Abstract
The genetic basis of resilience, defined as better cognitive functioning than predicted based on neuroimaging or neuropathology, is not well understood. Our objective was to identify genetic variation associated with executive functioning resilience. We computed residuals from regression models of executive functioning, adjusting for age, sex, education, Hachinski score, and MRI findings (lacunes, cortical thickness, volumes of white matter hyperintensities and hippocampus). We estimated heritability and analyzed these residuals in models for each SNP. We further evaluated our most promising SNP result by evaluating cis-associations with brain levels of nearby (±100 kb) genes from a companion data set, and comparing expression levels in cortex and cerebellum from decedents with AD with those from other non-AD diseases. Complete data were available for 750 ADNI participants of European descent. Executive functioning resilience was highly heritable (H2=0.76; S.E. = 0.44). rs3748348 on chromosome 14 in the region of RNASE13 was associated with executive functioning resilience (p-value=4.31×10−7). rs3748348 is in strong linkage disequilibrium (D′ of 1.00 and 0.96) with SNPs that map to TPPP2, a member of the α-synuclein family of proteins. We identified nominally significant associations between rs3748348 and expression levels of three genes (FLJ10357, RNASE2, and NDRG2). The strongest association was for FLJ10357 in cortex, which also had the most significant difference in expression between AD and non-AD brains, with greater expression in cortex of decedents with AD (p-value=7×10−7). Further research is warranted to determine whether this signal can be replicated and whether other loci may be associated with cognitive resilience.
Keywords: Memory, Executive functioning, Alzheimer’s disease, Psychometrics, Resilience, GWAS
Background
Neurodegenerative conditions are common in elderly populations and represent a distinct burden on individuals, families, and the economy (Lewin Group and Alzheimer’s Association 2003; Centers for Disease Control and Prevention and Alzheimer’s Association 2007). Magnetic resonance imaging (MRI) studies indicate that structural lesions such as lacunes (Longstreth et al. 1998; Longstreth et al. 2002) and white matter hyperintensities (Longstreth et al. 1996; Ikram et al. 2007) are common in community-dwelling older adults. While there is no question that these structural lesions are associated with diminished cognition, there remains significant variability in cognition beyond that explained by indicators of brain pathology or neuroimaging. The term “cognitive reserve” has been used to describe the concept of buffering the impact of pathology on brain performance (Satz 1993; Stern 2002).
We use the term “resilience” rather than “reserve,” though our notion of resilience is very similar to the notion of reserve proposed by Reed and colleagues, who evaluated associations with residual variability in a measure of episodic memory after controlling for demographic and MRI structural findings (Reed et al. 2010). In that paper, hypotheses regarding the relationship between that residual and several external criteria were robustly confirmed (Reed et al. 2010). Reed and colleagues address both positive reserve (which we call resilience) and negative reserve, referring to scores much worse than predicted. We use “resilience” specifically to refer to the top end of what Reed and colleagues called “reserve”—that is, specifically people whose performance was better than that predicted on the basis of imaging, demographic, and clinical characteristics
There is an extensive literature on factors influencing cognitive reserve. Much of this literature focuses on lifestyle (Scarmeas and Stern 2003) or cognitive (Hertzog et al. 2009) factors. Currently, it is not known whether genetic variation may be associated with resilience, or whether resilience is entirely due to environment.
Our goal was thus to use extensive imaging, genetic, and neuropsychological data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) to determine whether there may be genetic variability that is associated with resilience in executive functioning.
Methods
Alzheimer’s Disease Neuroimaging Initiative (ADNI)
Data used for this study were obtained from the ADNI database (http://adni.loni.ucla.edu/). The ADNI was initiated in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessments can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. Michael W. Weiner, MD, VA Medical Center and University of California-San Francisco is the Principal Investigator of this initiative. This $60 million, multiyear public-private partnership involves many coinvestigators from a broad range of academic institutions and private corporations. More than 800 participants, aged 55 to 90, have been recruited from across more than 50 sites in the US and Canada. This includes approximately 200 patients diagnosed with early AD followed for 2 years. Longitudinal imaging data, including structural 1.5 Tesla MRI scans, were collected on the full sample. Neuropsychological and clinical assessments were collected at baseline, and at follow-up visits of six-to-twelve month intervals. Other available data used in the present analysis included APOE genotype and genome-wide SNP data obtained in the full ADNI sample, as outlined in (Saykin et al. 2010). Further information about ADNI can be found in (Weiner et al. 2010) and at http://www.adni-info.org. The study was conducted after Institutional Review Board approval at each site. Written informed consent was obtained from all study participants, or their authorized representatives.
1.5T MRI neuroimaging
All participants received 1.5 Tesla structural magnetic resonance imaging (MRI). The neuroimaging methods utilized by ADNI have been described in detail previously (Jack et al. 2008) utilizing calibration techniques to maintain consistent protocols across scanners and sites. Raw dicom data of T1-weighted MP-RAGE scans acquired from 1.5 Tesla scanners at baseline visits from all participants were obtained via the ADNI database (http://www.loni.ucla.edu/ADNI/). Only MRI assessments with an overall quality control of “Pass” were included in these analyses. Images were processed using Freesurfer software (http://surfer.nmr.mgh.harvard.edu), an atlas-based approach that has been validated for use in subjects with a great deal of morphologic variability (Desikan et al. 2006). White matter hyperintensities (WMH) were detected on coregistered T1-, T2-, and PD-weighted images using an automated method described previously (Schwarz et al. 2009; Carmichael et al. 2010). WMH were detected in Minimal Deformation Template (MDT) space at each voxel based on corresponding PD, T1, and T2 intensities there, the prior probability of WMH there, and the conditional probability of WMH there based on the presence of WMH at neighboring voxels. The resulting map of WMH voxels across the brain is summarized by an estimate of total WMH volume. WMH volumes estimated with this method agreed strongly with WMH volumes estimated from fluidattenuated inversion recovery (FLAIR) MRI in a large, diverse elderly sample (Schwarz et al. 2009). In our analyses we used presence of one or more lacunes, cortical volume (summed across entorhinal cortex, fusiform, pars triangularis, caudal middle frontal, superior frontal, medial orbitofrontal, rostral, middle frontal, and lateral orbitofrontal, controlling for total brain volume), volume of bilateral hippocampus (controlling for total brain volume), and the natural log of white matter hyperintensity volume.
Neuropsychological tests
ADNI administers an extensive neuropsychological battery to participants at each study visit, including several measures of memory and executive function.
Memory tasks administered include immediate and delayed recall of Logical Memory prose passages from the Wechsler Memory Scale-Revised (Weschler 1987), a word list learning task from the ADAS-Cog and its delayed recall and recognition (Mohs et al. 1997), the Rey Auditory Verbal Learning Test (Rey 1964), and the recall task from the Mini-Mental State Examination (Folstein et al. 1975). Executive functioning tasks include parts A and B of the Trail Making test (Reitan 1958), a clock drawing task (Goodglass and Kaplan 1983), animal and vegetable category fluency (Morris et al. 1989), digit span backwards from the Wechsler Memory Scale-Revised (Weschler 1987), and the digit-symbol substitution task from the Wechsler Adult Intelligence Scale-Revised (Weschler 1981).
