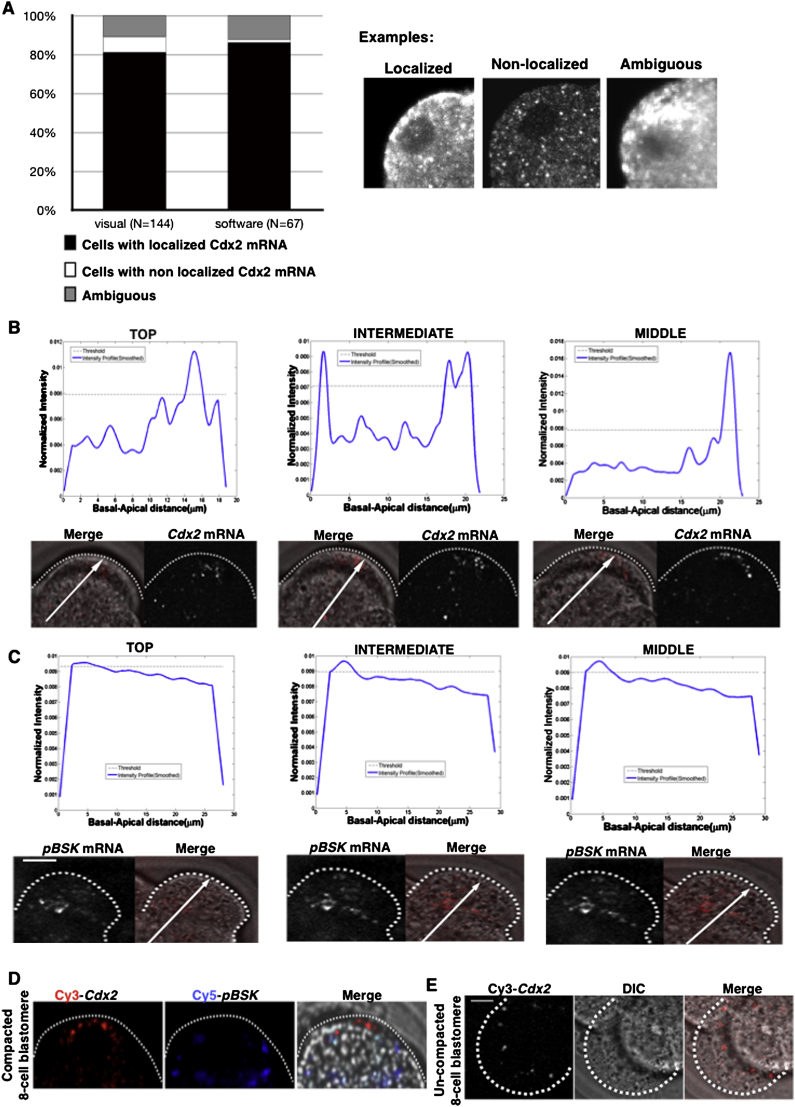
Localization of Endogenous and Exogenous Cdx2 mRNA in Eight-Cell-Stage Embryos, Related to Figure 1
(A) Quantification of blastomeres showing apical localization of endogenous Cdx2 mRNA by FISH in eight-cell-stage embryos, using visual observation and using the image analysis software. In the latter method, blastomeres were categorized based on the angle (θ) between the basal-apical axis of the blastomere and the polarization vector of the FISH staining as: showing apical localization for θ ≤ 45 deg, ambiguous for 45 < θ < 90 deg, and not showing apical localization for θ > = 90 deg. Note, the two methods being equally good in detecting apically localized Cdx2 transcripts. Examples of the three categories are shown on the right.
(B and C) Fluorescence intensity plots along the apical-basal axis derived for top, intermediate, and middle focal planes from confocal images of full-length Cdx2 mRNA (B) and control RNA (C), injected into compacted eight-cell-stage blastomeres. Notice the prevalent peak at the position corresponding the apical side of blastomere. For the most cortical plane the cell curvature leads to the appearance of two peaks one at the projected basal side and one at the apical side.
(D) Coinjection of full-length wt Cdx2 mRNA and RNA transcribed from the pBluescript vector, labeled with two different fluorescent dyes into compacted eight-cell blastomeres results in partitioning of the two RNAs similarly to single injections (see Figure 1). Scale bar, 5 μm.
(E) Injection of full-length wt Cdx2 mRNA to precompacted eight-cell blastomere does not result in the apical localization of the transcripts.
Extended Experimental Procedures.
Image Analysis
For quantification of spatial distribution of Cdx2 transcripts represented as the polarization index (PI) of FISH signal, the algorithm described before (Park et al., 2012) was adopted and implemented in JRuby as an ImageJ plugin or in Matlab using MathWorks. Polar coordinates for individual cells were expressed as a single point defined by vector PI and angle θ on a polar graph (lower part of panel). PI vector is defined by the center of gravity of the cell and the center of gravity of the FISH signal. This vector shows an angular displacement, θ (the angular coordinate), from the apical-basal (polar) axis. The radial co-ordinate (PI) can be alternatively expressed along the apical-basal (polar) axis as the value BA. Positive and negative BA values mean that the polarization vector is directed toward the apical or basal side of the embryo, respectively (Figure 1).
Fluorescence intensity plots were derived using Matlab. Fluorescent signal was summed within a cell normal to the basal-apical axis and each profile was normalized to give a total intensity of 1, before being smoothed using a moving average filter of ten pixels (Figure S1). Threshold values were determined and applied by taking the mean pixel intensity of the profile plus one standard deviation, to remove spurious data points.
Deconvolution of the acquired time-lapse images was performed with ‘Huygens Professional’ (Scientific Volume Imaging), adopting a signal/noise per channel ratio of 10 with automatic search for background option, to yield micrographs used in figures.
Drug Treatments
The microtubule cytoskeleton was depolymerized by addition of Nocodazole (Sigma) to KSOM culture media (final concentration of 30 μM) and embryos cultured in drops under paraffin oil, for 30 min. The actin network was disrupted by addition of Cytochalasin D (Sigma) at a final concentration of 0.5 μg/μl and embryos cultured for 60 min. To study the effect of aPKC inhibition, 8-cell embryos were incubated for 1h in M2 medium containing 60nM GÖ6983 (Sigma). To induce compaction of the 4-cell embryos, these were incubated for 30-60 min in the presence of 200 μM diacylglycerol analog, DiC8 (Sigma). Embryos were fixed immediately after the drug treatment (see below). Control embryos, for each drug treatment, were cultured in the appropriate concentration, for the drug treatment, of DMSO as a vehicle control.
qRT-PCR on Outside and Inside Cells
Embryos were collected into M2 medium containing 4mg/ml BSA 76h post hCG (8-cell stage) and observed to have undertaken eight- to 16-cell-stage divisions, before being incubated in M2 with a fluorescently labeled microsphere suspension diluted to 1:50 for 30 s. Embryos were subsequently disaggregated in divalent cations-free M2 medium with careful and gentle pipetting. Three to four outside (fluorescently labeled) and inside (unlabelled) cells from individual embryos were isolated under the fluorescent dissecting scope, released into a cell lysis buffer (PicoPure RNA isolation kit, Arcturus) and frozen on dry ice. RNA was isolated according to the manufacturer’s protocol and qRT-PCR performed as described earlier (Jedrusik et al., 2008).
RNA FISH Probes
Probes for Cdx2 were made using PCR-generated DNA template derived using Cdx2F1/Cdx2R1T7 primer pairs for anti-sense probe and Cdx2F1T7/Cdx2R1 primers for the sense probe (Cdx2F1—TCGCCACCATGTACGTGAGCTACCT; Cdx2R1—TTCAGACCACGGGAGGGGTCACTG; Cdx2F1T7—TAATACGACTCACTATAGGGATGTACGTGAGCTACCTTC; Cdx2R1T7—TAATACGACTCACTATAGGGAGGGGTCACTGGGTGACAG). A probe for Gata3 was generated using pBluescript plasmid containing EcoRI/EcoRI fragment of mouse Gata3 (generous gift of Dr. Katrin Ottersbach, Cambridge, UK) linearized with XbaI as a template. A probe for Dab2 was prepared using the pOTB7 plasmid containing full-length human Dab2 cDNA (kindly provided by Dr Folma Buss, Cambridge, UK) linearized with EcoRI. A probe for Sox2 was generated using a 430 bp cDNA fragment cloned into pBSK plasmid as a template.
Antibodies for Immunofluorescence
Primary antibodies used for immunofluorescence: mouse anti α-tubulin monoclonal antibody (SIGMA, T9026, 1:500), rabbit anti-aPKC-ζ polyclonal IgG (Santa-Cruz Biotechnology, Inc. sc-216, 1:200), mouse anti-Cdx2 (Biogenex, 1:200), goat anti-Sox17 (R&D Systems, 1:400), rabbit anti-Nanog (ReproCELL, 1:400), rabbit anti-FLOPED (1 μg/ml; generous gift from Jurrien Dean laboratory). Secondary antibodies: Alexa Fluor® 488 donkey anti-rabbit IgG (Invitrogen, A21206, 1:500), Alexa Fluor® 568 donkey anti-rabbit (Invitrogen, A10042, 1:500), Alexa Fluor® 647 donkey anti-rabbit IgG (Invitrogen, A31573, 1:500), Alexa Fluor® 488 goat anti-mouse IgG (Invitrogen, A 11029 1:500), Alexa Fluor® 647 donkey anti-mouse IgG (Invitrogen, A31571, 1:500), Alexa Fluor® 568 goat anti-rat IgG (Invitrogen, A11077, 1:500). Actin was visualized by incubation of embryos with Texas Red-X phalloidin (Invitogen, T7471) and Alexa Fluor® 647 phalloidin (Invitrogen, A22287) at a dilution of 1:50 in PBS for 30min.
Sequence Analysis
The secondary structure of mRNA was predicted and visualized using the RNAfold program (http://rna.tbi.univie.ac.at/) using default parameters (Mathews et al., 1999; Zuker, 2003). Sequences of Cdx2 gene from various species were obtained from the UCSC Genome Bioinformatics Site (http://genome.ucsc.edu/) and aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/). Evolutionary conservation was visualized using BoxShade 3.21 (http://www.ch.embnet.org/software/BOX_form.html).
