Abstract
As part of the visual cycle, the retinal chromophore in both rod and cone visual pigments undergoes reversible Schiff base hydrolysis and dissociation following photobleaching. We characterized light-activated retinal release from a short-wavelength sensitive cone pigment (VCOP) in 0.1% dodecyl maltoside using fluorescence spectroscopy. The half-time (t1/2) of retinal release from VCOP was 7.1 s, 250-fold faster than rhodopsin. VCOP exhibited pH-dependent release kinetics, with the t1/2 decreasing from 23 s to 4 s with pH 4.1 to 8, respectively. However, the Arrhenius activation energy (Ea) for VCOP derived from kinetic measurements between 4° and 20°C was 17.4 kcal/mol, similar to 18.5 kcal/mol for rhodopsin. There was a small kinetic isotope (D2O) effect in VCOP, but less than that observed in rhodopsin. Mutation of the primary Schiff base counterion (VCOPD108A) produced a pigment with an unprotonated chromophore (⌊max = 360 nm) and dramatically slowed (t1/2 ~ 6.8 min) light-dependent retinal release. Using homology modeling, a VCOP mutant with two substitutions (S85D/ D108A) was designed to move the counterion one alpha helical turn into the transmembrane region from the native position. This double mutant had a UV-visible absorption spectrum consistent with a protonated Schiff base (⌊max = 420 nm). Moreover, VCOPS85D/D108A mutant had retinal release kinetics (t1/2 = 7 s) and Ea (18 kcal/mol) similar to the native pigment exhibiting no pH-dependence. By contrast, the single mutant VCOPS85D had a ~3-fold decrease in retinal release rate compared to the native pigment. Photoactivated VCOPD108A had kinetics comparable to a rhodopsin counterion mutant, RhoE113Q, both requiring hydroxylamine to fully release retinal. These results demonstrate that the primary counterion of cone visual pigments is necessary for efficient Schiff base hydrolysis. We discuss how the large differences in retinal release rates between rod and cone visual pigments arise, not from inherent differences in the rate of Schiff base hydrolysis, but rather from differences in the non-covalent binding properties of the retinal chromophore to the protein.
Vision in most vertebrates is functionally duplex [1]. Scotopic and photopic vision, mediated by rods and cones respectively, have disparate sensitivity, spectral tuning, temporal properties and recovery rates following light exposure [2-4]. This division also occurs amongst the homologous phototransduction proteins which contribute to differences in cellular physiology, biochemistry and phototransduction, although the molecular basis underlying these differences is still emerging. A primary focus has been the visual pigments, which consist of a seven transmembrane alpha helical bundle (opsin) and an 11-cis retinal chromophore covalently attached via a Schiff base linkage [5, 6]. The visual pigments determine the spectral sensitivity of a particular photoreceptor and share roughly 50% amino acid sequence identity [7]. The X-ray structures of rhodopsin [8] and related photobleaching conformations [9-12] have permitted a unique opportunity to understand the activation of GPCRs through the combination of computational and a large repertoire of experimental approaches (recently reviewed [13]).
The primary isomerization event and subsequent photoreactions have been intensively studied [14-17]. Crystallography, UV/visible, ESR and FTIR spectroscopy have led to a model of the photoactivation pathway in which the steric strain of all-trans retinal drives movement of extracellular loop EL2, transmembrane helices H5-H6 and the disruption of ionic interactions in the transmembrane bundle, termed the ionic lock [13]. This framework has been applied to other members of the GPCR class [18], and thus forms a unifying principle for the activation of these proteins by ligands. However, the molecular basis for functional differences between the visual pigments is not yet understood.
There are four visual pigment properties that contribute to the distinctive photoresponses of rods and cones. First, the wavelength of light (λmax) to which the pigment is maximally sensitive varies from the UV [19] to the far red [20]. Spectral tuning is regulated by the protonation state of the Schiff base [21, 22], non-covalent interactions distributed between multiple amino acid side chains with the polyene chain of retinal [23] and structural water in the retinal binding pocket [8, 24]. Second, the relative instability of the dark-adapted pigment influences the rate of thermal isomerization and thus activation of the phototransduction cascade in the dark, thus setting the absolute noise level [25]. Thus, cones express a more labile visual pigment and exhibit much higher dark noise than rods [26] . In fact, rods transgenically expressing a cone opsin have elevated dark noise in either transgenic Xenopus or mice [27]. Following photoactivation, all-trans retinal is eventually released from opsin after Schiff base hydrolysis [28, 29] reducing of pigment concentration and visual sensitivity. The third property of visual pigments that contribute to rod-cone specialization is the lifetime of the light-activated conformation R* (coinciding with meta II) which sets a constraint on the maximum number of transducins that can be activated. In fact, photoreceptors with transgenes having shorter meta II lifetimes produce smaller photoresponse amplitudes [30, 31]. In rods, R* activity is terminated by downstream mechanisms before the Schiff base is hydrolyzed [32]. By contrast, the R*-meta II lifetime in cone pigments is much shorter, and this may contribute to the smaller photoresponse of the cones [33]. To reset the phototransduction cascade during response recovery and dark adaptation, regeneration of visual pigment occurs via formation of a new covalent bond with 11-cis retinal [28]. Fourth, rod and cone pigments have significantly different regeneration rates [34]. This contributes to variation between photoreceptor types in the overall adaptive changes to and recovery from steady light [2, 35, 36]. In summary, the biochemical differences between visual pigments are consistent with the hypothesis that visual pigments contribute to the differences in photoresponse sensitivity and kinetics between rods and cones, including their adaptation to background light.
The molecular determinants of the photochemical and biochemical properties of visual pigments have been investigated by site-specific mutagenesis of evolutionarily conserved amino acid residues. A primary focus has been the amino acids in the environment of the retinal-Schiff base linkage [13], particularly in comparative studies between chicken rhodopsin and a green-sensitive cone pigment [37]. The Schiff base has been extensively studied in rhodopsin, where it has been shown to have multiple functions, including Suppression of constitutive activity of opsin, Facilitation of regeneration Stabilization of Schiff base linkage and photosimoerization spectral tuning as mentioned above and role in determination of photointermediate lifetime [21] These studies identified a single amino acid (RhoE122, using bovine rhodopsin numbering) that influences the rate of meta II decay and lifetime of the light-activated conformation R*. In the Xenopus short-wavelength cone pigment (VCOP), mutation of the Schiff base counterion (Asp108) causes a deprotonation of the Schiff base and stabilization of the R* conformation [38]. Counterion mutations show similar behaviors in UV-sensitive cone pigments, despite the fact that the Schiff base is normally deprotonated in the dark [39]. Additional amino acids, in particular a highly conserved proline (Pro189) in the extracellular loop 2, are important in the thermal stability of both dark-adapted and light-activated cone pigments [40]. These results emphasize the central importance of the chromophore-protein linkage for inactivating the light-activated conformation, and its significant contribution to the differences between rod-cone visual pigments. However, mechanistic insight is needed to explain the role of the cone pigment Schiff base counterion in retinal release.
