Abstract
Background
Glioblastoma multiforme (GBM) is a heterogeneous, highly aggressive primary brain tumor with strongly variable patient survival. Because reliable prognostic biomarkers are lacking, we investigated the relation between telomerase-associated parameters and the disease course.
Methods
Telomerase-associated parameters were determined in 100 GBM tissues and associated with clinical characteristics and overall survival. Expressions of telomere length, telomerase activity (TA), and human telomerase reverse transcriptase (hTERT) were analyzed by quantitative PCR, telomeric repeat amplification protocol assay, and reverse transcriptase–PCR, respectively. Mutation status of isocitrate dehydrogenase (IDH)1 was determined by direct sequencing, and O6-methylguanine DNA methyltransferase (MGMT) promoter methylation by methylation-specific PCR.
Results
Of 100 GBM tissues, 61 were positive for both hTERT mRNA and TA, with a highly significant correlation between both parameters (linear regression, P < .0001). Telomere length determination revealed a significant difference between the hTERT/TA-positive and -negative subgroups, with markedly longer telomeres in the hTERT/TA-negative cohort (unpaired Student's t-test, P = .0001). Accordingly, significantly shorter telomeres were detected in GBM tissues derived from older patients (>60 y at diagnosis, P < .0001). While no association of telomere parameters with MGMT promoter status was found, all tumors with IDH1 mutation (6/100) were negative for both hTERT expression and TA and harbored significantly longer telomeres. Patients with tumors lacking hTERT expression/TA showed a significant survival benefit (Kaplan–Meier test, both P < .01), which, however, was based exclusively on the younger patient subgroup (≤60 y, both P < .005; >60 y, both ns).
Conclusions
Telomerase activation is not an independent prognostic parameter in GBM but predicts aggressive tumor behavior solely in a younger patient cohort.
Keywords: telomerase, glioblastoma, age, prognosis, survival
Human telomerase is a ribonucleoprotein polymerase that elongates telomeres by adding hexameric 5′-TTAGGG-3′ tandem repeats to the chromosomal ends. In normal somatic cells, telomeres shorten with each round of cell division. If telomeres reach a critical length, cells are directed into senescence and apoptosis.1,2 Most cancer cells escape telomere shortening by activating the enzyme telomerase, which leads to unlimited proliferation capacity and immortalization.3 Reactivation of telomerase was observed in more or less all types of human malignant tumors4 but was not detected in adjacent normal somatic cells, thus making it a favorable tumor-specific biomarker.
Glioblastoma multiforme (GBM), the most frequent primary brain tumor, is characterized by high aggressiveness based on local diffuse infiltration. Despite improved therapeutic strategies, prognosis remains poor, with a median survival still <15 months.5 The modest overall survival of GBM patients associated with a near 100% recurrence incidence mirrors the highly aggressive behavior of these tumors. Glioblastomas show distinct heterogeneity at both the microscopic and molecular levels, which is mostly reflected in the individually varying clinical courses. However, currently only a very few reliable biomarkers exist to predict the course of disease or the treatment response and outcome of GBM patients. Mutations of isocitrate dehydrogenase (IDH)1 (more prevalent in secondary glioblastoma) are associated with better survival, whereas promoter methylation of the O6-methylguanine DNA methyltransferase (MGMT) DNA repair gene predicts enhanced therapy response to the alkylating agent temozolomide.5–9
As has been reported for other solid tumor types, previous reports concerning gliomas have shown that either telomerase activity (TA) or expression of its catalytic subunit human telomerase reverse transcription (hTERT) might correlate with grade of malignancy.10–13 Additionally, telomerase activation was found to be associated with adverse prognosis,14 while a less aggressive course of disease was found in patients with GBM characterized by the alternative mechanism of telomere lengthening (ALT).15 In the current study, we comparatively investigated TA, hTERT mRNA expression, and length of telomeres in GBM tumors and their relation to patient characteristics and clinical outcome. On the basis of our single-center patient collective, we found that the prognostic quality of telomerase-associated parameters is limited by a significant interaction with patient age. Consequently, the worse prognosis associated with telomerase-positive GBM is restricted to the younger subgroup (≤60 y at diagnosis).
Materials and Methods
Clinical Data and Tumor Collection
GBM specimens (n = 100) were collected from consecutive patients who underwent surgery at the Department of Neurosurgery of the Wagner Jauregg Hospital in Linz, Austria, between 1997 and 2010. This study was approved by the local ethics committee, and written informed consent was obtained from all patients.
Reverse Transcriptase–Polymerase Chain Reaction
Expression of hTERT mRNA was determined by reverse transcriptase (RT)–PCR analysis. Total cellular RNA was extracted from tumor tissue and RT-PCR was performed with the OneStep RT-PCR Kit (Qiagen) using oligonucleotide primer sets specific for hTERT (sense 5′-CGGAGGAGTGTCTGGAGCAA-3′, antisense 5′-GGATGAAGCGGAGTCTGGA-3′; product size, 144 bp). Amplification was performed in a thermal cycler (iCycler, Bio-Rad) under the following conditions: 94°C for 30 s, 56°C for 40 s, and 72°C for 40 s. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was amplified (358 bp) as a housekeeping gene as described by Berger et al.16 For semiquantitative evaluation, 30 cycles were chosen for hTERT and 22 cycles for GAPDH. Amplification products were separated by acrylamide gel electrophoresis, stained with ethidium bromide, and quantified by scanning densitometry (Chemi Doc, Quantity One Quantitation software, Bio-Rad). Expression levels (arbitrary units) were calculated relative to GAPDH mRNA amplified concomitantly. In selected cases, hTERT mRNA expression levels were verified by quantitative real-time PCR using the Rotor-Gene SYBR Green RT-PCR Kit (Qiagen) and QuantiTect primer assay (Hs_TERT_1_SG, Qiagen) on a real-time PCR cycler (Rotor Gene Q, Qiagen). PCR was performed according to the manufacturer's instructions with slight modification. In brief, 50 ng of RNA isolated from tumor tissue was assessed by quantitative real-time PCR for data acquisition under the following conditions: 55°C for 10 min, 95°C for 5 min, 45 cycles: 95°C for 5 s, 60°C for 10 s, and 76°C for 15 s. Sample setup was performed in duplicate and no-template controls were included in each experimental setup. Data for ΔCT were calculated relative to GAPDH, which served as an internal control for the quality of cDNA (sense 5′-CAATGACCCCTTCATTGACC-3′, antisense 5′-GATCTCGCTCCTGGAAGATG-3′). Melting curve analysis and gel documentation confirmed the specificities of the fluorescence signals.
