Abstract
Recently, the small molecule 968 was found to block the Rho GTPase-dependent growth of cancer cells in cell culture and mouse xenografts, and when the target of 968 was found to be mitochondrial enzyme glutaminase (GLS1) it revealed a surprising link between Rho GTPases and mitochondrial glutamine metabolism. Signal transduction via the Rho GTPases, together with NFκB, appears to elevate mitochondrial glutaminase activity in cancer cells, thereby helping cancer cells satisfy their altered metabolic demands. Here, we review what is known about the mechanism of glutaminase activation in cancer cells, as well as compare the properties of two distinct glutaminase inhibitors, and discuss recent findings that shed new light on how glutamine metabolism might affect cancer progression.
Keywords: Rho, GTPase, signaling, glutamine, glutaminase, metabolism, cancer, Warburg
Rho GTPAses and cancer
Rho-family GTPases stimulate signaling activities that influence a broad array of cellular activities ranging from actin cytoskeletal rearrangements to cell polarity, migration, and cell cycle progression [1]. Although members of the Rho family have been classically linked to different regulatory roles that impact the actin cytoskeletal architecture, it has become increasingly evident that they play fundamental roles in several aspects of cancer progression and tumorigenesis [2]. For example, our laboratory and others have shown that the hyper-activation of different Rho GTPases – Rho, Rac, and Cdc42 – results in cellular transformation, either through mutations or by deregulation of their guanine nucleotide exchange factors (e.g., members of the Dbl family of oncogenic proteins) [3]. Cells expressing constitutively active forms of Rho GTPases can grow under conditions of serum deprivation and in the absence of a substratum (i.e., anchorage-independent growth), and they induce tumor formation when introduced into immunocompromised mice [4,5]. The overexpression of Rho GTPases has been demonstrated in human tumors including colon, lung, advanced stage breast cancers, testicular germ cell tumors, upper and lower urinary tract cancers, and ductal pancreatic adenocarcinoma [6–9]. Two members of the family, RhoA and RhoC, have been linked to the progression of malignancy and, in particular, metastasis [10–13]. Moreover, the expression of DLC1 (for deleted in liver cancer), a Rho GTPase-activating protein (Rho-GAP), is suppressed in liver cancer tissue and a variety of other cancers [14,15]. Thus, these various findings make the Rho GTPases and their regulatory proteins particularly intriguing targets for anticancer therapies.
Our laboratory has spent a great deal of effort trying to delineate the signaling activities downstream of Rho GTPases, particularly Cdc42, that influence cell growth and oncogenic transformation. Studies using a constitutively active (‘fast-cycling’) Cdc42 mutant, capable of spontaneously exchanging GDP for GTP, showed that activated forms of Cdc42 can influence the signaling lifetimes of EGFRs by negatively regulating the interactions between these receptors and c-Cbl, a signaling adaptor protein that functions as an E3 ubiquitin ligase [16,17]. Thus, cells expressing hyperactivated mutants of Cdc42 show enhanced EGFR levels because the ability of these mutants to form a complex with the Cdc42/Rac-signaling partner Cool-1/β-Pix (cloned-out of library 1/β Pak-interactive exchange factor),and c-Cbl sequesters Cbl away from EGFRs.
Studies using activated mutants of Cdc42 and the related GTPase Rac have also shown that these proteins can trigger a signaling pathway that results in the activation of mTORC1 and p70 S6 kinase, thereby influencing RNA processing to help accommodate the biosynthetic requirements of rapidly proliferating and transformed cells [18,19]. Given the many signaling targets of the various Rho GTPases, and the host of cellular responses they influence, there has been every reason to anticipate that members of this family will contribute to malignant transformation in a number of ways, aside from their well-known involvement in the migration and invasive activities of cancer cells.
Indeed, consistent with this expectation, we recently made an exciting discovery that highlights a new role for Rho GTPases in cancer progression through a previously unappreciated connection to cellular metabolism [20]. Specifically, we have found that fibroblasts transformed by oncogenic Dbl, or by activated fast-cycling mutants of Cdc42, Rac, or RhoC, exhibited significantly elevated activity of mitochondrial glutaminase, an enzyme that plays a key role in glutamine metabolism by catalyzing the hydrolysis of glutamine to glutamate and ammonia. Similarly, we found that glutaminase activity was markedly upregulated in different human cancer cell lines, including MDA-MB-231 and SKBR3 breast cancer cells.
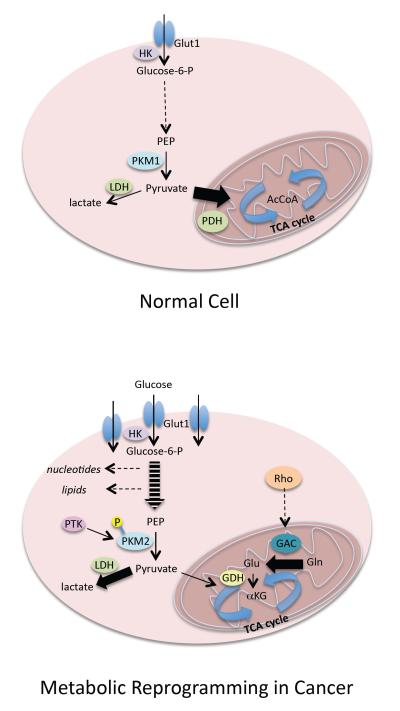
The importance of cellular metabolism in cancer development goes back to the early observations of Warburg that tumor cells exhibit enhanced glycolytic activity (i.e., the ‘Warburg effect’) [21], and this phenomenon has recently been receiving a great deal of renewed interest [22–24]. Two major events characterize the metabolic reprogramming of cancer cells (Figure 1). The first involves changes in glycolysis that result in the increased production of lactic acid (i.e., the Warburg effect), and the expression and activity of many of the enzymes in this pathway are increased in cancer cells. However, the activity of the penultimate enzyme in the pathway, pyruvate kinase, is attenuated because of the tyrosine phosphorylation that occurs at a key regulatory site within a specific isoform of the enzyme (M2) that is specifically expressed in cancer cells [23–25]. The pyruvate that is generated by the phosphorylated PKM2 is then largely converted to lactic acid (by lactate dehydrogenase) instead of entering the mitochondria where it would ultimately be converted to acetyl CoA, and then citrate, to initiate the TCA cycle.