In the present study, we investigate the mechanism of retinal release from light-activated VCOP in comparison to rhodopsin. We use the intrinsic fluorescence of opsin and its quenching by bound retinal to directly monitor retinal interactions with opsin [41]. We demonstrate that retinal release in VCOP is ~250-fold faster than in rhodopsin at neutral pH but the Arrhenius activation energy of Schiff base hydrolysis is similar between them, implying a conserved reaction mechanism. We show that the counterion (Asp108) is essential for rapid release following photoactivation and that movement of the counterion to a nearby position restores the rate to that of the native pigment. Furthermore, we show that the retinal chromophore release rate from rod and cone visual pigments is the same in the absence of a counterion, indicating that the primary counterion is the catalytic residue required for Schiff base hydrolysis.
Materials and Methods
Visual pigment expression and purification
The epitope-tagged VCOP plasmids used for site-directed mutagenesis and protein expression have been described previously [38]. The mutants were expressed in COS1 cells by transient transfection, purified by immunoaffinity chromatography and quantified by UV-visible spectroscopy. Except for RhoE113Q, pigments were eluted in buffer Y1 [50 mM HEPES, 140 mM NaCl, and 3 mM MgCl2 (pH 6.6)] with 0.1% N-dodecyl-β-D-maltoside and 20% glycerol. RhoE113Q was eluted in the modified buffer Y1 in pH 8.0 with 0.1% N-dodecyl-β-D-maltoside and 20% glycerol to deprotonate the Schiff base.
Fluorescence spectroscopy
The procedure was modified from Farrens et al. [41]. A FluroMax-3 (Jobin Yvon, Inc., Edison, NJ), fitted with a UV bandpass filter (U-360, Edmund Optics, Barrington, NJ) mounted in the sample chamber to eliminate light with wavelength >410 nm to prevent pigment from bleaching, was used to measure fluorescence changes. Protein samples (200 μl of 0.5 μM) were suspended in buffer A (0.1% N-dodecyl-β-D-maltoside, in 10mM MES/Na2HPO4) at three different pH (4.1, 6.0 and 8.0). Fluorescence was recorded from samples in a 0.2 ml quartz cuvette thermally controlled at 4°C, 6.7°C, 10°C, 12.5°C, 15°C, 18°C, and 20.4°C by a circulating water bath (C6 Launda, Germany). Excitation wavelength was 295 nm (slit width 0.25 nm) and emission at 330 nm (slit width 12 nm). There was noticeable pigment bleaching from the excitation beam provided in the FluoroMax-3 instrument, so a neutral density filter was introduced to minimize pigment loss during the experiment. This reduced bleaching so that no correction of the fluorescence kinetic trace was needed (i.e. Single exponential fits to the initial phase of the responses were sufficient). Data from VCOP samples were integrated for 80 ms at 250 ms intervals, with the emission shutter constantly open. Data from rhodopsin samples were integrated for 2 s at 10 s intervals, with emission shutter closed in between collection periods. Samples were photoactivated using an external light source (66906 Research Arc Source, Newport Corporation, Irvine, CA) containing a 200W Ozone-free Hg/Xe Arc lamp (6292, Newport Corporation, Irvine, CA). An IR cutoff filter (C53-711, Edmund Optics, Barrington, NJ) was used light was delivered to the cuvette via a quartz fiber optic light pipe (C38-955, Edmund Optics, Barrington, NJ). Complete photoactivation of the samples were accomplished by illumination for either 500 ms (VCOP) or 25 s (rhodopsin). The half time value of fluorescence change (t1/2) was measured as previously described [41, 42]. All fits in rhodopsin showed an r2 value > 0.95, while in VCOP an r, r2 values >0.80 and 0.65, respectively.
Fluorescence measurement in D2O
Water in purified samples was replaced by D2O (Cambridge Isotope Laboratory, Inc., MA) at 4°C by repetitive dilution and concentration using a Centricon YM-30 concentrator (Millipore, IN). Fluorescence measurements were performed at 10°C on protein diluted in buffer A at pH 4.1, 6.0, or 8.0 prepared with D2O.
Homology Models
Homology models for VCOPS85D/D108A and VCOPS85D were generated according to the procedures as previously described [43-45].
Results
Rate of retinal release of VCOP
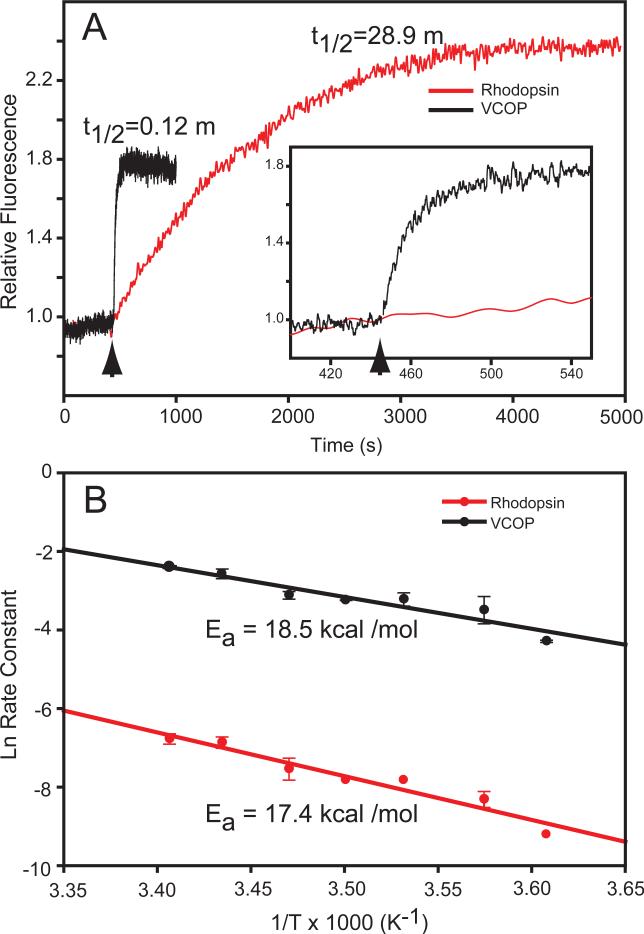
VCOP has a λmax of 427 nm when incubated with 11-cis retinal (Fig. 1A) which arises from a protonated Schiff base linkage [23, 46]. Illumination of the pigment causes a conversion in seconds to an unprotonated Schiff base with a λmax of 360 nm [47]. We measured the rate of retinal release following light-activation at 10°C and pH 6.0. The fluorescence increased rapidly following a bright flash and recovered slightly before reaching a steady state (Fig. 2A). The fluorescence change was monophasic over the first 25 s (Fig. 2A inset). A single exponential fit to the initial rising phase produced a half-time for the increase in fluorescence of t½=0.12 m. Changes in ionic strength from 0-300 mM NaCl had only a minor effect on the retinal release rates, ranging from 7.1 ± 2.0 s to 14.5 ±3.9 s respectively (data not shown). In similar experiments, rhodopsin exhibited a much slower monophasic increase following a bright flash, with a t½=28.9 min, in agreement with previous reports using this assay [41]. Thus, retinal release from VCOP was 240-fold faster than in rhodopsin under these conditions.