We used modern psychometric theory methods applied to item-level data from the ADNI neuropsychological battery to develop composite scores separately for memory (ADNI-Mem) and executive functioning (ADNI-EF). For complete details regarding the development of ADNI-Mem and ADNI-EF, please refer to the companion papers in this volume (Crane et al. 2012 submitted; Gibbons et al. 2012 submitted). For executive functioning, we found that a bi-factor model had the best fit to the data. We extracted factor scores for the general factor defined by all of the items from Mplus v5 (Muthén and Muthén 2006); this factor score is the ADNI-EF score. For memory, we used a longitudinal single factor model to account for different versions of the ADAS-Cog and of the Rey AVLT. We used parameters from that model to generate scores at each study visit, also using Mplus (Muthén and Muthén 2006). Further details are spelled out in our companion papers (Crane et al. 2012 submitted; Gibbons et al. 2012 submitted).
Derivation of phenotypes of interest
Several demographic and imaging-derived factors are associated with executive functioning, including age, educational attainment, atrophy, lacunes, and white matter hyperintensities (Mungas et al. 2005). We developed a regression model with executive functioning as the outcome variable and several predictors: age, sex, education (up to 8 years, 9–11 years, 12 years, 13–15 years, 16 years and up), Hachinski score, presence of lacunes, residual hippocampal volume (after adjusting for intracranial volume), residual cortical volume (after adjusting for intracranial volume), and the log of white matter hyperintensity (WMH) volume. The residual (standardized) from this regression model was our phenotype of interest for the first genome-wide association study (GWAS) analysis. This residual represents executive functioning that is beyond that predicted by the independent variables we included in our regression models. At one extreme these residuals are from people whose executive functioning is much better than that predicted by their demographic characteristics, Hachinski score, and brain structure as assessed with MRI (i.e., resilience), while at the other extreme these residuals are from people whose executive functioning is much worse than that predicted by the same set of independent variables.
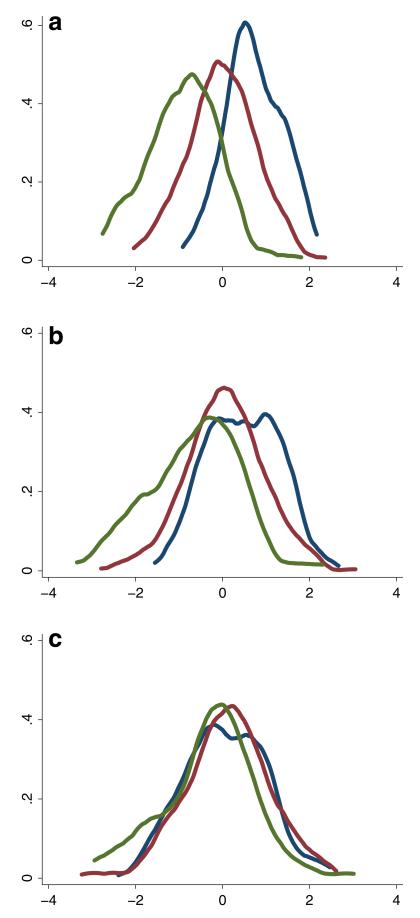
An important concern with ADNI was that the data came from a highly selected study cohort. Three highly selected groups were recruited: controls with normal cognition, people with MCI, and people with early AD, all of whom had low Hachinski scores (4 or less at baseline). Executive functioning was correlated with diagnostic group (polyserial correlation ρ=0.67, SE=0.02), as shown in the kernel density plot shown in Fig. 1a. In these figures, the blue curves represent the distribution of scores from people with normal cognition, red represents people with MCI, and green represents people with AD. Our executive functioning residuals were much less correlated with diagnostic group, as shown in Fig. 1b (ρ=0.27, SE=0.06). We were still concerned that diagnostic group differences might be driving our findings. We therefore added memory to our regression model, and the residuals from this model were even less tightly correlated with diagnostic group, as shown in Fig. 1c (ρ=0.08, SE=0.04). This residual was our phenotype of interest for our second GWAS analyses.
Fig. 1.
Kernel density plots* of a executive functioning, b residual for executive functioning when adjusting for demographics, Hachinski score and brain imaging parameters and c residual for executive functioning when adjusting for memory, demographics, Hachinski score and brain imaging parameters * Blue curves represent scores for people with normal cognition, red represents people with MCI, and green represents people with AD
Genotyping and quality control
The ADNI-I sample was genotyped using the Human610-Quad BeadChip (Illumina, Inc., San Diego, CA), resulting in 620,901 SNP and CNV biomarkers. The genotyping protocol followed the manufacturer’s instructions and is explained in detail in (Saykin et al. 2010).
APOE was genotyped at the time of screening. The two previously established APOE genotype SNPs (rs429358, rs7412) that characterize the ε2/ε3/ε4 alleles were not available on the Human610-Quad BeadChip array. Methods for genotyping this allele in ADNI have been published (Potkin et al. 2009). We downloaded APOE genotype from the ADNI database and added them to the genome-wide genotype data before we assessed sample quality.
Standard quality control procedures were performed on the ADNI genotype data using PLINK (ver. 1.07) (Purcell et al. 2007) as described in (Shen et al. 2010). Samples were excluded based on the following criteria: (1) call rate <90 %, (2) ambiguous sex identification, (3) identity check with PI_HAT >0.5 (3 sibling pairs were identified and one from each pair was randomly excluded). Markers were excluded based on the following criteria: (1) call rate <90 %, (2) Hardy-Weinberg equilibrium test in controls <10−6, (3) minor allele frequency (MAF)<5 %.
These analyses are restricted to non-Hispanic Caucasian participants. We used HapMap phase 3 release 2 data from 988 founders from 11 known populations (The International HapMap Project 2003) to identify non-Hispanic Caucasian participants from ADNI. We performed multi-dimensional scaling analysis on combined data from ADNI and HapMap using PLINK. ADNI participants were selected if they were grouped together with the HapMap CEU (Utah residents with Northern and Western European ancestry from the Centre d’Etude du Polymorphisme Humain (CEPH) collection) or TSI (Toscani in Italy) participants. If ADNI participants were separately grouped from any of the HapMap populations, ADNI’s ethnicity and race information were used. After the QC procedure, 750 participants and 531,096 single nucleotide polymorphisms (SNPs) were selected for the subsequent analyses and the genotyping rate in the remaining samples was ≥0.995.
Heritability
We estimated the heritability for our phenotype of interest (executive functioning residual after adjusting for memory, demographics, Hachinski score, brain imaging parameters) without adjusting for principal components using the software package GCTA v0.92.9 (Yang et al. 2011). In this approach, heritability is defined as a ratio of variances, by expressing the proportion of the phenotypic variance that can be attributed to variance of genotypic values (Visscher et al. 2008).