Figure 1.
Cancer cells undergo metabolic re-programming. The glycolytic pathway in normal cells (a) is used to generate pyruvate that enters the citric acid (TCA) cycle in the mitochondria. In cancer cells (b), many of the enzymes of the glycolytic pathway are upregulated and/or activated, although M2 pyruvate kinase activity, which catalyzes the penultimate step in the pathway, is inhibited by tyrosine phosphorylation. Pyruvate produced by PKM2 is preferentially converted to lactate (as catalyzed by lactate dehydrogenase). Elevated glutamine metabolism, through the up-regulation and/or activation of GAC (which converts glutamine to glutamate) and GDH (converting glutamate to α-ketoglutarate), is then essential for ‘feeding’ the TCA cycle in cancer cells. Abbreviations: Glut1, glucose transporter 1; HK, hexokinase; PEP, phosphoenolpyruvate; PKM, pyruvate kinase M; LDH, lactate dehydrogenase; PDH, pyruvate dehydrogenase; AcCoA, acetyl CoA; PTK, protein tyrosine kinase; GAC, glutaminase C; GDH, glutamate dehydrogenase; αKG, α-ketoglutarate. Dashed lines represent multistep pathways; heavy lines represent preferred and/or accelerated pathways.
This decrease in the amount of pyruvate available for the TCA cycle necessitates a second major change in cancer metabolism. Specifically, in response to changes in glycolysis, cancer cells exhibit greatly elevated glutamine metabolism (sometimes referred to as ‘glutamine addiction’), as catalyzed by the conversion of glutamine to glutamate by glutaminase, and the subsequent conversion of glutamate to α-ketoglutarate by glutamate dehydrogenase. The enhanced production of α-ketoglutarate helps to ‘feed’ the TCA cycle (Figure 1). Metabolic flux experiments using 13C-NMR have demonstrated that although proliferating cancer cells exhibit a pronounced Warburg effect, their TCA cycle remains intact and is necessary to replenish metabolic intermediates for the production of NADPH for fatty acid synthesis, to provide the carbon necessary for nucleotide synthesis and the production of asparagine and arginine, and to serve as a major anaplerotic source of oxaloacetate [22]. Cancer cells take advantage of their elevated glutamine metabolism to help drive the TCA cycle, and thus our discovery that glutaminase activity is significantly increased in response to the hyperactivation of Rho GTPases provides interesting new insights into how the demands of cancer cells for accelerated glutamine metabolism are met.
Mammals contain two distinct but structurally related genes that encode glutaminase enzymes, with one form being highly expressed in liver (referred to as liver-type glutaminase or GLS2) and a second gene found in kidney and a number of other tissues, including many cancer cells, that is referred to as kidney-type glutaminase or GLS1 [26–28]. GLS1 expresses two splice variants, KGA and GAC, and GAC is the predominant form expressed in transformed cells and cancers. GLS1 is the target of a small molecule inhibitor, 968, that we discovered which breaks the signaling connection between Rho GTPases and glutamine metabolism [20]. In the sections outlined below, we describe how this connection was initially discovered, as well as what we know thus far about the signals from Rho GTPases that activate glutaminase and what might be some interesting outcomes of these metabolic changes in cancer cells.
New roads for Rho: signaling to the mitochondria
The connection between Rho GTPases (specifically RhoA, RhoC, and Cdc42) was first uncovered while screening for small molecules that effectively inhibit the Rho-dependent transformation of fibroblasts in cell culture [20]. The effort began by assaying NIH 3T3 cells stably expressing the Dbl oncogene, an activator of Rho and Cdc42, because these cells are very efficient in their ability to form foci. The goal was to identify small molecules that could reverse this transformed phenotype. One molecule, the bromo-phenanthridinone molecule 968, was striking in its ability to block Dbl-induced cellular transformation, yet showed no effects on the growth of normal fibroblasts [20]. Although the initial assumption was that 968 acted by interfering with the guanine nucleotide binding capabilities of different Rho GTPases, it soon became apparent that 968 was not acting directly on the GTPases, nor was it directly affecting their regulators (i.e., GEFs, GAPs, or GDIs) or known effector proteins. Therefore, affinity chromatography utilizing a region on 968 critical for its inhibitory activity was used in an attempt to ‘fish’ out its protein target.