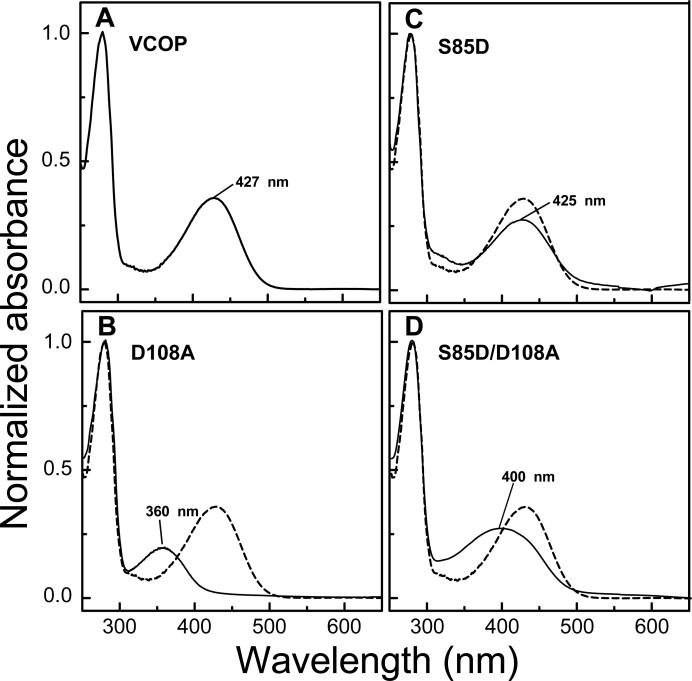
Figure 1.
UV/Visible spectroscopy of VCOP substitution mutants. Visual pigments were purified in dodecyl maltoside following addition of 11-cis retinal. UV/Vis spectra were obtained for the wild type VCOP (A), primary counterion mutant VCOPD108A (B), VCOPS85D (C) and the counterion replacement double mutant, VCOPS85D/D108A (D). The λmax (± 1 nm) is indicated for each pigment and the VCOP spectrum is included (dotted line) for comparison.
Figure 2.
Fluorescence increases and activation energies after photobleaching. (A) The kinetics of the fluorescence increase at 330 nm of VCOP (black) and rhodopsin (red) after illumination with a 2 s flash of white light (arrow). The time to reach half-maximal fluorescence intensity (t1/2) was 28.9 min and 0.12 min, respectively, at 10°C in a solution containing 0.1% dodecyl maltoside, pH 6.0. The inset shows an expanded time scale of the fluorescence increase of VCOP. For recording fluorescence, the excitation was at 295 nm and emission at 330 nm. Fluorescence was normalized to the initial value (set to 1). (B) Arrhenius plot of the natural log of the rates of fluorescence increase in illuminated rhodopsin and VCOP samples between 4° to 20°C at pH 6.0. Activation energies (Ea), as indicated in the figure, were calculated from the negative reciprocal of the slope of the best fit linear regression line (solid lines).
The rates of retinal release were determined at seven different temperatures ranging from 4° to 20°C at pH 6.0. Arrhenius plots were utilized to estimate the the activation energy of Schiff base hydrolysis (Fig. 2B). The slopes from the plots obtained by linear regression produced an activation energy of 18.5 kcal/mol for rhodopsin and 17.4 kcal/mole for VCOP. These results show that while the rates of retinal release are dramatically different, the activation energies for retinal release, which includes both Schiff base hydrolysis and all-trans dissociation, are very similar for both cone and rod visual pigments.
Alteration of the Schiff base counterion
Computational models suggest that Ser85 and Asp108 form an electrostatic network around the Schiff base in VCOP [48]. Mutations of Ser85 and Asp108 not only caused spectral shifts of absorbance maxima, but also destabilized photoactivation intermediates [38, 48]. We investigated whether Ser85 could stabilize the protonated Schiff base in the absence of the primary counterion D108. The single counterion mutant VCOPD108A has an unprotonated Schiff base with a λmax of 353 nm (Fig. 1B, see also [38]). Addition of another Asp in the Schiff base environment (VCOPS85D) causes a minor blue shift in λmax to 425 nm and an small accompanying broadening of the absorbance spectrum (Fig. 1C). Position 85 is in a helix adjacent to Asp108 and approximately one turn of an alpha helix toward the cytoplasmic face. This approach has been taken previously with bovine rhodopsin [49, 50]. Thus, the mutant VCOPS85D/D108A has a potential counterion in a different location compared to the wild type protein. This double mutant pigment has a λmax at 400 nm and a broad absorbance spectrum with significant intensity >425 nm. The anomalous absorbance spectrum suggests inhomogeneous broadening and is not consistent with an unprotonated Schiff base [51]. Thus, removal of the counterion from its normal position and reintroduction nearby produces a pigment with a stabilized protonated Schiff base, although the chromophore environment is not entirely restored to that of a normal visual pigment.
A Schiff base counterion is required for rapid retinal release
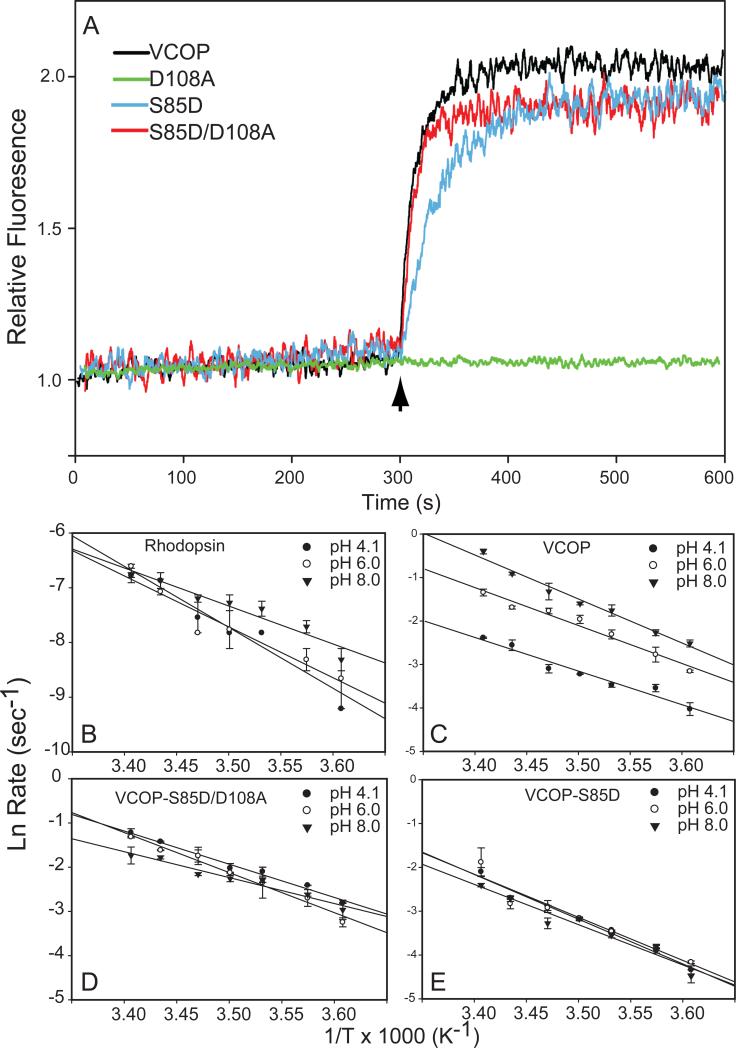
We investigated the role of the Schiff base counterion in retinal release by measuring the kinetics of light-induced fluorescence quenching for several counterion mutants at 10°C and pH 6.0. We used 350 nm illumination to photobleach VCOPD108A since it has an absorbance maximum at 353 nm. For this mutant, no change in fluorescence was detected (Fig. 3). Next, we tested the mutant with the counterion reintroduced toward the cytoplasmic face, VCOPS85D/D108A. After irradiation with white light for 0.5 s, VCOPS85D/D108A exhibited a rate similar to VCOP, with a half time of 6.9 s. Introduction of a second Asp in the Schiff base environment (VCOPS85D) slowed rate to 22.1 s half time value. Thus, changing the primary counterion to Ser85 did not alter the retinal release rate while removal or increasing electrostatic stabilization around the Schiff base slowed release. In the primary counterion mutant, we did not observe any fluorescence changes after more than 15 min (Fig. 3), even though the light-activated VCOPD108A is able to stimulate GTP exchange on transducin [38]. We also introduced a Glu acid in place of the Asp at position 108, and observed retinal release similar to wild-type kinetics (Suppl. Fig. 1).