Telomerase Activity Assay
TA was analyzed using a telomerase detection kit (TRAPeze, Chemicon International) according to the manufacturer's instructions. Briefly, 6–10 mg of tumor tissue was lysed in 100 µL CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate) lysis buffer, followed by incubation on ice for 30 min and centrifugation at 12 000 × g for 20 min at 4°C. The protein concentration of extracts was determined using the bicinchoninic acid protein assay (Pierce) with bovine serum albumin as a standard. Protein (500 ng) was subjected to the TRAPeze assay. Pre-incubation at 30°C for 30 min was performed to allow telomerase-mediated extension of a substrate oligonucleotide. Extended products were amplified by 33 cycles of a 3-step PCR (94°C for 30 s, 59°C for 30 s, 72°C for 1 min). PCR products were separated on 12% polyacrylamide gel electrophoresis stained with ethidium bromide and visualized under UV illumination (ChemiDoc, Bio-Rad). A heat-inactivated negative control of each sample and internal standards were included in each experiment. The GBM cell line T98G (American Type Culture Collection) served as a positive control (CRL1690).
Telomere Length Measurement by Quantitative PCR
Relative telomere length was determined as published.17 In brief, DNA was extracted from frozen tumor tissues with the QIAmp DNA Blood Mini Kit (Qiagen) according to the manufacturer's instructions. All samples were analyzed in duplicate on the Rotor Gene Q (Qiagen). Each reaction included 20 ng of DNA, 1× Maxima SYBR Green/ROX qPCR Master Mix (Fermentas), 100 nM telomere forward primer (CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT), and 100 nM telomere reverse primer (GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT).18,19 A single copy gene, 36B4 (forward primer 5′-CAGCAAGTGGGAAGGTGTAATCC-3′, reverse primer 5′-CCCATTCTATCATCAACGGGTACAA-3′, also 100 nM each per reaction), which encodes the acidic ribosomal phosphoprotein P0, was used as an amplification control for every sample assayed, as already described.19 Cycling conditions for both products were 10 min at 95°C, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. Finally, melting curve analysis was performed. A standard curve was generated to measure the content of the telomeric sequence in kilobase pairs (kb) for each sample using known quantities of a synthesized 84mer oligonucleotide composed of 14 TTAGGG repeats.17 DNA from an osteosarcoma cell line (SA-OS), which is demonstrably ALT positive, was used as a long telomere control in each experimental setup.20 Total telomeric length in kb per human diploid genome per sample was calculated relative to SA-OS set as 1.
DNA Modification and Methylation-Specific PCR
DNA was extracted from frozen GBM tumor tissue using the QIAmp DNA Mini Kit (Qiagen), and DNA bisulfite conversion was performed with the Epitect Bisulfite Kit (Qiagen) according to the manufacturer's instructions. Methylation-specific PCR was performed with primers specific for either methylated or unmethylated DNA as described previously.8
IDH1 Mutation Analysis
The genomic region of the IDH1 gene exon 4 containing the R132 codon was amplified in duplicate using the AmpliTaq Gold DNA Polymerase Kit (Applied Biosystems) according to the supplier's instructions. Primers for IDH1 amplification were published by Hartmann et al.21 For a better reverse sequence, we modified the reverse primer by adding the M13r sequence (marked in italic letters): forward primer CGGTCTTCAGAGAAGCCATT, reverse primer CAGGAAACAGCTATGACGCAAAATCACATTATTGCCAAC. Following PCR cleanup using the ExoSAP-IT Kit (Affymetrix), PCR products were sequenced on a 3130 Genetic Analyzer (Applied Biosystems) following standard procedures. As an initial screening, all samples were sequenced in the forward direction. Reverse sequences were made only of the second duplicate of samples positive for the R132 mutation to confirm the results. Sequence analysis and fragment assembly were performed using Ridom TraceEditPro version 1.1 software.
Statistical Analysis
Overall survival was defined as the period between time of surgery and death. Patients alive were censored with the date of the last follow-up. Survival probabilities were estimated by means of the Kaplan–Meier method, and survival rates were compared using the log-rank test. To describe the unadjusted effects of covariates on overall survival, univariate Cox proportional hazards regression models were used. For multivariate survival analyses, the Cox regression models were adjusted for age (dichotomized by mean age), sex, performance status (dichotomized by mean KPS: 80 ± 19, range: 40–100), therapy (surgery vs surgery plus any therapy), IDH1 mutation status, and the corresponding telomerase-associated parameters. P-values were all 2-sided and were considered statistically significant at <.05. All statistical analyses were performed using the PASW Statistics 18.0 package (Predictive Analytics Software/SPSS).
