Abstract
Background
Low-grade gliomas (LGGs) are rare brain neoplasms, with survival spanning up to a few decades. Thus, accurate evaluations on how biomarkers impact survival among patients with LGG require long-term studies on samples prospectively collected over a long period.
Methods
The 210 adult LGGs collected in our databank were screened for IDH1 and IDH2 mutations (IDHmut), MGMT gene promoter methylation (MGMTmet), 1p/19q loss of heterozygosity (1p19qloh), and nuclear TP53 immunopositivity (TP53pos). Multivariate survival analyses with multiple imputation of missing data were performed using either histopathology or molecular markers. Both models were compared using Akaike's information criterion (AIC). The molecular model was reduced by stepwise model selection to filter out the most critical predictors. A third model was generated to assess for various marker combinations.
Results
Molecular parameters were better survival predictors than histology (ΔAIC = 12.5, P< .001). Forty-five percent of studied patients died. MGMTmet was positively associated with IDHmut (P< .001). In the molecular model with marker combinations, IDHmut/MGMTmet combined status had a favorable impact on overall survival, compared with IDHwt (hazard ratio [HR] = 0.33, P< .01), and even more so the triple combination, IDHmut/MGMTmet/1p19qloh (HR = 0.18, P< .001). Furthermore, IDHmut/MGMTmet/TP53pos triple combination was a significant risk factor for malignant transformation (HR = 2.75, P< .05).
Conclusion
By integrating networks of activated molecular glioma pathways, the model based on genotype better predicts prognosis than histology and, therefore, provides a more reliable tool for standardizing future treatment strategies.
Keywords: biomarker, brain tumor, cancer pathways, prognosis
The median survival of gliomas varies from a few months to >20 years, mainly depending on tumor World Health Organization (WHO) grade (I–IV).1,2 Compared with the most malignant subtype, glioblastoma multiforme (GBM; WHO grade IV), low-grade gliomas (LGGs; WHO grade II) with their 3 different histologic types (diffuse astrocytoma [A], oligoastrocytoma [OA], and oligodendroglioma [OG]) are rare, and the median survival times are long.3,4 LGGs progress in an infiltrative manner and develop into malignant tumors (WHO grades III and IV). Grade IV tumors deriving from LGGs are designated secondary GBM and represent a small subset of GBM (∼5%), compared with the more frequent primary GBM (∼95%), which are considered to have developed de novo. With an incidence in Switzerland of 0.63 cases/100,000 adults per year (0.26 for A, 0.10 for OA, and 0.27 for OG), LGGs are rare and represent ∼15% of all gliomas.4 Patients with astrocytic tumors have a worse prognosis than patients with oligodendroglial or mixed oligoastrocytic histology:5,6 Median survival is 5.6 years for A, 6.6 years for OA, and 11.6 years for OG.4
Two molecular alterations characteristic of glioma have a particularly high prevalence in LGG: MGMT gene promoter methylation (MGMTmet)7 and IDH1/IDH2 mutations (IDHmut).8 IDH1 mutations have been discovered in GBM, with all mutations affecting codon 132, with the R132H mutation being the most frequent. To a lesser extent, IDH2 mutations at codon 172 can occur in a fraction of IDH1 wild-type gliomas.9 Taken together, 41% of gliomas were found to carry an IDH1 mutation, whereas 2% had an IDH2 mutation in a mutually exclusive manner.9,10 IDH1/2 gene mutations were mostly observed in LGGs (70%–80%) and in secondary GBM (85%), compared with primary GBM (3%–7%).8,9 IDH genes encode isocitrate dehydrogenase, which normally catalyzes the conversion of isocitrate into α-ketoglutarate (αKG) as part of the Krebs' cycle. The observed mutations lead to a novel catalytic function that converts αKG into 2-hydroxyglutarate (2HG).11 The oncogenic role of 2HG accumulation is attributable to impaired demethylation of genomic DNA, leading to an extensive methylome and transcriptome remodelling.12–14 This provides a molecular basis for the cosegregation observed between IDHmut and MGMTmet15 and for the response of MGMTmet to treatment with alkylating agent temozolomide.16
Although IDHmut and MGMTmet are frequent in LGG, regardless of histologic phenotype, TP53 mutations (TP53mut) mainly occur in diffuse astrocytomas, where they are also associated with a younger age of onset and a shorter survival.17 Combined loss of heterozygosity of 1p/19q (1p19qloh) is prevalent in oligodendrogliomas and is an indicator of longer survival.18 TP53mut and 1p19qloh are mutually exclusive.3,19,20 Whether there is a cause-effect relationship between TP53mut or 1p19qloh and astrocytic or oligodendrocytic histology remains unclear. During the course of gliomagenesis, IDHmut and MGMTmet are considered to be early events, followed by the acquisition of either TP53mut or 1p19qloh.20 However, because of its MGMTmet background and its association with longer survival, the role of 1p19qloh in LGG response to radiotherapy and temozolomide remains unclear.21 For these reasons, the treatment of individuals with LGG has not been universally standardized yet.
Here, we study the relevance of LGG prognostic markers on adult patient outcome in separate multivariate survival analyses. Thereby, we consider either histology (A, OA, and OG) or molecular marker status (IDH,15 MGMT,22 1p/19q,18,23 and TP5317) along with clinical characteristics of patients.
Materials and Methods
Patient Demographic Characteristics and Data Collection
Patients were recruited in a continuous geographic area consisting of the cantons Bern, Basel, Solothurn, and Jura. Demographic, clinical (i.e., epilepsy, focal neurological deficit, and tumor size) and survival data (i.e., overall survival [OS] and malignant transformation-free survival) were collected for these patients. Archived tissue specimens were selected by 2 independent neuropathologists according to a histologic diagnosis of a WHO grade II tumor (diffuse astrocytoma, oligodendroglioma, or oligoastrocytoma). The study was approved by the local Ethics Committee of Basel.