Dynamics of Cdx2 Transcript Localization
The finding that Cdx2 mRNA localizes apically in the late eight-cell embryo raised several questions: (1) What are the dynamics of spatial localization of Cdx2 transcripts? (2) Do Cdx2 transcripts partition asymmetrically during differentiative cell-fate-directing divisions? (3) What is the mechanism behind the Cdx2 transcripts localization? (4) What might be the role of asymmetrically localized Cdx2 transcripts? To address these questions, we first had to establish an assay to follow localization of Cdx2 mRNA in living mouse embryos. To that end, we synthesized fluorescently labeled Cdx2 and control mRNAs, and injected them into individual eight-cell embryo blastomeres (Figures 1E and 1F) that had compacted and were therefore expected to undergo apical-basal polarization (Johnson and Ziomek, 1981). We found that labeled control mRNAs transcribed from the pBluescript plasmid backbone (n = 15; Figure 1F), from green fluorescent protein (GFP, n = 12; Figure 3H), or from Nanog complementary DNA (cDNA; not shown) showed no regional localization in any focal plane. In contrast, labeled full-length Cdx2 mRNA with intact 5′ and 3′ untranslated regions (UTRs) showed significant accumulation in the apical region of cells (54%, n = 52; χ2 test, p < 0.001; Figure 1E; Movie S1). In the remaining cells, transcripts were observed either in a perinuclear location (25%) or dispersed throughout the cytoplasm (21%). Quantitation of the fluorescence intensity along the apical-basal axis of each cell at the focal plane of its maximum intensity revealed an apical concentration of the signal in Cdx2 but not in control mRNA injected embryos (Figures S1B and S1C). This apical concentration of Cdx2 mRNA was also observed on other focal planes. Finally, we coinjected Cdx2 and pBluescript RNAs labeled with two different fluorescent dyes, and this confirmed their differential localization (n = 7; Figure S1D).
Figure 3.
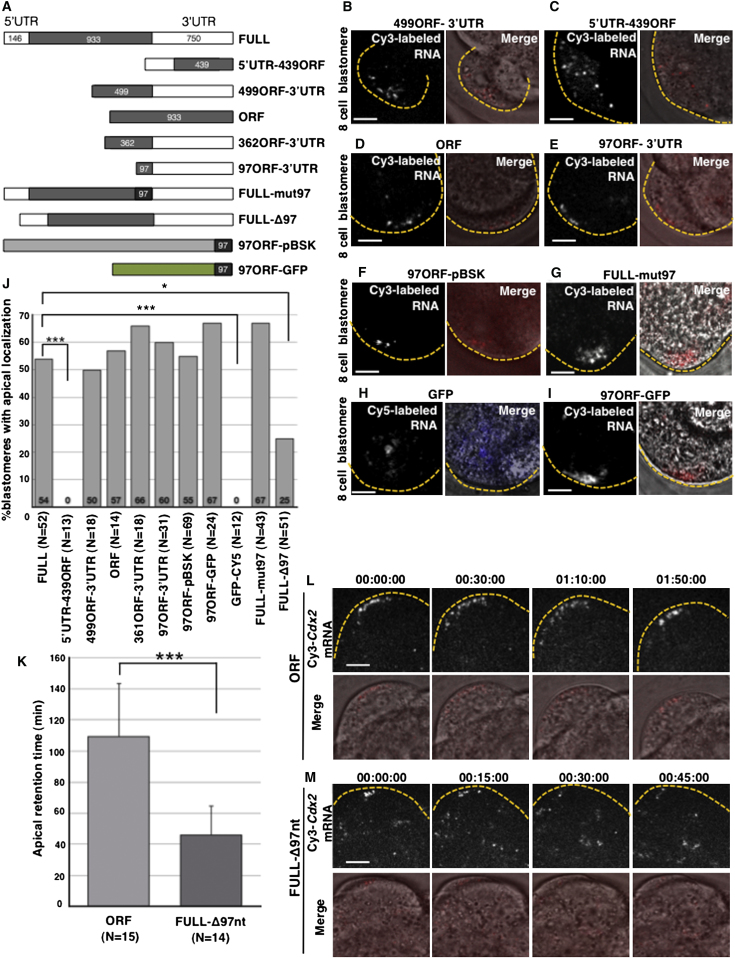
A Cdx2 mRNA Localization Element Resides in the Last 97 nt of the Coding Sequence
(A) Schematic representation of the Cdx2 and fusion constructs along with their controls used for injections.
(B–I) Representative images of localization patterns observed for the truncated forms of Cdx2 mRNA. Scale bar, 10 μm.
(J) Quantification of the injection experiments and numbers of injected blastomeres. Statistically significant differences compared with the control are marked with asterisks (χ2 test, ∗p < 0.05, ∗∗∗p < 0.001).
(K) Quantification of average (±SEM) apical persistence time of ORF and Δ97 Cdx2 transcripts (t test, p < 0.001).
(L and M) Stills from representative movies of labeled Cdx2 ORF RNA (L; n = 15) and Δ97 form (M; n = 14) injected into compacted blastomeres of an eight-cell embryo. Time points (hr:min:s) starting 40 min postinjection are indicated above the panels. Scale bar, 10 μm.
To determine whether the above three broad locations of Cdx2 mRNA represented final or intermediate positions of transcripts, we needed to follow their spatial dynamics in live embryos. To that end, we injected labeled full-length Cdx2 mRNA into 34 individual cells in a second cohort of compacted eight-cell embryos, and carried out confocal time-lapse microscopy beginning 1.5 hr after injection. At the beginning of the time-lapse series, 16 out of 34 injected cells already showed apical localization of Cdx2 transcripts. Of the remaining embryos, 11 had transcripts first enriched in the perinuclear space (Figure 1G, time points 0 and 20 min; Movie S1) that within the next 20–30 min translocated toward the apical region and spread along the apical cell surface (Figure 1G, time points 40 and 50 min). In only seven out of the 34 cells (20%) we could not detect any specific localization of Cdx2 mRNA. Similar behavior of Cdx2 mRNA was observed in newly formed outside cells at the 16-cell stage (Figure 1H; Movie S2). In agreement with the FISH results, Cdx2 mRNA injected into eight-cell blastomeres assessed prior to embryo compaction did not show any particular localization (n = 17; Figure S1E). Thus, the proportion of cells in which exogenous Cdx2 mRNA showed transient perinuclear or stable apical localization (80%, n = 86) is significant (χ2 test, p < 0.001 in comparison with pBluescript plasmid backbone) and similar to the proportion that displayed apical localization of endogenous Cdx2 mRNA detected by FISH at the late eight-cell stage (81%, n = 144, χ2 test, p < 0.001 in comparison with uncompacted embryos). Thus, exogenous Cdx2 mRNA, like endogenous mRNA, acquires a polarized distribution along the apical-basal axis preceding cell divisions leading to ICM and TE lineage segregation.
Partitioning of Cdx2 Transcripts at the Eight- to 16-Cell-Stage Transition
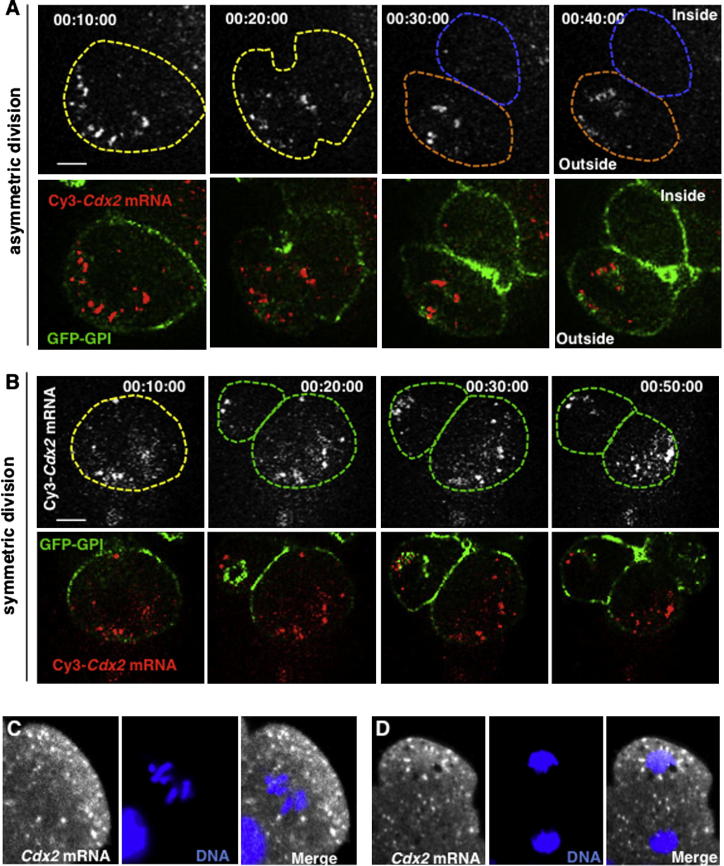
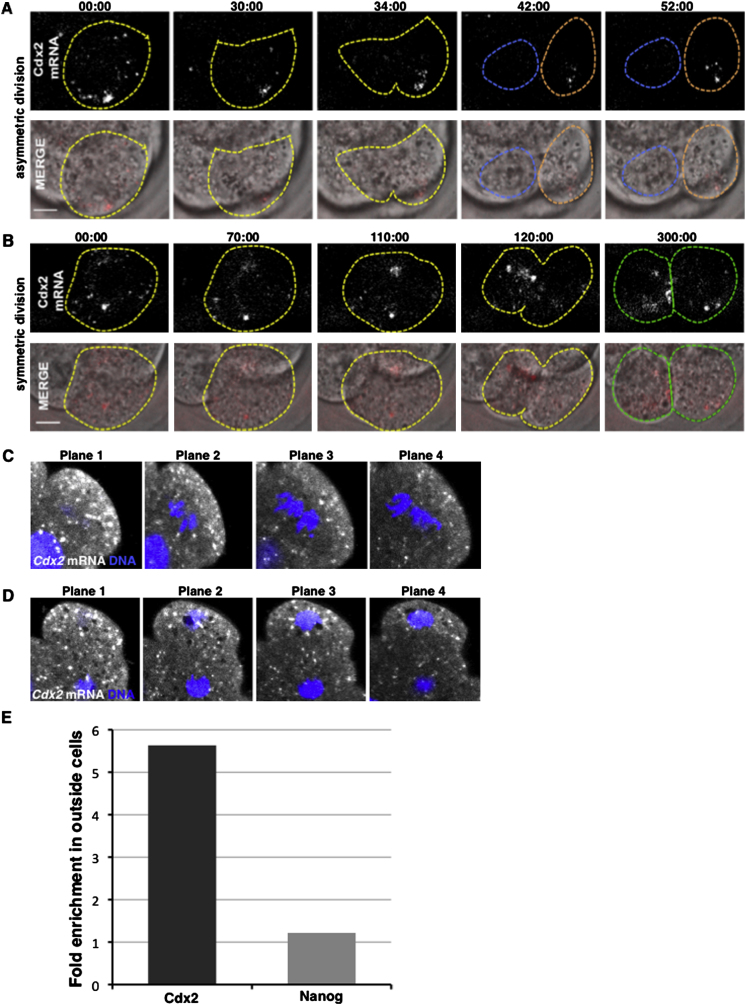
It is known that localizations of mRNAs can lead to an asymmetric inheritance by daughter cells upon cell division in many model systems. To determine whether this might be so in the mouse embryo, we followed the segregation of Cdx2 transcripts during divisions that generate cells of distinct fates. To that end, we injected fluorescent Cdx2 mRNA and imaged its distribution by time-lapse microscopy. We found that in differentiative divisions, in which cleavage furrows are perpendicular to the apical-basal axis and one daughter inherits the majority or entirety of the outer-facing apical domain, Cdx2 transcripts were inherited by the cell with the predominant apical domain (n = 10/10; Figure 2A; Figure S2A; Movie S3). In contrast, during conservative divisions, which generate outer daughter cells with apical domains equivalent in size, Cdx2 transcripts were inherited by both daughters in similar amounts (n = 11/11; Figure 2B; Figure S2B; Movie S4). This apical localization of transcripts could appear temporarily disrupted during mitosis but was then reestablished within 30–40 min of cytokinesis. We also attempted to catch embryos at the eight- to 16-cell transition to examine the distribution of endogenous Cdx2 by FISH. We found that at prometaphase, Cdx2 transcripts were enriched in apical regions, and at anaphase/telophase they were enriched in the cytoplasm destined for the outside rather than the inside daughter cell (17/34; Figures 2C and 2D; Figures S2C and S2D). The enrichment of Cdx2 transcripts in outside versus inside cells could also be detected by quantitative PCR (qPCR) following the cell division (Figure S2E). These results suggest that more Cdx2 transcripts become distributed to cells that initiate differentiation into TE rather than to cells that develop as pluripotent ICM.