Results
Patient Characteristics
A total of 100 GBM surgical specimens were analyzed; patient characteristics are outlined in Table 1. Ninety-five tumors were diagnosed as primary GBM (no preceding low-grade lesion), 3 as secondary GBM (clinical report of a preceding low-grade lesion), and 1 each as gliosarcoma and giant cell GBM. The patient group was composed of 35 women and 65 men with a mean age of 60 ± 14 years (range: 15–80). Median overall survival time for the total was 14.2 months. Subsequent to surgery, patients received radiotherapy (n = 4) or were treated with radiotherapy and chemotherapy either combined (n = 53) or adjuvant (n = 18). Twenty-five patients did not receive further therapy because of low performance status. For MGMT promoter methylation analysis, 99 DNA extracts from tumor specimens were available and investigated by methylation-specific PCR. Methylated MGMT gene promoter sequences were detected in 67 cases (68%), while in 32 patients (32%) the promoter was completely unmethylated. This relatively high percentage of methylated cases is somewhat surprising but comparable to GBM cohorts published by other groups.22–24 Additionally, in several cases at least partial methylation of the MGMT promoter was confirmed by pyrosequencing (data not shown). Additionally, IDH1 mutation sequencing was performed in all tumors included in the study. Six patients harbored IDH1 mutations, with 5 displaying the R132H (CGT→CAT) mutation and 1 the R132S (CGT→AGT) mutation. Concerning the clinical course of disease, this subgroup included 4 primary and 2 secondary GBM.
Table 1.
Characteristics of glioblastoma patients
| Characteristic | n (%) |
|---|---|
| Age, y Mean 60, range 15–85 | |
| ≤60 | 41 (41) |
| >60 | 59 (59) |
| Sex | |
| Female | 35 (35) |
| Male | 65 (65) |
| Karnofsky Performance Score (n = 98) | |
| <80 | 37 (38) |
| ≥80 | 61 (62) |
| Histology | |
| Primary GBM | 95 (95) |
| Secondary GBM | 3 (3) |
| Giant cell GBM | 1 (1) |
| Gliosarcoma | 1 (1) |
| Therapeutic intervention | |
| Surgery | 25 (25) |
| Surgery/radiotherapy | 4 (4) |
| Surgery/radiotherapy/chemotherapy | 71 (71) |
| MGMT promoter methylation status (n = 99) | |
| Methylated | 67 (68) |
| Unmethylated | 32 (32) |
| IDH1 | |
| Mutation | 6 (6) |
| No mutation | 94 (94) |
Telomerase Status Predicts Telomere Length in Human GBM
TA and hTERT mRNA expression values were analyzed in all 100 GBM specimens (Fig. 1A and B; representative samples are shown). Sixty-one percent (61/100) were positive for both TA and hTERT mRNA expression, whereas in 2 cases hTERT expression was negative despite detectable TA, and in another 2 cases hTERT expression did not result in positive TA. Both parameters correlated with high significance (linear regression, P < .0001; Supplementary Fig. S1A).
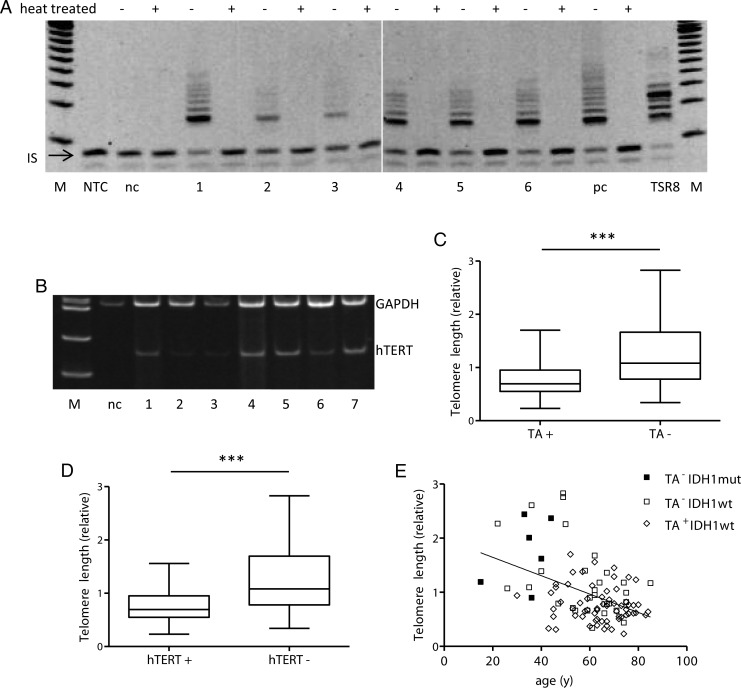
Fig. 1.
Telomerase status and its relation to telomere length of the GBM specimen. Representative examples are given. (A) Telomerase activity as measured by the telomeric repeat amplification protocol (TRAP) assay. M, 20-bp size marker; nc, negative control; 1–5, GBM tumor tissues; pc, positive control (GBM cell line CRL1690); TSR8 quantitation control template; IS, internal standard. For each sample (500 ng protein extract), the TRAP assay was performed with (+) and without (−) heat treatment. (B) hTERT (144 bp) mRNA expression was detected by RT-PCR and amplification products were calculated relative to GAPDH (358 bp), which served as a housekeeping gene. M, 100-bp size marker; nc, negative control; 1–7, GBM tumor tissues. Normal brain tissue obtained from epilepsy surgery served as negative control for both experiments. (C and D) Telomere lengths in relation to TA ± (C) and hTERT ± expression status (D) are shown by box plots. (E) Correlation between patient age and telomere length and relation to IDH1 and TA status (±) as indicated. Abbreviations: wt, wild-type; mut, mutated.
Using quantitative PCR, we determined relative telomere lengths in 98 of 100 tumor cases with sufficient high quality DNA available. Telomerase-positive GBM (62/98) harbored significantly shorter telomeres (mean 0.769) compared with 36 TA-negative tumors (mean 1.297) (Student's t-test; P < .0001; Fig. 1C). Accordingly, hTERT-positive tumor samples (59/98) had shorter telomeres (mean 0.767) compared with hTERT-negative (39/98) tumors (mean 1.259) (Student's t-test; P < .0001; Fig. 1D).
