Abstract
Although microRNA-135b (miR-135b) is known to be associated with cancer, with recent work showing that it is massively induced in the pulmonary tissues of mice challenged with nanoparticles suggests a critical role for this microRNA in mediating inflammatory response. In this study, we investigated the expression and function of miR-135b in mice exposed to cigarette smoke or nontypeable Haemophilus influenzae (NTHi). Exposure to both cigarette smoke and NTHi elicited robust lung inflammation, but increased miR-135b expression was observed only in the lungs of cigarette smoke–exposed mice. Using IL-1R 1 knockout mice, we show that miR-135b expression is IL-1R1 dependent. A series of in vitro experiments confirmed the role of IL-1R1 in regulating miR-135b expression. In vitro activation of the IL-1R1 pathway in mouse embryonic fibroblast (NIH3T3) and lung epithelial (FE1) cells resulted in increased miR-135b, which was blocked by IL-1R1 antagonists or small interfering RNA–mediated silencing of IL-1R1 expression. Overexpression of mature miR-135b in NIH3T3 cells (pEGP-mmu-mir-135b) resulted in the suppression of endogenous levels of IL-1R1 expression. pEGP-mmu-miR-135b cells transiently transfected with luciferase reporter vector containing the 3′UTR of mouse IL-1R1 showed reduced luciferase activity. Finally, we demonstrate that miR-135b targets IL-1–stimulated activation of Caspase-1, the IL-1R1 downstream activator of IL-1β leading to suppressed synthesis of the active form of IL-1β protein. These results suggest that miR-135b expression during cigarette smoke–induced inflammation is regulated by IL-1R1 in a regulatory feedback mechanism to resolve inflammation.
Introduction
The lungs are continuously exposed to a range of infectious and noninfectious environmental agents. A complex array of innate and adaptive immune inflammatory processes protects the host against these potentially harmful agents. These defense mechanisms are mediated through the coordinated activation and repression of several cytokines, chemokines, matrix proteases, and others. Accumulating evidence suggests a critical role for microRNAs (miRNAs) in regulating this response (reviewed in Ref. 1). Mature miRNA are short (20–25 nucleotides), noncoding, single-stranded RNA that directly regulate the expression of protein-coding genes by RNA interference-mediated mRNA degradation or by translational inhibition. miRNA can also target global methylation or transcription factors, leading to indirect regulation of transcription (reviewed in Ref. 2). Physiologic processes that are mediated by miRNA include cellular development, differentiation, growth and proliferation, and cell death. Deregulated miRNA expression in response to endogenous or external stimuli can lead to widespread gene expression changes, which in turn is shown to affect normal cellular function (3). Aberrant miRNA expression is associated with pathologic conditions, such as cardiovascular disorders (4), lung injury (5), inflammatory diseases (6), and cancer (7).
We recently reported a significant increase in the expression of several miRNA in mice following pulmonary exposure to nanosized titanium dioxide particles (8) and carbon black particles (9). Among those significantly altered, miR-135b was the most upregulated (>60- and 40-fold above nonexposed controls for the nanosized titanium dioxide and carbon black models, respectively). Particle-exposed mice exhibited significant changes in the expression of inflammatory cytokines and chemokines; however, mRNA levels of known or predicted miR-135b targets were not affected (8, 9).
In the current study, we used in vivo and in vitro models of inflammation to investigate whether miR-135b expression is associated with specific types of inflammation, and we identified direct targets of miR-135b during inflammation. We studied induction of miR-135b following exposure to mainstream cigarette smoke or inoculation with nontypeable Haemophilus influenzae (NTHi) in both standard inbred laboratory strains and IL-1R1 knockout mice. Because IL-1R1 mediated–signaling is involved in modulating particle-induced (10–12) and cigarette smoke–induced inflammation (13, 14), miR-135b expression in response to the IL-1R1 agonist IL-1α was investigated in mouse embryonic fibroblast (NIH3T3) and mouse lung epithelial (FE1) cell lines. The effects of miR-135b overexpression on IL-1R1 agonist-induced inflammatory response were explored, and direct targets of miR-135b during inflammation were identified.
Materials and Methods
Materials
DMEM:Nutrient Mixture F-12, bovine serum, FBS, penicillin-streptomycin, puromycin, and TRIzol reagents were purchased from Invitrogen (Carlsbad, CA). Mouse NIH3T3 cells were obtained from Dr. Mike Wade (Health Canada, Ottawa, ON, Canada). The FE1 cell line is derived from MutaMouse lung epithelial cells (15).
mirVana miRNA isolation kits were purchased from Applied Biosystems (Carlsbad, CA). The pEGP-mir–null control vector and pEGP-mmu-mir-135b expression vector were purchased from Cell Biolabs (San Diego, CA). Fugene 6 transfection reagent was purchased from Roche (Laval, QC, Canada). Accell Smart Pool IL-1R1 and Accell Nontargeting Pool were purchased from Thermo Fisher Scientific (Lafayette, CO). The pLuc-3′UTR vector, pIL-1R1-3′UTR-Luc and pCaspase-1-3′UTR-Luc target reporter vectors were purchased from Signosis (Sunnyvale, CA). QuickChange II Site-Directed Mutagenesis Kit was obtained from Agilent (Mississauga, ON, Canada). The Dual-Luciferase Reporter Assay System was obtained from Promega (Madison, WI). RNeasy Mini Kit and miScript PCR System were purchased from Qiagen (Mississauga, ON, Canada). RT2 First Strand Kit, RT2 SYBR Green PCR Master Mix and Mouse Inflammatory Cytokines and Receptors/Mouse TLR Signaling Pathway PCR Arrays were from SABiosciences (Frederick, MD).
Murine epidermal growth factor was purchased from Sigma (St. Louis, MO). Bradford protein assay kit, Bio-Plex Cell Lysis Kit and Bio-Plex Pro Mouse Cytokine Assays were purchased from Bio-Rad (Mississauga, ON, Canada). Mouse IL-1R1 agonists IL-1α, IL-1β, antagonist IL-1R1a, and the IL-1β and Cxcl12 Immunoassays were purchased from R&D Systems (Minneapolis, MN). Anti–Caspase-1 Ab was purchased from Abcam (Cambridge, MA). Anti β-actin Ab was acquired from Cell Signaling (Danvers, MA), anti-GFP Ab from Clontech (Mountain View, CA) and all other Abs were obtained from Santa Cruz (Santa Cruz, CA).
Animal exposure
Female BALB/c mice (6–8 wk old) were purchased from Charles River Laboratories (Montreal, QC, Canada). IL-1R1 knockout mice (C57BL/6 background) were purchased from the Jackson Laboratories (Bar Harbor, ME). All mice were kept under a 12-h light–dark cycle in autoclaved cages and bedding, with unlimited access to autoclaved food and water. All doses and time points for the exposure were chosen based on previous experiments (16, 17). Mice were exposed to cigarette smoke using the SIU-48 system (Promech Lab, Vintrie, Sweden). Mice were acclimatized for 3 d prior to the exposure. Mice were exposed to 12 3R4F reference cigarettes (Tobacco and Health Research Institute, University of Kentucky, Lexington, KY) without filters for 50 min, twice daily, for a total of 4 d and euthanized 3–4 h after exposure. Control mice were exposed to room air only. For NTHi infection, C57BL/6 mice were challenged intranasally with 106 CFU of NTHi and euthanized 12 h after infection. Control mice were exposed to PBS. Animals were anesthetized with isoflurane and euthanized by exsanguination. Animal exposures and sample collection followed the guidelines of the Canadian Council on Animal care, and procedures and were approved by the McMaster University Animal Research Ethics Board.