Figure 3.
Fluorescence increases and activation energies of VCOP counterion mutants. (A) Fluorescence increases for VCOP (black), the primary counterion mutant VCOPD108A (green), VCOPS85D (blue) and the counterion replacement mutant VCOPS85D/D108A (red) were normalized individually to the initial fluorescence value (set to 1). Photobleaching was initiated after recording baseline fluorescence by a 0.5 s flash of white light (arrow). The data were fit to a single exponential fit and the half-time to reach the maximum level was calculated. The t1/2 values were VCOP, 7.1 secs; VCOPD108A 6.8 min; VCOPS85D 22.1 s; and VCOPS85D/D108A, 6.9 s. (B-E) Arrhenius plots of the rates of fluorescence increases in illuminated pigment samples between 4° to 20°C at pH 4.1 (closed circles), 6.0 (open circles) and 8.0 (inverted triangles). Activation energies are given in Table 1. Linear regression was used to generate the best fit lines (solid lines).
pH dependence and isotope effect of Schiff base hydrolysis
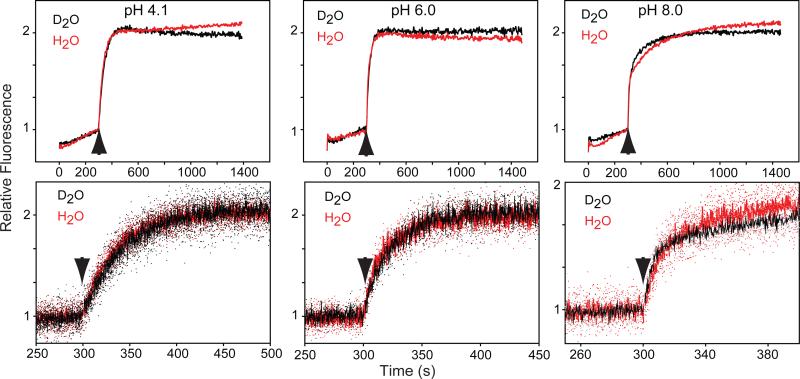
The chromophore environment is more accessible to solvent (e.g. H2O or NH2OH) in VCOP and other cone visual pigments [36, 46]. This suggests that solvent may contribute catalytically in the hydrolysis of the Schiff base in cone visual pigments. Thus, we investigated the effects of pH on the rate and activation energy of retinal release rate at three different pHs, pH 4.1, 6.0 and 8.0. As a control, we performed experiments on rhodopsin purified from bovine retina. The rate of retinal release in rhodopsin is not dependent upon pH, although the activation energy decreases with increasing pH (Table 1 and [42]). By contrast, at 10°C, the rate of retinal release from VCOP increased significantly as pH increased (Fig. 4 and Table 1). Overall, the activation energy decreased ~16% (Table 1). Retinal release was measured in VCOP samples in which H2O was replaced by D2O (Fig. 4) at 10°C. Retinal release exhibited a similar trend to faster rates at elevated pD. At pD 4.1 and 6.0, the half times of retinal release was slightly increased 1.2-fold in D2O (Table 1). At pD 8.0, the increase was slightly higher, at 1.6-fold. Thus, in wild type VCOP, the rate of retinal release shows a modest isotope effect, indicating proton transfer events may participate in Schiff base hydrolysis but that they are not the dominant catalytic moiety.
Table 1.
Retinal release rate and activation energies for Rho and VCOP (10°C)
| pH | t½ (S) | Ea (kcal/mole) | |
|---|---|---|---|
| 4.1 | 1730 | 22.1 | |
| Rhodopsin | 6.0 | 1730 | 18.5 |
| 8.0 | 1730 | 15.9 | |
| 4.1 | 22.9 (27.6) | 15.4 | |
| VCOP (D2O) | 6.0 | 7.1 (8.6) | 17.3 |
| 8.0 | 4.0 (6.2) | 19.9 | |
| 4.1 | 5.6 | 14.9 | |
| VCOPS85D/D108A | 6.0 | 6.9 | 18.0 |
| 8.0 | 6.9 | 11.7 | |
| 4.1 | 21.4 | 20.3 | |
| VCOPS85D | 6.0 | 22.1 | 19.5 |
| 8.0 | 22.8 | 18.3 | |
Figure 4.
Kinetics of fluorescence increases of VCOP samples in H2O (red) or D2O (black) following photobleaching (arrow) at 10°C, pH 4.1 (left), pH 6.0 (middle) or pH 8.0 (right). Bottom panels are an expanded time base of the top panels. Half-times are given in Table 1.
The rate of retinal release of the VCOPS85D/D108A mutant does not show a pH dependence in contrast to wild type protein (Table 1). However, the activation energy shows a complex behavior, with a peak at 6.0 and lower values at other pH (Table 1). In the VCOPS85D mutant, neither retinal release rate nor the activation energy was affected by pH (Table 1). However, compared to VCOP, there was a 3-fold increase in the rate and an elevated activation energy. In summary, these results show that mutations that alter hydrogen interactions with the Schiff base significantly effect the pH-dependence of retinal release.
Comparison of counterion mutants RhoE113Q and VCOPD108A
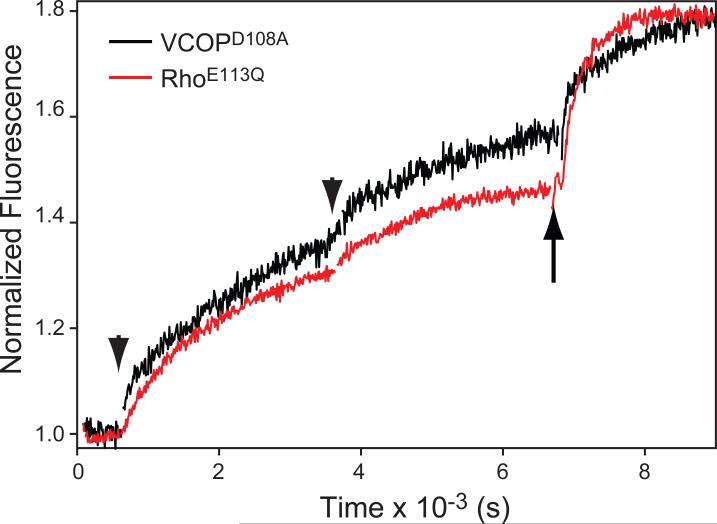
We compared the retinal release on extended time frames from a rod and cone visual pigments lacking s Schiff base counterion, RhoE113Q and VCOPD108A. Since protonation state of Schiff base in Rho-E113Q mutant is determined by the pH [52-54], we diluted the rhodopsin mutant in pH 8.0 buffer to deprotonate the Schiff base. For both counterion mutants, we used UV light, higher temperatures (20°C) and extended photobleaching (25 s) to activate the unprotonated visual pigment. Remarkably, both RhoE113Q and VCOPD108A showed very similar retinal release kinetics (Fig. 5). We used three irradiation treatments, after which hydroxylamine was added to completely bleach the pigments. The average rate of retinal release for RhoE113Q and VCOPD108A was 6.8 min. These observations strongly suggest that the primary counterion acts as the primary catalytic residue necessary for efficient Schiff base hydrolysis in both rod and cone visual pigments.