Genome-wide association analysis
We further analyzed population substructure due to genetic ancestry using Eigenstrat v4.0 (Price et al. 2006). Although we found that adjustment for substructure made little difference in the obtained results (see Appendix 2; Table 7), we included the first three principal components from Eigenstrat v4.0 in the final analyses shown here. First, we ran a GWAS on the residual for executive functioning which accounted for demographics, Hachinski score and brain imaging parameters to examine the main effect of each SNP using PLINK (Purcell et al. 2007). We followed up with a GWAS on the residual for executive functioning which accounted for all of those factors as well as ADNI-Mem. Bonferroni correction was applied and SNPs with corrected p-value <0.05 (uncorrected p-value <9.42×10−8, i.e. 0.05/531,096) were considered significant. Manhattan and Quantile-quantile (Q-Q) plots were generated in R (R Development Core Team 2005) using the results file from PLINK. Linkage disequilibrium (LD) plots were generated in Haploview v4.2 (http://www.broadinstitute.org/haploview/haploview) (Barrett et al. 2005). The LD plot was created by using a region ±250 kb from the peak SNP. Regional association results for the top SNPs obtained from the GWAS analysis were plotted (see Appendix 3) using Locus Zoom v1.1 (Pruim et al. 2010).
We followed up with three more GWAS analyses to check if the top signal (peak SNP) was driven by individuals whose executive functioning was better than predicted (i.e. resilience). We divided the residual for executive functioning (which was adjusted for memory, demographic characteristics, Hachinski score, and brain imaging parameters) into five quintiles where Q5 contains residual scores for those whose executive functioning was better than predicted by brain imaging parameters, demographics and Hachinski score, and Q1 contains residual scores for those whose executive functioning was worse than predicted by brain imaging parameters, Hachinski score and demographics. The three GWAS analyses we performed were a) Q5 vs. Q1, b) Q1 vs. Q2/Q3/Q4 combined and c) Q5 vs. Q2/Q3/Q4 combined. All these analyses were also adjusted for the first three principal components.
We performed additional sensitivity analyses of the top 20 SNPs in which we directly modeled the SNP effect on executive functioning, controlling for the covariates.
Association between SNPs and gene expression
We (N E-T) reviewed a brain expression data set for rs3748348. Gene expression levels are endophenotypes with a substantial underlying genetic component (Ertekin-Taner 2011) that can be reliably detected with relatively small sample size. We measured gene expression levels from temporal cortex of 399 subjects and cerebellar tissue from 374 subjects. 340 subjects had both measurements. All subjects were participants in the published Mayo LOAD GWAS (Carrasquillo et al. 2009) as part of the autopsy-based series. All subjects had neuropathologic evaluation by a single person. All AD (n=202 for temporal cortex and 197 for cerebellum) subjects had diagnosis of definite AD according to the NINCDS-ADRDA criteria (McKhann et al. 1984) and had Braak scores of ≥4.0. All control (non-AD, n=197 for temporal cortex and 177 for cerebellum) subjects had Braak scores of ≤2.5, but many had brain pathology unrelated to AD. Complete methods are described in (Allen et al. 2012 in press). Briefly, we extracted total RNA and determined its quantity and quality. We measured transcript levels and performed appropriate quality control. Genome-wide SNP data were also available for these individuals. We evaluated data for the three SNPs in the same linkage disequilibrium block as rs3748348 looking for cis-associations of brain levels of the five genes within 100 kb of rs3748348. We used regression models with minor allele dosage as the predictor and expression levels as endophenotypes, adjusting for APOE ε4 dosage (0, 1, or 2), age at death, sex, plate, RIN, and adjusted RIN squared, which was defined as the squared difference between RIN and mean RIN. RIN refers to the RNA Information Number, an indicator of the quality of the RNA (see http://gcf.pbrc.edu/docs/Agilent/RNA%20Integrity%20Analysis.pdf).
Results
We restricted our analyses to self-identified non-Hispanic whites (n=750) who had complete data for memory and executive functioning. All the included participants had complete genetic and MRI data. Demographic and clinical data for the cohort analyzed are shown in Table 1.
Table 1.
Demographic characteristics of participants included in the analyses
| Characteristic | Baseline diagnosis |
Total | ||
|---|---|---|---|---|
| NC | MCI | AD | ||
| # of participants | 207 | 365 | 178 | 750 |
| Sex: (M/F) | 113/94 | 235/130 | 97/81 | 445/305 |
| Age: Mean (S.D.) | 76.2 (4.9) | 75.0 (7.4) | 75.7 (7.5) | 75.5 (6.9) |
| Education | 16.2 (2.7) | 15.7 (3.0) | 14.9 (3.0) | 15.6 (3.0) |
| APOE ε4 (any/none) | 55/152 | 199/166 | 116/62 | 370/380 |
| ADNI-Mem: Mean (S.D.) | 0.97 (0.54) | −0.09 (0.59) | −0.85 (0.56) | 0.02 (0.87) |
| ADNI-EF: Mean (S.D.) | 0.72 (0.65) | −0.02 (0.79) | −0.92 (0.83) | −0.03 (0.96) |
| Hachinski score : Mean (S.D.) | 0.56 (0.7) | 0.61 (0.71) | 0.67 (0.71) | 0.61 (0.71) |
We found the heritability for our phenotype of interest to be quite high (H2=0.76 with a standard error of 0.44). The elevated standard error for the estimate is likely due to the small sample size. We performed similar analyses of the raw executive functioning scores (i.e., not the residuals but the scores themselves) and found that the heritability of executive functioning was much lower (H2=0.20, standard error=0.36).
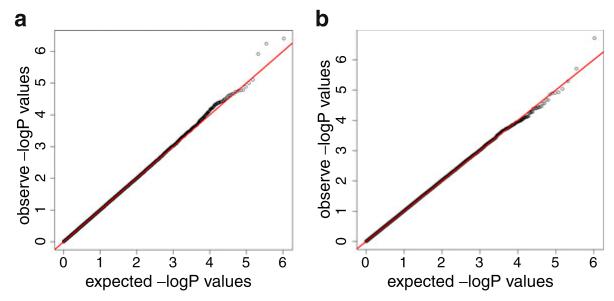
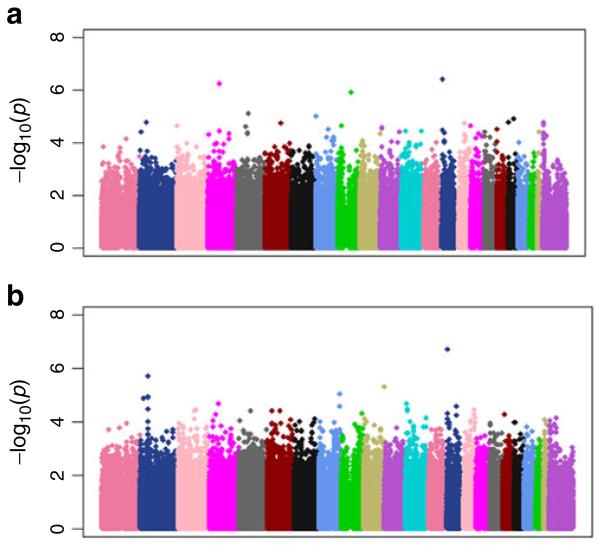
A Q-Q plot showed that our first phenotype of interest (residual for executive functioning adjusting for demographic characteristics, Hachinski score and brain imaging parameters) had little or no risk of confounding due to population stratification (Fig. 2a); the genomic inflation factor (λ) was 1.00. There were no SNPs which met the genome-wide significance threshold (9.42×10−8) likely due to the limited sample size. We identified the peak SNP rs3748348 (Chr 14) with a highly suggestive p-value of 4.31×10−7. The Manhattan plot for this phenotype is shown in Fig. 3a. We followed-up with a GWAS of the residual for executive functioning adjusting for ADNI-Mem in addition to demographic characteristics, Hachinski score, and the same brain imaging parameters. This Q-Q plot is shown in Fig. 2b; λ was 1.01. Although none of the SNPs met stringent genome-wide significance, we identified the same SNP (rs3748348) with an even more suggestive p-value of 2.12×10−7. The Manhattan plot for this phenotype is shown in Fig. 3b.