Formalin Fixation, Histopathology
Tumor samples were fixed with 4% buffered formalin and embedded into paraffin as previously described.24 Diagnosis based on H&E staining and routine immunohistochemistry (i.e., GFAP, MAP2c, MIB-1) was reviewed by 2 independent board-certified neuropathologists.
DNA Extraction
DNA was extracted from tissue sections containing at least 70% tumor cells using the QIAGEN (Hilden, Germany) QIAamp® FFPE Tissue Kit according to the manufacturer's instructions.
LOH Studies
LOH analysis was performed as described previously18 using microsatellite markers on chromosome regions 1p36 (D1S468, D1S1612, D1S228, and D1S214) and chromosome 19q13 (D19S219, D19S412, and D19-HRC).
Pyrosequencing
The genomic regions encompassing codons R132 of IDH1 and R172 of IDH2 were analyzed using pyrosequencing, as previously described,25 with use of the primers IDH1-Py-Reverse-5′TGATCCCCATAAGCAT-3′ with nucleotide dispensation order CGACTGACACTATCGAT and IDH2-Py-Forward-5′-AGCCCATCACCATTG-3′ with the nucleotide dispensation order TGCGATCGATCGCACGCA.
MGMT Promoter Methylation
MGMT promoter methylation status analysis was performed with a quantitative primer extension-based assay providing percentage values of methylated DNA in the tumor sample.26
TP53 Immunostaining
Immunostaining for TP53 was performed using monoclonal antibody DO-7 (Dako IR616) on the automatic staining system Roche Ventana BenchMark XT (Ventana Medical Systems, Tucson, AZ), according to the manufacturer's protocol. Tissue samples were counterstained with hematoxylin. TP53 nuclear positivity was defined by immunohistochemistry of vital tumor areas, excluding perinecrotic areas, which often show some degree of (hypoxia-associated) TP53 immunoreactivity. Cutoff of TP53 immunopositivity was defined from ≥20% of positive cells, whereas tumors with ≤19% were considered as negative. Magnification was 100×. Quantitative assessment was performed by 2 board-certified neuropathologists.
Statistics
We used the package mice27 of the statistical software package R28 to multiply impute incomplete multivariate data by chained equations (see Appendix for details). A set of m = 50 imputations was created per missing value.
OS among patients with LGG was analyzed using Cox proportional-hazards survival models. Thereby, models were fitted to each of the 50 imputed datasets, yielding 50 analyses per model. The results were then combined to derive pooled hazard ratio (HR) estimates, standard errors, and confidence intervals with use of Rubin's rules.29 By taking into account the variability in results among the imputed datasets, the pooled analysis reflects the uncertainty about the missing values.
OS was modeled by using either histology (A, OA, and OG) or molecular factors (IDHmut, 1p19qloh, MGMTmet, and TP53pos), along with age, sex, epilepsy, and focal neurological deficit as predictors. Because of a large amount of missing data (159 of 210), tumor size was considered in parallel analyses (Appendix 2). To evaluate the most important molecular predictors and to mitigate the problem of multicollinearity among molecular factors, we used stepwise model selection based on Akaike's information criterion (AIC) to reduce the molecular model. Because this did not completely resolve multicollinearity issues, we classified patients into 5 groups, based on common combinations of mutations and replaced individual molecular factors by combined molecular factors. To assess the relative goodness of fit of different models, we compared AIC by paired t tests (a pair of values for each of m = 50 imputed data set).
Moreover, to compare the predictive accuracy of models we used time-dependent receiver operating characteristic (ROC) curves and graphically displayed the corresponding areas under the curve (AUC), according to Heagerty and Zheng.30 We checked for violations of the proportional-hazard assumption with use of the method proposed by Grambsch and Therneau.31 All aforementioned statistical analyses were done using R (version 2.15.128).
Associations were calculated using Fisher's exact test (GraphPad Software).
Results
Multivariate Analysis Considering Histology
Of the 210 LGGs analyzed, 92 were diagnosed as A, 63 as OA, and 53 as OG. Mean age was 41.9 years, and 60.0% of the cohort was male. With use of OS as the end point, 45.2% died during the observation period (Table 1). We first performed a multivariate survival analysis focused on the effect of histology, including age, sex, epilepsy, and focal neurological deficit as covariates. Age at diagnosis showed a negative impact on OS (HR for death = 1.04; P< .001), with a 4% increase for every additional year of age at diagnosis. Consistent with the median OS of each histologic type, the HR associated with OA and OG histology using A as the reference were of 0.71 and 0.39, respectively (not significant and P< .01, respectively) (Table 2).
Table 1.