Bronchoalveolar lavage and differential cell counting
Bronchoalveolar lavage (BAL) fluid was collected by instilling the left lobe of the lungs with 0.25 ml of ice-cold 1× PBS, followed by 0.2 ml of 1× PBS. Total cell numbers were counted using a hematocytometer. Cytospins were prepared for differential cell counts and stained with Hema 3 (Biochemical Sciences). A total of 500 cells were counted per cytospin to identify mononuclear cells, and neutrophils (16).
Tissue processing
The left lobe of the lungs was snap frozen in liquid nitrogen and stored at −80°C. For molecular analysis, the frozen left lung lobe was sliced randomly into several sections and used for RNA extraction and miRNA extraction.
Total RNA and miRNA extraction from tissues
Total RNA was isolated from the lung sections (n = 3–5 per group) using TRIzol reagent and purified using RNeasy Mini Kit. The mirVana miRNA Isolation Kit was used to prepare total RNA enriched with small RNA species from randomly selected left lung sections. RNA quality was confirmed by ultraviolet spectrophotometry and using an Agilent bioanalyzer (Agilent Technologies).
Cell culture conditions
NIH3T3 cells or FE1 cells were cultured in DMEM containing 10% bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin or 1:1 DMEM:F12 nutrient mixture supplemented with 2% FBS, 100 U/ml penicillin G, 100 μg/ml streptomycin sulfate, and 1 ng/ml murine epidermal growth factor and incubated at 37°C, 95% humidity, and 5% CO2. Approximately 1.6 × 105 cells were seeded on six-well polystyrene culture plates and incubated overnight. To induce an inflammatory response, cells were treated with 10 ng/ml IL-1α and collected 1 h later. IL-1R1 inhibition experiments were conducted by pretreating cells with 2 μg/ml IL-1R1a for 16 h before adding IL-1α. Cells were washed twice with PBS, trypsinized, and centrifuged at 1000 rpm for 5 min to collect the cell pellets.
For small interfering RNA (siRNA) experiments, 25,000 (NIH3T3) cells were plated on 12-well dishes and incubated overnight at 37°C with 5% CO2. The next day, cells were transiently transfected with Accell Smart Pool IL-1R1 and Accell Nontargeting Pool (negative control) according to the manufacturer’s protocol. Cells were treated with IL-1α 72 h after the transfection. Total RNA was isolated from the cell lysates as described above, and IL-1R1 mRNA and miR-135b expression were analyzed.
Establishment of NIH3T3 cells stably expressing mmu-miR-135b or mir-135b-mutant
NIH3T3 cells were stably transfected with either pEGP-mmu-miR-135b or pEGP-null expression vector using Fugene 6 transfection reagent. Surviving clones were selected with puromycin. To create a mutant miR-135b, QuickChange II site-directed mutagenesis kits (forward primer: 5′-CTCTGCTGTGGCCTTCATTCCTATGTG-3′; and reverse primer: 5′-CACATAGGAATGAAGGCCACAGCAGAG-3′) were used to selectively delete 8 bp from the seed region of miR-135b contained in pEGP-mmu-miR-135b. The resultant PCR product was transformed into XL1-Blue super competent cells. Surviving clones were sequenced to confirm the deletion mutants. NIH3T3 cells expressing pEGP-mmu-miR-135b mutants were established as described above.
Real-time PCR
The Qiagen miScript PCR system was used to analyze miR-135b expression in all samples. For each sample (n = 3–5 per group), 1 μg of total RNA enriched with small RNA species was polyadenylated and then converted to cDNA using an oligo(dT) primer with a universal tag and miScript Reverse Transcription mix. Real-time (RT-PCR) was performed in duplicate for each sample, using a primer complementary to the universal tag and a miScript primer specific for miR-135b. Amplified product was detected using SYBR Green and a CFX real-time detection system. Expression levels of miR-135b were normalized to expression levels of small nuclear RNAs RNU1A1. A Student t test was used to test for statistical significance.
Pathway-specific PCR arrays
Approximately 800 ng of total RNA per sample (n = 3 per group) was reverse transcribed using an RT2 First Strand Kit. Reverse transcription and RT-PCRs were performed using RT2 SYBR Green PCR Master Mix in 96-well PCR arrays designed for the evaluation of mouse inflammatory cytokines and receptors (no. PAMM-011D; SABiosciences) using a CFX real-time detection system. Threshold cycle values were averaged. Relative gene expression was determined according to the comparative Ct method and normalized to the Hprt and β-actin housekeeping genes. Fold changes and statistical significance were calculated using online PCR array data analysis software (SABiosciences). All RT-PCR data are deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus database under accession number GSE43688 and can be accessed via http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43688.
Western blot and ELISA analysis
Protein extracts were prepared from control and treated cells using a Bio-Plex Cell Lysis Kit and quantified using a Bradford Protein Assay Kit. For Western blotting, 30 μg of total protein was immunoblotted on a 12% SDS-PAGE gel and analyzed using Abs against Caspase-1 and β-actin (housekeeping protein used for normalizing). Signals were detected using ECL plus reagent. Band intensities were determined by averaging the densitometric readings from three independent experiments. Actin levels in the samples were used to normalize Caspase-1 levels.
An IL-1β immunoassay was used to measure total IL-1 β in cellular lysates according to the manufacturer’s protocols (R&D Systems). Briefly, 50 μl of prediluted standards or diluted cell lysates (100 μg) were loaded onto a microplate precoated with mouse IL-1β Ab and incubated for 2 h. The unbound IL-1β was removed by washing and 100 μl IL-1β Ab was added to each well and incubated at 500 rpm for 2 h at room temperature. Plates were washed, and 100–200 μl of substrate was added to each well and incubated in the dark for 30 min. The reaction was stopped by adding 50–100 μl of stop solution. The OD for each well was determined at 450 nm using a microtiter plate spectrophotometer with the correction wavelength at 540 or 570 nm.
Luciferase reporter assay
Luciferase reporter assays were performed using the pLuc-3′UTR vector alone, pIL-1R1-3′UTR-Luc or pCaspase-1-3′UTR-Luc. pEGP-mir–null, pEGP-miR-135b, and pEGP-miR-135b mutant cells were transiently transfected with 100 ng of reporter constructs together with 25 ng pRL-CMV (an internal control reporter vector used to cotransfect cells) and 375 ng ssDNA. Cells were harvested 48 h after transfection and assayed for reporter activity using the Dual-Luciferase Reporter Assay System.