Figure 5.
Comparison of the fluorescence increase of the bovine RhoE113Q counterion mutant with VCOPD108A at 20°C. The former is at pH 8.0 to deprotonate the Schiff base, and the latter is at pH 7.0. Two flashes were sequentially applied (arrowheads) The t1/2 measured after after the first flash was 6.8 min for both samples. A final concentration of 10 mM hydroxylamine was added (arrow) to fully release all-trans retinal. Fluorescence was normalized to the initial value (set to 1).
Discussion
We have studied the fluorescence change during the decay of the meta II intermediate in VCOP and rhodopsin. We have found that the chromophore is more solvent accessible in the former, leading to a pH-dependent retinal release rate and activation energy of Schiff base hydrolysis. This study shows that the retinal release rate in VCOP is ~250-fold faster than in rhodopsin, which explains the faster decay of meta II in cone compared to rod pigments as reported by Govardovskii [55]. Meta II is the principal intermediate that couples to the G-protein transducin, starting the phototransduction cascade. R* is terminated in two ways: via phosphorylation-arrestin binding (see review in [32, 56]), and the release of all-trans retinal to produce aproprotein. Previous experiments have shown that cone meta II has lower thermal stability than rhodopsin meta II [34, 37, 47, 57, 58]. The former has a short life time (~1 s), while the latter a much longer life time (>30 min). The destabilized meta II in cone opsin may partially explain the physiological difference in late recovery between rods and cones. In humans, it takes five or more times longer for rods to recover following bright flashes compared to cones [2, 59]-arising from the intrinsic difference between rhodopsin and cone opsin. Our results suggest that the hydrolysis of the Schiff base per se is similar in the two pigment classes. But, we find support for the hypothesis that the molecular basis for these different properties resides primarily in the release of retinal from the light-activated visual pigments.
Mechanism of Schiff base hydrolysis
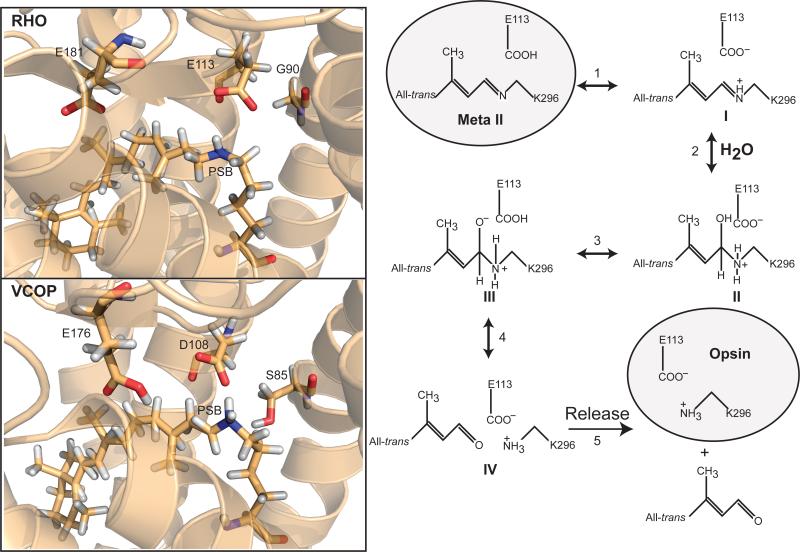
Following the mechanism for Schiff base hydrolysis discussed by Cooper et al [60], we propose a central catalytic role for the counterion in cone visual pigments (Fig. 6). In this model, Meta II has a deprotonated retinylidene Schiff base, protonated Glu113/Asp108 and undergoes a reversible and transient protonation, with the proton donor being Asp108. Note that this state is distinct from meta I, which also has a similar configuration of Schiff base and counterion protonation states. There is evidence for such a state in rhodopsin, which may either be meta IIb [61], meta III [62] or a related post-meta II photoproduct. There is no direct evidence for such a species produced from photoactivation of cone visual pigments. However, given the similar activation energies for retinal release, strong sequence homology between all visual pigments and the known mechanisms for formation and hydrolysis of model Schiff base compounds [60], it is reasonable to postulate that the formation of this intermediate [I] is the first step in hydrolysis. In [I], the unprotonated Schiff base may be attacked by a water molecule (step 2) catalyzed by a near-by acidic residue (primary counterion), to form the protonated carbinolamine intermediate with a conformational change of the Schiff base from trigonal to tetrahedral, forming intermediate [II]. Proton abstraction by the acidic residue (step 3) produces the unstable intermediate [III] that decomposes (step 4), resulting in non-covalently bound all-trans retinal, intermediate [IV]. Finally, the all-trans chromophore can dissociate from the protein (step 5), and leaving the apoprotein with a deprotonated Glu113/Asp108 ion-paired with Lys291.
Fig. 6.
Left, Three-dimension models of selected amino acids in the binding pockets in rhodopsin (RHO) and VCOP were prepared using PyMOL[68]. Structural water has been omitted for clarity. Amino acid numbering is based upon individual protein sequences. The primary counterion is E113 for rhodopsin and D108 for VCOP. Right, The proposed mechanism for Schiff base hydrolysis and retinal release. Two shaded conformations have been established experimentally, while the proposed intermediates are numbered I-IV. See Discussion for more details.
Experiments presented in this paper support the proposed mechanism. The rate of retinal release in VCOP was increased by elevated pH, favoring meta II conformation. Since the cone opsin has a more open chromophore environment [46], acidic conditions may retard the proton transfer and formation of [I] in step 1 and/or the proton transfer in step 3, leading to a slower retinal release. This also supports the idea that there is rapid exchange of solvent into the binding pocket, compared to rhodopsin. A kinetic isotope effect has been observed for rhodopsin and model Schiff base compounds [63, 64]. However, we were unable to observe a similar effect for VCOP, finding a ratio of <1.2-fold compared to the 2.3-2.5-fold observed for rhodopsin. While we can not rule out the existence of non-exchangeable water molecules bound in the retinal binding pocket of VCOP, the sensitivity of the pigment to hydrolysis in the dark (by hydroxylamine and more slowly by water suggests this is not likely. It is important to note that the rates of retinal release for VCOP are much faster than rhodopsin, and this may make it more difficult to observe an isotope effect. The Schiff base in VCOP may not readily reform after hydrolysis, since it leaves the binding pocket so quickly. In rhodopsin, a substantial reverse reaction apparently occurs [60], possibly due to sterically unfavorable conditions for release [63]. Moreover, hydrolysis of model Schiff compounds in detergent were studied under equilibirum conditions [60]. The similar activation energies, sequence homology and catalytic role of the counterion all suggest that the catalytic mechanism of hydrolysis is quite similar in VCOP and rhodopsin. Thus, the lack of an observable kinetic isotope effect may reflect the absence of a significant reverse (Schiff base reforming) step in cone pigments due to the rapid dissociation of retinal from the protein, compared to rhodopsin.