Fig. 2.
Quantile-quantile (Q-Q) plots for a residual for executive functioning when adjusting for demographics, Hachinski score and brain imaging parameters, and b residual for executive functioning when adjusting for memory, demographics, Hachinski score and brain imaging parameters
Fig. 3.
Manhattan plots for a residual for executive functioning when adjusting for demographics, Hachinski score and brain imaging parameters and b residual for executive functioning when adjusting for memory, demographics, Hachinski score and brain imaging parameters. The X-axis refers to points along the genome (separated by chromosome) at which each of the several hundred thousand single nucleotide polymorphisms (SNPs; represented by a dot) evaluated are located. The y axis refers to the negative logarithm of the p-value for a test of association between each SNP and the quantitative trait
We followed up with three more GWAS analyses to determine whether this signal was driven by individuals with better or worse executive functioning than predicted. As shown in Table 2, the signal for rs3748348 was driven by the higher end—people whose executive functioning was better than predicted—and not by the lower end—people whose executive functioning was worse than predicted—The purpose of these analyses was to determine whether our findings were due to resilience (i.e., driven by people whose executive function was greater than that predicted) or by the converse. We relied on the overall analyses including all participants as our primary analysis, and did not try to interpret SNP findings from these additional analyses.
Table 2.
Comparison of the top 10 SNPs across different analyses in terms of p-values, coefficients and position
| CHR | SNP | Minor Allele | MAF | Final residual model |
Q1 vs. Q5 analysis |
Q1 vs. Q2/Q3/Q4 analysis |
Q5 vs. Q2/Q3/Q4 analysis |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | p-value | Position | β | p-value | Position | β | p-value | Position | β | p-value | Position | ||||
| 14 | rs3748348 | A | 0.40 | 0.28 | 2.12×10−7 | 1 | 0.37 | 5.86×10−7 | 1 | 0.78 | 0.0781 | 40579 | 0.55 | 2.48×10−5 | 7 |
| 2 | rs1609361 | C | 0.44 | 0.25 | 2.10×10−6 | 2 | 0.4 | 6.87×10−7 | 2 | 0.68 | 0.0057 | 2758 | 0.62 | 0.0005 | 248 |
| 10 | rs7897741 | C | 0.12 | −0.36 | 5.91×10−6 | 3 | 2.92 | 0.0001 | 31 | 1.71 | 0.0040 | 1914 | 1.59 | 0.0530 | 28134 |
| 2 | rs16987794 | C | 0.14 | 0.32 | 1.10×10−5 | 4 | 0.48 | 0.0022 | 1004 | 0.71 | 0.1108 | 58308 | 0.65 | 0.0125 | 6331 |
| 8 | rs13251704 | G | 0.24 | −0.27 | 1.16×10−5 | 5 | 1.84 | 0.0028 | 1272 | 1.35 | 0.0524 | 27066 | 1.35 | 0.0766 | 40961 |
| 2 | rs2698011 | A | 0.44 | 0.23 | 1.29×10−5 | 6 | 0.43 | 4.12×10−6 | 3 | 0.68 | 0.0066 | 3236 | 0.66 | 0.0023 | 1177 |
| 2 | rs2715077 | T | 0.44 | 0.23 | 1.37×10−5 | 7 | 0.44 | 6.46×10−6 | 5 | 0.71 | 0.0155 | 7787 | 0.64 | 0.0012 | 626 |
| 12 | rs7306879 | G | 0.27 | −0.25 | 1.42×10−5 | 8 | 2.17 | 8.63×10−5 | 24 | 1.27 | 0.0895 | 46725 | 1.65 | 0.0026 | 1335 |
| 2 | rs2698029 | A | 0.44 | 0.23 | 1.44×10−5 | 9 | 0.44 | 6.44×10−6 | 4 | 0.72 | 0.0182 | 9135 | 0.64 | 0.0010 | 501 |
| 4 | rs4865353 | C | 0.44 | 0.23 | 2.23×10−5 | 10 | 0.57 | 0.0015 | 600 | 0.92 | 0.5669 | 302273 | 0.62 | 0.0006 | 314 |
In our sensitivity analyses, we performed separate models for the 20 most salient SNPs in which ADNI-EF was the dependent variable and the SNP was an independent variable along with the same covariates. Results were indistinguishable from the associations reported here for the SNP effects on the residuals (Appendix Table 8).
We identified nominally significant associations with FLJ10357, RNASE2 and NDRG2 levels (see Table 3). The strongest association was for FLJ10357 in temporal cortex, where the minor allele was associated with higher brain levels of this mRNA. Dr. Ertekin-Taner also evaluated transcript levels for brains of people with AD and people without AD. Levels of FLJ10357 showed the greatest differences in expression levels (p-value=10−7); see Table 4.
Table 3.
Association between minor alleles at three selected SNPs and mRNA expression in cerebellum and cortex of brains from people with AD and non-AD pathologya
| SNP | Allele | Probe | Symbol | Cerebellum |
Cortex |
||||
|---|---|---|---|---|---|---|---|---|---|
| n | β | p-value | n | β | p-value | ||||
| rs1243459 | A | ILMN_1705743 | FLJ10357 | 363 | −0.020 | 0.27 | 381 | 0.040 | 0.24 |
| ILMN_2276811 | METT11D1 | −0.020 | 0.95 | ||||||
| ILMN_1670535 | NDRG2 | −0.001 | 0.08 | 0.010 | 0.43 | ||||
| ILMN_2361603 | NDRG2 | −0.030 | 0.64 | 0.020 | 0.03 | ||||
| ILMN_1762871 | RNASE13 | −0.006 | 0.17 | ||||||
| ILMN_1730628 | RNASE2 | 0.030 | 0.07 | −0.140 | 0.02 | ||||
| ILMN_1688295 | ZNF219 | −0.080 | 0.63 | 0.010 | 0.40 | ||||
| rs3748348 | A | ILMN_1705743 | FLJ10357 | 365 | 0.040 | 0.07 | 382 | 0.002 | 0.96 |
| ILMN_2276811 | METT11D1 | 0.020 | 0.11 | ||||||
| ILMN_1670535 | NDRG2 | 0.030 | 0.08 | −0.020 | 0.22 | ||||
| ILMN_2361603 | NDRG2 | 0.006 | 0.64 | −0.020 | 0.03 | ||||
| ILMN_1762871 | RNASE13 | −0.030 | 0.19 | ||||||
| ILMN_1730628 | RNASE2 | 0.030 | 0.45 | 0.130 | 0.05 | ||||
| ILMN_1688295 | ZNF219 | 0.010 | 0.50 | −0.006 | 0.74 | ||||
| rs9624 | T | ILMN_1705743 | FLJ10357 | 365 | 0.010 | 0.53 | 382 | 0.150 | 0.002 |
| ILMN_2276811 | METT11D1 | 0.010 | 0.39 | ||||||
| ILMN_2361603 | NDRG2 | 0.030 | 0.04 | 0.010 | 0.27 | ||||
| ILMN_1670535 | NDRG2 | 0.000 | 0.98 | −0.030 | 0.26 | ||||
| ILMN_1762871 | RNASE13 | 0.050 | 0.12 | ||||||
| ILMN_1730628 | RNASE2 | −0.050 | 0.44 | −0.060 | 0.48 | ||||
| ILMN_1688295 | ZNF219 | 0.010 | 0.45 | 0.010 | 0.51 | ||||
β coefficient refers to the adjusted strength of association between the number of copies of the minor allele and the level of expression for each probe. The adjustment model is specified in the Methods section
Table 4.