Figure 2.
Cdx2 mRNA Localization at the Eight- to 16-Cell Transition
(A and B) Live imaging of exogenous Cy3-labeled Cdx2 mRNA distribution during asymmetric (A) and symmetric (B) divisions. Time points are presented in hr:min:s format.
(C and D) FISH detection of endogenous Cdx2 mRNA during the eight- to 16-cell transition in mitotic cells (C) and during telophase (D). Scale bar, 10 μm.
Figure S2.
Inheritance of Cdx2 mRNA during Eight- to 16-Cell Division, Related to Figure 2
(A and B) Time-lapse imaging of asymmetric (A) and symmetric divisions (B). Time-points (in hh:mm:ss format) were as indicated above the panels. Upper row shows the images taken in the red channel that results from the fluorescence of the Cy3-labeled Cdx2 mRNA. Lower panels represent the merge of the DIC and red channels. Note that outside cells inherits majority of the labeled mRNA during differentiative cell division, while both cells arising from conservative division inherit comparable amounts of the RNA. Cell boundaries highlighted in dashed lines. Scale bar, 10 μm.
(C and D) Localization of Cdx2 mRNA detected by FISH (white) in the eight-to-16 transiting cell. Four different focal planes are shown. DNA (blue) is stained with DAPI.
(E) Relative amounts of Cdx2 and Nanog mRNAs measured by qRT-PCR in outside and inside cells isolated after the eight-to-16 cell division. Bars represent the average fold enrichment of particular mRNA in outside cells (14 samples of three to four cells) as compared with inside cells (nine samples of three to four cells).
Cdx2 Transcript Localization Depends on a cis Element within the Coding Region
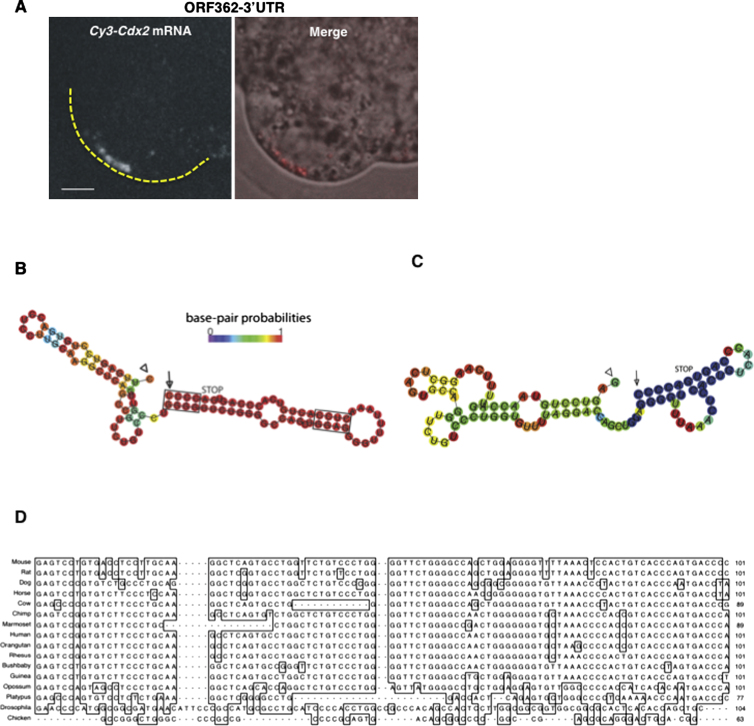
It is known that specific sequence elements within targeted mRNAs are able to direct them to subcellular localizations. These cis-acting elements (also known as localization elements or zipcodes) frequently reside in the 3′ untranslated region (UTR) of a targeted mRNA but can also be found within the 5′UTR or open reading frame (ORF) (Jambhekar and Derisi, 2007). To identify the element required for localization of Cdx2 mRNA, we first generated a series of truncated forms of the Cdx2 gene and followed their localization (Figure 3A). We found that whereas truncated RNA missing the 5′UTR and part of the ORF (499ORF-3′UTR) could localize to the apical region (n = 9/18, χ2 test, p = 0.994 compared with the wild-type (WT) mRNA; Figures 3B and 3J), truncated RNA missing the 3′UTR and part of the ORF (5′UTR-439ORF) could not, and was always distributed throughout the cytoplasm (n = 13/13; χ2 test, p < 0.001 compared with the WT mRNA; Figures 3C and 3J). Because RNA containing only the ORF construct also localized apically (n = 8/14, χ2 test, p = 1 compared with the WT mRNA; Figures 3D and 3J), we argued that a localization element must reside within the ORF of Cdx2, and, more specifically, within the last 499 nt. To define the localization element more tightly, we generated two further constructs comprising the 3′UTR and shorter segments (326 or 97 nt) of the 3′ part of the ORF (362ORF-3′UTR and 97ORF-3′UTR). Both of these RNAs distributed apically (n = 12/18, χ2 test, p = 0.502 [Figures 3J and S3A], and n = 19/31, χ2 test, p = 0.665 [Figures 3E and 3J], respectively, compared with the WT mRNA), suggesting that the 97 nt element at the 3′ of the ORF is sufficient to drive the localization.
To confirm the functionality of this element, we fused it to either RNA transcribed from the pBluescript plasmid or from GFP cDNA and determined whether it could direct localization of these unrelated RNAs. We found that the 97 nt element was able to confer apical localization upon these RNAs (n = 38/69, χ2 test, p = 1 [Figures 3F–3H], and n = 16/24, χ2 test, p = 0.422 [Figure 3I], respectively, compared with the full-length transcript). Together, these results suggest that a region of the Cdx2 transcript comprising the last 97 nt of the ORF is sufficient for localization. Moreover, because the 97 nt element in these last two constructs was placed out of the reading frame, translation of polypeptide from this element does not seem to play a role in apical localization.
Some RNA localization elements form hairpin stem loops (Hamilton and Davis, 2007), which led us to consider whether this might be the case for the 97 nt Cdx2 localization signal. We found that the lowest free-energy structure predicted by the mfold program (Extended Experimental Procedures) contains two stem loops, one of which (comprising the last 51 nt) has a high probability of base-pairing (Figure S3B) and contains runs of purines in the predicted stem that are similar to those previously reported to be important for localization signal activity (Bullock et al., 2010). To test whether this predicted structure plays a role, we introduced ten mutations that should abolish a hairpin loop but preserve the amino acid sequence (Figure S3C). This mutated RNA (FULL-mut97) was still able to localize apically (n = 29/43, χ2 test, p = 0.178 compared with WT mRNA; Figures 3G and 3J), suggesting that this putative structure is not required.
Figure S3.
Localization Element of Cdx2 mRNA, Related to Figure 3
(A) Fluorescently labeled RNA comprising 362 bases from 3′ of Cdx2 ORF and the whole 3′UTR localizes apically upon injection to compacted eight-cell blastomere (n = 12/18).
(B and C) RNA secondary structure prediction of the (B) WT and (C) mutated form of the 97 bases element from the 3′ end of the Cdx2 coding sequence (plus 4 additional bases after stop codon); arrow head and arrow mark the 5′ and 3′ end of RNA, respectively; base-pair probabilities are color coded. Runs of purins in WT version are boxed.
(D) The 97 bases long 3′ terminal part of Cdx2 coding sequence (including codon stop) is highly conserved in live-bearing, especially placental, mammals but not in monotremes or lower vertebrates and invertebrates.
Finally, to verify that the 97 nt sequence is necessary for Cdx2 mRNA localization, we created a full-length Cdx2 construct deleted only for the 97 nt element (Δ97nt). We found that the localization of labeled Δ97nt RNA was significantly reduced to 25% of injected cells (n = 13/51, χ2 test, p < 0.05; Figure 3J) in comparison with WT mRNA, but not totally abolished. This prompted us to examine the dynamics of localization of the Δ97nt form by time-lapse microscopy. This revealed that the Δ97nt form was transported apically (n = 14; Figures 3L and 3M; Movies S5 and S6). However, its average time of persistence at the apical region was significantly decreased to 46 min relative to 109 min for the WT ORF construct (n = 15, t test, p < 0.001; Figure 3K). Together, these results indicate that the 97 nt element is sufficient to drive the apical localization of Cdx2 transcripts and is also required for anchorage and/or stability of the transcripts at the apical cortex. Interestingly, the 97 nt at the 3′ terminal part of Cdx2 coding sequence is highly conserved in placental mammals (Figure S3D).