Association of Telomere-Related Parameters and Patient Characteristics
The relations among TA, hTERT mRNA expression, and telomere length and patient characteristics are outlined in Table 2. Statistical analyses (χ2 test) revealed significant association between age and both hTERT expression and telomere length, while the association for TA reached only borderline significance. Similar results were obtained using linear regression models, in which telomere length (Fig. 1E) and hTERT (Supplementary Fig. S2A) were strongly associated with age (P < .0001, r = 0.44 and r = 0.37, respectively), and a weaker correlation was found for TA (P = .021, r = 0.22) (Supplementary Fig. S2B). Telomere lengths in tumors of patients aged ≤60 years were at mean 1.6-fold longer compared with the older patient subgroup (Student's t-test, P < .0001) (Supplementary Fig. S2C). This significant association between telomere length and age was based solely on the TA-negative GBM panel (linear regression, P < .005, r = 0.47), while no correlation was found in telomerase-positive tumors (P = .23) (Supplementary Fig. S2D). A worse KPS was significantly associated with both hTERT expression and higher TA, whereas only a trend toward shorter telomere length was observed (Table 2). Accordingly, for those patients eligible for any treatment subsequent to surgery, a significantly lower hTERT expression and a tendency toward reduced TA were noted. Mutations in IDH1 (n = 6) were distinctly linked to enhanced telomere length and lack of TA (Table 2). The 6 tumors harboring IDH1 mutations belonged to the group of younger patients (≤60 y), whereas the majority of TA-positive and IDH1 wild-type tumors were derived from older patients and had shorter telomeres (<1) (Fig. 1E). Moreover, 5 TA-negative samples from patients aged <60 years at diagnosis showed extremely long telomeres (>2), which might indicate ALT.15 No significant association was observed between telomerase-related parameters and MGMT promoter methylation (Table 2).
Table 2.
Characteristics of patients according to hTERT mRNA, TA, and telomere length
| n Patients (%) | hTERT mRNA Expression, n patients (%) |
P* | TA, n patients (%) |
P* | n Patients (%) | Telomere Length, n patients (%) |
P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristic | n = 100 | Positive n = 63 (63) | Negative n = 37 (37) | Positive n = 63 (63) | Negative n = 37 (37) | n = 98 | >1 n = 34 (35) | <1 n = 64 (65) | |||
| Age, y | |||||||||||
| ≤60 | 41 (41) | 19 (46) | 22 (54) | 21 (51) | 20 (49) | 41 (42) | 20 (49) | 21 (51) | |||
| >60 | 59 (59) | 44 (75) | 15 (25) | .006* | 42 (71) | 17 (29) | .06 | 57 (58) | 14 (25) | 43 (75) | .018* |
| Sex | |||||||||||
| Female | 35 (35) | 23 (66) | 12 (34) | 22 (63) | 13 (37) | 34 (35) | 13 (38) | 21 (62) | |||
| Male | 65 (65) | 40 (61.5) | 25 (38.5) | .83 | 41 (63) | 24 (37) | 1 | 64 (65) | 21 (33) | 41 (67) | .82 |
| KPS | n = 98 | n = 96 | |||||||||
| <80 | 37 (38) | 30 (81) | 7 (19) | 29 (78) | 8 (22) | 36 (38) | 8 (22) | 28 (78) | |||
| ≥80 | 61 (62) | 31 (51) | 30 (49) | .003* | 32 (52) | 29 (48) | .01* | 60 (62) | 25 (42) | 35 (58) | .08 |
| Any therapy | |||||||||||
| Yes | 75 (75) | 42 (56) | 33 (44) | 41 (57) | 29 (40) | 70 (71) | 26 (37) | 44 (63) | |||
| No | 25 (25) | 21 (84) | 4 (16) | .016* | 22 (79) | 6 (21) | .07 | 28 (29) | 8 (29) | 6 (71) | .23 |
| MGMT promoter status | n = 99 | ||||||||||
| Methylated | 67 (68) | 41 (61) | 26 (39) | 41 (61) | 26 (39) | 66 (67) | 24 (36) | 42 (64) | |||
| Unmethylated | 32 (32) | 22 (69) | 10 (31) | .51 | 22 (69) | 10 (31) | .51 | 32 (33) | 10 (31) | 22 (69) | .66 |
| IDH1 | |||||||||||
| Mutated | 6 (6) | 0 (0) | 6 (100) | 0 (0) | 6 (100) | 6 (6) | 5 (83) | 1 (17) | |||
| Wild-type | 94 (94) | 63 (67) | 31 (33) | .003* | 63 (67) | 31 (33) | .002* | 92 (94) | 29 (32) | 63 (68) | .02* |
*P-values indicating significance
Telomerase Status Predicts Survival in Human GBM
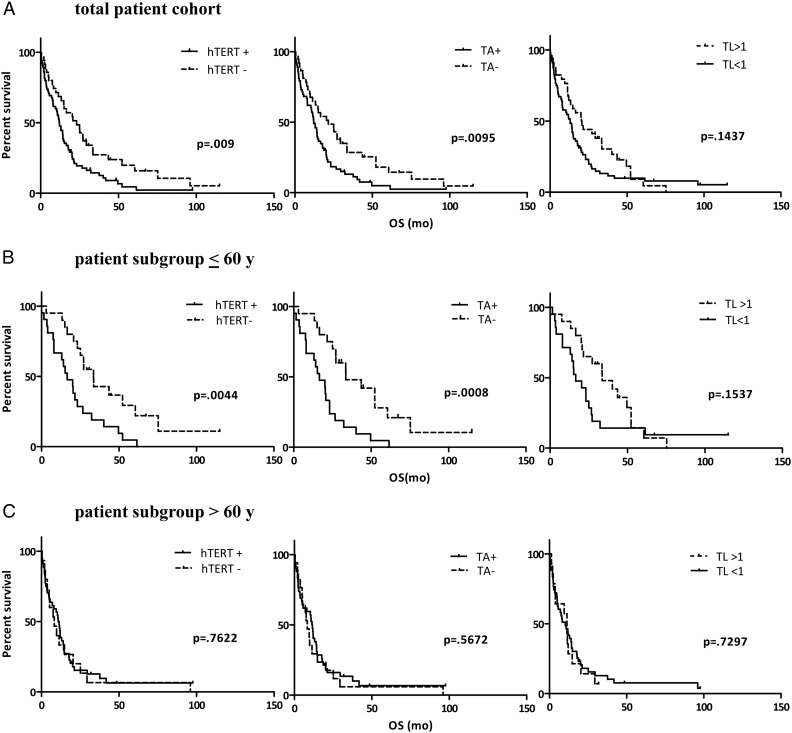
To evaluate the effect of telomerase activation on disease progression, we set hTERT expression, TA, and telomere lengths in relation to GBM patient survival time (Fig. 2A). Kaplan–Meier estimates revealed a significant survival benefit for patients lacking hTERT expression or TA. The survival benefit of patients with longer telomeres did not reach statistical significance. Closer inspection revealed 2 “uncommon” long-term survivors with primary GBM harboring short telomeres (<1) but lack of telomerase. Exclusion of these 2 patients rendered the survival differences according to telomere length statistically significant (P = .03; data not shown).