The primary counterion is necessary for rapid retinal release, as VCOPD108A has very retarded kinetics. Substitution of an Asp in place of Ser85 restores near normal retinal release in the counterion mutant. Given the proximity of these residues to the Schiff base (Fig. 6) in the homology models, these results indicate that either the proton donor in step 1 or proton acceptor in step 3 (or both) is the primary counterion. Although It is possible that two different amino acid side chains participate in each of these steps, the more likely scenario, given the behavior of VCOPS85D/D108A, is that the primary counterion is the catalytically relevant residue as illustrated in Fig. 6. Our homology model places Asp85 2.6 Å deeper in the transmembrane bundle than Ala108, locating the counterion in a more hydrophobic environment. This new environment would expel solvent more strongly, and potentially stabilize the pK’ of the counterion thus rendering it less pH-sensitive. Additional support for the dual role of the counterion comes from the additional restraints imposed upon water molecules by both Asp108 and Asp85 (in VCOPS85D). This mutant would require more free energy to alter the structure during Schiff base hydrolysis, leading to a slower rate of reaction in step 1. This is in fact observed in the retinal release measurements (Fig. 3 and Table 1).
A remarkable prediction of this model is that the main catalytic power for Schiff base hydrolysis resides in the protonation of the counterion for both rod and cone visual pigments. When the primary counterion is neutralized, the reaction in step 1 is drastically slowed down due to the absence of a proton donor. In such counterion mutants, the hydrolysis of the Schiff base is still mediated through a water. However, in a hydrophobic environment, the protonation of the Schiff base is not favorable and thus formation of the carbinolamine intermediate would be very slow, leading to a very slow hydrolysis rate. Since the formation of a Schiff base is expected to proceed by the reverse of the same reaction, reformation would also be expected to be significantly reduced in the counterion mutants. Thus, the reformation of Meta II from noncovalently bound retinal in rhodopsin would not occur, and the proposed kinetic trap [63] would be removed. Thus, we predict that the rate limiting step in retinal release for both rod and cone pigments in the absence of a primary counterion, would become Schiff base hydrolysis. The slow rate of hydrolysis can be dramatically accelerated by hydroxylamine which protonates the deprotonated Schiff base to initiate the nucleophilic attack in step 1. Deprotonated hydroxylamine absorbs the proton from the unstable carbinolamine intermediate, and completes step 2, and step 3. This is supported in the retinal release experiments described here (Fig. 5).
Conclusions
The primary counterion stabilizes the protonated Schiff base linkage in dark-adapted rod and cone visual pigments but is essential for normal Schiff base hydrolysis. Visual pigments without a counterion have extremely low rates of retinal release. Thus, even UV sensitive visual pigments which have an unprotonated Schiff base [39, 65] and thus do not require a counterion to neutralize the retinylidene linkage, still require one for Schiff base hydrolysis and retinal release. Presumably the reverse reaction, also known as pigment generation,will proceed through the reverse sequence of steps (Fig. 6) and thus also require a primary counterion for efficient pigment formation. These experiments provide support for the primary role of the counterion at position 113 (rhodopsin numbering) in Schiff base hydrolysis-retinal release, and presumably in pigment formation in all vertebrate visual pigments. These results also are consistent with a number of previous reports on the primary role of the counterion in retinal-Schiff base chemistry [63, 66, 67]. Furthermore, our results provide evidence for the original suggestion that the counterion in bovine rhodopsin was necessary for efficient Schiff base hydrolysis [52]. The similar energetics of the Schiff base hydrolysis and catalytic mechanism for both rod and cone visual pigments strongly support the hypothesis that the intrinsic differences in retinal release between light-activated rod and cone pigments resides in the different dissociation rates for all-trans retinal. This may be due to differences in chromophore interactions with retinal binding pocket side chains or in the accessibility of non-covalently bound chromophore to pass through the seven transhelical bundle. Finally, we note that these experiments were performed in dodecy maltoside solutions, not in the native outer segment membrane. Future experiments should be directed toward examining how the native membrane could influence the release of all-trans retinal or the binding of 11-cis retinal to form visual pigment.
Supplementary Material
Acknowledgements
We thank K. Babu for helpful advice on pigment expression and B. Hajjari for providing stimulating discussions.
This work was supported in part by the National Institutes of Health grants EY-11256 and EY-12975 to B.E.K., and GM-34548 to R.R.B., Research to Prevent Blindness (Unrestricted Grant to SUNY UMU Department of Ophthalmology), the Harold S. Schwenk Sr. Distinguished Chair funds for support of specialized instrumentation at the University of Connecticut and the Lions of CNY.
Abbreviations
- VCOP
Xenopus short-wavelength sensitive cone pigment
- Ea
Arrhennius activation energy
Footnotes
Supplemental Information
The kinetics of fluorescence increase following illumination are presented for additional mutants VCOPD108E and VCOPS85C that alter the retinal Schiff base environment. The supplemental material may be accessed free of charge online at http://pubs.acs.org.