Cdx2 Transcript Localization Requires an Intact Cytoskeleton
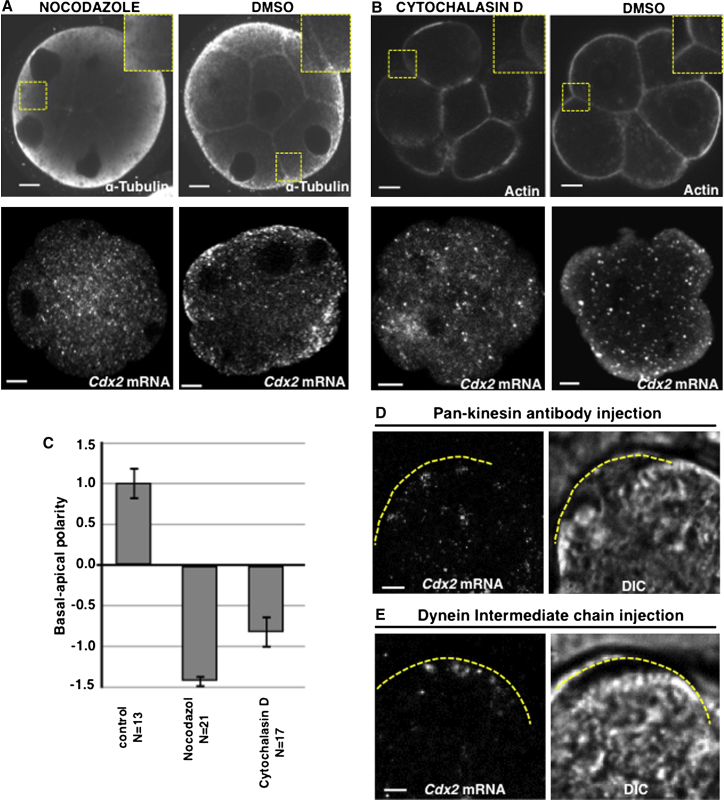
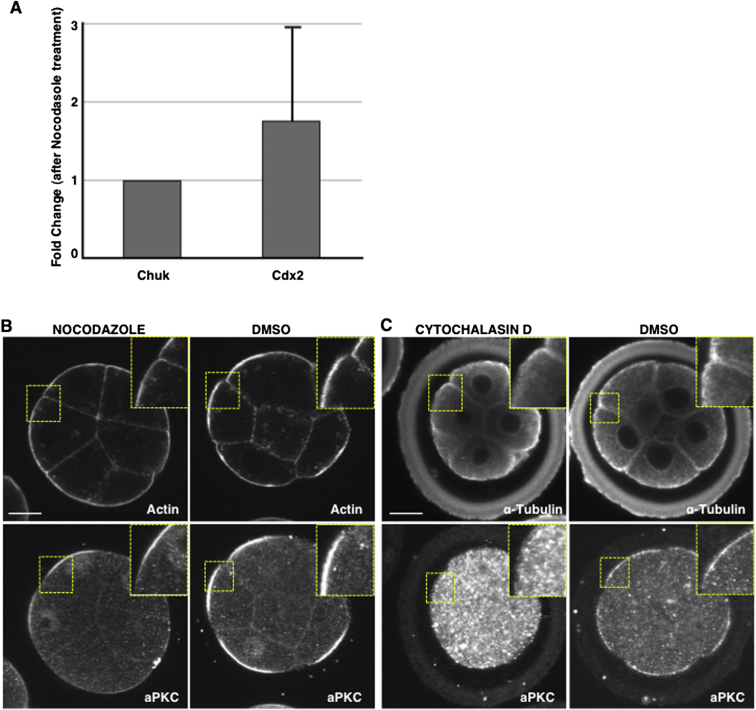
The actin and microtubule networks participate in the transport and anchoring of transcripts at target sites in many model systems (López de Heredia and Jansen, 2004). We therefore wished to determine whether this is also the case for Cdx2 mRNA. To disrupt the microtubule network, we used nocodazole and first confirmed that a 30 min treatment of eight-cell embryos was sufficient for this purpose (n = 20/20; Figures 4A and 4C) as previously reported (Johnson and Maro, 1984). We found that this treatment prevented the apical localization of Cdx2 transcripts in all compacted eight-cell embryos (n = 0/18 embryos, θ = 166° ± 3° and BA = −1.42 ± 0.08), whereas vehicle-treated controls were unaffected (n = 6/7 embryos, θ = 33° ± 12° and BA = 1.01 ± 0.20, χ2 test, p < 0.001). This was not due to decreased stability of the Cdx2 transcript, because the nocodazole did not significantly change the Cdx2 mRNA level determined by qRT-PCR (pairwise fixed reallocation randomization test, p (H1) = 0.12; Figure S4).
Figure 4.
Localization of Cdx2 mRNA Requires an Intact Cytoskeleton
(A and B) Embryos treated with nocodazole (A) and cytochalasin D (B) and with the vehicle control (DMSO). FISH detection of endogenous Cdx2 mRNA in eight- to 16-cell-stage embryos (lower panels in A [18 embryos] and B [12 embryos]). Immunostaining for α-tubulin (upper panels in A; n = 14). F-actin staining with phalloidin (upper panels in B; n = 15). Scale bar, 15 μm.
(C) Quantification of the polarization, represented as average BA value (±SEM), in individual cells of embryos treated with nocodazole and cytochalasin D.
(D and E) Localization of labeled Cdx2 mRNA following injection of anti-pan-kinesin (D; n = 0/20) or dynein intermediate chain (E; n = 4/7) antibodies. Scale bar, 5 μm.
See also Figure S4.
Figure S4.
Effect of Cytoskeleton Disruption on Cdx2 mRNA Expression and aPKC Localization, Related to Figure 4
(A) Quantitative RT- PCR was performed to embryos treated with 30 mM Nocodazole for 30 min and to the DMSO vehicle control group. Normalization was done against the gene Chuk. The Fold change between the Nocodazole treated group and the untreated group is shown. There is no significant difference in Cdx2 transcripts level between the treated and the untreated group (Pair Wise Fixed Reallocation Randomization Test, P(H1) = 0.12). Data represent the average of two biological replicates.
(B and C) Embryos treated with Nocodazole (B) and Cytochalasin D (C) and with the vehicle control (DMSO). F-actin staining with phalloidin (upper panels in (B) and immunostaining for α-tubulin (upper panels in (C). Immunostaining for aPKC (lower panels in (B) and (C); Note - the differences in aPKC staining of control embryos in (B) and (C) result from different fixation protocols used in the two experiments. In panel (C), methanol fixation was used to preserve microtubules and this resulted in non-specific staining of the zona. Scale bar, 15 μm.
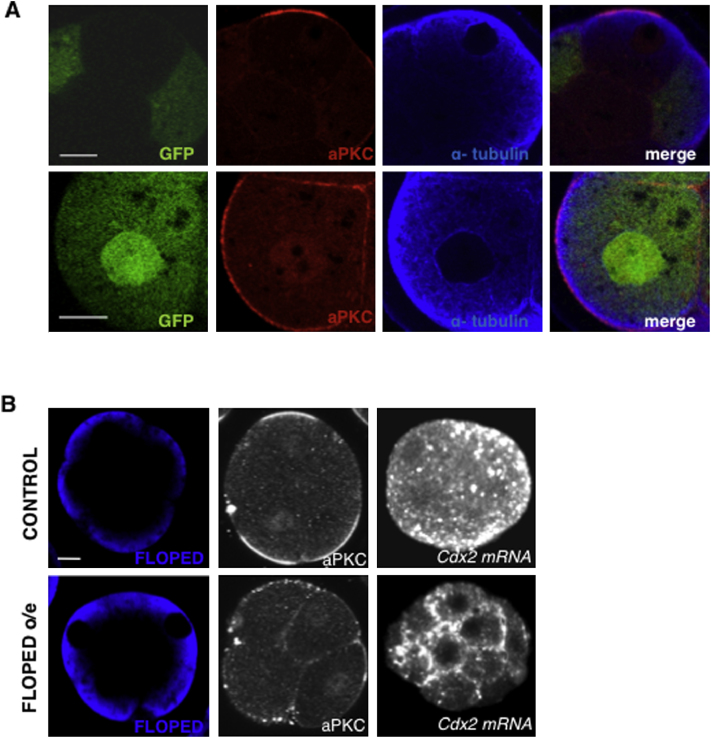
We next established that a 60-min treatment with cytochalasin-D resulted in partial depolymerization of the filamentous actin, as was evident from the disorganization of cortical actin, some embryo decompaction, and a decrease in overall cytoplasmic staining (n = 15/15; Figure 4B). This partial depolymerization of actin was sufficient to disrupt the apical localization of Cdx2 transcripts in all embryos examined (n = 0/12 embryos, θ = 140° ± 13° and BA = −0.82 ± 0.2), whereas control embryos were not affected (n = 12/12 embryos, θ = 33° ± 12° and BA = 1.01 ± 0.20; χ2 test, p < 0.001). The disruption of the actin cytoskeleton also caused some perturbation in microtubule organization, and vice versa, and both of these treatments caused mislocalization of atypical protein kinase C (aPKC) from apical poles (n = 15/15 and n = 17/18 embryos, respectively, for both χ2 tests, p < 0.001 compared with untreated embryos; Figures S4B and S4C).
Prompted by the fact that depolymerization of microtubules had a strong effect on Cdx2 transcript localization, we wished to determine whether a microtubule-dependent molecular motor could be involved. To that end, we injected compacted eight-cell blastomeres with either anti-pan-kinesin or anti-dynein intermediate chain antibodies, and 30–60 min later with labeled Cdx2 mRNA. None of the blastomeres injected with the pan-kinesin antibodies showed apical localization of transcripts 1.5–2 hr after injections (n = 0/20, χ2 test, p < 0.001 in comparison with Cdx2 mRNA injection in control embryos; Figure 4D), whereas antibodies against dynein had no observable effect (n = 4/7, Fisher’s exact test, p < 0.05 in comparison with Cdx2 mRNA injection in control embryos; Figure 4E). We confirmed the successful injection of antibodies by immunostaining (data not shown). Together, these results suggest that Cdx2 transcript localization requires either one or both of the microtubule and actin networks, which are interdependent at this developmental stage, and the activity of a motor protein of the kinesin superfamily.