Results
BAL cellular profiles and expression of miR-135b in lungs following exposure to mainstream cigarette smoke or NTHi inoculation
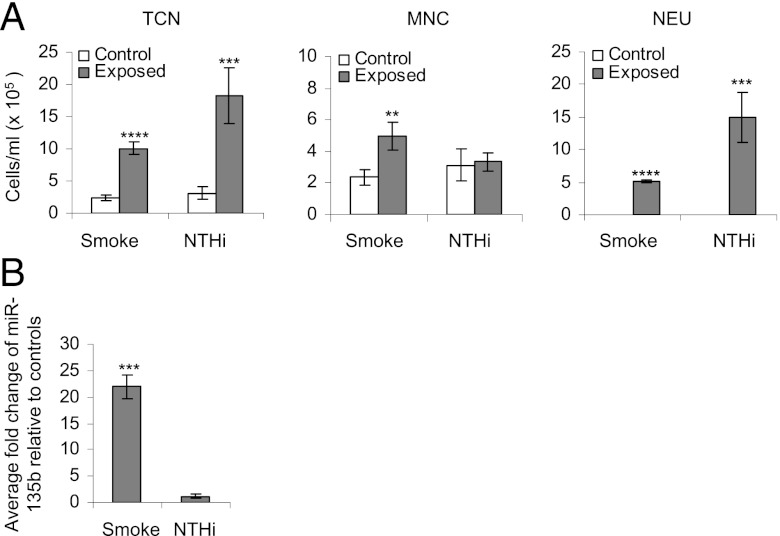
BAL cellular profiles were investigated in BALB/c mice that were exposed to cigarette smoke for 4 d or inoculated with NTHi for 12 h alongside sham-treated controls (exposed to room air). Sham-treated mice did not exhibit any inflammation (Fig. 1A). Total cellular count increased significantly in mice exposed to either cigarette smoke or NTHi. Inflammation in cigarette smoke–exposed mice was characterized by a significant increase in both mononuclear cells and neutrophils, whereas neutrophilia were predominant in NTHi-treated mice.
FIGURE 1.
(A) Bronchoalveolar lavage cellular profiles. Open bars (controls), gray bars (exposed to 4 d cigarette smoke or inoculated with NTHi). The average of five mice is presented. Statistical analysis was performed using a two-way ANOVA with a Bonferroni post hoc test. **p < 0.005, ***p < 0.0005, ****p < 0.00005. (B) RT-PCR validation of miR-135b in lung tissues exposed to cigarette smoke for 4 d or inoculated with NTHi. Data are presented as average fold changes over matched controls (n = 5 per group ± SEM). Expression values were normalized to small nuclear RNA RNUA1. ***p < 0.05, Student t test. MNC, Mononuclear cells; NEU, neutrophils; TCN, total cell number.
A significant 22-fold increase in miR-135b levels was observed in the lungs of BALB/c mice exposed to cigarette smoke compared with matched controls (Fig. 1B). MiR-135b expression was unaffected in the lungs of mice inoculated with NTHi (Fig. 1B). To confirm that the observed lack of miR-135b expression in NTHi-inoculated lungs is not simply due to differences in timing or duration of exposure, we exposed mice to heat-inactivated NTHi for 12 h as described above, or on 4 consecutive days to match the time of cigarette smoke exposure (Supplemental Fig. 1). Comparatively less inflammation was found in mouse lungs exposed to heat-inactivated NTHi. A 3.4-fold increase in miR-135b levels above controls was observed in lungs after treatment of mice with heat-inactivated NTHi for 4 consecutive days (Supplemental Fig. 1B). This increase is negligible relative to the 22-fold increase observed in the smoke-exposed mice (Fig. 1B). Currently, it is not clear whether heat-inactivated and live NTHi induce inflammation via the same mechanisms. Therefore, additional experiments are needed to interpret these results meaningfully. For now, we can hypothesize that different molecular mechanisms trigger cigarette smoke– or NTHi-induced inflammation, and that miR-135b expression is predominantly regulated by the mechanisms that control cigarette smoke–induced inflammation.
miRNA-135b expression is regulated by IL-1R1 during cigarette smoke–induced inflammation
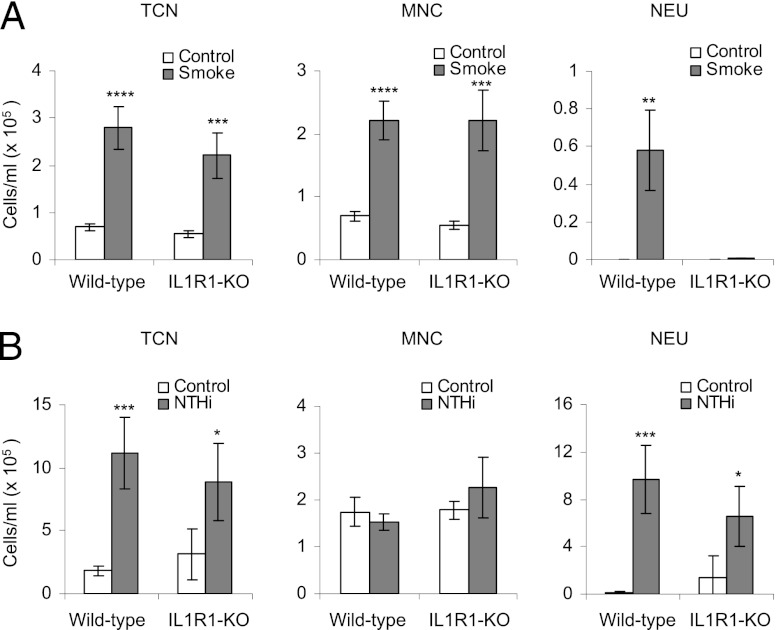
Cigarette smoke–induced pulmonary inflammation has been shown to be dependent on activation of the IL-1R1 pathway (13). Because miR-135b expression is specifically induced after exposure to cigarette smoke and not in response to NTHi, we used IL-1R1 knockout mice to examine whether miR-135b expression is regulated by IL-1R1 during cigarette smoke–induced inflammation. We exposed IL-1R1 knockout mice and C57BL/6 wild-type mice to cigarette smoke for 4 d and compared them with mice inoculated with NTHi for 12 h. Analysis of BAL cellular infiltrate profiles revealed that IL-1R1 knockout mice exposed to cigarette smoke had increased BAL total cell numbers and mononuclear cells (consisting primarily of macrophages), consistent with wild-type controls. However, neutrophilia were completely absent in the BAL of IL-1R1 knockout mice exposed to cigarette smoke (Fig. 2A). In contrast, the absence of IL-1R1 had no effect on NTHi-induced neutrophilic inflammation (Fig. 2B), suggesting that the IL-1R1 pathway is specific to cigarette smoke–induced inflammatory response in this experiment.
FIGURE 2.
(A) Bronchoalveolar lavage cellular profiles of wild-type and IL-1R1 knockout mice exposed to cigarette smoke for 4 d. (B) Wild-type and IL-1R1 knockout mice exposed to NTHi for 12 h. The average for five mice per group is presented. Statistical analysis was performed using a two-way ANOVA with a Bonferroni post test. *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.00005. MNC, Mononuclear cells; NEU, neutrophils; TCN, total cell number.
To investigate whether IL-1R1 deficiency affects miR-135b expression following cigarette smoke exposure, we measured miR-135b levels in lung tissues of wild-type and IL-1R1 knockout mice exposed to cigarette smoke for 4 d. Despite clear evidence of inflammation in IL-1R1–deficient mice after cigarette smoke exposure (Fig. 2A), miR-135b levels in the lungs of these mice were unaffected (Fig. 3). In contrast, IL-1R1 wild-type mice showed close to 20-fold induction, which was similar to the fold induction observed in BALB/c mice exposed to cigarette smoke. Thus, miR-135b is specific to inflammation types involving IL-1R1, and its expression could be directly or indirectly regulated by IL-1R1.
FIGURE 3.
RT- PCR validation of miR-135b in lung tissues of wild-type and IL-1R1 knockout mice exposed to cigarette smoke for 4 d. Data are presented as average fold changes over matched controls (n = 5 per group ± SEM). Expression values were normalized to small nuclear RNA RNUA1. *p < 0.05, Student t test.