The results were completely unanticipated: the downstream component of Rho signaling in transformation that was being targeted by 968 was a specific isoform of the mitochondrial enzyme glutaminase, designated GAC [20]. The connection between Rho proteins and GAC was verified by knocking down the Rho proteins and determining that this disrupted mitochondrial glutaminase activity, similar to what was observed by treating cells with 968. Most importantly, the inhibitory effects of 968 could be reversed by eliminating the need for glutaminolysis during cellular transformation by supplying the cells with a soluble product of that reaction, dimethyl α-ketoglutarate [20]. Thus, although the original goal was to identify molecules that function as inhibitors of Rho GTPases, in the end, these efforts led to an even more intriguing discovery. By blocking the Rho-dependent metabolic reprogramming of glutamine metabolism in cancer, 968 strategically targets an outcome of Rho GTPase-signaling that is specifically required for the growth of cancer cells.
GAC activation in cancer cells
How is glutaminase activity being regulated in response to signals emanating from Rho GTPases? One mechanism by which cancer cells can modulate GLS1 is via a c-Myc-dependent microRNA. Specifically, it has been shown in B-lymphoma and prostate cancer cells that c-Myc suppresses the expression of miR-23 which, in turn, increases GLS1 expression and activity [29]. However, across a number of cells lines we have not observed a direct one-to-one correlation between GAC expression patterns and activity levels. For example, the highly aggressive breast cancer cell line MD-MBA- 231 exhibits both higher GAC protein and activity levels compared with normal human mammary epithelial cells (HMECs), whereas a less aggressive breast cancer cell line, SKBR3, shows lower GAC protein levels compared with HMECs, but significantly higher activity [20]. Thus, there appears to be more to the regulation of GAC than simply changes in protein expression levels.
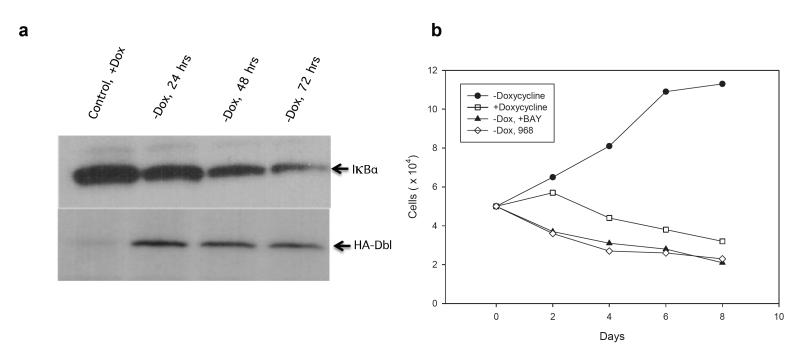
A critical element of the signaling downstream of Rho GTPases that leads to the elevated glutaminase activity observed in Rho-transformed cells, as well as in many cancer cell lines, is activation of the NFκB transcriptional program. Prior to this finding, it had been known for some time that Rho GTPase-signaling is a potent upstream stimulus for initiating NFκB nuclear translocation and the expression of a multitude of proteins associated with malignancy [30,31]. Indeed, studies in a MEF cell line expressing a tetracycline-inducible oncogenic form of Dbl demonstrate the rapid onset of IκBα degradation with the concomitant activation and nuclear translocation of NFκB following Dbl expression ([32]; see Figure 2a). Microarray analysis of these cells immediately after oncogenic induction reveals a marked increase in a host of metastatic markers (e.g., tenascin), including upregulation of the glucose transporter and the microvesicle-associated protein tissue transglutaminase ([32] and see below). Moreover, the onset of elevated Rho GTPase-signaling in Dbl-expressing cells markedly increases glutamine dependence for cell proliferation and acquired sensitivity to glutaminase inhibition by 968 (Figure 2b). The proliferation of these cells and their induced dependence on glutaminase activity to support this increased growth rate are also reversed by directly inhibiting NFκB-signaling with the small molecule BAY 11-7082.
Figure 2.
Rho GTPase signaling and NFκB activation. a. Oncogenic HA-tagged Dbl inducible mouse embryonic fibroblasts (MEFs) show a time dependent loss of IKBα activity, nuclear translocation of NFκB and initiation of NFκB-induced transcription. b. Proliferation of induced and uninduced HA-Dbl MEFs in low serum (1%) and the inhibition of growth in the presence of 0.5 μM of the NFκB inhibitor BAY 11-7082 or 5 μM of the glutaminase inhibitor 968.
Although NF-κB is a necessary component of the Rho GTPase-dependent activation of GAC, it does not appear to do so by influencing the expression levels of the enzyme. Specifically, the decrease in glutaminase activity that is observed after treating cells with the NF-κB inhibitor BAY 11-7082 is not accompanied by a concomitant decrease in GAC protein levels [20]. This observation presents the possibility that the mechanism by which Rho proteins signal to activate GAC may involve either the post-translational modification(s) of GAC and/or a regulation of protein–protein interactions that are necessary for glutaminase activity in vivo. In this scenario, the role of NF-κB may be to regulate the expression of protein(s) necessary for the contextual modification of GAC in cancer cells to achieve enzymatic activity.
There are other clues to suggest that the modification of GAC in cancer cells may play a role in regulating its enzymatic activity. For example, GAC isolated from cancer cells displays a ‘basal’ activity that greatly exceeds that of recombinant, purified GAC preparations. GLS1 is often referred to in the literature as a ‘phosphate-dependent glutaminase’ because high millimolar concentrations of phosphate are required as a necessary cofactor that activates purified preparations of the enzyme. The cancer cell-derived GAC, however, has significant activity when assayed in the absence of phosphate. Indeed, we routinely observe a basal activity in cancer cells that can approach 75% of the phosphate-stimulated activity. Additionally, this basal activity can be reversed by treating GAC isolated from transformed cells with alkaline phosphatase, which removes phosphate moieties from tyrosine, serine, and threonine residues. Alkaline phosphatase does not affect the phosphate-stimulated activity [20], suggesting a potential role for phosphorylation in generating the basal GAC activity.