Differences in expression in brains of people with AD and people with non-AD pathology
| Probe | Symbol | β | Cerebellum |
Cortex |
|||
|---|---|---|---|---|---|---|---|
| S.E. | p-value | β | S.E. | p-value | |||
| ILMN_1705743 | FLJ10357 | 0.04 | 0.03 | 0.24 | 0.27 | 0.05 | 7.01×10−7 |
| ILMN_2276811 | METT11D1 | 0.03 | 0.02 | 0.17 | |||
| ILMN_2361603 | NDRG2 | 0.01 | 0.02 | 0.38 | 0.01 | 0.01 | 0.41 |
| ILMN_1670535 | NDRG2 | 0.01 | 0.02 | 0.72 | −0.11 | 0.03 | 3.14×10−4 |
| ILMN_1762871 | RNASE13 | −0.02 | 0.03 | 0.52 | |||
| ILMN_1730628 | RNASE2 | 0.04 | 0.07 | 0.55 | 0.08 | 0.10 | 0.38 |
| ILMN_1688295 | ZNF219 | 0.001 | 0.02 | 0.95 | −0.01 | 0.02 | 0.64 |
Discussion
We identified a single SNP, rs3748348, associated with resilience in executive functioning among participants in ADNI. This association was stronger when we additionally conditioned on episodic memory, which dramatically reduced differences in phenotype levels across diagnostic group (e.g. normal vs. MCI vs. AD). This association was driven by people whose executive functioning was better than that predicted by neuroimaging markers of brain pathology.
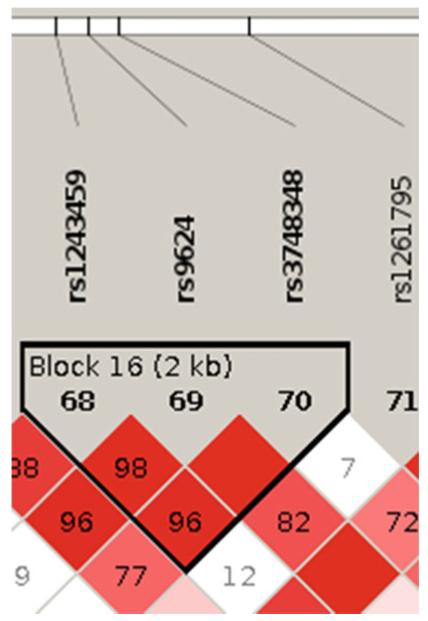
rs3748348 is found on chromosome14 and was on the genome-wide SNP chip used for ADNI participants. rs3748348 is within 3′ UTR the gene RNASE13 (ribonuclease, RNase A family, 13). It is in high linkage disequilibrium with two other SNPs, rs1243459 (D′=0.96) and rs9624 (D′=1.00) (see Fig. 4). rs1243459 and rs9624 are associated with TPPP2, tubulin polymerization-promoting protein 2. There are five genes within 100 kb: NDRG2, RNASE13, RNASE2, TPPP2, and FLJ10357. Of these, only NDRG2 has been studied for neurobiological activity. NDRG2 is one of a family of proteins in the N-myc downstream-regulated gene (NDRG) family. NDRG2 is involved in cell differentiation and stress response. It has been studied mostly in the context of malignancy, including gliomas. NDRG2 is expressed widely in the central nervous system where it is localized to astrocytes, and its expression is induced in response to ischemic or toxic stress. A few studies have investigated NDRG2 in the context of neurodegenerative diseases where it appears to be expressed in astrocytes in diseased regions of Parkinson’s disease and corticobasal degeneration, and upregulated in AD where it is perhaps localized to pyramidal neurons. A search of the phosphoproteome in frontal cortex from individuals with frontotemporal lobar degeneration identified NDRG2 and glial fibrillary acidic protein with apparent increased phosphorylation. Though there is a literature on the neurobiological activity of TPPP1, there is not for its homolog TPPP2, which is expressed in fetal but not adult brain. The neurobiological activities, if any, of the other candidates identified have not been reported. We did not find any SNP nominally associated with our phenotype close to this locus (See Appendix C).
Fig. 4.
The block of interest in the Linkage disequilibrium (LD) plot. Blocks of high LD are outlined as triangles and numbered as indicated in the figure. Shading reflects differences in pairwise LD (whiter r2 = low LD; redder r2 = near-perfect LD). Numbers in diamonds are estimates of pairwise coefficients (D′), expressed in percentages
Although we found nominally significant associations with FLJ10357, RNASE2 and NDRG2 when measuring gene expression levels in brain for our top SNP, we note that the expression data were derived from a convenience sample of participants from research studies who died and came to autopsy. It is uncertain whether the same results would be found using only controls without any neurodegenerative conditions.
We found the phenotype to be highly heritable, though this result had a relatively large standard error. A larger sample size would lead to a lower standard error for the estimate of the variance explained by the SNPs and hence a lower standard error for the heritability estimate but it should not systematically affect the current estimate since it is unbiased (Visscher et al. 2010). In our GWAS analysis, although the effect of individual SNPs was small, our results show that common SNPs in total may explain around 75 % of the phenotypic variability.
We developed our phenotype of interest by regressing out factors known to be associated with executive functioning, including demographic characteristics and MRI findings. This strategy produced a highly heritable phenotype. The raw executive functioning score was much less heritable. This general strategy may prove to be useful in other settings.
An important characteristic and potential limitation of genetic analyses of ADNI participants is that they were a clinically ascertained sample who were self- and/or clinician-selected for possible participation in ADNI. We attempted to account for this after confirming that memory scores were highly correlated with disease status, as would be expected. Indeed, when we further regressed our executive functioning scores by adjusting for memory scores, the resulting residuals were barely associated with diagnostic group (see Fig. 1). Nevertheless, replication of these findings in a separate sample is necessary. Studies of population-based and a less highly selected group of study participants would be ideal for future analyses.
The p-value for rs3748348 for the association with executive functioning resilience (2.12×10−7) did not quite meet the very stringent threshold typically employed for evaluating genome-wide significance (p<10−8). This is not surprising, given the relatively small sample size (n=750) available for these analyses. These results should therefore be considered preliminary, and as in any association study, in need of replication in larger independent data sets.