IL-1R1 agonists induce miR-135b expression in vitro
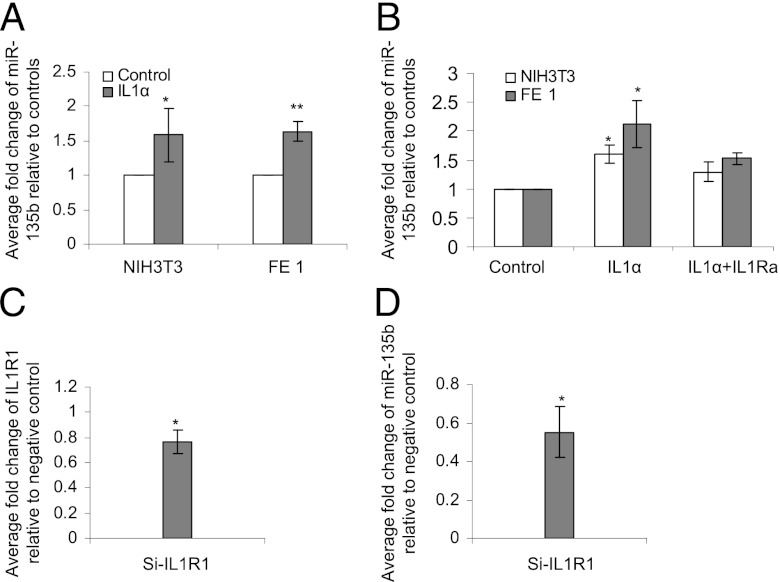
To investigate the role of IL-1R1–mediated induction of miR-135b during inflammation, we conducted several in vitro experiments. First, we exposed NIH3T3 and FE1 lung epithelial cell monolayers to varying concentrations of the IL-1R1 agonist IL-1α. The expression of miR-135b was analyzed at different time points following the stimulation. Treatment with IL-1α–induced miR-135b expression in both NIH3T3 and FE1 cells within 60 min (Fig. 4A). miR-135b induction was blocked by the IL-1R1 antagonist IL-1R1a in both cell lines (Fig. 4B). Cells treated with vehicle control showed only weak endogenous expression of miR-135b. The responses were similar in both cell types, suggesting that inflammatory response to IL-1α stimulation was also largely similar. Because FE1 cells are resistant to DNA transfection, the subsequent experiments were conducted using NIH3T3 cells.
FIGURE 4.
(A) RT-PCR validation of miR-135b in NIH 3T3 cells and FE1 cells treated with 10 ng/ml IL-1α and (B) RT-PCR validation of miR-135b in NIH3T3 cells and FE1 cells treated with IL-1α for 30 min or pretreated for 16 h with the inhibitor IL-1Ra followed by treatment with IL-1α. Expression values were normalized to small nuclear RNA RNUA1. *p < 0.05, Student t test. (C) RT- PCR validation of IL-1R1 mRNA in NIH 3T3 cells transfected with siRNA against IL-1R1 (Si-IL-1R1) or negative control (siNTC). Data represent results from three independent experiments. (D) RT-PCR validation of miR-135b in NIH 3T3 cells transiently transfected with siRNA against IL-1R1 (Si-IL-1R1) followed by treatment with IL-1 α. Values represent average fold change of miR-135b relative to cells transfected with negative control (siNTC) and treated with IL-1α. Data represent results from three independent experiments. *p < 0.05, **p < 0.005, Student t test.
To confirm the involvement of IL-1R1 in controlling miR-135b expression, we transiently transfected NIH3T3 cells with siRNA against IL-1R1 and stimulated the cells with the IL-1R1 agonist IL-1α. The overall expression of IL-1R1 was reduced by 25% in NIH3T3 cells transfected with siIL-1R1 compared with small interfering nontemplate control (SiNTC)–transfected cells (Fig. 4C). The small effect of the siRNA-mediated suppression of IL-1R1 might be due to the low efficiency of transient transfection. We then investigated the effect of silencing IL-1R1 on miR-135b expression following IL-1α challenge. Fig. 4D shows that, compared with SiNTC-transfected cells, siIL-1R1–transfected cells showed a 45% reduction in miR-135b levels after IL-1α challenge, confirming that IL-1R1 expression is important for miR-135b induction.
As shown in Fig. 4A–C, the extent of upregulation in miR-135b in vitro is subtle compared with the robust response observed in vivo. This discrepancy could be due to the cell type chosen for in vitro studies. The specific cell type in lungs in which miR-135b is expressed is unknown. In addition, the in vivo inflammatory response is a complex phenomenon involving multiple biologic pathways and cellular interactions. This biologic complexity is not possible to reproduce in a monolayer and could dampen the observed responses. Identification of the precise cell types in lungs that express miR-135b will clarify the reasons behind this discrepancy.
In vitro overexpression of miR-135b alters endogenous expression of inflammatory mediators including IL-1R1 in NIH3T3 cells
To explore the role of miR-135b in the inflammatory process, NIH3T3 clones stably expressing miR-135b (pEGP-mmu-miR-135b) or vector alone (pEGP-miR–null) were established. Clones that expressed nearly equal amounts of GFP protein (data not shown) were used in the experiments. Using PCR arrays designed for the evaluation of mouse inflammatory cytokines and receptors, the endogenous expression of 70 different inflammatory modulators was analyzed. Compared with pEGP-miR-null–expressing cells, pEGP-mmu-miR-135b cells overexpressing miR-135b by 2198-fold showed significantly altered endogenous mRNA levels of various inflammatory modulators, including C-C chemokines Ccl7 (−2.6-fold) and Ccl8 (−7.5-fold), C-X-C chemokine Cxcl12 (−2.4-fold) and Cxcl11 (2.1-fold), and IL-1R1 (−1.8-fold) and SPP1 (4.1-fold; Table I). Complete RT-PCR array results are presented in Supplemental Table I. The results imply that excess physiologic levels of miR-135b leading to suppressed expression of proinflammatory genes might impair cellular response to external inflammatory stimuli, and that these genes can be directly or indirectly targeted by miR-135b during inflammation.
Table I. RT-PCR analysis of inflammatory cytokines and receptors in pEGP-miR-135b overexpressing cells relative to controls.
| Gene | pEGP-miR-135b/pEGP-miR–null |
|
|---|---|---|
| FC | p Value | |
| Spp1 | 4.15 | 0.036 |
| Cxcl11 | 2.15 | 0.030 |
| Il1r1 | −1.86 | 0.017 |
| Cxcl12 | −2.46 | 0.005 |
| Ccl7 | −2.66 | 0.037 |
| Ccl8 | −7.55 | 0.050 |
Genes with p < 0.05 are shown.
FC, Fold change over matched controls.