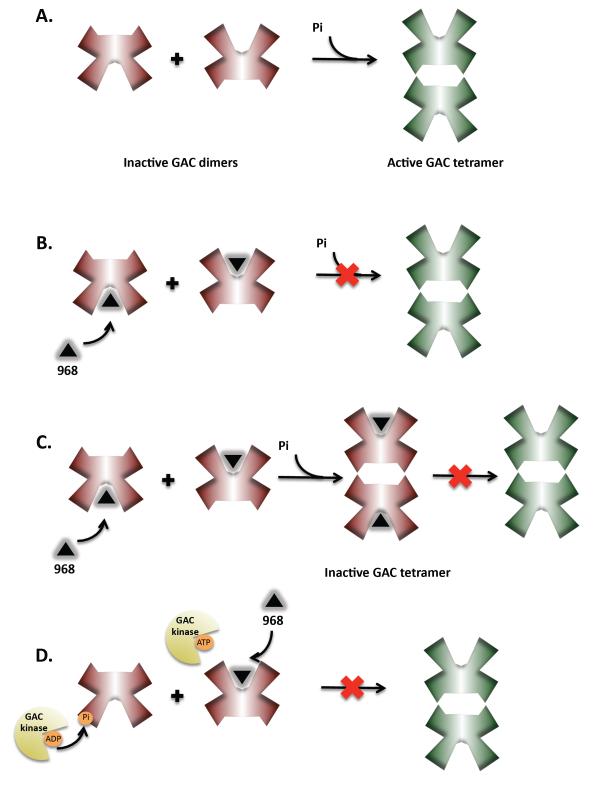
Identifying phosphorylated residues on GAC, as well as determining the GAC kinase, will no doubt be important advances in our understanding of how Rho proteins regulate this metabolic enzyme. Glutaminase exists as a homodimer in its inactive state and transitions to a tetramer in the active state (see Figure 4A). In vitro, phosphate activates GAC by stimulating a change in the oligomeric state of the enzyme from an inactive dimer to an active tetramer [26]. The phosphorylation of GAC in vivo may lead to a similar transition as to that which is induced by phosphate in vitro. It is also possible that in addition to, or in conjunction with, phosphorylation, other regulatory events (i.e., additional post-translational modifications and/or GAC binding partners) may play roles in activating GAC. Indeed, given the critical role that GAC is playing in cancer cells, a complex mode of regulation for this enzyme can almost certainly be anticipated.
Figure 4.
Models for GAC activation and inhibition by 968. a. GAC activation - GAC exists as a dimer in its inactive state and the transition to a tetramer is required for activation. This transition to an active state occurs in response to inorganic phosphate in vitro and in cancer cells is thought to require post-translational modification (e.g. phosphorylation). 968 binds to the inactive GAC dimer and prevents its activation. b. One way this might be achieved is by blocking the dimer to tetramer transition. c. Alternatively, after 968 binds to dimeric GAC the enzyme can still transition to the tetrameric state but cannot become activated. d. In vivo, post-translational modification of GAC might serve either to stimulate the dimer to tetramer transition, or confer activity to the tetramer. By disrupting the ability of GAC to become modified, 968 prevents the activation of GAC in cells.
Small molecule inhibitors and glutaminase
As mentioned above, the connection between Rho GTPases and mitochondrial glutaminase was uncovered while determining the mechanism by which the small molecule 968 inhibited cellular transformation. 968 functions as an allosteric GAC inhibitor; that is, it does not compete with either glutamine or glutamate for binding to the enzyme [20]. When considering GAC as a therapeutic target for treating cancer, the use of allosteric inhibitors of glutaminase is essential. In the 1990s, preclinical studies using glutamine mimetics to inhibit tumorogenesis showed that although these compounds were effective against cancer growth, there were toxic side effects that indicated the blanket inhibition of glutamine metabolism was not a viable therapeutic strategy [33].