Fig. 2.
Telomerase-associated parameters compared with overall survival. Kaplan–Meier survival curves for patient subgroups according to the indicated telomerase-associated parameters (hTERT expression, TA, and telomere length [TL]) are shown for the whole GBM patient cohort (n = 100) (A), in the patient subgroup aged ≤60 y (n = 41) (B, upper panel), and in the subgroup of patients aged >60 y (n = 59) (B, lower panel). Abbreviation: OS, overall survival.
Prognostic Significance of Telomerase-Associated Parameters in GBM Patients
To evaluate the prognostic quality of telomerase-associated parameters, univariate and multivariate Cox regression analyses were performed. Univariate analysis (Table 3) confirmed the association of both hTERT expression and TA with shorter patient survival time, whereas data for telomere lengths did not reach statistical significance. Additionally, well-known prognostic factors such as age, performance status, and therapy were significantly associated with overall survival of GBM patients. Moreover, IDH1 mutations were linked to a significantly less aggressive clinical course.
Table 3.
Univariate survival analysis
| Variable | HR* | 95% Confidence Interval | P |
|---|---|---|---|
| Age | 2.13 | 1.38–3.28 | .001* |
| Sex | 1.03 | 0.67–1.59 | .90 |
| KPS (median) | 3.87 | 2.43–6.15 | <.001* |
| Treatmenta | 8.41 | 4.99–14.16 | <.001* |
| hTERT mRNA expression | 1.90 | 1.23–2.96 | .004* |
| TA | 1.78 | 1.14–2.76 | .011* |
| Telomere length | 0.70 | 0.45–1.09 | .11 |
| MGMT promoter methylation | 1.27 | 0.81–1.99 | .30 |
| IDH1 mutation status | 4.59 | 1.43–14.64 | .010* |
*Hazard ratio (HR) for death.
aTreatment grouped according to surgery only vs surgery plus subsequent therapy.
Surprisingly, when the data were analyzed in the multivariate Cox regression model (Table 4), no telomerase-associated parameter was found to have independent prognostic power. While data for KPS and therapy remained statistically significant, age and IDH1 mutation status were not significantly linked to patient survival in the multivariate setting. Consequently, we calculated the interaction of the telomerase-associated parameters with the other prognostic factors derived from the univariate survival analysis by including all respective interaction terms in the multivariate model. While all combinations lacked significance (data not shown), a strong interaction was found between age and both hTERT expression and TA (Table 4, lower part).
Table 4.
Multivariate survival analysis
| hTERT mRNA Expression |
Telomerase Activity |
Telomere Length |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| HR* | 95% Confidence Interval (CI) | P | HR* | 95% CI† | P | HR* | 95% CI† | P | |
| Variable without interaction term | |||||||||
| Age, ≤60/>60 y | 1.15 | 0.69–1.91 | .60 | 0.87 | 0.53–1.45 | .60 | 0.89 | 0.53–1.49 | .66 |
| Sex | 0.73 | 0.44–1.24 | .24 | 0.74 | 0.44–1.24 | .25 | 0.72 | 0.42–1.21 | .21 |
| KPS ≥80/>80 | 2.07 | 1.13–3.79 | .018* | 2.08 | 1.13–3.82 | .019* | 2.11 | 1.14–3.90 | .017* |
| Treatment | 5.03 | 2.49–9.93 | .001* | 5.02 | 2.55–9.89 | .001* | 5.18 | 2.60–10.29 | .001* |
| IDH1 mutation status | 2.80 | 0.81–9.70 | .10 | 2.81 | 0.81–9.76 | .10 | 2.80 | 0.82–9.56 | .101 |
| hTERT mRNA expression | 1.02 | 0.61–1.71 | .93 | ||||||
| Telomerase activity | 1.03 | 0.62–1.70 | .91 | ||||||
| Telomere length | 0.93 | 0.57–1.52 | .80 | ||||||
| Variable with interaction term | |||||||||
| hTERT mRNA Expression × Age | .033* | ||||||||
| Telomerase Activity × Age | .024* | ||||||||
| Telomere Length × Age | .22 | ||||||||
*Adjusted hazard ratio (HR) for death.
Telomerase Activity Is Associated With Shorter Survival in Younger Patients
Given the significant interaction between age and telomerase-associated parameters, we performed age-related subgroup analyses by subdividing our patients into 2 age cohorts: ≤60 years (n = 41) and >60 years (n = 59). Kaplan–Meier survival estimates revealed a significant survival benefit for patients aged ≤60 years whose tumors lacked either hTERT expression or TA (P = .0044 or P = .0008, respectively; Fig. 2B). In contrast, not even a trend toward altered overall survival according to hTERT/TA status was observed in the subgroup of patients aged >60 years. Regarding telomere length, only an insignificant trend toward better survival of patients harboring longer telomeres (>1) was observed in the younger patient subgroup.