Our focus on a residualized cognitive outcome, “resilience,” is not the usual approach for GWAS. Conventional GWAS of unresidualized cognitive outcomes could potentially identify determinants of resilience, because on average resilience will be associated with better cognitive function and less risk of impairment. However, resilience emerges in the context of pathology, such as vascular disease or neurodegeneration. In a conventional analysis, desirable cognitive outcomes may therefore reflect either high resilience or an absence of pathology that impacts cognitive impairment. Because the physiologic processes contributing to diseases (e.g., Alzheimer’s disease, white matter hyperintensities, MRI-defined lacunes) may be distinct from the processes providing resilience, conducting separate genetic studies using residualized outcomes as indicators of resilience may provide a complimentary approach to identifying important biological signals associated with late life cognition.
It is important to further delineate the biological substrate underlying cognitive resilience. If a GWAS could identify targets potentially modifiable with medications (i.e. “druggable targets”), this could facilitate development of new interventions to help reduce or perhaps even eliminate the personal and societal devastation associated with neurodegenerative conditions in old age. Understanding the genetic architecture of cognitive resilience offers an opportunity to further understand the aging brain and its ability to accommodate neurodegenerative processes.
In summary, we performed a GWAS of resilience in executive functioning, defined as the residual term in regression models predicting executive functioning on the basis of imaging and demographic characteristics, and identified a very interesting signal associated with TPPP2. This promising result warrants confirmation in other large populations.
Acknowledgment
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation. Data management and the specific analyses reported here were supported by NIH grant R01 AG029672 (Paul Crane, PI), R13 AG030995 (Mungas), and P50 AG05136 (Raskind), from the NIA as well as NIA R01 AG19771 (Saykin) and P30 AG10133 (Saykin/Ghetti).
APPENDIX A: Regression results for the phenotypes of interest
Table 5.
Regression results for executive functioning residual when adjusting for demographics, Hachinski score and MRI variables
| β | p-value | 95 % CI | |
|---|---|---|---|
| Sex | −0.03 | 0.65 | (−0.17, 0.10) |
| Age | 0.01 | 0.01 | (0.01, 0.02) |
| Education | 0.20 | < 0.01 | (0.15, 0.24) |
| White matter hyperintensities | −0.04 | 0.04 | (−0.07, −0.01) |
| Residual hippocampal volume | 0.45 | < 0.01 | (0.33, 0.58) |
| Residual cortical volume | 0.01 | < 0.01 | (0.01, 0.02) |
| Presence of lacunes | −0.03 | 0.80 | (−0.25, 0.19) |
| Constant | 0.33 | 0.56 | (−0.78, 1.44) |
Table 6.
Regression results for executive functioning residual when adjusting for ADNI-Mem, demographics, Hachinski score and MRI variables
| β | p-value | 95 % CI | |
|---|---|---|---|
| ADNI-Mem | 0.70 | < 0.01 | (0.63, 0.77) |
| Sex | −0.14 | 0.01 | (−0.25,−0.03) |
| Age | −0.01 | 0.28 | (−0.01, 0.01) |
| Education | 0.11 | < 0.01 | (0.07, 0.15) |
| White matter hyperintensities | −0.03 | 0.04 | (−0.07,−0.01) |
| Residual hippocampal volume | −0.13 | 0.03 | (−0.24,−0.01) |
| Residual cortical volume | 0.01 | < 0.01 | (0.01, 0.02) |
| Presence of lacunes | 0.01 | 0.91 | (−0.17, 0.19) |
| Constant | 1.52 | < 0.01 | (0.61, 2.42) |
APPENDIX B: Sensitivity GWAS analyses results
Table 7.
Comparison of GWAS results of our phenotype of interest when a) not adjusting for principal components, b) adjusting for the first three principal components and c) adjusting for the first three principal components and APOE status
| Top 20 SNPs for GWAS w/o PCs |
Top 20 SNPs for GWAS with PC1, PC2 & PC3 |
Top 20 SNPs for GWAS with PC1, PC2, PC3 and APOE |
||||||
|---|---|---|---|---|---|---|---|---|
| CHR | SNP | p-value | CHR | SNP | p-value | CHR | SNP | p-value |
| 14 | rs3748348 | 1.92×10−7 | 14 | rs3748348 | 2.12e-07 | 14 | rs3748348 | 2.13×10−7 |
| 2 | rs1609361 | 1.96×10−6 | 2 | rs1609361 | 2.099e-06 | 2 | rs1609361 | 2.15×10−6 |
| 10 | rs7897741 | 5.03×10−6 | 10 | rs7897741 | 5.907e-06 | 10 | rs7897741 | 6.04×10−6 |
| 8 | rs13251704 | 9.10×10−6 | 2 | rs16987794 | 1.103e-05 | 2 | rs16987794 | 1.12×10−5 |
| 2 | rs2698011 | 1.16×10−5 | 8 | rs13251704 | 1.163e-05 | 8 | rs13251704 | 1.18×10−5 |
| 2 | rs2698029 | 1.26×10−5 | 2 | rs2698011 | 1.294e-05 | 2 | rs2698011 | 1.31×10−5 |
| 2 | rs2715077 | 1.26×10−5 | 2 | rs2715077 | 1.366e-05 | 12 | rs7306879 | 1.39×10−5 |
| 2 | rs16987794 | 1.38×10−5 | 12 | rs7306879 | 1.422e-05 | 2 | rs2715077 | 1.40×10−5 |
| 12 | rs7306879 | 2.10×10−5 | 2 | rs2698029 | 1.438e-05 | 2 | rs2698029 | 1.48×10−5 |
| 4 | rs4865353 | 2.13×10−5 | 4 | rs4865353 | 2.23e-05 | 4 | rs4865353 | 2.28×10−5 |
| 8 | rs13258868 | 2.75×10−5 | 14 | rs2365682 | 2.585e-05 | 14 | rs2365682 | 2.63×10−5 |
| 14 | rs2365682 | 2.77×10−5 | 12 | rs11055126 | 3.007e-05 | 12 | rs11055126 | 3.06×10−5 |
| 2 | rs2698024 | 3.39×10−5 | 8 | rs13258868 | 3.1e-05 | 8 | rs13258868 | 3.15×10−5 |
| 12 | rs11055126 | 3.50×10−5 | 5 | rs13183341 | 3.412e-05 | 5 | rs13183341 | 3.48×10−5 |
| 3 | rs9851461 | 3.65×10−5 | 6 | rs9344230 | 3.519e-05 | 6 | rs9344230 | 3.49×10−5 |
| 15 | rs4456480 | 3.66×10−5 | 12 | rs12316661 | 3.754e-05 | 6 | rs1737038 | 3.79×10−5 |
| 12 | rs12316661 | 3.94×10−5 | 6 | rs1737038 | 3.756e-05 | 12 | rs12316661 | 3.81×10−5 |
| 3 | rs6809436 | 3.98×10−5 | 2 | rs2698024 | 3.81e-05 | 2 | rs2698024 | 3.89×10−5 |
| 6 | rs1737038 | 4.02×10−5 | 3 | rs9851461 | 4.16e-05 | 15 | rs4456480 | 4.05×10−5 |
| 6 | rs9344230 | 4.03×10−5 | 15 | rs4456480 | 4.186e-05 | 3 | rs9851461 | 4.08×10−5 |
PC Principal component
Table 8.