Demographic characteristics and biomarker statuses of studied population with LGG
| Numeric | |||||
|---|---|---|---|---|---|
| Observed data |
Imputed data | ||||
| Mean | Range | Median | n | Mean | |
| Demographic data | |||||
| age (years) | 41.9 | 18.0–75.9 | 40.5 | 210 | 41.9 |
| Survival | |||||
| days to death | 2509.5 | 35–8432 | 2019 | 210 | 2509.5 |
| days to malignant transformation | 2144.6 | 13–8239 | 1525 | 210 | 2144.6 |
| Tumor size | |||||
| tumor volume (cm3) | 92.9 | 1.3–287.6 | 71.3 | 51 | 118.9 |
| Biomarkers | |||||
| MGMT promoter methylation (%) | 48.3 | 0–100 | 60 | 136 | 46.6 |
| TP53 positivity (%) | 10.6 | 0–60 | 1 | 145 | 11.5 |
| Categorial† | |||||
| Observed data | Imputed data | ||||
| % | Number | n | % | ||
| Demographic data | |||||
| male patients | 60.0 | 126 | 210 | 60.0 | |
| Survival | |||||
| death | 45.2 | 95 | 210 | 45.2 | |
| malignant transformation | 32.9 | 69 | 210 | 32.9 | |
| Clinical presentation | |||||
| epilepsy | 76.9 | 133 | 173 | 76.5 | |
| focal neurological deficit | 29.9 | 46 | 154 | 29.8 | |
| Treatment | |||||
| chemotherapy | 35.2 | 57 | 162 | 37.2 | |
| Histology | |||||
| astrocytoma (A) | 44.2 | 92 | 208 | 44.4 | |
| oligoastrocytoma (OA) | 30.3 | 63 | 208 | 30.2 | |
| oligodendroglioma (OG) | 25.5 | 53 | 208 | 25.4 | |
| Biomarkers | |||||
| IDH1 major mutation (R132H) | 70.3 | 130 | 185 | 68.7 | |
| IDH1 minor mutations (R132X) | 7.0 | 13 | 185 | 7.1 | |
| IDH2 mutations (R172X) | 3.8 | 7 | 185 | 4.5 | |
| all IDH mutations (IDHmut) | 81.1 | 150 | 185 | 80.3 | |
| IDH wild-type (IDHwt) | 18.9 | 35 | 185 | 19.7 | |
| MGMT methylation (>0%) (MGMTmet) | 69.9 | 95 | 136 | 67.6 | |
| TP53 positivity (≥20%) (TP53pos) | 24.1 | 35 | 145 | 27.1 | |
| 1p/19q combined allelic loss (1p19qloh) | 24.4 | 39 | 160 | 23.4 | |
| Biomarker combinations | |||||
| IDHmut + MGMTmet | 20.0 | 21 | 105 | 24.6 | |
| IDHmut + MGMTmet + 1p19qloh | 24.8 | 26 | 105 | 20.8 | |
| IDHmut + MGMTmet + TP53pos | 13.3 | 14 | 105 | 16.6 | |
| IDHwt | 33.3 | 35 | 105 | 19.7 | |
| other combinations | 8.6 | 9 | 105 | 18.3 | |
Columns 1–4 refer to observed data, column 5 shows the mean across m = 50 imputed data sets, where n = 210.
†Columns 1–3 refer to observed data, column 4 shows the mean percentage across m = 50 imputed data sets, where n = 210.
Table 2.
Hazard ratios associated with LGG histological and molecular markers
| Overall survival | Hazard ratio | lower 0.95 | higher 0.95 | t-value | P |
|---|---|---|---|---|---|
| Histology model | |||||
| mean Akaike's information criterion (AIC): 861.9 | |||||
| age* | 1.04 | 1.02 | 1.05 | 4.12 | .000 |
| gender (m vs. f) | 1.15 | 0.75 | 1.76 | 0.64 | .519 |
| epilepsy | 0.89 | 0.51 | 1.55 | −0.42 | .674 |
| focal neurological deficit | 1.42 | 0.84 | 2.41 | 1.30 | .195 |
| OA vs. A | 0.71 | 0.43 | 1.18 | −1.32 | .187 |
| OG vs. A | 0.39 | 0.22 | 0.70 | −3.17 | .002 |
| Molecular model | |||||
| mean AIC: 850.0 | |||||
| IDHmut° | 0.64 | 0.27 | 1.54 | −0.99 | .321 |
| IDH2mut vs. IDH1mut | 1.38 | 0.41 | 4.67 | 0.52 | .602 |
| IDH1R132H vs. IDHminor | 1.11 | 0.49 | 2.55 | 0.26 | .798 |
| age* | 1.04 | 1.02 | 1.06 | 4.35 | .000 |
| gender (m vs. f) | 1.23 | 0.77 | 1.95 | 0.87 | .384 |
| epilepsy | 1.08 | 0.59 | 1.99 | 0.25 | .799 |
| focal neurological deficit | 1.55 | 0.89 | 2.71 | 1.54 | .125 |
| 1p19qloh vs. wild-type | 0.58 | 0.26 | 1.31 | −1.31 | .191 |
| TP53pos* | 1.02 | 1.00 | 1.04 | 2.22 | .027 |
| MGMTmet* | 0.99 | 0.98 | 1.00 | −1.52 | .130 |
| Reduced molecular model | |||||
| mean AIC: 845.8 | |||||
| age* | 1.04 | 1.02 | 1.06 | 4.70 | .000 |
| IDHmut° | 0.54 | 0.27 | 1.09 | −1.72 | .086 |
| TP53pos* | 1.02 | 1.01 | 1.04 | 2.66 | .008 |
| MGMTmet* | 0.99 | 0.98 | 1.00 | −1.94 | .053 |
| Combined molecular model | |||||
| mean AIC: 845.3 | |||||
| age* | 1.04 | 1.02 | 1.05 | 4.44 | .000 |
| gender (m vs. f) | 1.27 | 0.80 | 2.01 | 1.02 | .307 |
| epilepsy | 1.05 | 0.57 | 1.94 | 0.16 | .875 |
| focal neurological deficit | 1.56 | 0.90 | 2.71 | 1.60 | .111 |
| IDHmut/MGMTmet° | 0.33 | 0.16 | 0.68 | −2.97 | .003 |
| IDHmut/MGMTmet/1p19qloh° | 0.18 | 0.07 | 0.45 | −3.68 | .0002 |
| IDHmut/MGMTmet/TP53pos†° | 0.88 | 0.45 | 1.74 | −0.36 | .722 |
| others° | 0.76 | 0.38 | 1.51 | −0.79 | .429 |
| Malignant transformation-free survival | |||||
| Combined molecular model | |||||
| age* | 1.00 | 0.98 | 1.02 | 0.12 | .905 |
| gender (m vs. f) | 0.76 | 0.46 | 1.27 | −1.04 | .299 |
| epilepsy | 1.07 | 0.53 | 2.17 | 0.20 | .841 |
| focal neurological deficit | 2.00 | 1.06 | 3.78 | 2.15 | .032 |
| IDHmut/MGMTmet° | 1.15 | 0.42 | 3.18 | 0.28 | .780 |
| IDHmut/MGMTmet/1p19qloh | 0.58 | 0.18 | 1.88 | −0.90 | .366 |
| IDHmut/MGMTmet/TP53pos† | 2.75 | 1.03 | 7.37 | 2.02 | .044 |
| others° | 3.21 | 1.15 | 8.93 | 2.23 | .026 |
*continuous variables.