Discussion
Activation of a telomere maintenance mechanism (TMM) is one key prerequisite for malignant transformation preventing telomere attrition and thus allowing immortalization. Additionally, TMMs might also have value as prognostic markers in several cancer types.25 In the current study, we investigated hTERT expression, TA, and telomere length in GBM tissues and their relation to clinicopathological parameters and patient survival. Our results confirm that hTERT expression and resulting TA are detectable in a subgroup of GBM tumors (∼60%) characterized by short telomeres and enhanced patient age. Both hTERT expression and TA as parameters for telomerase activation were associated with significantly shortened patient survival, corroborating the prognostic quality of these markers. However, in multivariate analysis, none of the telomerase-associated markers had independent prognostic value in the entire patient cohort. Subgroup analysis showed that age was the interacting factor. Consequently, better prognosis of telomerase-negative patients was confined to patients of younger age, thus excluding a general and independent prognostic value for telomerase-associated parameters in GBM patients.
Our single center–derived data suggest that ∼60% of GBM patients harbor tumor tissues characterized by activated telomerase based on the significant correlation between hTERT mRNA expression and TA. Additionally, telomerase expression inversely correlated with telomere length, indicating that telomerase in GBM keeps telomeres at minimal effective dimensions. Generally, telomerase activation is detected in ∼90% of all human cancer types, including solid tumors and hematological malignancies.3,4,26,27 Thus, in comparison with other solid tumors, a substantial proportion of GBM cases do not exhibit telomere stabilization by telomerase activation. Variable proportions of telomerase-positive GBM ranging between 26% and 89% have been reported in previous studies. However, in all cases, telomerase activation increased with tumor grade.10,14,15,28–32 Lack of any known TMM in other tumor types is generally restricted to low-grade malignancies.3,11,33 In contrast, even in primary GBM a considerable number of tumors lack any sign of telomerase activation, as was also found in the current study. Notably, only a minor proportion of these tumors are characterized by an alternative telomere stabilization mechanism (ie, ALT),15,34,35 posing the question of how telomerase- and ALT-negative tumors stabilize their telomeres. A high prevalence of ALT-positive tumors was found in anaplastic astrocytoma,34 suggesting that secondary GBM might represent predominantly the telomerase-negative subgroup. However, in our patient population only 3% harbored secondary GBM based on the clinical course of disease and accompanied by IDH1 mutations. In these cases, and also in 3 additional ones with IDH1 mutations presenting primarily as GBM without preceding low-grade lesions, we detected very long telomeres indicative of ALT. Although this study did not directly investigate ALT, telomere lengths suggested that in addition to the 6 IDH1 mutant patients, 5 others might harbor this TMM. This would indicate ∼11% ALT-positive tumors, thus leaving ∼30% of tumors without any TMM. Interestingly, we have observed that establishment of stable cell lines from the investigated GBM tissues was limited exclusively to tumors with detectable hTERT expression (manuscript in preparation). This suggests that telomerase-negative GBM cells are not fully immortalized and might depend on so far uncharacterized signals from the tumor microenvironment to avoid telomere shortening–mediated senescence in vivo.
The association of telomerase activation with glioma patient prognosis has been investigated in several studies with variable results. Some reports failed to find a significant association between telomerase activation and patient prognosis.36,37 However, the majority of studies observed a significantly worse survival for patients harboring telomerase-positive gliomas, thus corroborating the findings in our patient cohort.14,15,35,38,39 In some cases, Cox proportional hazard models even revealed an independent prognostic function of telomerase expression, especially in studies involving substantial proportions of low-grade and anaplastic astrocytomas.14,39 In the study by Hiraga et al,29 a significantly worse survival was found exclusively for patients with telomerase-positive, low-grade, and anaplastic gliomas but not GBM. In contrast, 2 studies from the Royds group consistently found a major and independent negative effect of telomerase expression on patient survival, whereas ALT-positive tumors were associated with favorable outcome.15,35 These inconsistent results might be explained by different detection methods for telomerase activation and by the investigation of different patient collectives. Regarding the effect of the detection methods, in the current study, we comparatively analyzed hTERT mRNA expression and TA in relation to telomere length. The highly significant association found between these completely different parameters supports the reliability of our analysis. Considering the study cohort, we report here, prospectively accumulated single-center data including patients with a diagnosis of GBM without further stratification regarding performance status and patient age. In contrast, those studies reporting an independent prognostic value for telomerase activation are based on multicenter-retrospective data collections 15,35 and stratify on patients with good preoperative performance score.15 Also, in our study, telomerase activation was associated with a distinctly worse prognosis based on Kaplan–Meier survival and univariate Cox regression analysis. However, both hTERT expression and TA failed to have independent prognostic power in multivariate analysis. This suggests interaction of telomerase parameters with another prognostic factor in our GBM patient cohort. In accordance with previous studies, univariate analysis showed that age, KPS, treatment, and IDH1 mutation status were significantly associated with overall survival. Of these parameters, patient age, KPS, and IDH1 mutation status were significantly related to telomerase activation in GBM tissues. Patients >60 years of age at diagnosis were characterized not only by lower KPS but also by significantly higher telomerase activation. This is in accordance with previous studies reporting an effect of patient age on the proportion of telomerase-positive GBM.12,14 However, when we included interaction terms between telomerase-associated parameters and these clinicopathological features, only age was significantly linked to telomerase status. Hence, subgroup analysis according to patient age revealed that better prognosis for patients with telomerase-negative tumors is confined to the patient cohort <60 years of age at diagnosis. In contrast, not even a trend was seen in the older patient subgroup.