Results from singlestep GWAS analysis for ADNI-EF when a) not adjusting for principal components, b) adjusting for the first three principal components
| Single-step GWAS for ADNI-EF without PC |
Single-step GWAS for ADNI-EF with PC |
||||
|---|---|---|---|---|---|
| CHR | SNP | p-value | CHR | SNP | p-value |
| 14 | rs3748348 | 1.42×10−7 | 14 | rs3748348 | 1.56×10−7 |
| 2 | rs1609361 | 1.53×10−6 | 2 | rs1609361 | 1.64×10−6 |
| 10 | rs7897741 | 3.90×10−6 | 10 | rs7897741 | 4.70×10−6 |
| 8 | rs13251704 | 7.38×10−6 | 8 | rs13251704 | 9.64×10−6 |
| 2 | rs2698011 | 9.23×10−6 | 2 | rs16987794 | 9.95×10−6 |
| 2 | rs2715077 | 1.01×10−5 | 2 | rs2698011 | 1.02×10−5 |
| 2 | rs2698029 | 1.02×10−5 | 2 | rs2715077 | 1.09e-05 |
| 2 | rs16987794 | 1.22×10−5 | 2 | rs2698029 | 1.16×10−5 |
| 12 | rs7306879 | 1.87×10−5 | 12 | rs7306879 | 1.23×10−5 |
| 4 | rs4865353 | 1.96×10−5 | 4 | rs4865353 | 2.00×10−5 |
| 14 | rs2365682 | 2.47×10−5 | 14 | rs2365682 | 2.40×10−5 |
| 2 | rs2698024 | 2.90×10−5 | 12 | rs11055126 | 2.64×10−5 |
| 15 | rs4456480 | 2.98×10−5 | 12 | rs12316661 | 3.00×10−5 |
| 12 | rs11055126 | 3.04×10−5 | 5 | rs13183341 | 3.04×10−5 |
| 12 | rs12316661 | 3.20×10−5 | 2 | rs9344230 | 3.13×10−5 |
| 8 | rs13258868 | 3.20×10−5 | 2 | rs2698024 | 3.25×10−5 |
| 3 | rs9851461 | 3.37×10−5 | 15 | rs4456480 | 3.29×10−5 |
| 6 | rs1737038 | 3.48×10−5 | 6 | rs1737038 | 3.42×10−5 |
| 2 | rs9344230 | 3.55×10−5 | 8 | rs13258868 | 3.65×10−5 |
| 5 | rs13183341 | 3.75×10−5 | 3 | rs9851461 | 3.98×10−5 |
PC Principal component
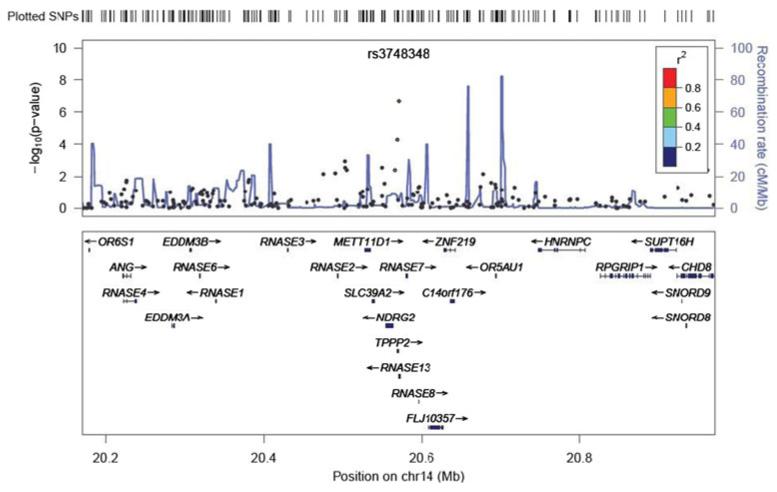
APPENDIX C: Regional association plot
Fig. 5.
Regional association plot of the locus (rs3748348) nominally associated with resilience
Footnotes
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Contributor Information
Shubhabrata Mukherjee, Department of Medicine, University of Washington, Box 359780, 325 Ninth Avenue, Seattle, WA 98104, USA.
Sungeun Kim, Center for Neuroimaging, Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, USA.
Laura E. Gibbons, Department of Medicine, University of Washington, Box 359780, 325 Ninth Avenue, Seattle, WA 98104, USA
Kwangsik Nho, Center for Neuroimaging, Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, USA.
Shannon L. Risacher, Center for Neuroimaging, Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, USA
M. Maria Glymour, Department of Society, Human Development, and Health, Harvard School of Public Health, Boston, MA, USA.
Christian Habeck, Cognitive Neuroscience Division, The Taub Institute for Research on Aging and Alzheimer’s Disease, Columbia University, New York, NY, USA.
Grace J. Lee, Mary S. Easton Center for Alzheimer’s Disease Research at UCLA, Department of Neurology, David Geffen School of Medicine at UCLA, Los Angeles, CA, USA
Elizabeth Mormino, Helen Wills Neuroscience Institute, University of California Berkeley, Berkeley, CA, USA.
Nilüfer Ertekin-Taner, Departments of Neurology and Neuroscience, Mayo Clinic Florida, Jacksonville, FL, USA.
Thomas J. Montine, Department of Pathology, University of Washington, Seattle, WA, USA
Charles DeCarli, Department of Neurology and Center for Neuroscience, University of California at Davis, Sacramento, CA, USA.