IL-1R1 is targeted by miR-135b
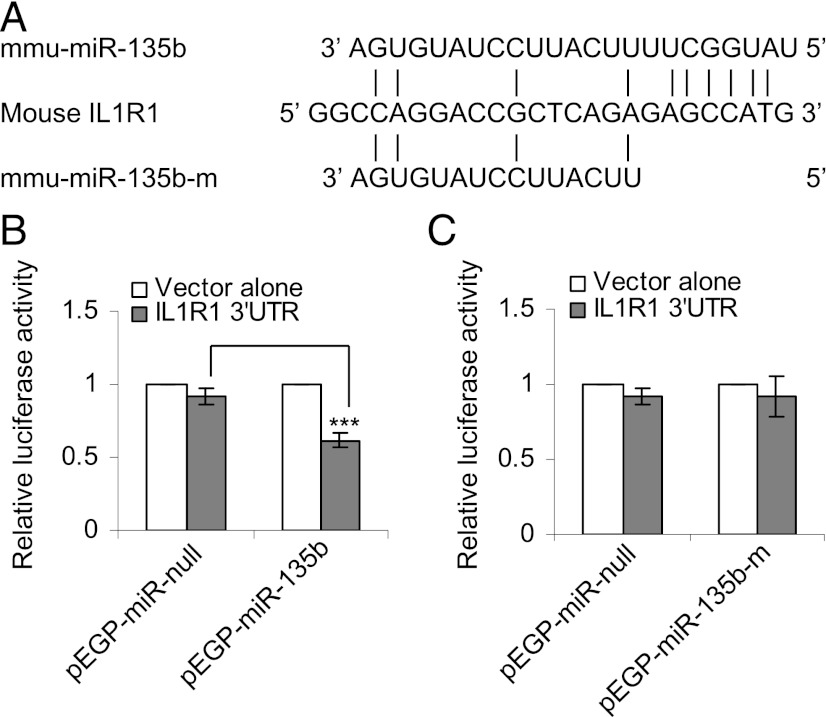
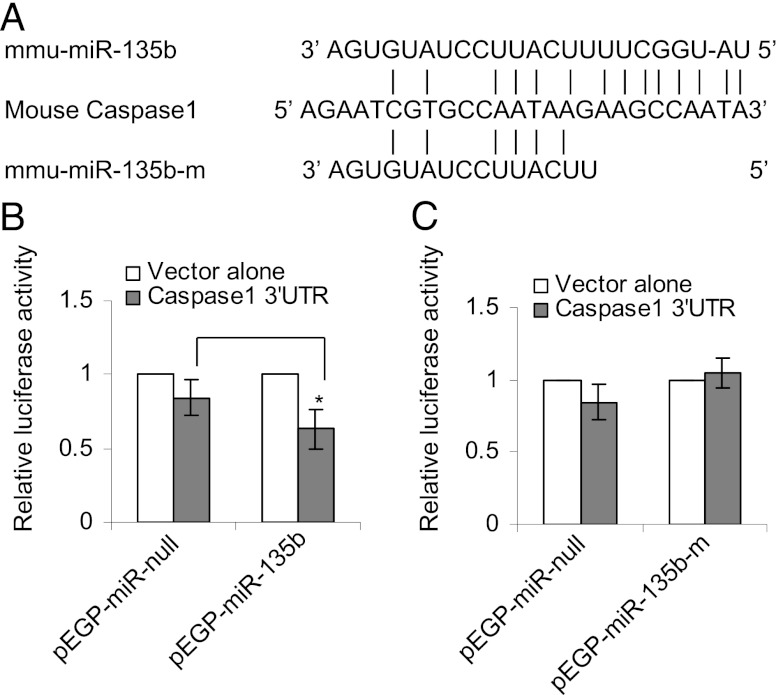
Cigarette smoke–induced inflammation is mediated by the IL-1R1/IL-1α pathway (13). Endogenous expression of the IL-1R1 gene is negatively affected in pEGP-mmu-miR-135b cells (Table I). Target-prediction software (e.g., TargetScan, Pictar) did not predict that any of the aforementioned inflammation-responsive genes are targets of miR-135b, nor are these curated targets for miR-135b. However, sequence alignment of the 3′UTR of mouse IL-1R1 with the seed region of miR-135b revealed six matching base pairs (Fig. 5A). Interestingly, human IL-1R1 is a predicted target of hsa-miR-135b with seven matching base pairs in the seed region (the typical cutoff used by prediction software in determining miRNA targets). To investigate whether mouse IL-1R1 is targeted by miR-135b, pEGP-miR–null and pEGP-miR-135b cells were transiently transfected with pLuc-3′UTR vector containing the 3′UTR of mouse IL-1R1. There was a 30–40% reduction in the luciferase activity in pEGP-miR-135b cells compared with pEGP-miR–null cells (Fig. 5B). To confirm that the observed decrease in luciferase activity was mediated by miR-135b, 8 bp in the seed region of miR-135b were deleted by site-directed deletion, and pEGP-miR-135b-mutant cells were established. miR-135b devoid of a seed region failed to inhibit luciferase activity in the pEGP-miR-135b-mutant cells (Fig. 5C), suggesting that mouse IL-1R1 is likely targeted by miR-135b in a negative regulatory feedback loop.
FIGURE 5.
(A) Alignment of the 3′UTR of IL-1R1 with the seed region of mature murine miR-135b and mutant miR-135b, in which the 6 bp in the seed region are deleted. (B) Relative luciferase activity in pEGP-miR–null (controls) and pEGP-miR-135b (overexpressing miR-135b) cells transfected with pLuc-3′UTR vector alone or pIL-1R1–3′UTR-Luc. (C) Relative luciferase activity in pEGP-miR–null (controls) and pEGP-miR-135b-m (overexpressing mutant form of miR-135b) transfected with pLuc-3′UTR vector alone or pIL-1R1-3′UTR-Luc. ***p < 0.0005.
miR-135b abundance suppresses Caspase-1 and IL-1β expression following IL-1α stimulation
To understand the effects of miR-135b overexpression on the IL-1α–induced inflammatory signaling cascade, we compared the expression profiles of 84 inflammatory modulators in pEGP-miR–null and pEGP-miR-135b cells following IL-1α treatment using a mouse inflammatory cytokine and receptor RT-PCR array. pEGP-miR–null and pEGP-miR-135b cells treated with IL-1α showed a dramatic increase in the expression of several chemokines and cytokines including Cxcl1, Ccl2, Cxcl5, and Ccl7 compared with the vehicle-treated controls (Table II), suggesting the initiation of an IL-1R1/IL-1α–induced inflammatory response. To explore the effects of miR-135b overexpression on this pathway, we compared the mRNA expression between the IL-1α–treated pEGP-miR–null and pEGP-miR-135b cells. Compared with the treated null cells, no significant decreases in the mRNA levels of Cxcl1, Ccl2, Cxcl5, and Ccl7 were found in pEGP-miR-135b cells. However, mRNA levels of Ccl20, IL-11, Ccl8, and Caspase-1 were significantly reduced in pEGP-miR-135b cells compared with the treated pEGP-miR–null cells (Table II). Complete RT-PCR array results are presented in Supplemental Table II. Caspase-1 is a downstream target of the IL-1R1 pathway and is a component of a multimeric protein complex known as the NLRP3 inflammasome, which consists of NALP (NACHT-leucine-rich repeat– and PYD-containing protein), and the adaptor protein ASC (apoptosis-associated specklike protein containing a caspase recruitment domain). Inflammasome-activated Caspase-1 catalyzes the cleavage of precursor IL-1β to its mature form, thereby triggering the inflammatory cascade. Sequence alignment of the 3′UTR of mouse Caspase-1 with the seed region of miR-135b revealed six matching base pairs (Fig. 6A). To confirm that Caspase-1 is a true target of miR-135b, pEGP-miR–null and pEGP-miR-135b cells were transiently transfected with pLuc-3′UTR vector containing the 3′UTR of mouse Caspase-1. A 30–40% reduction in the luciferase activity in pEGP-miR-135b cells was observed relative to pEGP-miR–null cells (Fig. 6B). This decrease in luciferase activity was not observed in pEGP-miR-135b-mutant cells (Fig. 6C), suggesting that Caspase-1 is targeted by miR-135b.