°vs. IDH wild-type.
†≥20%.
Acquisition of Molecular Marker Status
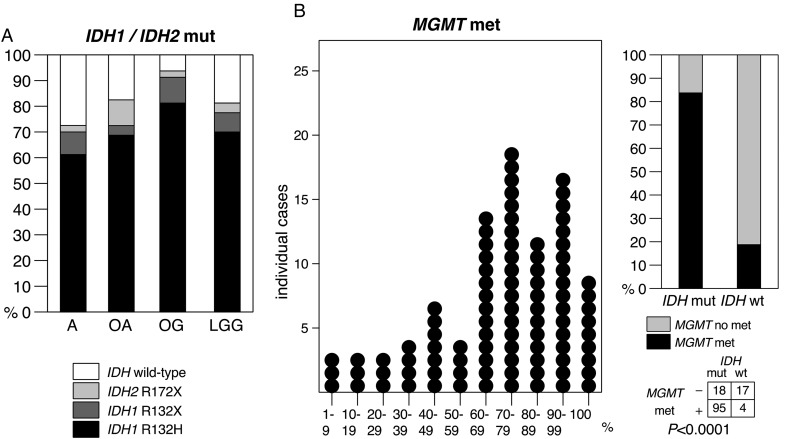
Eighty-one percent of LGG had an IDH1 or an IDH2 mutation. Seventy percent showed the IDH1 major mutation R132H, 7 had another IDH1 mutation, and 4 had an IDH2 mutation (Table 1). Frequencies of IDHmut in A, OA, and OG were of 72%, 83%, and 94%, respectively (Fig. 1A). MGMTmet (>0% methylation) was found in nearly 70% of LGGs and ranged up to 100%, with a median degree of methylation of 60% (Fig. 1B). When restricted to the IDHmut LGG population, 84% were MGMTmet. As previously reported,15 IDHmut was strongly positively associated with MGMTmet (P< .001; Fig. 1B).
Fig. 1.
Distribution of biomarkers in the studied population with LGG. (A) Frequencies of IDH1 and IDH2 mutations by tumor histology; (B) distribution plot of percentages of methylated MGMT promoter in individual IDH-mutant LGG (left), association between IDH mutation (IDH1 or IDH2), and MGMT promoter methylation (right).
Analysis for 1p and 19q allelic loss revealed that 24% (39 of 160) of LGGs and 30% (37 of 121) of IDHmut LGGs had combined 1p19qloh, with the highest prevalence in OG (65%; 26 of 40). Immunodetection of TP53-positive cell nuclei ranged up to 60%. TP53-positive cells were detected in 55% (80 of 146) of all tumors and 60% (65 of 109) of IDHmut LGG.
Multivariate Analysis Considering Molecular Biomarkers
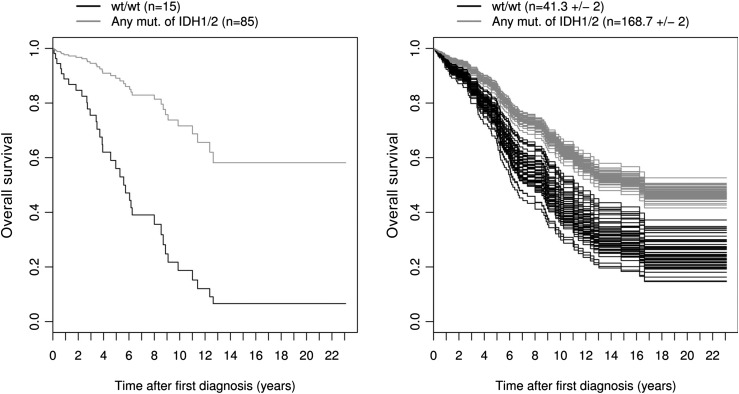
We then focused on the effects of molecular markers IDH, MGMT, 1p/19q, and TP53 on OS, where age, sex, epilepsy, and focal neurological deficit were used as covariates. MGMTmet and TP53pos were treated as continuous variables (percentages), similar to patient age. By comparing the histology and molecular models using AIC) values, the molecular model had a significantly lower AIC value than did the histology model (mean AIC, 861.9 vs. 850.0; ΔAIC = 11.9; P< .001). Moreover, the AUC of time-dependent ROC curves was consistently higher for the molecular model than for the histology model for prognosis beyond 500 days (Fig. 4B). The AUC for the molecular model (and the genotype group model) is always larger than 0.7 after 250 days. This means that, on any day t for t >250, the probability that a patient who dies on day t has a greater risk of dying than a patient who survives beyond day t is at least 0.7.30 This analysis therefore shows that molecular markers represent stronger predictors for survival among patients with LGG than does histology. There was a negative impact of TP53 immunohistochemistry (HR = 1.02; P< .05) and a positive but nonsignificant impact of IDHmut status (HR = 0.64) (Fig. 2), similar to 1p19qloh status (HR = 0.58) (Table 2). These nonsignificant HRs of IDHmut and 1p19qloh status can be explained by the strong association between the molecular markers used in the same model.