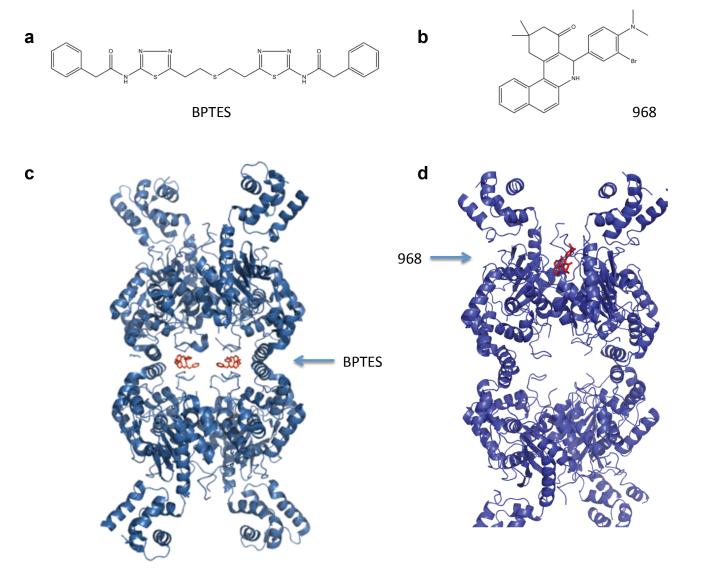
In addition to 968, BPTES [bis-2-(5-phenylacetamido-1,2,4-thiadiazoyl-2-yl)ethyl sulfide] is another allosteric inhibitor of GLS1 that has been described in the literature [28]. 968 and BPTES are structurally distinct (Figures 3a and b) and several lines of evidence indicate the two molecules act through different mechanisms. We do not find that BPTES and 968 are competitive with one another in their ability to inhibit GAC in vitro, suggesting they bind different sites on the enzyme. BPTES specifically binds to and inhibits the phosphate-stimulated activation of GLS1, but it has only weak affinity for GLS2 (IC50 of 60 nM and 33 μM, respectively [35]). Biochemical studies indicate that BPTES traps GLS1 in an inactive tetrameric state [28,34], and these studies have been confirmed by two independent x-ray crystallographic structures of BPTES bound to both GLS1 isoforms (Figure 3c) [35,36]. The x-ray structures reveal that two BPTES molecules bind at the helical dimer-dimer interface of the GLS1 tetramer and induce changes in a loop proximal to the catalytic site, thus trapping the enzyme in an inactive state. 968, by contrast, is largely ineffective at inhibiting GAC once it has been activated by phosphate [37] and thus, must be added before the phosphate to inhibit purified, recombinant GAC. This suggests that 968 functions by binding to the inactive conformation of GAC and preventing it from adopting an active conformational state [20,37]. This might be achieved either by 968 binding to the inactive dimer and preventing the formation of a higher oligomeric species, or by 968 binding to the inactive dimer and preventing the activation of the tetramer upon formation (See Figure 4b and 4c). Molecular docking and mutagenesis studies suggest that 968 binds in a hydrophobic pocket lying between the N- and C-termini at the monomer– monomer interface of the GAC dimer (Figure 3d) [37].
Figure 3.
Comparison of two GLS1 allosteric inhibitors, BPTES and 968. a. Chemical structure of BPTES. b. Chemical structure of 968. c. X-ray crystallographic structure of human GLS1 bound to BPTES, adapted from [35,36]. Two BPTES molecules are bound at the dimer–dimer interface of the GLS1 tetramer. d. Molecular docking model of 968 to GAC, adapted from [37]. One molecule of 968 is proposed to bind into a hydrophobic pocket formed by the N- and C-termini at the monomer–monomer interface of the GAC dimer.
There exists apparent differences in how 968 functions to inhibit GAC in vitro versus in vivo, which can be explained by differences in how the enzyme is activated under these different conditions (i.e., millimolar phosphate versus post-translational modification). We have consistently observed that lower concentrations of 968 are required to inhibit cellular GAC activity as compared to the recombinant enzyme (IC50: 5 μM in vivo vs. 17 μM in vitro). Approximately 48 hours is required to observe an effect of 968 on cellular proliferation and glutaminase activity, which is in contrast to the rapid effects observed with BPTES. Additionally, when assaying either endogenous mitochondrial GAC or ectopically expressed GAC immunoprecipitated from cells that have been treated with 968, basal enzyme activity is inhibited compared to untreated cells even though the small molecule inhibitor should have been diluted out during sample preparation. Together, these observations suggest that only the inactive (and presumably unmodified) pool of GAC is susceptible to the effects of 968 and that protein turnover is necessary in order for 968 to bind to and prevent the activation of a significant portion of cellular GAC, and that this GAC has a “memory” of its exposure to the inhibitor. Thus, 968 may be functioning in cells to block the activating modifications on the enzyme described above, making the inhibitor unique in its ability to disrupt GAC activity under specific signaling contexts (Figure 4d). Whereas BPTES is expected to inhibit both GLS1 isoforms independent of how they are being activated in cells, 968 inhibits the aberrant Rho-dependent signaling activation of GAC that occurs in cancer cells. These observations are likely to explain how 968 functions as a potent inhibitor of cancer cell growth, while having little or no effect on normal cell proliferation.
Unexpected connections between Rho GTPases, metabolism, and microvesicles
The altered metabolism exhibited by cancer cells is crucial for the ability of these cells to maintain an accelerated rate of growth and to evade apoptotic challenges, two major hallmarks of cancer [38,39]. Altered metabolism does so, in part, by fulfilling the enhanced energetic demands of cancer cells as they rapidly progress through the cell cycle. The re-programming of a cancer cell’s metabolism also contributes to the development of the malignant state in a second important way, by increasing the production of nucleotides, amino acids, fatty acids, and lipids. Each of these molecular building blocks is used by cells to generate daughter cells, and so, it is generally believed that the metabolic changes that cancer cells undergo are uniquely suited to promote their growth.