Table II. RT- PCR analysis of inflammatory cytokines and receptors in pEGP-miR-135b overexpressing cells compared with controls (pEGP-miR–null) following IL-1α treatment.
| pEGP-miR-135b/pEGP-miR–null (1h IL1a) |
||
|---|---|---|
| Symbol | FC | p Value |
| Ccl1 | 4.14 | 0.00 |
| C3 | 3.82 | 0.00 |
| Cxcl5 | 2.98 | 0.00 |
| Il18 | 2.92 | 0.00 |
| Itgb2 | 1.88 | 0.00 |
| Ccl17 | 1.67 | 0.03 |
| Cxcl1 | 1.61 | 0.00 |
| Cx3cl1 | 1.56 | 0.00 |
| Ccl25 | 1.46 | 0.02 |
| Mif | 1.42 | 0.02 |
| Il13ra1 | 1.41 | 0.01 |
| Il1a | 1.40 | 0.05 |
| Ccl9 | 1.37 | 0.01 |
| Il15 | 1.35 | 0.06 |
| Aimp1 | 1.27 | 0.01 |
| Spp1 | 1.26 | 0.04 |
| Abcf1 | 1.21 | 0.04 |
| Bcl6 | −1.12 | 0.06 |
| Tollip | −1.18 | 0.03 |
| Ccl11 | −1.25 | 0.07 |
| Ccl3 | −1.35 | 0.07 |
| Lta | −1.44 | 0.08 |
| Cxcl12 | −1.76 | 0.00 |
| Ccl20 | −1.86 | 0.04 |
| Il11 | −2.10 | 0.00 |
| Casp1 | −2.51 | 0.07 |
| Ccl8 | −4.23 | 0.00 |
| Il2rg | −6.53 | 0.00 |
Genes with p < 0.1 are shown.
FC, Fold change over matched controls.
FIGURE 6.
(A) Alignment of the 3′UTR of Caspase-1 with the seed region of mature murine miR-135b and mutant miR-135b, in which the 6 bp in the seed region are deleted. (B) Relative luciferase activity in pEGP-miR–null (controls) and pEGP-miR-135b (overexpressing miR-135b) cells transfected with pLuc-3′UTR vector alone or pCaspase-1-3′UTR-Luc. (C) Relative luciferase activity in pEGP-miR–null (controls) and pEGP-miR-135b-m (overexpressing mutant form of miR-135b) transfected with pLuc-3′UTR vector alone or pCaspase-1-3′UTR-Luc. *p < 0.05.
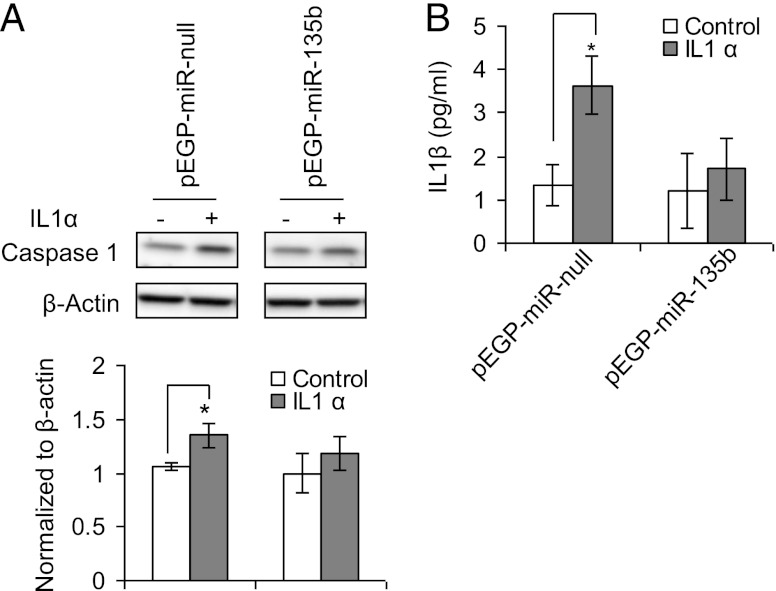
To investigate the effects of reduced Caspase-1 mRNA expression on IL-1α–treated pEGP-miR-135b cells, we analyzed the protein levels of activated Caspase-1 and the mature form of IL-1β. Levels of both Caspase-1 and IL-1β were significantly lower in pEGP-miR-135b cells than pEGP-miR–null cells (Fig. 7A, 7B).
FIGURE 7.
(A) Western blot images of Caspase 1 (upper panel) in pEGP-miR–null (controls) and pEGP-miR-135b (overexpressing miR-135b) with or without treatment with IL-1 α. The lower panel shows the normalized (to actin) band intensities of Caspase-1 levels. (B) Total levels of IL-1β in pEGP-miR–null (controls) and pEGP-miR-135b (overexpressing miR-135b) cells treated with IL-1α. *p < 0.05.
Our results demonstrate that pulmonary expression of miR-135b is increased during cigarette smoke–induced acute inflammation in an IL-1α/IL-1R1–dependent manner. In addition, we show that miR-135b directly targets IL-1R1 in a negative regulatory feedback, as well as targeting its downstream mediators Caspase-1 and IL-1β, perhaps in an effort to resolve the inflammation.
Discussion
Previously, we reported increased expression of miR-135b following acute exposure to nanoparticles of titanium dioxide and carbon black (8, 9). Exposure to these particles is associated with pulmonary inflammation. In this study, we demonstrate increased expression of miR-135b in vivo in response to cigarette smoke–associated inflammation. A number of studies have shown increased expression of miR-135b in certain types of tumors (18, 19), suggesting an association between miR-135b expression and cancer. It is widely accepted that inflammatory processes play a critical role in tumorigenesis. A broad spectrum of inflammatory mediators is often observed in the tumor microenvironment; however, it is unclear whether increased expressions of miR-135b in cancers and during inflammation are interrelated. In this study, we establish several lines of evidence to support that miR-135b expression is regulated by IL-1R1 during IL-1R1/IL-1α–mediated inflammation, and that IL-1R1 is also a direct target of miR-135b, implying that these molecules self-regulate in a negative feedback loop.
IL-1 is an inflammatory cytokine with an important role in the initiation and propagation of inflammatory processes (20). IL-1R1 mediates sterile inflammation induced by silica crystals (11), asbestos (10), monosodium urate crystals (12), and cigarette smoke (13, 14). The IL-1R–mediated signaling cascade is involved in neutrophilic inflammation. Because nanoparticle and cigarette smoke–induced inflammation is neutrophilic in nature, we hypothesized that an increase in miR-135b expression could be related to neutrophilia. Of interest, we observed little (Supplemental Fig. 1) or no increase (Fig. 1B) in miR-135b expression in response to NTHi inoculation, suggesting that not all neutrophilic signaling pathways lead to the induction of miR-135b. Because IL-1R1 signaling is involved in neutrophilic inflammation, and mice deficient in IL-1R1 do not exhibit neutrophilic inflammation after exposure to cigarette smoke, we evaluated miR-135b expression in the lungs of cigarette smoke–exposed, IL-1R1–deficient mice. Exposure to cigarette smoke for 4 d induced inflammation in IL-1R1–deficient mice that were devoid of neutrophils. However, no significant changes in miR-135b expression was observed in the smoke–exposed mice relative to controls (Figs. 2, 3), suggesting that miR-135b expression is associated with an IL-1R1–induced neutrophilic type of inflammation.