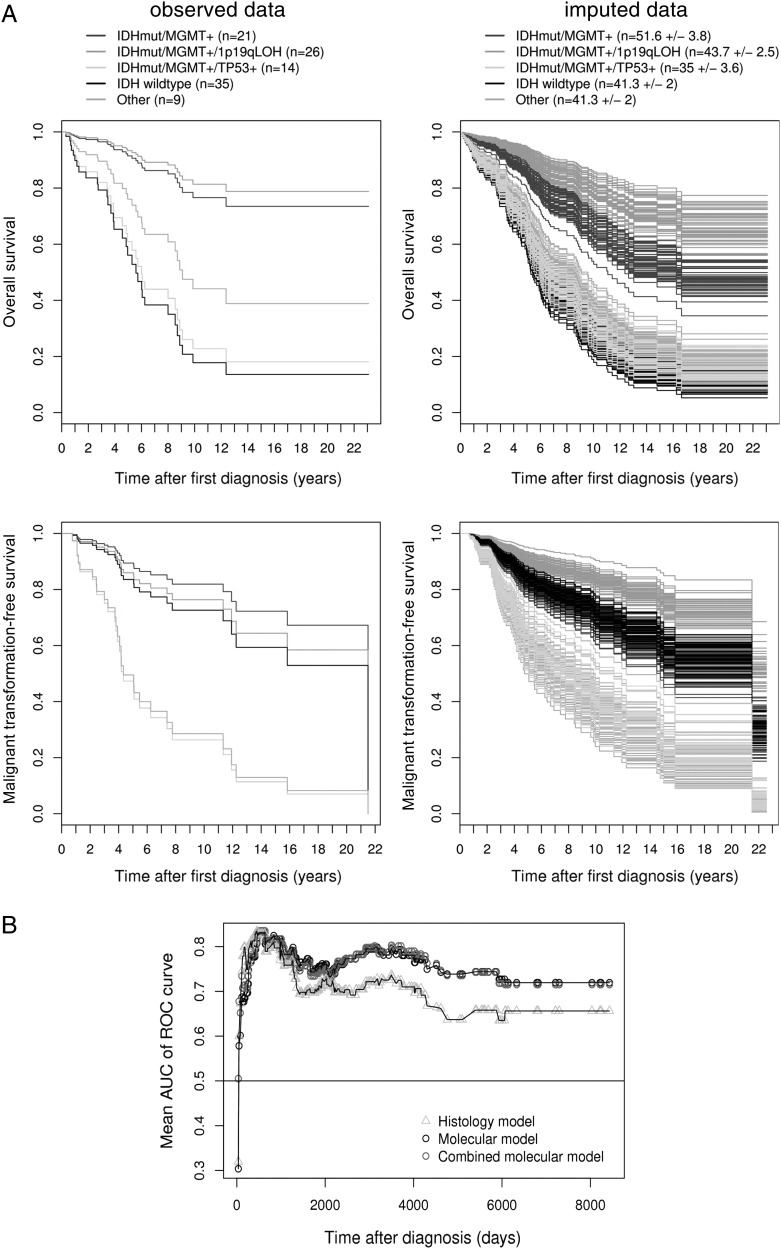
Fig. 4.
(A) Cox proportional-hazards survival curves by group from the combined molecular model for overall survival (top) and malignant transformation free survival (bottom). The left panel shows the survival curves for the observed data (n = 105). The right panel shows the survival curves for all m = 50 imputed data sets (n = 210). For the multiply imputed data, the mean number of observations is given for each category (± SD), because this number varies. Note that the curves were drawn for patients with a median age (40.5 years), with male sex and epilepsy, but without neurodeficits (most frequent combination). (B) Time-dependent AUC of the ROC curve for the histology model, the molecular, and the combined molecular model. Each AUC value shown in the graph represents a mean from m = 50 imputed data sets.
Fig. 2.
Cox proportional-hazards survival curves by IDH1/2 status from the reduced molecular model for overall survival. The left panel shows the survival curves for the observed data (n = 100). The right panel shows the survival curves for all m = 50 imputed data sets (n = 210). For the multiply imputed data, the mean number of observations is given for each category (±SD), because this number varies. Note that the curves were drawn for patients with median values for the numeric predictors age (40.5 years), TP53 positivity (1%), and MGMT methylation (60%).
Stepwise model selection using AIC led to a reduced molecular model, including age and molecular markers IDHmut, TP53pos, and MGMTmet as the best predictors for survival among patients with LGG (AIC = 845.8). The percentage of MGMTmet had a significant impact on OS (HR = 0.99; P = 0.053), because the hazard of death decreased by 1%, with an increased degree of MGMTmet by 1%. Percentage of TP53pos cells had a negative impact on survival (HR = 1.02; P< .01), because the hazard of death increased by 2%, with an increased TP53pos by 1%. Therefore, the degrees of MGMTmet and TP53pos both gradually affected survival among patients with LGG (Table 2).