The reasons for this age-dependent effect are unknown. One might hypothesize that GBM in older patients is generally characterized by worse clinical outcome possibly overruling the effect of TMM. Accordingly, the proportion of patients not treated by standard chemo/radiotherapy based on low KPS is almost confined to the older patient subgroup. However, no significant interaction was detected between telomerase-associated parameters and either KPS or therapy in multivariate analysis, making this explanation rather unlikely. In our study, telomere lengths in tumor tissues shortened significantly with patient age, specifically in telomerase-negative GBM tissues, as was also observed previously by Hakin-Smith et al.15 This might reflect the prevalence of IDH1-mutated, ALT-positive tumors in the younger, telomerase-negative patient subgroup.35,40 On the other hand, telomeres in healthy tissues shorten with age,41 suggesting that activation of TMM in GBM is an earlier step during malignant progression in the older subgroup compared with the younger subgroup. Together, these observations suggest that general differences might exist between carcinogenic processes depending on patient age that might be reflected in clinical outcome and prognostic quality of TMM. Thus IDH1-mutant, ALT-positive GBM might represent a less aggressive subentity congruent molecularly with the proneural subtype35,42 and clinically with secondary GBM associated with younger patient age. However, telomerase per se might also transmit oncogenic functions.43–45 Our in vitro culture experiments outlined above suggest that telomerase might generally be more efficient in driving glioma cell immortalization. In corroboration, only 1 ALT-positive GBM cell line established under specific stem cell culture conditions has been reported so far.46 Alternatively, hTERT was associated with telomere-independent, growth-promoting functions.45 In addition, the tumorigenic potential of telomerase was attributed to the induction of hTERT expression mediated by growth factor in vascular endothelial cells, thus promoting neo-angiogenesis in GBM.43,44
Summarizing, our data suggest that the activation of telomerase in GBM tissues predicts an unfavorable outcome solely in younger GBM patients. These data are derived from an unselected single-center collective without any effect of defined inclusion criteria as used in clinical studies. Thus, telomerase-associated parameters might have limited value as independent prognostic markers concerning GBM in clinical routine. The molecular background of these age-related effects needs to be investigated in depth.
Supplementary Material
Funding
This work was supported by the Forschungsfond der Österreichischen Krebshilfe OÖ and the OÖ Kinderkrebshilfe Forschungsverein, by the Initiative Krebsforschung of the Medical University Vienna, and by the Jubiläumsfonds der Österreichischen Nationalbank.
Supplementary Material
Acknowledgments
The authors thank Kerstin Schauer for skillful technical assistance.
Conflict of interest statement. No potential conflicts of interest were disclosed.
References
- 1.Blackburn EH. Structure and function of telomeres. Nature. 1991;350(6319):569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 3.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33(5):787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 4.Soria JC, Vielh P, el-Naggar AK. Telomerase activity in cancer: a magic bullet or a mirage? Adv Anat Pathol. 1998;5(2):86–94. doi: 10.1097/00125480-199803000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 6.Ducray F, El Hallani S, Idbaih A. Diagnostic and prognostic markers in gliomas. Curr Opin Oncol. 2009;21(6):537–542. doi: 10.1097/CCO.0b013e32833065a7. [DOI] [PubMed] [Google Scholar]
- 7.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 8.Spiegl-Kreinecker S, Pirker C, Filipits M, et al. O6-methylguanine DNA methyltransferase protein expression in tumor cells predicts outcome of temozolomide therapy in glioblastoma patients. Neuro Oncol. 2010;12(1):28–36. doi: 10.1093/neuonc/nop003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27(34):5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 10.Langford LA, Piatyszek MA, Xu R, Schold SC, Jr, Shay JW. Telomerase activity in human brain tumours. Lancet. 1995;346(8985):1267–1268. doi: 10.1016/s0140-6736(95)91865-5. [DOI] [PubMed] [Google Scholar]
- 11.Maes L, Van Neste L, Van Damme K, et al. Relation between telomerase activity, hTERT and telomere length for intracranial tumours. Oncol Rep. 2007;18(6):1571–1576. doi: 10.3892/or.18.6.1571. [DOI] [PubMed] [Google Scholar]
- 12.Shervington A, Patel R. Differential hTERT mRNA processing between young and older glioma patients. FEBS Lett. 2008;582(12):1707–1710. doi: 10.1016/j.febslet.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 13.Shervington A, Patel R, Lu C, et al. Telomerase subunits expression variation between biopsy samples and cell lines derived from malignant glioma. Brain Res. 2007;1134(1):45–52. doi: 10.1016/j.brainres.2006.11.093. [DOI] [PubMed] [Google Scholar]
- 14.Boldrini L, Pistolesi S, Gisfredi S, et al. Telomerase activity and hTERT mRNA expression in glial tumors. Int J Oncol. 2006;28(6):1555–1560. doi: 10.3892/ijo.28.6.1555. [DOI] [PubMed] [Google Scholar]
- 15.Hakin-Smith V, Jellinek DA, Levy D, et al. Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet. 2003;361(9360):836–838. doi: 10.1016/s0140-6736(03)12681-5. [DOI] [PubMed] [Google Scholar]
- 16.Berger W, Elbling L, Micksche M. Expression of the major vault protein LRP in human non-small-cell lung cancer cells: activation by short-term exposure to antineoplastic drugs. Int J Cancer. 2000;88(2):293–300. [PubMed] [Google Scholar]
- 17.O'Callaghan N, Dhillon V, Thomas P, Fenech M. A quantitative real-time PCR method for absolute telomere length. BioTechniques. 2008;44(6):807–809. doi: 10.2144/000112761. [DOI] [PubMed] [Google Scholar]