Recently, Matsuyama et al. (21) reported that miR-135b is a downstream mediator of NPM-ALK/STAT3 signaling, and that miR-135b modulates immune response by targeting the Th2 master regulators STAT6 and GATA3. These authors found that the inhibition of miR-135b results in the suppression of Th17 cell–associated molecules, such as IL-17, in ALCL cells (21). IL-17 is a proinflammatory cytokine that induces production of inflammatory cytokines (e.g., IL-1β, IL-6, IL-8, G-CSF, GM-CSF) and chemokines (e.g., Ccl2, Ccl7, Ccl20, Cxcl1, Cxcl2, Cxcl5) (22). The secretion of some of these cytokines and chemokines is dependent on miR-135b in cocultured Karpas 299 and WI 38 human fibroblast cells, respectively (21). IL-1R1 signaling has been recognized as a vital step in the early differentiation and expansion of Th17 cells (reviewed in Ref. 23). We observed a similar increases in miR-135b expression in both FE1 (lung epithelial) and NIH3T3 (fibroblasts) cells following IL-1R1 stimulation by its agonist IL-1α (Fig. 4), which also resulted in a dramatic induction of several inflammatory modulators, including the Th17/IL-17 responders (Table II). To understand the underlying role of miR-135b during inflammation, we generated stable NIH3T3 cells overexpressing miR-135b. These cells exhibited more than a 2000-fold increase in miR-135b levels compared with cells transfected with null vectors. miR-135b overexpression resulted in suppressed endogenous expression of a subset of inflammation-associated molecules, including IL-1R1 itself (Table I). Interestingly, the mRNA levels of IL-17 downstream targets Ccl2, Cxcl1, Cxcl5, Ccl20, and Ccl7 were increased in miR-135b–overexpressing cells, a result that correlates well with Matsuyama et al. (21). These findings suggest that IL-1R1–regulated expression of miR-135b immediately after ligand stimulation is involved in the early initiation and expansion of the IL-1R1–mediated inflammatory signaling cascade, supporting the assertion that miR-135b has a regulatory role in the IL-1 signaling pathway.
In contrast to the induction of inflammation producing cytokines, the expression of Caspase-1 (a proinflammatory caspase) was suppressed in NIH3T3 cells overexpressing miR-135b after treatment with IL-1α. Activated Caspase-1 is required for the cleavage and activation of IL-1β, which is a pyrogenic cytokine that responds to infection, injury, and immunologic challenge. To understand the implications of Caspase-1 suppression on IL-1β activation, we analyzed the levels of activated Caspase-1 and IL-1β following IL-1α challenge. Although there was no statistically significant change in the overall cleaved form of Caspase-1, the levels of the processed form of IL-1β were significantly lower in treated miR-135b–overexpressing cells compared with treated null cells (Fig. 7). However, there was no change in the mRNA levels of IL-1β, suggesting that miR-135b could be controlling IL-1R1–downstream expression of IL-1β via regulation of Caspase-1. It is well established that IL-1α and IL-1β function in different aspects of the inflammatory response. For example, IL-1α is produced by neutrophils, whereas IL-1β is produced by macrophages; they are produced at different stages of inflammation, and both recruit a distinct set of myeloid cells. Thus, IL-1α and IL-1β each produce unique inflammatory responses. IL-1α acts as an alarm signal for initiating inflammation, and IL-1β propagates the signal by inducing a suite of inflammatory cascade (24). In our in vitro experiments, siRNA-mediated inhibition of IL-1R1 reduced the expression of miR-135b significantly in NIH3T3 cells challenged with IL-1α (Fig. 4). These results collectively indicate that miR-135b is directly regulated by IL-1R1, which probably serves a dual function: 1) to propagate the alarm signal that was initiated by IL-1α leading to recruitment of neutrophils, chemokines, and cytokines (Table II), including the Th-17/IL-17 responders during the acute phase of inflammation, and 2) curtailing inflammation before it becomes chronic by indirectly targeting IL-1β via Caspase-1. This hypothesis is supported by the observation that IL-1R1 and Caspase-1 are directly targeted by miR-135b in vitro (Figs. 5, 6).
We then explored whether any of the changes observed downstream of IL-1R1, such as Caspase-1 and activated IL-1β, in the NIH 3T3 cell model are also observed in smoke-exposed mouse lungs. We assessed the levels of Caspase-1, IL-1β, and miR-135b in lung tissues exhibiting inflammation following acute exposure to cigarette smoke (exposed for 4 d to cigarettes without filters for 50 min twice daily for a total of 4 d [present study]) or subchronic (exposed for 50 min twice daily, for 5 d/wk for 8 wk, and euthanized 3–4 h after exposure, described in Ref. 16). If our hypothesis that miR-135b acts to curtail inflammation before it becomes chronic is correct, then we expected to see an increase or no effect on activated levels of IL-1β and Caspase-1 in the lungs of mice exposed to smoke acutely. In support of this hypothesis, we note elevated expression of activated Caspase-1 and IL-1β in these acutely exposed mouse lungs (Supplemental Fig. 2B, 2C). A similar increase in Caspase-1 and IL-1β levels in lung tissues showing acute inflammation following a smoking regimen similar to that in our study was reported by Botelho et al. (13). Using various knockout models, they also showed that this elevated expression of IL-1β is independent of Caspase-1 activation, suggesting that Caspase-1 or IL-1β are not the primary targets of miR-135b during cigarette smoke–induced acute inflammation. However, in contrast, we observed a significant reduction in activated levels of Caspase-1 and IL-1β (Supplemental Fig. 2B, 2C) in mouse lungs exposed subchronically to cigarette smoke in the presence of high levels of miR-135b (Supplemental Fig. 2A). Although it is difficult to prove that the observed reduction in Caspase-1 and IL-1β levels is directly related to higher miR-135b expression in these samples (Supplemental Fig. 2A), this observation indirectly supports our hypothesis that miR-135b is acting on Caspase-1 and IL-1β during postacute phases in an effort to stop the progression of inflammation into the chronic phase.
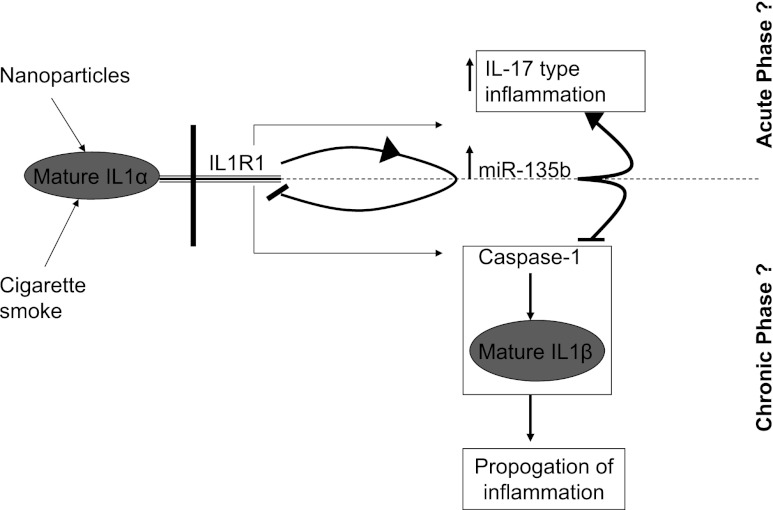
In conclusion, we show that miR-135b hosted by the LEM domain–containing 1 gene is induced in vivo in response to immunologic challenges such as cigarette smoke in an IL-1R1–dependent manner. Induced expression of miR-135b acts to signal the presence of danger and initiates the inflammation process by stimulating Th17 cells to produce IL-17–responding cytokines and chemokines. Once the initial danger is signaled and the initial phase of inflammation is well underway, miR-135b can bind to its own regulator (IL-1R1) in a negative feedback loop, as well as its downstream effector Caspase-1, in an attempt to stop further expansion of the inflammatory process by other potent cytokines such as IL-1β (Fig. 8). Although, our results provide clear evidence for in vivo regulation of miR-135b expression during inflammation via the IL1-R1 pathway, it remains to be shown whether Caspase-1 is indeed targeted in vivo by miR-135b, in addition to its true biologic implications under inflammatory conditions. Our results demonstrate that miR-135b serves a complex role as both a pro- and anti-inflammatory agent that shapes immune responses and contributes to the unique inflammatory phenotype elicited by the IL-1 signaling pathway. Our results also suggest that our previous nanoparticle-induced inflammation is similar to the cigarette smoke–induced inflammation in this study, and that responses to these exposures are regulated by common molecular mechanisms that involve the IL-1R1–miR-135b pathway.