Determination of a Cutoff Value for TP53 Immunopositivity
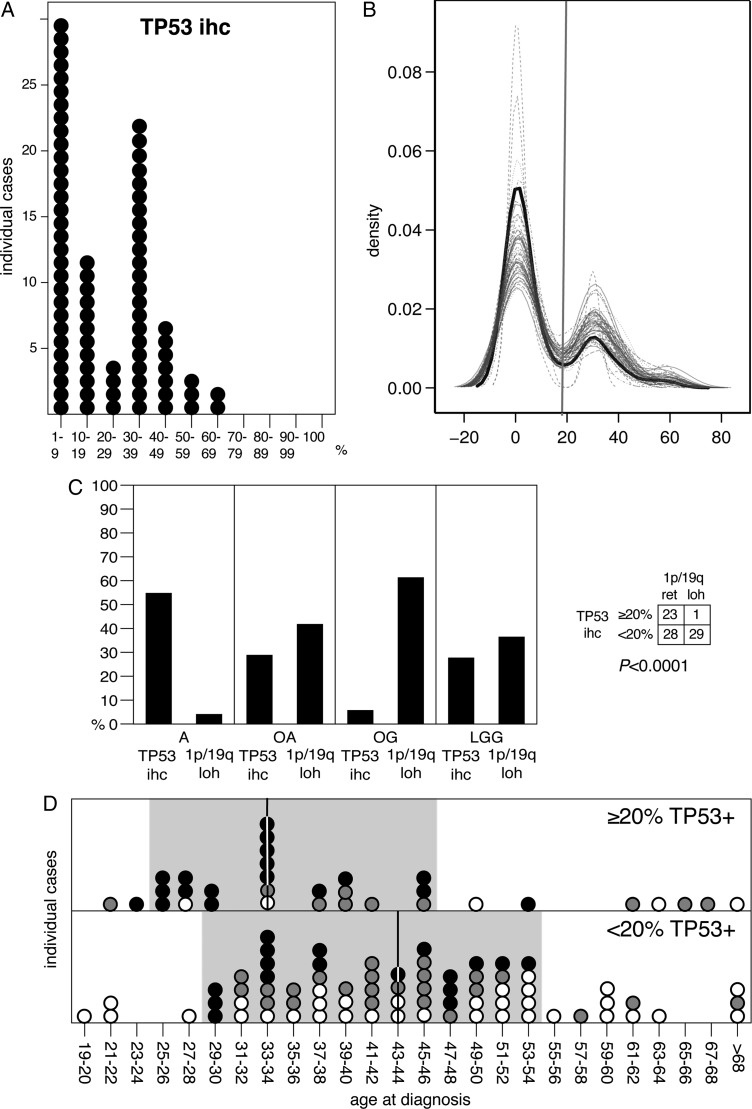
Tumor labeling by immunohistochemistry (IHC) for TP53 has been proposed as a potential indicator of tumors with TP53 mutations.32–34 Because of their variabilities, percentages of TP53 immunopositivity estimates were plotted by bins of 10%. Percentages of TP53-stained cells in individual tumors showed a bimodal distribution, peaking at 1%–9% and 30%–39% (Fig. 3A). Kernel density estimates for marginal distribution of the observed data suggested a 20% cutoff between both peaks (Fig. 3B), which is in agreement with the one empirically applied to select tumors potentially carrying TP53 mutations (TP53mut).
Fig. 3.
TP53 data. (A) distribution plot of percentages of TP53 immunopositivity in individuals with LGG; (B) kernel density estimates for the marginal distributions of the observed data (black line) and the m = 50 densities from the imputed data (gray lines), only numeric imputed variables are shown; (C) frequencies of 1p/19q loss and TP53 immunopositivity (cutoff: ≥20%) in IDH-mutant tumors by histology (left) and tested association between TP53 immunopositivity and 1p/19q loss (right); (D) distribution plot of age at diagnosis for IDH-mutant LGG depending on TP53 immunopositivity (≥20%). Vertical bars indicate median value and grey background highlights sample clustering. Filled circles: A, grey circles: OA, open circles: OG.
Considering this ≥20% threshold, we found decreasing frequencies of IDHmut LGG with TP53pos from A to OG and an inverse correlation between 1p19qloh and TP53pos (Fig. 3). This is consistent with the literature and reflects the mutual exclusion observed between both events.3,19,20,35 We also found a median age at diagnosis of 9.9 years younger for patients with TP53pos tumors (33.9 vs. 43.8 years; Fig. 3).17 These observations support that TP53pos is a suitable indicator for identifying putative TP53mut and underline the potential of routine TP53 IHC as a time- and material-saving, cost-efficient alternative method for evaluating TP53 status.
Combination of Molecular Biomarkers
Furthermore, molecular markers were tested in combination for their prognostic relevance, dividing patients into groups with common molecular combinations. MGMT status was considered either as methylated (>0%) or as nonmethylated, and the 20% cutoff was applied for TP53 immunopositivity, as described above. Against LGG with wild-type IDH, the combination IDHmut/MGMTmet (with neither 1p19qloh nor TP53pos) is associated with OS (HR = 0.33; P< .01). Although the triple combination IDHmut/MGMTmet/1p19qloh had an even more favorable impact (HR = 0.18; P< .001), the triple combination IDHmut/MGMTmet/TP53pos canceled the positive effect (HR = 0.88). However, the latter had a significant negative impact on malignant transformation-free survival (HR = 2.75; P< .05) (Table 2 and Fig. 4A). With regard to the full molecular model, time-dependent AUC of the ROC curves of the combined molecular model was consistently higher than for the histology model for prognosis beyond 500 days (Fig. 4B). Combination of biomarkers status therefore appears to be a powerful tool, virtually as powerful as the molecular model, to predict survival among patients with LGG.
Multivariate Analyses Considering Tumor Volumes
Tumor volume is a strong predictor of survival among patients with LGG.36 Because of the small number of available tumor size data, an analysis that included tumor size has been performed in parallel. Nevertheless, addition of this parameter did not alter our main results (Appendix 2) and our conclusions.
Discussion
Although some studies have shown no impact of IDH1 or IDH2 status on survival among patients with LGG or grade II astrocytoma, others have revealed a positive correlation between IDH mutations and OS.15,19,37,38 Compared with those performed on all types and grades of gliomas,15 we focused on the glioma types with the highest frequencies of IDH mutations.9 Because of the particularly long survival among patients with LGG, we performed a retrospective study based on LGG of all histological glioma subtypes.