Andrew J. Saykin, Center for Neuroimaging, Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, USA
Paul K. Crane, Department of Medicine, University of Washington, Box 359780, 325 Ninth Avenue, Seattle, WA 98104, USA
References
- Allen M, Zou F, Chai HS, Younkin CS, Crook J, Pankratz VS, et al. Novel late-onset Alzheimer’s disease loci variants associate with brain region expression. Neurology. 2012 doi: 10.1212/WNL.0b013e3182605801. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. doi:10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Carmichael O, Schwarz C, Drucker D, Fletcher E, Harvey D, Beckett L, et al. Longitudinal changes in white matter disease and cognition in the first year of the Alzheimer disease neuroimaging initiative. Archives of Neurology. 2010;67(11):1370–1378. doi: 10.1001/archneurol.2010.284. doi:10.1001/archneurol.2010.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer’s disease. Nature Genetics. 2009;41(2):192–198. doi: 10.1038/ng.305. doi:10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Alzheimer’s Association . The healthy brain initiative: a national public health road map to maintaining cognitive health. Alzheimer’s Association; Chicago: 2007. [Google Scholar]
- Crane PK, Carle A, Gibbons LE, Insel P, Mackin RS, Gross A, et al. Development and assessment of a composite score for memory in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Brain Imaging and Behavior. 2012 doi: 10.1007/s11682-012-9186-z. doi:10.1007/s11682-012-9186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Lee I, Thundat T. Effect of nanometer surface morphology on surface stress and adsorption kinetics of alkanethiol self-assembled monolayers. Ultramicroscopy. 2006;106(8-9):795–799. doi: 10.1016/j.ultramic.2005.11.012. doi:10.1016/j.ultramic.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Ertekin-Taner N. Gene expression endophenotypes: a novel approach for gene discovery in Alzheimer’s disease. Molecular Neurodegeneration. 2011;6:31. doi: 10.1186/1750-1326-6-31. doi:10.1186/1750-1326-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. Mini-mental state. A practical method for grading the cognitive state of patients for the clinician. Journal of Psychiatric Research. 1975;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Gibbons LE, Carle AC, Mackin RS, Harvey D, Mukherjee S, Insel P, et al. A composite score for executive functioning, validated in Alzheimer’s Disease Neuroimaging Initiative (ADNI) participants with baseline mild cognitive impairment. Brain Imaging and Behavior. 2012 doi: 10.1007/s11682-012-9176-1. doi:10.1007/s11682-012-9176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodglass H, Kaplan D. The assessment of aphasia and related disorders. 2nd ed. Lea & Febiger; Philadelphia: 1983. [Google Scholar]
- Hertzog C, Kramer AF, Wilson RS, Lindenberger U. Enrichment effects on adult cognitive development: can the functional capacity of older adults be preserved and enhanced? Psychological Science in the Public Interest. 2009;9(1):1–65. doi: 10.1111/j.1539-6053.2009.01034.x. [DOI] [PubMed] [Google Scholar]
- Ikram MA, Vrooman HA, Vernooij MW, van der Lijn F, Hofman A, van der Lugt A, et al. Brain tissue volumes in the general elderly population The Rotterdam Scan Study. Neurobiology of Aging. 2007;29(6):882–890. doi: 10.1016/j.neurobiolaging.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr., Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging. 2008;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin Group. Alzheimer’s Association . Saving lives, saving money: dividends for Americans investing in Alzheimer’s research. Alzheimer’s Association; Washington, D.C.: 2003. [Google Scholar]
- Longstreth WT, Jr., Manolio TA, Arnold A, Burke GL, Bryan N, Jungreis CA, et al. Clinical correlates of white matter findings on cranial magnetic resonance imaging of 3301 elderly people. The Cardiovascular Health Study. Stroke; A Journal of Cerebral Circulation. 1996;27(8):1274–1282. doi: 10.1161/01.str.27.8.1274. [DOI] [PubMed] [Google Scholar]
- Longstreth WT, Jr., Bernick C, Manolio TA, Bryan N, Jungreis CA, Price TR. Lacunar infarcts defined by magnetic resonance imaging of 3660 elderly people: the Cardiovascular Health Study. Archives of Neurology. 1998;55(9):1217–1225. doi: 10.1001/archneur.55.9.1217. [DOI] [PubMed] [Google Scholar]
- Longstreth WT, Jr., Dulberg C, Manolio TA, Lewis MR, Beauchamp NJ, Jr., O’Leary D, et al. Incidence, manifestations, and predictors of brain infarcts defined by serial cranial magnetic resonance imaging in the elderly: the Cardiovascular Health Study. Stroke. 2002;33(10):2376–2382. doi: 10.1161/01.str.0000032241.58727.49. [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Mohs RC, Knopman D, Petersen RC, Ferris SH, Ernesto C, Grundman M, et al. Development of cognitive instruments for use in clinical trials of antidementia drugs: additions to the Alzheimer’s Disease Assessment Scale that broaden its scope. The Alzheimer’s Disease Cooperative Study. Alzheimer Dis Assoc Disord. 1997;11(Suppl 2):S13–21. [PubMed] [Google Scholar]
- Morris JC, Heyman A, Mohs RC, Hughes JP, van Belle G, Fillenbaum G, et al. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part I. Clinical and neuropsychological assessment of Alzheimer’s disease. Neurology. 1989;39(9):1159–1165. doi: 10.1212/wnl.39.9.1159. [DOI] [PubMed] [Google Scholar]
- Mungas D, Harvey D, Reed BR, Jagust WJ, DeCarli C, Beckett L, et al. Longitudinal volumetric MRI change and rate of cognitive decline. Neurology. 2005;65(4):565–571. doi: 10.1212/01.wnl.0000172913.88973.0d. doi:10.1212/01.wnl.0000172913.88973.0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthén L, Muthén B. Mplus users guide. Muthen and Muthen; Los Angeles: 2006. Version 4.1 ed. [Google Scholar]
- Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PLoS One. 2009;4(8):e6501. doi: 10.1371/journal.pone.0006501. doi:10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. doi:10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2005. [Google Scholar]
- Reed BR, Mungas D, Farias ST, Harvey D, Beckett L, Widaman K, et al. Measuring cognitive reserve based on the decomposition of episodic memory variance. Brain: A Journal of Neurology. 2010;133(Pt 8):2196–2209. doi: 10.1093/brain/awq154. doi:10.1093/brain/awq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitan RM. Validity of the Trail Making Test as an indicator of organic brain damage. Perceptual and Motor Skills. 1958;8:271–276. [Google Scholar]
- Rey A. L’examen clinique en psychologie. Presses Universitaires de France; Paris: 1964. [Google Scholar]
- Satz P. Brain reserve capacity on symptom onset after brain injury: a formulation and review of evidence for threshold theory. Neuropsychology. 1993;7(3):273–295. [Google Scholar]
- Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, et al. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimer’s & Dementia. 2010;6(3):265–273. doi: 10.1016/j.jalz.2010.03.013. doi: S1552-5260(10)00082-8 [pii]10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarmeas N, Stern Y. Cognitive reserve and lifestyle. Journal of Clinical and Experimental Neuropsychology. 2003;25(5):625–633. doi: 10.1076/jcen.25.5.625.14576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz C, Fletcher E, DeCarli C, Carmichael O. Fully-automated white matter hyperintensity detection with anatomical prior knowledge and without FLAIR. Information Processing in Medical Imaging. 2009;21:239–251. doi: 10.1007/978-3-642-02498-6_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. NeuroImage. 2010;53(3):1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. doi:S1053-8119(10)00064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern Y. What is cognitive reserve? Theory and research application of the reserve concept. Journal of the International Neuropsychological Society. 2002;8(3):448–460. [PubMed] [Google Scholar]
- The International HapMap Project Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. doi:10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Hill WG, Wray NR. Heritability in the genomics era-concepts and misconceptions. Nature Reviews. Genetics. 2008;9(4):255–266. doi: 10.1038/nrg2322. doi:10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Yang J, Goddard ME. A commentary on ‘common SNPs explain a large proportion of the heritability for human height’ by Yang et al. (2010) Twin Research and Human Genetics: The Official Journal of the International Society for Twin Studies. 2010;13(6):517–524. doi: 10.1375/twin.13.6.517. doi:10.1375/twin.13.6.517. [DOI] [PubMed] [Google Scholar]
- Weiner MW, Aisen PS, Jack CR, Jr., Jagust WJ, Trojanowski JQ, Shaw L, et al. The Alzheimer’s Disease Neuroimaging Initiative: progress report and future plans. Alzheimer’s & Dementia. 2010;6(3):202–211. e207. doi: 10.1016/j.jalz.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weschler D. Weschler Adult Intelligence Scale-Revised. Psychological Corporation; NY: 1981. [Google Scholar]
- Weschler D. WMS-R: Weschler Memory Scale - Revised manual. Psychological Corporation/HBJ; NY: 1987. [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. American Journal of Human Genetics. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. doi:10.1016/j.ajhg. 2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]