Cdx2 Transcript Localization Requires Cell Polarization
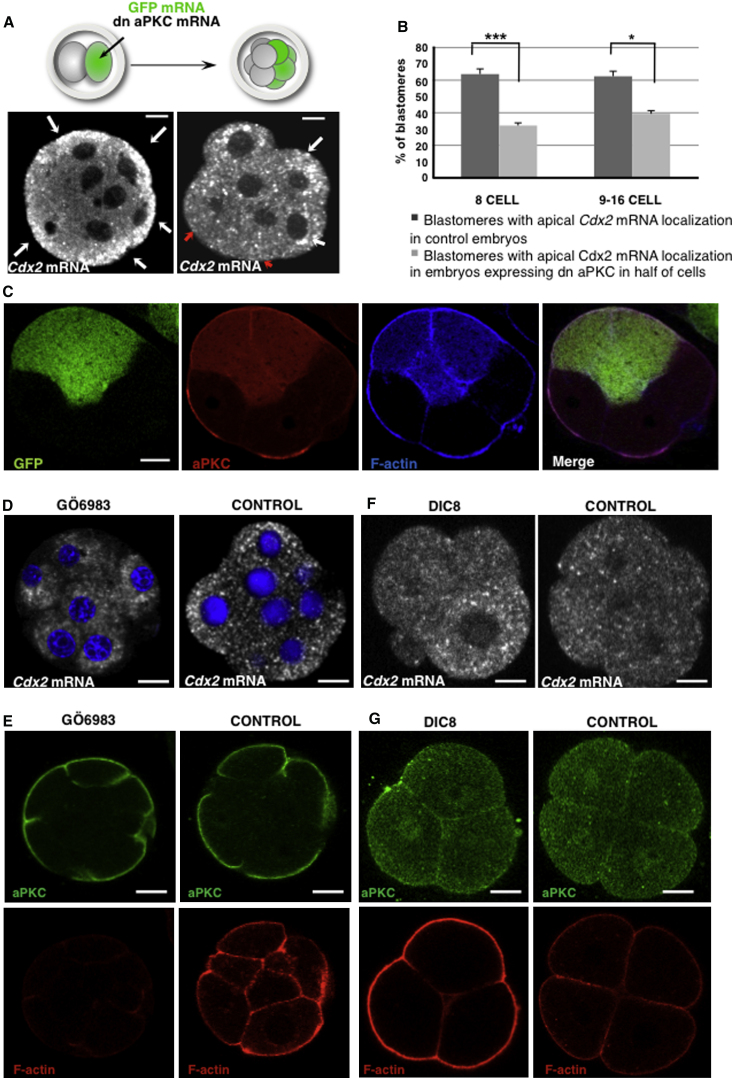
The apical enrichment of the Cdx2 transcripts upon apical-basal polarization suggested a possible role of cell polarization in this process. To determine whether this is the case, we first overexpressed the dominant-negative form of aPKC zeta (dn-aPKC), which disrupts cell polarization at the eight-cell stage (Plusa et al., 2005), together with GFP mRNA as a lineage marker. We coinjected these constructs into one cell at the late two-cell stage to restrict the effect to half of the embryo. The embryos were allowed to develop to the late eight-cell stage, and then Cdx2 mRNA was detected by FISH in one group and aPKC protein was detected by immunofluorescence in a second group (Figures 5A and 5B). Following downregulation of aPKC in half of the embryo, there were 2-fold fewer cells with apical Cdx2 transcripts than in control embryos (n = 18/56 cells, seven embryos, versus n = 51/80 cells, ten embryos; χ2 test, p < 0.001). Consistently, aPKC distribution was perturbed in cells descended from the injected blastomere in the second cohort of embryos (Figure 5C). Filamentous actin staining in the cytoplasm appeared stronger following treatment with dn-aPKC (Figure 5C), although there was no effect on the microtubule network (Figure S5).
Figure 5.
Disruption of Cell Polarity Abolishes Apical Localization of Cdx2 mRNA
(A–C) dn-aPkc mRNA along with lineage tracer (GFP mRNA) was injected into one blastomere of two-cell embryos that were then cultured until the late eight-cell stage.
(A) FISH detection of endogenous Cdx2 mRNA in control (left panel; n = 51/80, ten embryos) and dn-aPKC injected (right panel; n = 18/56, seven embryos) embryos. Cdx2 mRNA localized apically is indicated by white arrows, and its absence in some blastomeres is shown by red arrows. Scale bar, 5 μm.
(B) Quantification of the blastomeres that showed apical Cdx2 mRNA localization in the experiment shown in (A) and (B) (mean value ± SEM; Student’s t test; ∗p < 0.05, ∗∗∗p < 0.001).
(C) Effect of dn-aPKC (marked with GFP fluorescence, first panel) on localization of aPKC (second panel) and actin (third panel). Scale bar, 15 μm.
(D and E) Effect of aPKC inhibition with Gö6983 in compacted eight-cell blastomeres on Cdx2 mRNA localization (D; n = 8 embryos) and on actin (E, upper panel; n = 6 embryos) and aPKC (E, lower panel; n = 6 embryos) localization. Scale bar, 15 μm.
(F and G) Effect of premature compaction of four-cell embryos resulting from activation of PKCs with DiC8 on mRNA localization (F; n = 6 embryos), and on actin (G, upper panel; n = 6 embryos) and aPKC (G, lower panel; n = 6 embryos) localization. Scale bar, 15 μm.
See also Figure S5.
Figure S5.
Overexpression of dn-aPkc and Floped, Related to Figure 5
(A) Effect of overexpression of dominant-negative (dn) form of aPKC on microtubule cytoskeleton. Both, injected and un-injected eight-cell stage blastomeres are shown (upper panels). Blow up of the dn-aPKC expressing blastomeres. Notice, disruption of the aPKC staining but not difference in alpha-tubulin staining (lower panels).
(B) Effect of overexpression of Floped protein on localization of aPKC and Cdx2 mRNA in compacted eight-cell stage embryos.
In a second series of experiments, we confirmed the requirement for aPKC activity on Cdx2 mRNA localization by treating eight-cell embryos with Gö6983, an inhibitor of aPKC activity (particularly PKC zeta). A 60-min treatment with Gö6983 completely abolished Cdx2 mRNA localization (n = 0/8 embryos, θ = 136° ± 12° and BA = −0.46 ± 0.13, χ2 test, p < 0.01; Figure 5D) compared with vehicle only-treated control embryos (n = 6/6 embryos, θ = 18° ± 4° and BA = 1.33 ± 0.11, Fisher’s exact test, p < 0.001). The inhibitor did not affect the localization of aPKC but had a strong effect on actin, as evidenced by loss of phalloidin staining of cytoplasmic and cortical/microvilli (Figure 5E).
Finally, we wondered whether induction of premature embryo compaction would be sufficient for Cdx2 mRNA localization. We therefore induced compaction by treating four-cell embryos with DiC8, a diacylglycerol analog and an activator of PKCs, and monitored compaction by phalloidin staining and differential interference contrast (DIC) microscopy (data not shown). Such prematurely induced compaction did not lead to either premature apical localization of aPKC in all examined embryos (n = 0/6 embryos; Fisher’s exact test p < 0.05; Figure 5G) or localization of Cdx2 transcripts (n = 0/6 embryos, Fisher’s exact test p < 0.05; Figure 5F). These results indicate that Cdx2 mRNA localization requires aPKC kinase activity that is associated with cell polarization at the eight-cell stage.
Preventing Cdx2 Transcript Localization Leads to Expression of CDX2 Protein in ICM
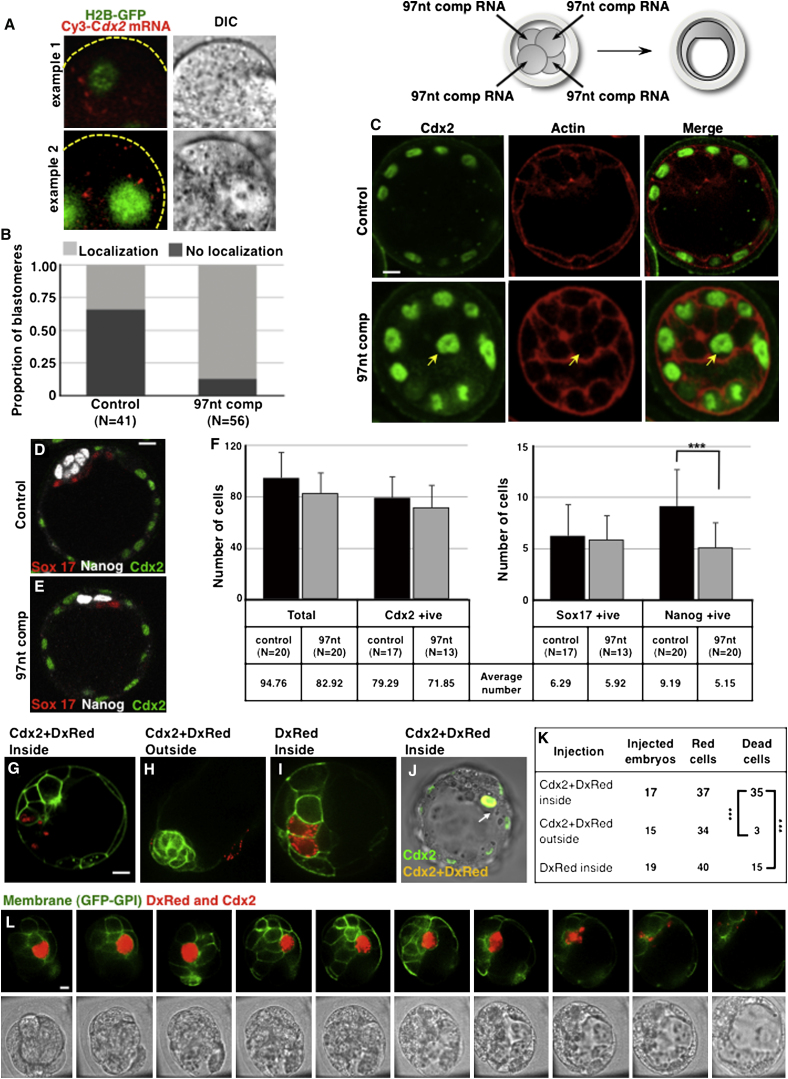
Having observed that Cdx2 mRNA is inherited mainly by outside cells upon cell division at the eight- to 16-cell transition, we wished to determine whether this is important for cell-fate specification. To address this, we needed to prevent Cdx2 mRNA localization and determine the effect on cell fate. To achieve this, we injected blastomeres with the 97nt RNA containing the Cdx2 localization element to compete for localization with endogenous Cdx2 transcripts. To determine the efficiency of this approach, we first coinjected a 10× molar excess of the 97nt RNA with labeled Cdx2 mRNA, and found a 5-fold reduction of cells showing apical Cdx2 mRNA in comparison with control cells injected with unrelated RNA of comparable size (χ2 test, p < 0.001; Figures 6A and 6B). Having established this proof of principle, we injected all blastomeres at the four-cell stage with 97nt (comp) RNA or with unrelated RNA (control), and cultured embryos until the blastocyst stage. We found that the embryos injected with control RNA expressed CDX2 only in the TE (n = 14 embryos), as expected. In contrast, in >75% of embryos injected with the 97nt RNA, CDX2 protein was clearly detectable in one or more inside cells (n = 10/13 embryos, χ2 test, p < 0.001; Figure 6C). Thus, interfering with the Cdx2 transcripts’ localization machinery results in CDX2-positive inside cells.