Percentages of MGMT Methylation and TP53 Positivity
Quantification of MGMTmet26 allowed the analysis of its impact not only as a binary but also as a continuous variable. Indeed, plotting of individual tumors by percentage of methylated DNA reveals a single-peaked distribution ranging from 4% to 100% with a median value of 60%, with progressive correlation between the degree of methylation and survival that may reflect variability in the sets of silenced genes.
The same approach was applied to TP53, where the percentage of TP53pos cells was assessed. We found a graded correlation between degree of TP53pos and shorter survival. Although with limited accuracy, this time- and material-saving method provides a reasonable approximation of the TP53mut status. Of interest, TP53pos, ranging from 1% to 60%, had 2 peaks, one between 1 and 9 and one between 30% and 39%, where the gap (20%) coincides with the cutoff empirically applied by pathologists to select TP53mut tumors from TP53 wild types. Consistently, the 20% cutoff was mutually exclusive of combined 1p19qloh (Fig. 3). This cutoff also consistently distinguishes a subset of patients with a median age of 33.9 years at diagnostics from those with a median age of 43.8 years (Fig. 3). This younger age is closer to the median age at diagnosis of TP53mut among patient with LGG and confirms that TP53pos status implies TP53mut.
IDH Mutations
Most previous studies have associated IDHmut with positive outcome on survival among patients with glioma.15,19,37,38 We have also shown that IDHmut has a significant positive impact on long-term OS among patients with LGG in combination with MGMTmet. Accumulation of the oncometabolite 2HG resulting from IDH mutation is regarded as a founder biochemical event in tumorigenesis that triggers methylation of genomic segments covering, among others, the MGMT gene.12,39 This is in agreement with the strong correlation between IDHmut and MGMTmet. IDHmut is believed to occur before either 1p19qloh or TP53mut,20 both of which occur in a mutually exclusive manner.3,19,20 Because of their respective prevalences in OG and A, cause-and-effects between 1p/19q allelic loss or TP53 mutation and oligodendroglial- or astrocytic differentiation remain unclear. Nevertheless, on the basis of the selective positive impact of IDHmut on survival, the groups of LGG that accumulate 2HG at early stages of development may be regarded as less aggressive with slower evolution than wild-type IDH LGG.
Molecular Biomarkers Versus Histology
Compared with the histology model, the molecular model has both a significantly lower AIC and a consistently higher AUC of time-dependent ROC curves after 500 days, meaning that the latter provides a better model to predict long-term survival among patients with LGG. This strongly suggests that a defined combination of genetic alterations, likely to be reflected in specifically activated cancer pathways, provides a better indicator than does histopathologic subclassification in WHO grade II tumors (A, OA, OG) for assessing patient outcome, including malignant transformation.
Stepwise model selection on the molecular model only revealed IDHmut, MGMTmet, and TP53pos as the most critical molecular prognostic factors in LGGs. That 1p19qloh did not remain in the reduced molecular model is most likely attributable to strong correlations with other markers. In fact, by grouping LGGs according to combinations of all molecular markers into 4 major subgroups, we found that (1) patients with tumors with IDHmut/MGMTmet/1p19qloh had the longest, those with IDHmut/MGMTmet had intermediate, and those with IDH wild-type had the lowest overall survival; and (2) patients with tumors with IDHmut/MGMTmet/TP53pos had the shortest time to malignant transformation.
Through a detailed retrospective study of LGG tissue samples from the past 20 years, we have shown that a combination of biomarkers, combined with demographic and clinical variables, represents a stronger predictor of survival among patients with LGG than does histological analysis. This suggests that such markers should be routinely assessed in parallel to histopathological examination to further stratify these WHO grade II tumors according to their individual prognosis. This could help to standardize treatment strategies and to identify activated cancer pathways that may be targets for future therapies.
Supplementary Material
Funding
This work was supported by the Swiss Cancer League (Grant KLS 2765-02-2011 to J.-L. B., L. M., S. F., E. V., and P. S.)
Conflict of interest statement: None declared.