FIGURE 8.
Regulation of miR-135b and its role during IL-1R1–mediated inflammation. Binding of mature IL-1α to its receptor IL-1R1 initiates a signal leading to transcription of miR-135b. MiR-135b indirectly regulates a cascade of cytokines and chemokines (IL-17 type) and inflammation. Excess production of miR-135b targets its own regulator IL-1R1 in a negative feedback loop and its downstream effectors, Caspase-1 and IL-1β, in an effort to curtail the progression of inflammation to a chronic phase.
This work was supported by Health Canada’s Genomics Research and Development Initiative.
The RT-PCR data presented in this article have been submitted to the National Center for Biotechnology Information's Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/query/acc.cgi?acc=GSE43688) under accession number GSE43688.
The online version of the article contains supplemental material.
- BAL
- bronchoalveolar lavage
- miR-135b
- microRNA-135b
- miRNA
- microRNA
- NTHi
- nontypeable Haemophilus influenzae
- siRNA
- small interfering RNA
- SiNTC
- small interfering nontemplate control.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Lindsay M. A. 2008. microRNAs and the immune response. Trends Immunol. 29: 343–351 [DOI] [PubMed] [Google Scholar]
- 2.Pasquinelli A. E. 2012. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 13: 271–282 [DOI] [PubMed] [Google Scholar]
- 3.Sayed D., Abdellatif M. 2011. MicroRNAs in development and disease. Physiol. Rev. 91: 827–887 [DOI] [PubMed] [Google Scholar]
- 4.Abdellatif M. 2012. Differential expression of microRNAs in different disease states. Circ. Res. 110: 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou T., Garcia J. G., Zhang W. 2011. Integrating microRNAs into a system biology approach to acute lung injury. Transl. Res. 157: 180–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oglesby I. K., McElvaney N. G., Greene C. M. 2010. MicroRNAs in inflammatory lung disease—master regulators or target practice? Respir. Res. 11: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lujambio A., Lowe S. W. 2012. The microcosmos of cancer. Nature 482: 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halappanavar S., Jackson P., Williams A., Jensen K. A., Hougaard K. S., Vogel U., Yauk C. L., Wallin H. 2011. Pulmonary response to surface-coated nanotitanium dioxide particles includes induction of acute phase response genes, inflammatory cascades, and changes in microRNAs: a toxicogenomic study. Environ. Mol. Mutagen. 52: 425–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourdon J. A., Saber A. T., Halappanavar S., Jackson P. A., Wu D., Hougaard K. S., Jacobsen N. R., Williams A., Vogel U., Wallin H., Yauk C. L. 2012. Carbon black nanoparticle intratracheal installation results in large and sustained changes in the expression of miR-135b in mouse lung. Environ. Mol. Mutagen. 53: 462–468 [DOI] [PubMed] [Google Scholar]
- 10.Dostert C., Petrilli V., Van Bruggen R., Steele C., Mossman B. T., Tschopp J. 2008. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science 320: 674–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hornung V., Bauernfeind F., Halle A., Samstad E. O., Kono H., Rock K. L., Fitzgerald K. A., Latz E. 2008. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torres R., Macdonald L., Croll S. D., Reinhardt J., Dore A., Stevens S., Hylton D. M., Rudge J. S., Liu-Bryan R., Terkeltaub R. A., et al. 2009. Hyperalgesia, synovitis and multiple biomarkers of inflammation are suppressed by interleukin 1 inhibition in a novel animal model of gouty arthritis. Ann. Rheum. Dis. 68: 1602–1608 [DOI] [PubMed] [Google Scholar]
- 13.Botelho F. M., Bauer C. M., Finch D., Nikota J. K., Zavitz C. C., Kelly A., Lambert K. N., Piper S., Foster M. L., Goldring J. J., et al. 2011. IL-1α/IL-1R1 expression in chronic obstructive pulmonary disease and mechanistic relevance to smoke-induced neutrophilia in mice. PLoS ONE 6: e28457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doz E., Noulin N., Boichot E., Guénon I., Fick L., Le Bert M., Lagente V., Ryffel B., Schnyder B., Quesniaux V. F., Couillin I. 2008. Cigarette smoke-induced pulmonary inflammation is TLR4/MyD88 and IL-1R1/MyD88 signaling dependent. J. Immunol. 180: 1169–1178 [DOI] [PubMed] [Google Scholar]
- 15.White P. A., Douglas G. R., Gingerich J., Parfett C., Shwed P., Seligy V., Soper L., Berndt L., Bayley J., Wagner S., et al. 2003. Development and characterization of a stable epithelial cell line from Muta Mouse lung. Environ. Mol. Mutagen. 42: 166–184 [DOI] [PubMed] [Google Scholar]
- 16.Botelho F. M., Gaschler G. J., Kianpour S., Zavitz C. C., Trimble N. J., Nikota J. K., Bauer C. M., Stämpfli M. R. 2010. Innate immune processes are sufficient for driving cigarette smoke-induced inflammation in mice. Am. J. Respir. Cell Mol. Biol. 42: 394–403 [DOI] [PubMed] [Google Scholar]
- 17.Gaschler G. J., Zavitz C. C., Bauer C. M., Stämpfli M. R. 2010. Mechanisms of clearance of nontypeable Haemophilus influenzae from cigarette smoke-exposed mouse lungs. Eur. Respir. J. 36: 1131–1142 [DOI] [PubMed] [Google Scholar]
- 18.Bandrés E., Cubedo E., Agirre X., Malumbres R., Zárate R., Ramirez N., Abajo A., Navarro A., Moreno I., Monzó M., García-Foncillas J. 2006. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer 5: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong A. W., Fulgham P., Jay C., Chen P., Khalil I., Liu S., Senzer N., Eklund A. C., Han J., Nemunaitis J. 2009. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 16: 206–216 [DOI] [PubMed] [Google Scholar]
- 20.Dinarello C. A. 1996. Biologic basis for interleukin-1 in disease. Blood 87: 2095–2147 [PubMed] [Google Scholar]
- 21.Matsuyama H., Suzuki H. I., Nishimori H., Noguchi M., Yao T., Komatsu N., Mano H., Sugimoto K., Miyazono K. 2011. miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood 118: 6881–6892 [DOI] [PubMed] [Google Scholar]
- 22.Yang X. O., Chang S. H., Park H., Nurieva R., Shah B., Acero L., Wang Y. H., Schluns K. S., Broaddus R. R., Zhu Z., Dong C. 2008. Regulation of inflammatory responses by IL-17F. J. Exp. Med. 205: 1063–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sha Y., Markovic-Plese S. 2011. A role of IL-1R1 signaling in the differentiation of Th17 cells and the development of autoimmune diseases. Self Nonself 2: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luheshi N. M., Rothwell N. J., Brough D. 2009. Dual functionality of interleukin-1 family cytokines: implications for anti-interleukin-1 therapy. Br. J. Pharmacol. 157: 1318–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]