Figure 6.
Mislocalization of Cdx2 mRNA in the Eight-Cell Embryo Results in CDX2 Protein in Inside Cells and a Decrease in Epiblast Cell Number
(A) Coinjection of excess of RNA containing competitor Cdx2 97 nt localization signal (97nt comp) with labeled Cdx2 mRNA abolishes apical localization of Cdx2 mRNA, which now localizes to basolateral regions.
(B) Quantification of cells showing apical localization of labeled Cdx2 mRNA upon coinjection of 97nt comp (competitor; n = 56) or control unrelated RNA of similar size (n = 41); the difference is statistically significant (χ2 test, p < 0.001).
(C–F) All cells of the four-cell embryo were injected with control or 97nt comp RNA and allowed to develop until the early (C) or mature blastocyst (D–F) stage.
(C) Representative images of embryos stained for Cdx2 (green) and actin (red). Yellow arrow marks an inside cell positive for Cdx2 in the embryo injected with 97nt comp RNA (n = 10/13); control injected embryos do not express Cdx2 in inside cells (n = 0/14).
(D and E) Examples of embryos injected with control (D) and 97nt comp (E) RNA stained for CDX2 (green), SOX17 (red), or NANOG (white).
(F) Quantification of the percentage (±SEM) of cells positive for Cdx2, Sox17, or Nanog in blastocyst (∼96-cell stage).
(G–L) Single inside (G, J, and L; n = 17) or outside (H; n = 15) cells of a 16-32 cell embryos injected with Cdx2 mRNA and DxRed (G, H, and J) or with DxRed alone (I; n = 19). Progeny of inside cells injected with Cdx2 mRNA shows nuclear CDX2 at blastocyst stage (H, arrow; green plus red = yellow). Scale bar, 10 μm.
(K) Summary of injection data (Fisher’s exact test, ∗∗∗p < 0.001).
(L) Time-lapse series of an embryo with one inside cell injected with Cdx2 mRNA and DxRed.
See also Figure S6.
Preventing CDX2 Transcript Localization Decreases the Number of Pluripotent Cells
In order to determine the developmental effects of Cdx2 transcript mislocalization, we performed an experiment similar to the one described above, but this time allowed embryos to develop further so that we could assess the precise cell lineage allocation (Figures 6D and 6E). We found that injection of the 97nt RNA reduced the number of epiblast (NANOG-expressing) cells significantly (t test, p < 0.001) compared with control embryos (average values of 5.2 versus 9.2 cells, respectively; n = 20). In contrast, development of the TE and primitive endoderm was not significantly affected (Figure 6F).
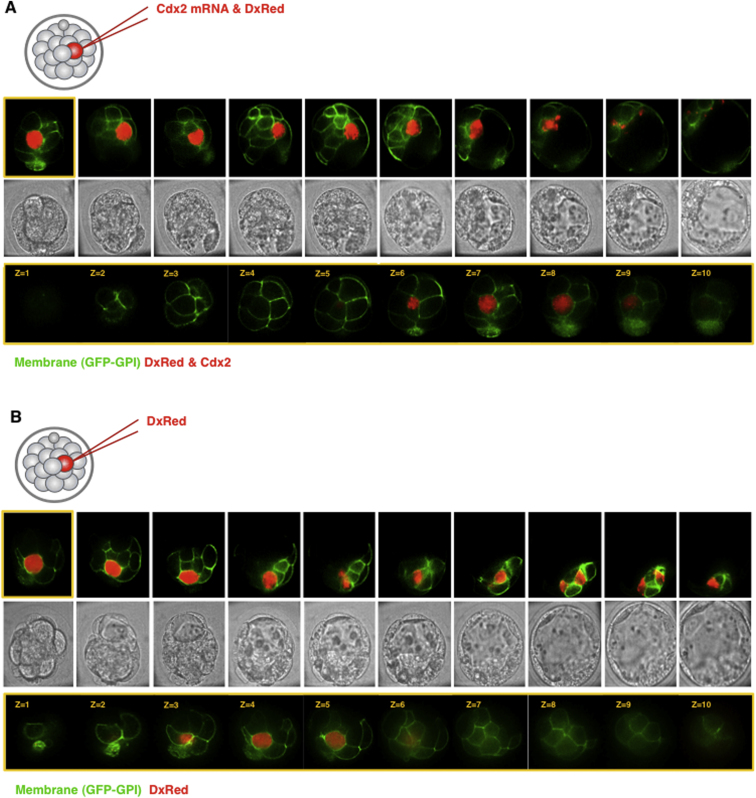
To address how mislocalization of CDX2 to inside cells can lead to fewer epiblast cells, we overexpressed CDX2 in inside cells (or outside cells as control) together with a lineage marker, Rhodamine Dextran (DxRed), to follow their fate directly by four-dimensional (4D) time-lapse microscopy (Figure S6). This revealed that inside cells injected with Cdx2 mRNA at the 16- to 32-cell stage developed normally at first and gave rise to 37 inside progeny (n = 17 embryos). However, as the embryos reached the mature blastocyst stage, 95% of CDX2-expressing cells underwent cell death (n = 35/37; Figures 6G, 6K, and 6L). The timing of the apoptosis of CDX2-positive cells coincided with the onset of apoptosis known to occur in normal development (Handyside and Hunter, 1986; Morris et al., 2010). The two CDX2-positive cells that survived gave rise to primitive endoderm (Figure 6J). In contrast, outside cells injected with Cdx2 mRNA gave rise to 34 cells, of which only three (9%) underwent apoptosis (n = 3/34; Figures 6H and 6K). To eliminate the possibility that injection of inside cells per se might lead to cell death, we injected inside cells with DxRed alone (n = 19 embryos). These cells gave rise to 40 inside progeny, 37% of which died (n = 15/40; Figures 6I and 6K). This suggests that the increased death of inside cells could be due in part to their greater sensitivity to injection per se, but most likely it reflects the known incidence of cell death within the ICM. This is because the rate of cell death in inside CDX2-positive cells was significantly greater than in both control groups of cells (Fisher’s exact test, two-tailed, p < 0.001; Figure 6K), indicating that a high CDX2 level is not well tolerated in the ICM. Together, our results suggest that preventing Cdx2 mRNA apical localization leads to a decrease in pluripotent cell numbers.
Figure S6.
Overexpression of Cdx2 in the Inside Cells of a Morula-Stage Embryo, Related to Figure 6
(A and B) Time-lapse series of a typical embryo with one inside cell injected with either Cdx2 mRNA and dextran red – DxRed – (A) or DxRed alone (B). Embryos are expressing membrane form of GFP (green) to visualize cell boundaries to track cell behavior.
Discussion
The results we present here indicate that transcripts for Cdx2, a critical factor in the first cell-fate decision in the mouse embryo, preferentially localize asymmetrically toward the apical region of blastomeres at the late eight-cell stage. Cdx2 mRNA localization depends on a 97 nt cis element within the ORF and requires intact actin and microtubule cytoskeletons as well as apical-basal cell polarization. We find that the asymmetric localization of Cdx2 mRNA leads to the acquisition of more Cdx2 by outside cells compared with inside cells. This asymmetry likely contributes to the elevated CDX2 expression that directs the development of outside cells into TE. But perhaps more importantly, it can serve to ensure that inside cells do not inherit Cdx2 transcripts and consequently CDX2 protein, thus enabling them to develop as the pluripotent cell lineage. Our findings therefore indicate that differentiative divisions are truly asymmetric because they lead to the asymmetric segregation of at least one factor that is critical for cell differentiation: Cdx2 mRNA. When cells divide conservatively, each daughter inherits similar levels of Cdx2 mRNA, and thus, from this perspective, these divisions can be considered symmetric. After the completion of cell division, Cdx2 transcripts relocalize to the apical regions of cells. This may be important for ensuring that in the second round of differential divisions, inside cells do not become enriched with CDX2.
Our studies of Cdx2 transcript localization dynamics indicate that the mechanism of their localization depends on a 97 nt element within the coding region of Cdx2. This is suggested by three observations: First, a 97 nt element from the 3′ end of the ORF is sufficient to position Cdx2 mRNA apically. Second, other RNAs tagged with this 97 nt element localize apically. Third, Cdx2 transcripts lacking this element show only transient localization, indicating that the 97 nt element provides anchorage/stability of the transcript in the apical region. Although localization elements are more usually placed within the UTRs, they are known to reside within the ORFs of targeted mRNAs in other model systems (Jambhekar and Derisi, 2007). This 97 nt is highly conserved in the Cdx2 genes of placental mammals, raising the possibility that Cdx2 mRNA might also localize in other species. Although it remains unknown which parts of the 97 nt element are essential for the apical anchorage, our mutational studies suggest that is not a predicted loop structure. Our results also suggest that another apical transporting sequence may lie outside of the 97 nt element, but that this sequence is unable to mediate the efficient anchorage or stability of the transcript at this site. Such binary elements mediating redundant and/or developmentally orchestrated functions in other localizing mRNAs have been reported (Simmonds et al., 2001).