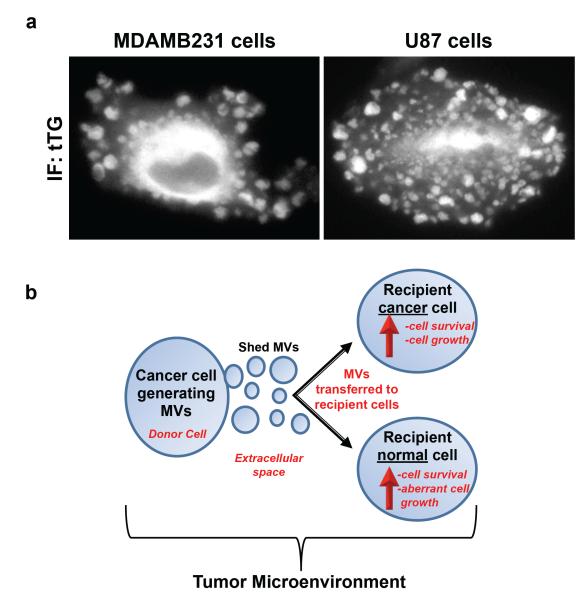
However, another intriguing and potentially important cellular process that may be intimately coupled to the altered metabolism of cancer cells involves the generation of microvesicles (MVs). MVs, which are also referred to as shedding vesicles, oncosomes, microparticles, and tumor-derived microvesicles, are a novel form of cell-to-cell communication that is garnering a tremendous amount of attention from clinical and basic cancer researchers [40–42]. They are unusually large vesicular structures (ranging from 0.2–3.0 μm in diameter) that are formed and shed directly from the plasma membranes of cancer cells via a poorly understood mechanism. Figure 5a shows immunofluorescent images of a human breast carcinoma cell, MDA-MB-231, and a human glioma cell, U87, both of which are heavily decorated with MVs on their surfaces, as detected by staining the cells for the MV marker tissue transglutaminase (tTG) [43,44]. Notably, these highly aggressive cancer cell lines constitutively generate MVs; ~35% of the MDA-MB-231 cells and ~25% of the U87 cells show detectable levels of MVs on their surfaces at any one time.
Figure 5.
Highly aggressive forms of human cancer cells constitutively generate microvesicles (MVs) that can be transferred to other cells that comprise the tumor microenvironment.. Cultures of serum-starved MDA-MB-231 breast carcinoma cells and U87 glioma cells were fixed and stained with an antibody against tissue transglutaminase (tTG) to label MVs. a. Representative images of the cells stained for tTG. b. A tumor microenvironment is composed of cancer cells, as well as normal cell lineages (i.e., fibroblasts). One cancer cell forms and sheds MVs from its surface into the extracellular space, and these MVs can then be transferred to another cancer cell, stimulating the growth and survival of the recipient cell. MVs can also be transferred to normal cells, conferring upon the recipient cells the transformed characteristics of a cancer cell.
Once MVs are shed from cancer cells, they can impact the behavior of neighboring cells, both normal and cancerous, that are present in the tumor microenvironment [43–48]. For example, MVs shed by one cancer cell can be taken-up by another cancer cell, stimulating the growth and promoting the survival of the recipient cell (Figure 5b) [45,46]. In addition, MVs can be transferred between the high grade MDA-MB-231 breast carcinoma cells or U87 glioma cells and normal (non-transformed) NIH-3T3 fibroblasts or MCF10A mammary epithelial cells, conferring upon the recipient cells the transformed characteristics of a cancer cell [43]. Thus, a cancer patient’s primary tumor burden might not be solely due to the clonal expansion of the cancer cells, but could also involve contributions from MVs through their ability to stimulate the growth and/or alter the signaling capabilities of normal cells .
Interestingly, a member of the Rho GTPase family has been linked to the generation of MVs. RhoA, signaling through Rho-associated, coiled-coil containing protein kinase (ROCK) to LIM kinase (LIMK) and cofilin, is both necessary and sufficient for the outward budding of MVs from plasma membranes in several different cancer cell lines [44]. Activation of the RhoA-cofilin pathway is well known to regulate actin cytoskeletal rearrangements [49], which is consistent with the idea that the dynamic morphological changes that accompany a cell generating MVs would involve such a signaling input.
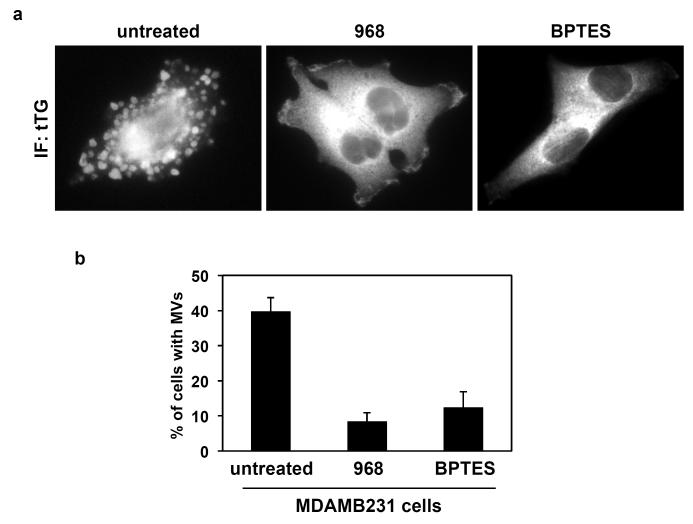
Given that the aberrant activation of Rho GTPases promotes the metabolic reprogramming of cancer cells and is critical for inducing the formation of MVs, we wondered whether there might be a connection between the altered metabolism exhibited by cancer cells and the biogenesis of MVs. By examining the images of the cancer cells shown in Figure 5a, one can begin to appreciate the demands placed upon cells that are generating MVs, including the expenditure of a significant amount of energy to form these structures, together with the need to rapidly replace the plasma membrane once the MVs are shed. The re-programming of the metabolic machinery in cancer cells can uniquely fulfill both of these demands [38,39], and so we asked whether treating MDA-MB-231 breast cancer cells with 968 or BPTES to inhibit glutaminase activity would influence the ability of these cells to generate MVs. Figures 6a and 6b show that exposing MDA-MB-231 cells to either of these inhibitors potently blocked the ability of these cells to form MVs. Although we are only at the earliest stages of this work and appreciate that much more remains to be done, it is still exciting to consider the possible connections that may exist between the Rho GTPases, metabolic changes that accompany the development of the malignant state, and the generation of MVs. If our preliminary findings bear out, then the energy and lipids generated through glutamine metabolism would not only be used to generate daughter cells, but they would also be important for the formation and shedding of MVs, strongly influencing the tumor microenvironment and the establishment of the pre-metastatic niche.