Supplementary Material
References
- 1.Kleihues P, Burger PC, Scheithauer BW. The new WHO classification of brain tumours. Brain Pathol. 1993;3:255–268. doi: 10.1111/j.1750-3639.1993.tb00752.x. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. doi:10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okamoto Y, Di Patre PL, Burkhard C, et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol. 2004;108:49–56. doi: 10.1007/s00401-004-0861-z. doi:10.1007/s00401-004-0861-z. [DOI] [PubMed] [Google Scholar]
- 4.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64:479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 5.Leighton C, Fisher B, Bauman G, et al. Supratentorial low-grade glioma in adults: an analysis of prognostic factors and timing of radiation. J Clin Oncol. 1997;15:1294–1301. doi: 10.1200/JCO.1997.15.4.1294. [DOI] [PubMed] [Google Scholar]
- 6.Pignatti F, van den Bent M, Curran D, et al. Group EOfRaToCRC. Prognostic factors for survival in adult patients with cerebral low-grade glioma. J Clin Oncol. 2002;20:2076–2084. doi: 10.1200/JCO.2002.08.121. [DOI] [PubMed] [Google Scholar]
- 7.Costello JF, Futscher BW, Tano K, et al. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J Biol Chem. 1994;269:17228–17237. [PubMed] [Google Scholar]
- 8.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. doi:1164382[pii]10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. doi:360/8/765[pii]10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. doi:10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 11.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. doi:nature08617[pii]10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. doi:S1535-6108(10)00108-X[pii]10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. doi:nature10860[pii]10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. doi:nature10866[pii]10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. doi:JCO.2009.21.9832[pii]10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 16.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. doi:352/10/997[pii]10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 17.Kim YH, Nobusawa S, Mittelbronn M, et al. Molecular classification of low-grade diffuse gliomas. Am J Pathol. 2010;177:2708–2714. doi: 10.2353/ajpath.2010.100680. doi:ajpath.2010.100680[pii]10.2353/ajpath.2010.100680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mariani L, Deiana G, Vassella E, et al. Loss of heterozygosity 1p36 and 19q13 is a prognostic factor for overall survival in patients with diffuse WHO grade 2 gliomas treated without chemotherapy. J Clin Oncol. 2006;24:4758–4763. doi: 10.1200/JCO.2006.05.9238. doi:JCO.2006.05.9238[pii]10.1200/JCO.2006.05.9238. [DOI] [PubMed] [Google Scholar]
- 19.Houillier C, Wang X, Kaloshi G, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75:1560–1566. doi: 10.1212/WNL.0b013e3181f96282. doi:75/17/1560[pii]10.1212/WNL.0b013e3181f96282. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe T, Nobusawa S, Kleihues P, et al. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. doi:S0002-9440(10)60974-1[pii]10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bromberg JE, van den Bent MJ. Oligodendrogliomas: molecular biology and treatment. Oncologist. 2009;14:155–163. doi: 10.1634/theoncologist.2008-0248. doi:theoncologist.2008-0248[pii]10.1634/theoncologist.2008-0248. [DOI] [PubMed] [Google Scholar]
- 22.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. doi:10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 23.Boulay JL, Miserez AR, Zweifel C, et al. Loss of NOTCH2 positively predicts survival in subgroups of human glial brain tumors. PLoS One. 2007;2:e576. doi: 10.1371/journal.pone.0000576. doi:10.1371/journal.pone.0000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bandi N, Zbinden S, Gugger M, et al. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–5559. doi: 10.1158/0008-5472.CAN-08-4277. doi:0008-5472.CAN-08-4277[pii]10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- 25.Setty P, Hammes J, Rothämel T, et al. A pyrosequencing-based assay for the rapid detection of IDH1 mutations in clinical samples. J Mol Diagn. 2010;12:750–756. doi: 10.2353/jmoldx.2010.090237. doi:jmoldx.2010.090237[pii]10.2353/jmoldx.2010.090237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vassella E, Vajtai I, Bandi N, et al. Primer extension based quantitative polymerase chain reaction reveals consistent differences in the methylation status of the MGMT promoter in diffusely infiltrating gliomas (WHO grade II-IV) of adults. J Neurooncol. 2011;104:293–303. doi: 10.1007/s11060-010-0490-4. doi:10.1007/s11060-010-0490-4. [DOI] [PubMed] [Google Scholar]
- 27.van Buuren S, Groothuis-Oudshoorn K. mice: Multivariate Imputation by Chained Equations in R. J Stat Software. 2011;45:1–67. [Google Scholar]
- 28.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 29.Rubin D. Multiple imputation for nonresponse in surveys. New York, USA: John Willey & Sons; 1987. [Google Scholar]
- 30.Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61:92–105. doi: 10.1111/j.0006-341X.2005.030814.x. doi:BIOM030814[pii]10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- 31.Grambsch P, Therneau T. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81:515–526. [Google Scholar]
- 32.Vojtěsek B, Bártek J, Midgley CA, et al. An immunochemical analysis of the human nuclear phosphoprotein p53. New monoclonal antibodies and epitope mapping using recombinant p53. J Immunol Methods. 1992;151:237–244. doi: 10.1016/0022-1759(92)90122-a. [DOI] [PubMed] [Google Scholar]
- 33.van Meyel DJ, Ramsay DA, Casson AG, et al. p53 mutation, expression, and DNA ploidy in evolving gliomas: evidence for two pathways of progression. J Natl Cancer Inst. 1994;86:1011–1017. doi: 10.1093/jnci/86.13.1011. [DOI] [PubMed] [Google Scholar]
- 34.Hagel C, Laking G, Laas R, et al. Demonstration of p53 protein and TP53 gene mutations in oligodendrogliomas. Eur J Cancer. 1996;32A:2242–2248. doi: 10.1016/s0959-8049(96)00259-6. [DOI] [PubMed] [Google Scholar]
- 35.Bigner SH, Matthews MR, Rasheed BK, et al. Molecular genetic aspects of oligodendrogliomas including analysis by comparative genomic hybridization. Am J Pathol. 1999;155:375–386. doi: 10.1016/S0002-9440(10)65134-6. doi:S0002-9440(10)65134-6[pii]10.1016/S0002-9440(10)65134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mariani L, Siegenthaler P, Guzman R, et al. The impact of tumour volume and surgery on the outcome of adults with supratentorial WHO grade II astrocytomas and oligoastrocytomas. Acta Neurochir (Wien) 2004;146:441–448. doi: 10.1007/s00701-004-0222-7. doi:10.1007/s00701-004-0222-7. [DOI] [PubMed] [Google Scholar]
- 37.Dubbink HJ, Taal W, van Marion R, et al. IDH1 mutations in low-grade astrocytomas predict survival but not response to temozolomide. Neurology. 2009;73:1792–1795. doi: 10.1212/WNL.0b013e3181c34ace. doi:73/21/1792[pii]10.1212/WNL.0b013e3181c34ace. [DOI] [PubMed] [Google Scholar]
- 38.Mellai M, Piazzi A, Caldera V, et al. IDH1 and IDH2 mutations, immunohistochemistry and associations in a series of brain tumors. J Neurooncol. 2011;105:345–357. doi: 10.1007/s11060-011-0596-3. doi:10.1007/s11060-011-0596-3. [DOI] [PubMed] [Google Scholar]
- 39.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. doi:S1535-6108(10)00527-1[pii]10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.