Our results indicate that the maintenance or stability of the Cdx2 mRNA at the apical region requires the integrity of the microtubule and actin cytoskeletons. Transcript localization also requires cell polarization because disruption of aPKC zeta, which is critical for cell polarity at this stage (Plusa et al., 2005), leads to the loss of Cdx2 mRNA’s apical enrichment. Additionally, we find that overexpression of a component of the subcortical maternal complex FLOPED (Li et al., 2008), which interferes with eight-cell embryo polarization, results in Cdx2 transcript relocalization to basolateral regions (Figure S5B). Whether this effect of cell polarity on mRNA localization is direct or indirect remains unknown. Importantly, we find that experimentally induced premature embryo compaction cannot induce the localization of Cdx2 transcripts, which suggests that correct cell polarization at the eight-cell stage, rather than compaction per se, is required.
We can detect asymmetric Cdx2 transcripts in ∼80% of cells in late eight-cell-stage embryos. This raises the question of what happens in the remaining 20% of cells. At least four possibilities can be considered. First, it is possible that polarization of Cdx2 mRNA does not arise simultaneously in all blastomeres. This would accord with the known lack of synchrony of preimplantation development. A second possibility is that Cdx2 transcript distribution does not polarize in all cells, which would lead to some inside cells inheriting Cdx2 mRNA. This would explain the fraction of inside cells reported to contain Cdx2 protein (Dietrich and Hiiragi, 2007). Third, Cdx2 mRNA localization might be prevented by disruption of the apical region through the process of cell injection (Morris, 2011; Yamanaka, 2011). We consider this possibility to be less likely, because we examined Cdx2 mRNA asymmetry 1.5–2 hr after injection, when polarity is likely to have been restored. Fourth, polarization of Cdx2 transcripts might not be detectable in blastomeres in which Cdx2 expression is very low. Indeed expression of CDX2 is heterogeneous at the eight-cell stage (Jedrusik et al., 2008; Ralston and Rossant, 2008).
It has been suggested that Cdx2 mRNA is provided not only zygotically but also maternally (Jedrusik et al., 2010; Wu et al., 2010), and that simultaneous depletion of both maternal and zygotic Cdx2 pools has a stronger effect on TE development than depletion of only the zygotic pool (Jedrusik et al., 2010). The maternal pool of Cdx2 appears not to be essential on its own (Blij et al., 2012). However, whether the maternal provision of Cdx2 message is superfluous or helpful in enhancing TE specification is not clear and awaits a thorough comparison of the developmental dynamics of embryos with both maternal and zygotic pools of Cdx2 eliminated versus embryos in which only the zygotic pool is removed. It is important to note that the work we present here does not distinguish between maternal and zygotic pools of Cdx2 mRNA.
The asymmetric localization of Cdx2 mRNA is likely to have important implications for the first cell lineage specification (Figure 7), for two main reasons. First, polarized localization of Cdx2 transcripts can contribute to increased levels of CDX2 protein in outside cells, which is critical for TE development (Strumpf et al., 2005; Jedrusik et al., 2008). Such initial asymmetry in Cdx2 mRNA would be further enhanced through the nuclear translocation of YAP, which together with TEAD4 promotes Cdx2 expression via transcriptional control (Yagi et al., 2007; Nishioka et al., 2008, 2009). Second, asymmetric localization and inheritance of Cdx2 mRNA is likely to contribute to the development of pluripotency of inside cells by ensuring low levels of CDX2 in these cells, which downregulates pluripotency gene expression (Niwa et al., 2005). In support of this, our results show that interference with Cdx2 mRNA localization leads to CDX2 protein synthesis in inside cells, and that this interferes with their development along the pluripotent lineage. We therefore suggest that assignment of TE identity and de facto ICM identity during the first fate decision draws upon more than one mechanism. It is perhaps for this reason that mammalian embryos can call upon regulative developmental mechanisms more effectively than their more deterministic metazoan relatives. In agreement with this, the preferential inheritance of Cdx2 transcripts by outside, TE-destined cells might provide an elegant cell-fate “insurance policy” that would help negate any inappropriate Hippo signaling input. Such inappropriate signaling could arise from cell divisions that are neither parallel nor perpendicular to the apical-basal axis (Sutherland et al., 1990) and affect the size of cell-to-cell contact domains on which Hippo signaling might depend. In conclusion, our results add support to the notion that cell polarity and CDX2-mediated cell fate are interdependent, and bring together earlier models proposed for cell-fate specification (Figure 7). At the same time, they raise a new possibility that the polarized localization of mRNA for other cell-fate factors at the eight-cell stage might contribute to cell-fate specification in mouse embryo.
Figure 7.
Working Model for How the Localization and Partitioning of Cdx2 Transcripts together with Differential Transcription Lead to Lineage Segregation
(Top) Symmetric cell division generates two outside cells (green) that inherit similar amounts of Cdx2 transcripts. Asymmetric division generates an outside daughter that inherits more Cdx2 transcripts (green) than the inside daughter (yellow).
(Bottom) Cell polarization leads to the asymmetric localization of Cdx2 transcripts and their differential inheritance, contributing to elevated levels of CDX2 in outside cells (as we show here). This CDX2 asymmetry is then reinforced by a transcriptional response to TEAD4 and the nuclear localization of YAP in outside cells (Yagi et al., 2007; Nishioka et al., 2008, 2009). In turn, the elevated CDX2 level reinforces cell polarization (Jedrusik et al., 2008) and represses the expression of pluripotency factors (Niwa et al., 2005) to promote differentiation into the TE lineage. In contrast, the reduced CDX2 level in inside cells is a consequence of (1) reduced inheritance of Cdx2 transcripts, (2) prevention of Cdx2 transcription as a result of the cytoplasmic retention of YAP, and (3) the repressive effects of OCT4 and NANOG that promote pluripotent fate.
Experimental Procedures
Embryo Collection and Culture
Embryos were collected from F1 (C57Bl6 × CBA) females mice superovulated with 10 IU of PMSG (Intervet) and 10 IU of human chorionic gonadotropin (HCG; Intervet) 48 hr later and mated with F1 or with EGFP-H2B (Hadjantonakis and Papaioannou, 2004) males. The embryos were recovered in M2 medium with 4 mg/mL BSA and cultured in KSOM medium under paraffin oil at 37.5°C in 5% CO2 as previously described (Bischoff et al., 2008). Eight-cell embryos were obtained by collecting four-cell embryos 56–61 hr after HCG injection and culturing either for a further 1–2 hr or for 10–12 hr after division to the eight-cell stage.
Whole-Mount RNA FISH
FISH was performed as previously described (Chazaud et al., 2006). Digoxigenin-labeled RNA probes were generated by direct in vitro transcription using the T7 MEGA-Script kit (Ambion). Details of the protocol are provided in Extended Experimental Procedures.
Synthesis of Fluorescently Labeled, Capped RNA and Microinjections
In vitro synthesis of mRNA was performed as previously described (Bullock and Ish-Horowicz, 2001). Linearized plasmid template was transcribed in a solution containing 0.4 mM ATP, 0.4 mM CTP, 0.36 mM UTP, 0.04 mM Cy3-UTP (Perkin Elmer), 0.12 mM GTP, and m7G(5′)ppp(5′)G cap analog (Ambion) using T3 (pBluescriptRN3P) or T7 (pBluescriptSK+) RNA polymerases (Roche) according to the manufacturer’s instructions. The resultant RNA was treated with DNase I (Ambion), purified using the MegaClear kit (Ambion), and precipitated with NH4OAc/EtOH. One or more blastomeres of compacted eight-cell embryos were injected at a concentration of 500 ng/μL. The integrity of the RNA was checked by electrophoresis before injection. Injection of fluorescently labeled Drosophila transcripts (kindly provided by Simon Bullock) adopted a dispersed distribution similar to that observed for our control RNAs. None of the synthetic RNAs we used impaired development.
Constructs and mRNA Injection
A full-length ORF Cdx2 cDNA clone containing the majority of the 3′ and 5′ UTRs was obtained from Geneservice (Cambridge, UK) and cloned into pBluescript SK + plasmid. Truncated forms of Cdx2 were obtained by subcloning of respective fragments. dnaPKC, Floped, and GFP mRNAs were prepared and injected into blastomeres as previously described (Zernicka-Goetz et al., 1997). The embryos were cultured until the eight-cell stage and then processed for FISH and immunostaining. To obtain statistically significant numbers, all experiments were performed on embryos from more than a single batch, and several independent experiments were conducted.
Cdx2 mRNA (50 ng/μl) was injected into cells of 16- to 32-cell embryos expressing membrane-bound GFP protein with DxRed as a lineage tracer. The embryos were imaged as described previously (Morris et al., 2010).
97nt comp RNA (total length of ∼200 nt) was transcribed from a DNA construct comprising the mRNA localization signal of Cdx2 in pBluescript vector. Control RNA consisted of ∼200 nt of a 5′ sequence of GFP mRNA. The RNAs were injected into all four-cell embryo cells, which were then allowed to develop until blastocyst stage.
Image Analysis
To calculate the PI of the FISH signal, a previously described algorithm (Park et al., 2012) was adopted and implemented in JRuby as an ImageJ plugin or in MATLAB using MathWorks (Extended Experimental Procedures). Fluorescence intensity plots were derived using MATLAB. All data analyses were performed on raw images.
4D Imaging
Time-lapse imaging of labeled RNA was performed using a spinning-disk confocal microscope (Marianas; Intelligent Imaging Innovations) with Slidebook (Intelligent Imaging Innovations) software and a camera (QuantEM 512SC; Yokogawa). Images were taken with 500 ms exposure in focal planes spaced 3.5 μm apart and using a 63 × Plan-Neofluar water immersion objective.
Immunofluorescence
Embryos were fixed in 4% paraformaldehyde in PBS containing 0.1% Tween20 for 30 min at 37°C or in 10% DMSO in methanol for 5 min at −20°C. The samples were washed in 0.1% Tween20 in PBS (PBST), permeabilized in 0.25% Triton X-100 in PBS for 20 min at room temperature, washed in PBST, and incubated in blocking solution (3% BSA in PBST) for 2 hr at room temperature as previously described (Jedrusik et al., 2008). The antibodies used are listed in Extended Experimental Procedures.