- 18.Callicott RJ, Womack JE. Real-time PCR assay for measurement of mouse telomeres. Comp Med. 2006;56(1):17–22. [PubMed] [Google Scholar]
- 19.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30(10):e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gan Y, Mo Y, Johnston J, Lu J, Wientjes MG, Au JL. Telomere maintenance in telomerase-positive human ovarian SKOV-3 cells cannot be retarded by complete inhibition of telomerase. FEBS Lett. 2002;527(1–3):10–14. doi: 10.1016/s0014-5793(02)03141-1. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 22.Brandes AA, Tosoni A, Cavallo G, et al. Temozolomide 3 weeks on and 1 week off as first-line therapy for recurrent glioblastoma: phase II study from Gruppo Italiano Cooperativo di Neuro-oncologia (GICNO) Br J Cancer. 2006;95(9):1155–1160. doi: 10.1038/sj.bjc.6603376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10(6):1871–1874. doi: 10.1158/1078-0432.ccr-03-0384. [DOI] [PubMed] [Google Scholar]
- 24.Karayan-Tapon L, Quillien V, Guilhot J, et al. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2009;97(3):311–322. doi: 10.1007/s11060-009-0031-1. [DOI] [PubMed] [Google Scholar]
- 25.Royds JA, Al Nadaf S, Wiles AK, et al. The CDKN2A G500 allele is more frequent in GBM patients with no defined telomere maintenance mechanism tumors and is associated with poorer survival. PLoS One. 2011;6(10):e26737. doi: 10.1371/journal.pone.0026737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim NW, Piatyszek MA, Prowse KR, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266(5193):2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 27.Shay JW, Wright WE. Telomerase therapeutics for cancer: challenges and new directions. Nat Rev Drug Discov. 2006;5(7):577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 28.Chong EY, Lam PY, Poon WS, Ng HK. Telomerase expression in gliomas including the nonastrocytic tumors. Hum Pathol. 1998;29(6):599–603. doi: 10.1016/s0046-8177(98)80009-9. [DOI] [PubMed] [Google Scholar]
- 29.Hiraga S, Ohnishi T, Izumoto S, et al. Telomerase activity and alterations in telomere length in human brain tumors. Cancer Res. 1998;58(10):2117–2125. [PubMed] [Google Scholar]
- 30.Le S, Zhu JJ, Anthony DC, Greider CW, Black PM. Telomerase activity in human gliomas. Neurosurgery. 1998;42(5):1120–1124. doi: 10.1097/00006123-199805000-00099. [DOI] [PubMed] [Google Scholar]
- 31.Morii K, Tanaka R, Onda K, Tsumanuma I, Yoshimura J. Expression of telomerase RNA, telomerase activity, and telomere length in human gliomas. Biochem Biophys Res Commun. 1997;239(3):830–834. doi: 10.1006/bbrc.1997.7562. [DOI] [PubMed] [Google Scholar]
- 32.Sano T, Asai A, Mishima K, Fujimaki T, Kirino T. Telomerase activity in 144 brain tumours. Br J Cancer. 1998;77(10):1633–1637. doi: 10.1038/bjc.1998.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valls C, Pinol C, Rene JM, Buenestado J, Vinas J. Telomere length is a prognostic factor for overall survival in colorectal cancer. Colorectal Dis. 2011;13(11):1265–1272. doi: 10.1111/j.1463-1318.2010.02433.x. doi:10.1111/j.1463-1318.2010.02433.x. [DOI] [PubMed] [Google Scholar]
- 34.Heaphy CM, Subhawong AP, Hong SM, et al. Prevalence of the alternative lengthening of telomeres: telomere maintenance mechanism in human cancer subtypes. Am J Pathol. 2011;179(4):1608–1615. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald KL, McDonnell J, Muntoni A, et al. Presence of alternative lengthening of telomeres mechanism in patients with glioblastoma identifies a less aggressive tumor type with longer survival. J Neuropathol Exp Neurol. 2010;69(7):729–736. doi: 10.1097/NEN.0b013e3181e576cf. [DOI] [PubMed] [Google Scholar]
- 36.Nakatani K, Yoshimi N, Mori H, et al. The significant role of telomerase activity in human brain tumors. Cancer. 1997;80(3):471–476. doi: 10.1002/(sici)1097-0142(19970801)80:3<471::aid-cncr15>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 37.Persson A, Englund E. Different assessments of immunohistochemically stained Ki-67 and hTERT in glioblastoma multiforme yield variable results: a study with reference to survival prognosis. Clin Neuropathol. 2008;27(4):224–233. doi: 10.5414/npp27224. [DOI] [PubMed] [Google Scholar]
- 38.Alonso MM, Fueyo J, Shay JW, et al. Expression of transcription factor E2F1 and telomerase in glioblastomas: mechanistic linkage and prognostic significance. J Natl Cancer Inst. 2005;97(21):1589–1600. doi: 10.1093/jnci/dji340. [DOI] [PubMed] [Google Scholar]
- 39.Fukushima T, Yoshino A, Katayama Y, Watanabe T, Kusama K, Moro I. Prediction of clinical course of diffusely infiltrating astrocytomas from telomerase expression and quantitated activity level. Cancer Lett. 2002;187(1–2):191–198. doi: 10.1016/s0304-3835(02)00357-9. [DOI] [PubMed] [Google Scholar]
- 40.Henson JD, Hannay JA, McCarthy SW, et al. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin Cancer Res. 2005;11(1):217–225. [PubMed] [Google Scholar]
- 41.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12(10):1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 42.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Falchetti ML, Mongiardi MP, Fiorenzo P, et al. Inhibition of telomerase in the endothelial cells disrupts tumor angiogenesis in glioblastoma xenografts. Int J Cancer. 2008;122(6):1236–1242. doi: 10.1002/ijc.23193. [DOI] [PubMed] [Google Scholar]
- 44.Falchetti ML, Pierconti F, Casalbore P, et al. Glioblastoma induces vascular endothelial cells to express telomerase in vitro. Cancer Res. 2003;63(13):3750–4. [PubMed] [Google Scholar]
- 45.Gorbunova V, Seluanov A. Telomerase as a growth-promoting factor. Cell Cycle. 2003;2(6):534–537. doi: 10.4161/cc.2.6.515. [DOI] [PubMed] [Google Scholar]
- 46.Silvestre DC, Pineda JR, Hoffschir F, et al. Alternative lengthening of telomeres in human glioma stem cells. Stem Cells. 2011;29(3):440–451. doi: 10.1002/stem.600. doi:10.1002/stem.600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.