References
- 1.Rodieck RW. The First Steps in Seeing. 1st edition Sinauer Associates, Inc.; Sunderland, MA: 1998. p. 562. [Google Scholar]
- 2.Lamb TD, Pugh EN. Dark adaptation and the retinoid cycle of vision. Prog. Retinal Eye Res. 2004;23:307–380. doi: 10.1016/j.preteyeres.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Lamb TD, Pugh EN. Phototransduction, dark adaptation, and rhodopsin regeneration the proctor lecture. Invest. Ophthalmol. Vis. Sci. 2006;47:5137–5152. doi: 10.1167/iovs.06-0849. [DOI] [PubMed] [Google Scholar]
- 4.Naarendorp F, Esdaille TM, Banden SM, Andrews-Labenski J, Gross OP, Pugh EN. Dark light, rod saturation, and the absolute and incremental sensitivity of mouse cone vision. J. Neurosci. 2010;30:12495–12507. doi: 10.1523/JNEUROSCI.2186-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakmar TP, Menon ST, Marin EP, Awad ES. Rhodopsin: insights from recent structural studies. Annu. Rev. Biophys. Biomol. Struct. 2002;31:443–484. doi: 10.1146/annurev.biophys.31.082901.134348. [DOI] [PubMed] [Google Scholar]
- 6.Palczewski K. G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yokoyama S. Molecular evolution of vertebrate visual pigments. Prog. Retinal Eye Res. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- 8.Okada T, Sugihara M, Bondar A-N, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 9.Choe H-W, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 10.Park JH, Scheerer P, Hofmann KP, Choe H-W, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 11.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe H-W, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 12.Ruprecht JJ, Mielke T, Vogel R, Villa C, Schertler GFX. Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 2004;23:3609–3620. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith SO. Structure and Activation of the Visual Pigment Rhodopsin. Annu. Rev. Biophys. 2010;39:1–23. doi: 10.1146/annurev-biophys-101209-104901. [DOI] [PubMed] [Google Scholar]
- 14.Birge RR. Nature of the primary photochemical events in rhodopsin and bacteriorhodopsin. Biochim. Biophys. Acta. 1990;1016:293–327. doi: 10.1016/0005-2728(90)90163-x. [DOI] [PubMed] [Google Scholar]
- 15.Kukura P, McCamant DW, Yoon S, Wandschneider DB, Mathies RA. Structural observation of the primary isomerization in vision with femtosecond-stimulated Raman. Science. 2005;310:1006–1009. doi: 10.1126/science.1118379. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Schoenlein RW, Peteanu LA, Mathies RA, Shank CV. Vibrationally coherent photochemistry in the femtosecond primary event of vision. Science. 1994;266:422–424. doi: 10.1126/science.7939680. [DOI] [PubMed] [Google Scholar]
- 17.Nakamichi H, Buss V, Okada T. Photoisomerization mechanism of rhodopsin and 9-cis-rhodopsin revealed by x-ray crystallography. Biophys J: Biophys Lett. 2007;92:L106–L108. doi: 10.1529/biophysj.107.108225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shukla AK, Sun J-P, Lefkowitz RJ. Crystallizing thinking about the β2-adrenergic receptor. Mol. Pharm. 2008;73:1333–1338. doi: 10.1124/mol.108.045849. [DOI] [PubMed] [Google Scholar]
- 19.Hunt DM, Carvalho LS, Cowing JA, Parry JWL, Wilkie SE, Davies WL, Bowmaker JK. Spectral tuning of shortwave-sensitive visual pigments in vertebrates. Photochem. Photobiol. 2007;83:303–310. doi: 10.1562/2006-06-27-IR-952. [DOI] [PubMed] [Google Scholar]
- 20.Amora TL, Ramos LS, Galan JF, Birge RR. Spectral tuning of deep red cone pigments. Biochemistry. 2008;47:4614–4620. doi: 10.1021/bi702069d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsutsui K, Shichida Y. Multiple functions of Schiff base counterion in rhodopsins. Photochem. Photobiol. Sci. 2010;9:1426–1434. doi: 10.1039/c0pp00134a. [DOI] [PubMed] [Google Scholar]
- 22.Andersen LH, Nielsen IB, Kristensen MB, El Ghazaly MOA, Haacke S, Nielsen MB, Petersen MA. Absorption of schiff-base retinal chromophores in vacuo. J. Am. Chem. Soc. 2005;127:12347–12350. doi: 10.1021/ja051638j. [DOI] [PubMed] [Google Scholar]
- 23.Kusnetzow A, Dukkipati A, Babu KR, Singh D, Vought BW, Knox BE, Birge RR. The photobleaching sequence of a short-wavelength visual pigment. Biochemistry. 2001;40:7832–7844. doi: 10.1021/bi010387y. [DOI] [PubMed] [Google Scholar]
- 24.Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo D-G, Yue WWS, Ala-Laurila P, Yau K-W. Activation of visual pigments by light and heat. Science. 2011;332:1307–1312. doi: 10.1126/science.1200172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kefalov V, Fu Y, Marsh-Armstrong N, Yau K-W. Role of visual pigment properties in rod and cone phototransduction. Nature. 2003;425:526–531. doi: 10.1038/nature01992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Y, Kefalov V, Luo D-G, Xue T, Yau K-W. Quantal noise from human red cone pigment. Nat. Neurosci. 2008;11:565–571. doi: 10.1038/nn.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wald G. Molecular basis of visual excitation. Science. 1968;162:230–239. doi: 10.1126/science.162.3850.230. [DOI] [PubMed] [Google Scholar]
- 29.Blazynski C, Ostroy SE. Pathways in the hydrolysis of vertebrate rhodopsin. Vision Res. 1984;24:459–470. doi: 10.1016/0042-6989(84)90043-9. [DOI] [PubMed] [Google Scholar]
- 30.Imai H, Kefalov V, Sakurai K, Chisaka O, Ueda Y, Onishi A, Morizumi T, Fu Y, Ichikawa K, Nakatani K, Honda Y, Chen J, Yau K-W, Shichida Y. Molecular properties of rhodopsin and rod function. J. Biol. Chem. 2007;282:6677–6684. doi: 10.1074/jbc.M610086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakurai K, Onishi A, Imai H, Chisaka O, Ueda Y, Usukura J, Nakatani K, Shichida Y. Physiological properties of rod photoreceptor cells in green-sensitive cone pigment knock-in mice. J. Gen. Physiol. 2007;130:21–40. doi: 10.1085/jgp.200609729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burns ME, Pugh EN. Lessons from Photoreceptors: Turning Off G-Protein Signaling in Living Cells. Physiology. 2010;25:72–84. doi: 10.1152/physiol.00001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tachibanaki S, Shimauchi-Matsukawa Y, Arinobu D, Kawamura S. Molecular Mechanisms Characterizing Cone Photoresponses. Photochem. Photobiol. 2007;83:19–26. doi: 10.1562/2006-02-28-IR-823. [DOI] [PubMed] [Google Scholar]
- 34.Imai H, Kuwayama S, Onishi A, Morizumi T, Chisaka O, Shichida Y. Molecular properties of rod and cone visual pigments from purified chicken cone pigments to mouse rhodopsin in situ. Photochem. Photobiol. Sci. 2005;4:667–674. doi: 10.1039/b416731g. [DOI] [PubMed] [Google Scholar]
- 35.Perry RJ, McNaughton PA. Response properties of cones from the retina of the tiger salamander. J. Physiol. (Lond) 1991;433:561–587. doi: 10.1113/jphysiol.1991.sp018444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kefalov VJ, Estevez ME, Kono M, Goletz PW, Crouch RK, Cornwall MC, Yau K-W. Breaking the Covalent Bond— A Pigment Property that Contributes to Desensitization in Cones. Neuron. 2005;46:879–890. doi: 10.1016/j.neuron.2005.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shichida Y, Imai H, Imamoto Y, Fukada Y, Yoshizawa T. Is chicken green-sensitive cone visual pigment a rhodopsin-like pigment? A comparative study of the molecular properties between chicken green and rhodopsin. Biochemistry. 1994;33:9040–9044. doi: 10.1021/bi00197a002. [DOI] [PubMed] [Google Scholar]