Acknowledgments
We are grateful to Simon Bullock, David Glover, Claire Chazaud, Rui Pires Martins, Alexander Bruce, Kitai Kim, Isabel Palacios, and Alejandra Gardiol for help and advice. This work was supported by a Wellcome Trust Senior Research Fellowship to M.Z.G. M.S. was funded by the Greek State Scholarships Foundation.
Published: January 31, 2013
Footnotes
Supplemental Information includes Extended Experimental Procedures, six figures, and six movies and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.01.006.
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information
Movie of a compacted eight-cell-stage blastomere injected with fluorescently labeled Cdx2 mRNA. The movie shows the green channel (membrane-GFP), the red channel (Cy3-Cdx2 mRNA), and the merge of the two channels. Timing is displayed in the upper-left corner.
Dynamics of Cdx2 mRNA localization immediately after cell division of the eight-cell blastomere. The movie shows the red channel (Cy3-Cdx2 mRNA) and the merge with DIC. Timing is displayed in the upper-left corner. An asterisk marks the timing and position of Cdx2 mRNA localization.
Cdx2 mRNA localization during a differentiative division of an eight-cell blastomere. The movie shows three different focal planes of the same dividing cell. Each focal plane is a merge of the green (membrane-GFP) and red (Cy3-Cdx2 mRNA) channels. Timing is displayed in the upper-right corner.
Cdx2 mRNA localization during a conservative division of eight-cell blastomeres. The movie shows one focal plane as a merge of the green (membrane-GFP) and red (Cy3-Cdx2 mRNA) channels. Timing is displayed in the upper-left corner.
Movies of fluorescently labeled Cdx2 ORF RNA injected into a compacted blastomere of an eight-cell embryo. Notice that the RNA persists at the cortex for the whole length of the movie. The timing is displayed in the upper-left corner.
Movies of fluorescently labeled Δ97bp RNA injected into a compacted blastomere of an eight-cell embryo. Notice that the RNA gets quickly removed from the cortex. Timing is displayed in the upper-left corner.
References
- Avilion A.A., Nicolis S.K., Pevny L.H., Perez L., Vivian N., Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff M., Parfitt D.E., Zernicka-Goetz M. Formation of the embryonic-abembryonic axis of the mouse blastocyst: relationships between orientation of early cleavage divisions and pattern of symmetric/asymmetric divisions. Development. 2008;135:953–962. doi: 10.1242/dev.014316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blij S., Frum T., Akyol A., Fearon E., Ralston A. Maternal Cdx2 is dispensable for mouse development. Development. 2012;139:3969–3972. doi: 10.1242/dev.086025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A.W., Zernicka-Goetz M. Developmental control of the early mammalian embryo: competition among heterogeneous cells that biases cell fate. Curr. Opin. Genet. Dev. 2010;20:485–491. doi: 10.1016/j.gde.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Bullock S.L., Ish-Horowicz D. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature. 2001;414:611–616. doi: 10.1038/414611a. [DOI] [PubMed] [Google Scholar]
- Bullock S.L., Ringel I., Ish-Horowicz D., Lukavsky P.J. A’-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat. Struct. Mol. Biol. 2010;17:703–709. doi: 10.1038/nsmb.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chazaud C., Yamanaka Y., Pawson T., Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Dietrich J.E., Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis A.K., Papaioannou V.E. Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol. 2004;4:33. doi: 10.1186/1472-6750-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton R.S., Davis I. RNA localization signals: deciphering the message with bioinformatics. Semin. Cell Dev. Biol. 2007;18:178–185. doi: 10.1016/j.semcdb.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handyside A.H., Hunter S. Cell division and death in the mouse blas- tocyst before implantation. Rouxs Arch. Dev. Biol. 1986;195:519–526. doi: 10.1007/BF00375893. [DOI] [PubMed] [Google Scholar]
- Holt C.E., Bullock S.L. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A., Derisi J.L. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 2007;13:625–642. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik A., Parfitt D.E., Guo G., Skamagki M., Grabarek J.B., Johnson M.H., Robson P., Zernicka-Goetz M. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev. 2008;22:2692–2706. doi: 10.1101/gad.486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik A., Bruce A.W., Tan M.H., Leong D.E., Skamagki M., Yao M., Zernicka-Goetz M. Maternally and zygotically provided Cdx2 have novel and critical roles for early development of the mouse embryo. Dev. Biol. 2010;344:66–78. doi: 10.1016/j.ydbio.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.H., Maro B. The distribution of cytoplasmic actin in mouse 8-cell blastomeres. J. Embryol. Exp. Morphol. 1984;82:97–117. [PubMed] [Google Scholar]
- Johnson M.H., Ziomek C.A. The foundation of two distinct cell lineages within the mouse morula. Cell. 1981;24:71–80. doi: 10.1016/0092-8674(81)90502-x. [DOI] [PubMed] [Google Scholar]
- Li L., Baibakov B., Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev. Cell. 2008;15:416–425. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Yang X., Wasser M., Cai Y., Chia W. Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell. 1997;90:437–447. doi: 10.1016/s0092-8674(00)80504-8. [DOI] [PubMed] [Google Scholar]
- López de Heredia M., Jansen R.P. mRNA localization and the cytoskeleton. Curr. Opin. Cell Biol. 2004;16:80–85. doi: 10.1016/j.ceb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Meignin C., Davis I. Transmitting the message: intracellular mRNA localization. Curr. Opin. Cell Biol. 2010;22:112–119. doi: 10.1016/j.ceb.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Melton D.A. Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature. 1987;328:80–82. doi: 10.1038/328080a0. [DOI] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Morris S.A. Cell fate in the early mouse embryo: sorting out the influence of developmental history on lineage choice. Reprod. Biomed. Online. 2011;22:521–524. doi: 10.1016/j.rbmo.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Morris S.A., Teo R.T., Li H., Robson P., Glover D.M., Zernicka-Goetz M. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc. Natl. Acad. Sci. USA. 2010;107:6364–6369. doi: 10.1073/pnas.0915063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman-Silberberg F.S., Schüpbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell. 1993;75:165–174. [PubMed] [Google Scholar]
- Nishioka N., Yamamoto S., Kiyonari H., Sato H., Sawada A., Ota M., Nakao K., Sasaki H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Nishioka N., Inoue K., Adachi K., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R.O., Ogonuki N. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Palmieri S.L., Peter W., Hess H., Schöler H.R. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev. Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- Park H.Y., Trcek T., Wells A.L., Chao J.A., Singer R.H. An unbiased analysis method to quantify mRNA localization reveals its correlation with cell motility. Cell Rep. 2012;1:179–184. doi: 10.1016/j.celrep.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusa B., Frankenberg S., Chalmers A., Hadjantonakis A.K., Moore C.A., Papalopulu N., Papaioannou V.E., Glover D.M., Zernicka-Goetz M. Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J. Cell Sci. 2005;118:505–515. doi: 10.1242/jcs.01666. [DOI] [PubMed] [Google Scholar]
- Ralston A., Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev. Biol. 2008;313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Russ A.P., Wattler S., Colledge W.H., Aparicio S.A., Carlton M.B., Pearce J.J., Barton S.C., Surani M.A., Ryan K., Nehls M.C. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Simmonds A.J., dosSantos G., Livne-Bar I., Krause H.M. Apical localization of wingless transcripts is required for wingless signaling. Cell. 2001;105:197–207. doi: 10.1016/s0092-8674(01)00311-7. [DOI] [PubMed] [Google Scholar]
- St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Strumpf D., Mao C.A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Sutherland A.E., Speed T.P., Calarco P.G. Inner cell allocation in the mouse morula: the role of oriented division during fourth cleavage. Dev. Biol. 1990;137:13–25. doi: 10.1016/0012-1606(90)90003-2. [DOI] [PubMed] [Google Scholar]
- Yagi R., Kohn M.J., Karavanova I., Kaneko K.J., Vullhorst D., DePamphilis M.L., Buonanno A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–3836. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y. Response: Cell fate in the early mouse embryo—sorting out the influence of developmental history on lineage choice. Reprod. Biomed. Online. 2011;22:525–527. doi: 10.1016/j.rbmo.2011.03.011. discussion 528. [DOI] [PubMed] [Google Scholar]
- Wu G., Gentile L., Fuchikami T., Sutter J., Psathaki K., Esteves T.C., Araúzo-Bravo M.J., Ortmeier C., Verberk G., Abe K., Schöler H.R. Initiation of trophectoderm lineage specification in mouse embryos is independent of Cdx2. Development. 2010;137:4159–4169. doi: 10.1242/dev.056630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernicka-Goetz M., Pines J., McLean Hunter S., Dixon J.P.C., Siemering K.R., Haseloff J., Evans M.J. Following cell fate in the living mouse embryo. Development. 1997;124:1133–1137. doi: 10.1242/dev.124.6.1133. [DOI] [PubMed] [Google Scholar]
Supplemental References
- Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie of a compacted eight-cell-stage blastomere injected with fluorescently labeled Cdx2 mRNA. The movie shows the green channel (membrane-GFP), the red channel (Cy3-Cdx2 mRNA), and the merge of the two channels. Timing is displayed in the upper-left corner.
Dynamics of Cdx2 mRNA localization immediately after cell division of the eight-cell blastomere. The movie shows the red channel (Cy3-Cdx2 mRNA) and the merge with DIC. Timing is displayed in the upper-left corner. An asterisk marks the timing and position of Cdx2 mRNA localization.
Cdx2 mRNA localization during a differentiative division of an eight-cell blastomere. The movie shows three different focal planes of the same dividing cell. Each focal plane is a merge of the green (membrane-GFP) and red (Cy3-Cdx2 mRNA) channels. Timing is displayed in the upper-right corner.
Cdx2 mRNA localization during a conservative division of eight-cell blastomeres. The movie shows one focal plane as a merge of the green (membrane-GFP) and red (Cy3-Cdx2 mRNA) channels. Timing is displayed in the upper-left corner.
Movies of fluorescently labeled Cdx2 ORF RNA injected into a compacted blastomere of an eight-cell embryo. Notice that the RNA persists at the cortex for the whole length of the movie. The timing is displayed in the upper-left corner.
Movies of fluorescently labeled Δ97bp RNA injected into a compacted blastomere of an eight-cell embryo. Notice that the RNA gets quickly removed from the cortex. Timing is displayed in the upper-left corner.