Figure 6.
Microvesicle (MV) formation in human cancer cells can be blocked using glutaminase inhibitors. Cultures of serum-starved MDA-MB-231 breast carcinoma cells were left untreated or were treated with either 10 μm 968 or 20 μm BPTES for 35 hours, at which time they were fixed and stained with an antibody against tissue transglutaminase (tTG) to label MVs. a. Representative images of the cells exposed to the indicated culturing condition. b. Quantification of MV production by MDA-MB-231 cells treated without or with the glutaminase inhibitors.
Concluding remarks
How might the results described in the preceding sections inform our understanding of tumor biology? Except for the very isolated examples where de-regulated guanine nucleotide exchange activity toward Rac, Rho, or Cdc42 results in tumorigenesis (i.e., LARG [50]), constitutively active, truncated forms of Rho GEFs do not underlie Rho-driven cancers. Similarly, the search for somatic mutations in Rho GTPases have for the most part failed to identify any changes that would give rise to hyperactivated Rho activity (i.e., ‘fast cycling’ or GTPase-defective, constitutively active mutants) and transformation [51], with the exception of the recent identification of a Rac1 mutation in a small percentage of sun-induced melanoma [52]. Nevertheless, the wild-type alleles of Rho GTPases are often either overexpressed in tumor cells and/or an increased fraction of these GTPases are in the active, GTP-bound state. As mentioned earlier, in many cancer cell lines and in tumor tissue samples this elevated Rho GTPase activity can likely be attributed to the loss of DLC1 [15,53]. The striking absence of DLC1 from a number of isolated cancers of the lung, breast, prostate, and liver raises the possibility that Rho GTPases, although not sufficient for full tumorigenicity, contribute to many cancer phenotypes by their influence on cytoskeletal remodeling, giving rise to the motility and invasiveness of metastatic cells, and by their NFκB-mediated alterations of the metabolic transcriptome. It will be important to gain a better understanding of the molecular basis of the changes that give rise to the observed alterations in metabolism in Rho GTPase-driven transformation, and in particular, how the enzymatic machinery of glutaminolysis might be shifted transcriptionally or post-translationally to bring about the observed changes in cellular growth and metabolism.
Outstanding Questions Box.
What are the signaling events emanating from the Rho GTPases that culminate in the activation of mitochondrial GAC? What is the role of NFκB?
What is the molecular mechanism responsible for the activation of GAC in cancer cells? If it involves phosphorylation, what sites on GAC are phosphorylated and what kinase(s) are responsible?
Can GAC be activated in cells as a consequence of Rho-independent signaling events?
Does the other GLS1 isoform, KGA, also support the glutamine addiction of cancer cells?
Where does 968 bind to GAC and how does this binding inhibit glutaminase activity?
What is the link between mitochondrial glutaminase activity and microvesicle biogenesis?
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- 1.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 2.Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093–2101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 3.Erickson JW, Cerione RA. Structural elements, mechanism, and evolutionary convergence of Rho protein-guanine nucleotide exchange factor complexes. Biochemistry. 2004;43:837–842. doi: 10.1021/bi036026v. [DOI] [PubMed] [Google Scholar]
- 4.Lin R, et al. Specific contributions of the small GTPases Rho, Rac and Cdc42 to Dbl transformation. J. Biol. Chem. 1999;274:23633–23641. doi: 10.1074/jbc.274.33.23633. [DOI] [PubMed] [Google Scholar]
- 5.Fort P. Small GTPases of the Rho family and cell transformation. Prog. Mol. Subcell. Biol. 1999;22:159–181. doi: 10.1007/978-3-642-58591-3_8. [DOI] [PubMed] [Google Scholar]
- 6.Suwa H, et al. Overexpression of the rhoC gene correlates with progression of ductal adenocarcinoma of the pancreas. Br. J. Cancer. 1998;77:147–152. doi: 10.1038/bjc.1998.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mira JP, et al. Endogenous, hyperactive Rac3 controls proliferation of breast cancer cells by a p21-activated kinase-dependent pathway. Proc. Natl. Acad. Sci. USA. 2000;97:185–189. doi: 10.1073/pnas.97.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritz G, et al. Rho GTPases in human breast tumors: expression and mutation analyses and correlation with clinical parameters. Br. J. Cancer. 2002;87:635–644. doi: 10.1038/sj.bjc.6600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamai T, et al. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin. Cancer Res. 2004;10:4799–4805. doi: 10.1158/1078-0432.CCR-0436-03. [DOI] [PubMed] [Google Scholar]
- 10.Kleer CG, et al. Characterization of RhoC expression in benign and malignant breast disease: a potential new marker for small breast carcinomas with metastatic ability. Am. J. Pathol. 2002;160:579–584. doi: 10.1016/S0002-9440(10)64877-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark EA, et al. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 12.Burbelo P, et al. Altered Rho GTPase signaling pathways in breast cancer cells. Breast Cancer Res. Treat. 2004;84:43–48. doi: 10.1023/B:BREA.0000018422.02237.f9. [DOI] [PubMed] [Google Scholar]
- 13.Valastyan S, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Xue W, et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22:1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lahoz A, Hall A. DLC1: a significant GAP in the cancer genome. Genes Dev. 2008;22:1724–1730. doi: 10.1101/gad.1691408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu WJ, et al. Activated Cdc42 sequesters c-Cbl and prevents EGF receptor degradation. Cell. 2003;114:715–725. doi: 10.1016/s0092-8674(03)00688-3. [DOI] [PubMed] [Google Scholar]