- 38.Babu KR, Dukkipati A, Birge RR, Knox BE. Regulation of phototransduction in short-wavelength cone visual pigments via the retinylidene Schiff base counterion. Biochemistry. 2001;40:13760–13766. doi: 10.1021/bi015584b. [DOI] [PubMed] [Google Scholar]
- 39.Kusnetzow AK, Dukkipati A, Babu KR, Ramos L, Knox BE, Birge RR. Vertebrate ultraviolet visual pigments: protonation of the retinylidene Schiff base and a counterion switch during photoactivation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:941–946. doi: 10.1073/pnas.0305206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuwayama S, Imai H, Hirano T, Terakita A, Shichida Y. Conserved proline residue at position 189 in cone visual pigments as a determinant of molecular properties different from rhodopsins. Biochemistry. 2002;41:15245–15252. doi: 10.1021/bi026444k. [DOI] [PubMed] [Google Scholar]
- 41.Farrens DL, Khorana HG. Structure and function in rhodopsin. Measurement of the rate of metarhodopsin II decay by fluorescence spectroscopy. J Biol Chem. 1995;270:5073–5076. doi: 10.1074/jbc.270.10.5073. [DOI] [PubMed] [Google Scholar]
- 42.Janz JM, Fay JF, Farrens DL. Stability of dark state rhodopsin is mediated by a conserved ion pair in intradiscal loop E-2. J. Biol. Chem. 2003;278:16982–16991. doi: 10.1074/jbc.M210567200. [DOI] [PubMed] [Google Scholar]
- 43.Chen M-H, Sandberg DJ, Babu KR, Bubis J, Surya A, Ramos LS, Zapata HJ, Galan JF, Sandberg MN, Birge RR, Knox BE. Conserved Residues in the Extracellular Loops of Short-Wavelength Cone Visual Pigments. Biochemistry. 2011;50:6763–6773. doi: 10.1021/bi101557m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandberg MN, Amora TL, Ramos LS, Chen M-H, Knox BE, Birge RR. Glutamic Acid 181 Is Negatively Charged in the Bathorhodopsin Photointermediate of Visual Rhodopsin. J. Am. Chem. Soc. 2011;133:2808–2811. doi: 10.1021/ja1094183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramos LS, Chen M-H, Knox BE, Birge RR. Regulation of photoactivation in vertebrate short wavelength visual pigments: protonation of the retinylidene Schiff base and a counterion switch. Biochemistry. 2007;46:5330–5340. doi: 10.1021/bi700138g. [DOI] [PubMed] [Google Scholar]
- 46.Starace DM, Knox BE. Cloning and expression of a Xenopus short wavelength cone pigment. Exp. Eye Res. 1998;67:209–220. doi: 10.1006/exer.1998.0507. [DOI] [PubMed] [Google Scholar]
- 47.Starace DM, Knox BE. Activation of transducin by a Xenopus short wavelength visual pigment. J. Biol. Chem. 272:1095–1100. doi: 10.1074/jbc.272.2.1095. [DOI] [PubMed] [Google Scholar]
- 48.Dukkipati A, Vought BW, Singh D, Birge RR, Knox BE. Serine 85 in Transmembrane Helix 2 of Short-Wavelength Visual Pigments Interacts with the Retinylidene Schiff Base Counterion. Biochemistry. 2001;40:15098–15108. doi: 10.1021/bi011354l. [DOI] [PubMed] [Google Scholar]
- 49.Zvyaga TA, Min KC, Beck M, Sakmar TP. Movement of the retinylidene Schiff base counterion in rhodopsin by one helix turn reverses the pH dependence of the metarhodopsin I to metarhodopsin II transition. J. Biol. Chem. 1993;268:4661–4667. [PubMed] [Google Scholar]
- 50.Zvyaga T, Fahmy K, Sakmar T. Characterization of rhodopsin-transducin interaction: a mutant rhodopsin photoproduct with a protonated Schiff base activates transducin. Biochemistry. 1994;33:9753–9761. doi: 10.1021/bi00198a046. [DOI] [PubMed] [Google Scholar]
- 51.Honig B, Greenberg A, Dinur U, Ebrey T. Visual-pigment spectra: implications of the protonation of the retinal Schiff base. Biochemistry. 1976;15:4593–4599. doi: 10.1021/bi00666a008. [DOI] [PubMed] [Google Scholar]
- 52.Sakmar TP, Franke RR, Khorana HG. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nathans J. Determinants of visual pigment absorbance: identification of the retinylidene Schiff's base counterion in bovine rhodopsin. Biochemistry. 1990;29:9746–9752. doi: 10.1021/bi00493a034. [DOI] [PubMed] [Google Scholar]
- 54.Zhukovsky EA, Oprian DD. Effect of carboxylic acid side chains on the absorption maximum of visual pigments. Science. 1989;246:928–930. doi: 10.1126/science.2573154. [DOI] [PubMed] [Google Scholar]
- 55.Golobokova EY, Govardovskii VI. Late stages of visual pigment photolysis in situ: cones vs. rods. Vision Res. 2006;46:2287–2297. doi: 10.1016/j.visres.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 56.Arshavsky VY, Lamb TD, Pugh EN. G proteins and phototransduction. Annu. Rev. Physiol. 2002;64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- 57.Imai H, Kojima D, Oura T, Tachibanaki S, Terakita A, Shichida Y. Single amino acid residue as a functional determinant of rod and cone visual pigments. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2322–2326. doi: 10.1073/pnas.94.6.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imai H, Imamoto Y, Yoshizawa T, Shichida Y. Difference in molecular properties between chicken green and rhodopsin as related to the functional difference between cone and rod photoreceptor cells. Biochemistry. 1995;34:10525–10531. doi: 10.1021/bi00033a026. [DOI] [PubMed] [Google Scholar]
- 59.Rushton WA. Rod/cone rivalry in pigment regeneration. J. Physiol. (Lond) 1968;198:219–236. doi: 10.1113/jphysiol.1968.sp008603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cooper A, Dixon S, Nutley M, Robb J. Mechanism of retinal Schiff base formation and hydrolysis in relation to visual pigment photolysis and regeneration: resonance Raman spectroscopy of a tetrahedral carbinolamine intermediate and oxygen-18 labeling of retinal at the metarhodopsin stage in photoreceptor membranes. J. Am. Chem. Soc. 1987;109:7254–7263. [Google Scholar]
- 61.Szundi I, Mah TL, Lewis JW, Jäger S, Ernst OP, Hofmann KP, Kliger DS. Proton transfer reactions linked to rhodopsin activation. Biochemistry. 1998;37:14237–14244. doi: 10.1021/bi981249k. [DOI] [PubMed] [Google Scholar]
- 62.Vogel R, Siebert F, Zhang X-Y, Fan G, Sheves M. Formation of Meta III during the decay of activated rhodopsin proceeds via Meta I and not via Meta II. Biochemistry. 2004;43:9457–9466. doi: 10.1021/bi049337u. [DOI] [PubMed] [Google Scholar]
- 63.Janz JM, Farrens DL. Role of the retinal hydrogen bond network in rhodopsin Schiff base stability and hydrolysis. J. Biol. Chem. 2004;279:55886–55894. doi: 10.1074/jbc.M408766200. [DOI] [PubMed] [Google Scholar]
- 64.Oseroff AR, Callendar RH. Resonance Raman spectroscopy of rhodopsin in retinal disk membranes. Biochemistry. 1974;13:4243–4248. doi: 10.1021/bi00717a027. [DOI] [PubMed] [Google Scholar]
- 65.Dukkipati A, Kusnetzow A, Babu KR, Ramos L, Singh D, Knox BE, Birge RR. Phototransduction by vertebrate ultraviolet visual pigments: protonation of the retinylidene Schiff base following photobleaching. Biochemistry. 2002;41:9842–9851. doi: 10.1021/bi025883g. [DOI] [PubMed] [Google Scholar]
- 66.Gross AK, Rao VR, Oprian DD. Characterization of rhodopsin congenital night blindness mutant T94I. Biochemistry. 2003;42:2009–2015. doi: 10.1021/bi020613j. [DOI] [PubMed] [Google Scholar]
- 67.Janz JM, Farrens DL. Assessing structural elements that influence Schiff base stability: mutants E113Q and D190N destabilize rhodopsin through different mechanisms. Vision Res. 2003;43:2991–3002. doi: 10.1016/j.visres.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 68.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.