- 17.Feng Q, et al. Cool-1 functions as an essential regulatory node for EGF receptor- and Src-mediated cell growth. Nat. Cell Biol. 2006;8:945–956. doi: 10.1038/ncb1453. [DOI] [PubMed] [Google Scholar]
- 18.Wilson KF, et al. Cdc42 stimulates RNA splicing via the S6 kinase and a novel S6 kinase target, the nuclear cap-binding complex. J. Biol. Chem. 2000;275:37307–37310. doi: 10.1074/jbc.C000482200. [DOI] [PubMed] [Google Scholar]
- 19.Ly TK, et al. Activation of the Ran GTPase is subject to growth factor regulation and can give rise to cellular transformation. J. Biol. Chem. 2010;285:5815–5826. doi: 10.1074/jbc.M109.071886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang JB, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 22.DeBerardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christofk HR, et al. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008a;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 24.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008b;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 25.Hitosugi T, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci. Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curthoys NP. Regulation of glutaminase activity and glutamine metabolism. Annu. Rev. Nutr. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- 27.Kenny J, et al. Bacterial expression, purification and characterization of rat kidney-type mitochondrial glutaminase. Protein Expr. Purif. 2003;31:140–148. doi: 10.1016/s1046-5928(03)00161-x. [DOI] [PubMed] [Google Scholar]
- 28.Robinson MM, et al. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) Biochem. J. 2007;406:407–414. doi: 10.1042/BJ20070039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perona R, et al. Activation of the nuclear factor-kappaB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 1997;11:463–475. doi: 10.1101/gad.11.4.463. [DOI] [PubMed] [Google Scholar]
- 31.Cammarano M, Minden A. Dbl and the Rho GTPases activate NF kappa B by I kappa B kinase (IKK)-dependent and IKK-independent pathways. J. Biol. Chem. 2001;276:25876–25882. doi: 10.1074/jbc.M011345200. [DOI] [PubMed] [Google Scholar]
- 32.McBrayer M, et al. in preparation.
- 33.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartwick EW, Curthoys NP. BPTES inhibition of hGA(124-551), a truncated form of human kidney-type glutaminase. J. Enzyme Inhib. Med. Chem. 2011 doi: 10.3109/14756366.2011.622272. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 35.DeLaBarre B, et al. Full-length human glutaminase in complex with an allosteric inhibitor. Biochemistry. 2011;50:10764–10770. doi: 10.1021/bi201613d. [DOI] [PubMed] [Google Scholar]
- 36.Thangavelu K, et al. Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism. Proc. Natl. Acad. Sci. USA. 2012;109:7705–7710. doi: 10.1073/pnas.1116573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katt WP, et al. Dibenzophenanthridines as inhibitors of glutaminase C and cancer cell proliferation. Mol. Cancer Ther. 2012;11:1269–1278. doi: 10.1158/1535-7163.MCT-11-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erickson JW, Cerione RA. Glutaminase: a hot spot for regulation of cancer cell metabolism? Oncotarget. 2010;1:734–740. doi: 10.18632/oncotarget.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muralidharan-Chari V, et al. Microvesicles: mediators of extracellular communication during cancer progression. J. Cell Sci. 2010;123:1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kharaziha P, et al. Tumor cell-derived exosomes: A message in a bottle. Biochim. Biophys. Acta. 2012;1826:103–111. doi: 10.1016/j.bbcan.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 42.Lee TH, et al. Microvesicles as mediators of intercellular communication in cancer – the emerging science of cellular ‘debris’. Semin. Immunopathol. 2011;33:455–467. doi: 10.1007/s00281-011-0250-3. [DOI] [PubMed] [Google Scholar]
- 43.Antonyak MA, et al. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc. Natl. Acad. Sci. USA. 2011;108:4852–4857. doi: 10.1073/pnas.1017667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B, et al. RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells. Oncogene. 2012 doi: 10.1038/onc.2011.636. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Nedawi K, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 46.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svensson KJ, et al. Hypoxia triggers a proangiogenic pathway involving cancer cell microvesicles and PAR-2-mediated heparin-binding EGF signaling in endothelial cells. Proc. Natl. Acad. Sci. USA. 2011;108:13147–13152. doi: 10.1073/pnas.1104261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Al-Nedawi K, et al. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc. Natl. Acad. Sci. USA. 2009;106:3794–3799. doi: 10.1073/pnas.0804543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rathinam R, et al. Role of Rho GTPases and their regulators in cancer progression. Front. Biosci. 2011;17:2561–2571. doi: 10.2741/3872. [DOI] [PubMed] [Google Scholar]
- 50.Booden MA, et al. Leukemia-associated Rho guanine nucleotide exchange factor promotes Gαq-coupled activation of RhoA. Mol. Cell Biol. 2002;22:4053–4060. doi: 10.1128/MCB.22.12.4053-4061.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rihet S, et al. Mutation status of genes encoding RhoA, Rac1, and Cdc42 GTPases in a panel of invasive human colorectal and breast tumors. J. Cancer Res. Clin. Oncol. 2001;127:733–738. doi: 10.1007/s004320100272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krauthammer M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan BZ, et al. DLC-1 gene inhibits human breast cancer cell growth and in vivo tumorigenicity. Oncogene. 2003;22:445–450. doi: 10.1038/sj.onc.1206064. [DOI] [PubMed] [Google Scholar]