Abstract
The anti-AIDS drug rilpivirine undergoes conformational changes to bind HIV-1 reverse transcriptase and retain potency against drug-resistance mutations. Our discovery that water molecules play an essential role in the drug binding is reported. Femtosecond experiments and theory expose molecular level dynamics of rilpivirine bound to HIV-1 reverse transcriptase. The two nitrile substituents (-CN), one on each arm of the drug, have vibrational spectra consistent with their protein environments being similar in crystals and in solutions. Two-dimensional vibrational-echo spectroscopy reveals a dry environment for one nitrile while unexpectedly the other is hydrogen-bonded to a mobile water molecule, not identified in earlier X-ray structures. Ultrafast nitrile-water dynamics are confirmed by simulations. A higher (1.51 Å) resolution X-ray structure indeed reveals a water-drug interaction network. Maintenance of a crucial anchoring hydrogen bond, despite the enlargement and structural variation of the binding pocket, may help retain the potency of rilpivirine against the pocket mutations.
Reverse transcriptase (RT) is an essential enzyme in the replication of the human immunodeficiency virus (HIV). HIV-1 RT is a 117 kDa heterodimer composed of two subunits, p66 and p51. Nonnucleoside RT inhibitors (NNRTIs) bind to a pocket of the p66 subdomain where drug-resistance mutations occur.1,2 The NNRTI rilpivirine (TMC278/Edurant), a potent diarylpyrimidine (DAPY) inhibitor of wild-type (WT) and NNRTI-resistant viruses, recently received FDA approval as an anti-AIDS drug. Previous studies have shown that rilpivirine could retain potency against mutant HIV-1 viruses through a combination of torsional flexibility, ability to reposition, and hydrogen bonding between drug linker atoms.1 However, a complete and detailed map of molecular interactions by which the inhibitor maintains its potency in mutated binding pockets has remained unclear.
Rilpivirine has distinct structural elements, in particular, the two nitrile (CN) groups which are required for the creation of an effective and potent drug.1 By a fortunate coincidence, these CN groups have readily observable characteristic vibrational spectra3 that allow them to serve as vibrational probes4–6 of structure and dynamics inside the binding pocket of RT7. The current work has successfully used these two CN groups to provide comparative information on the dynamics inside the pocket for rilpivirine in complexes with WT-RT and two clinically important double mutant RTs, namely M1-RT (RT51A; L100I/K103N-RT) and M2-RT (RT55A; K103N/Y181C-RT), which are known for significantly lowering the potency of other NNRTIs.1 The results are both unexpected and remarkable in that a special role for water in drug binding and mobility was discovered.
This work follows basically three lines of approach: linear vibrational spectra of the nitrile groups of the drug in crystal and solution to observe equilibrium features of the nitriles environments; two dimensional vibrational echo (2D-IR) spectra8–11 of the drug complexes to extract dynamic properties of the vibrational frequency distributions for the nitrile groups; and molecular dynamics (MD) simulations and density functional theory (DFT) computations to interpret the spectral features/changes of the nitrile groups in relation to the dynamic and structure of the RT/drug complex. The IR spectrum of the RT/drug complex shows two bands in the stretching vibrational region of nitrile7 and each is assigned to a single nitrile group, as strongly indicated by the similarity of the solution and single crystal IR spectra and by the single conformation of the protein backbone observed in the X-ray structure.1 2D-IR spectroscopy allows one to experimentally monitor and measure the equilibrium dynamics of the vibrational frequency distributions of nitriles in the drug due to fluctuations of its environment. The decays of the resulting frequency-frequency correlation functions (FFCF) cannot be detected from conventional IR spectra yet they contain essential information regarding the dynamics of the changing local environments and prompt strong inferences regarding the equilibrium fluctuations of the structure of the NNRTI binding pocket. Finally, the dynamics results are brought into relation with a very high-resolution (1.51 Å) X-ray crystal structure and classical MD simulations.
Results
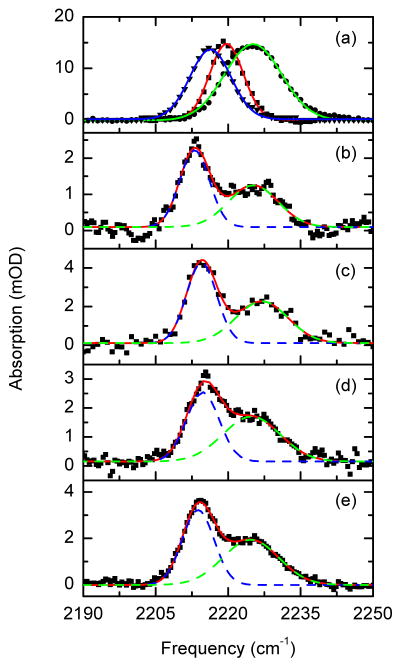
The drug inside the WT-RT7 shows two resolvable, but slightly overlapped, vibrational absorption bands in the same region (Figure 1b–c). Naturally, the two bands are attributed to the two nitrile groups of the drug.7 The first band is located at 2215 cm−1 and has a full width at half maximum (FWHM) of ~7 cm−1 and the second is centered at ~2225 cm−1 with FWHM ~12 cm−1. The ratio of the peak absorptivities of the low to the high frequency transition is ~2. While the overall IR spectral shape of the drug/enzyme complex for different mutants are similar (Figure 1d–e), a significant change in the width of the high frequency band and in the ratios of integrated transition dipoles (area) is observed (see Table S1 of Supplementary Information). The FWHM of the high frequency transition increased from ~12 cm−1 for the WT-RT to ~14 cm−1 for the M1-RT and to ~18 cm−1 for the M2-RT. Similarly, a change in the ratio of areas of the broad to narrow bands is observed in the different complexes: 1.10:1 in the WT-RT, 0.95:1 in the M1-RT, and 0.90:1 in the M2-RT. A key result concerns the vibrational spectra of the WT-RT/rilpivirine complex in a single crystal (Figure 1b). Vibrational spectra are very sensitive to the local environment of the vibrational mode so the similarity of the spectrum of the drug in the crystal (Figure 1b) and aqueous solution (Figure 1c) establishes that the local environment of the nitrile groups in the binding pocket, as deduced from the X-ray crystal structure, are maintained in solution. Remarkably the vibrational spectra (Figure 1a) of rilpivirine7 in bulk solvents display only a single, unresolved, and broad transition (FWHM ~8–14 cm−1) that shifts from 2216 cm−1 in dimethylsulfoxide to 2225 cm−1 in methanol and is always ~30 percent broader than any model compounds12,13 in the same solvent (see Table S1 of Supplementary Information).
Figure 1.
Experimental linear IR spectra of rilpivirine in solution and in complex to the different RT enzymes. (a) FTIR and fits of inhibitor in methanol (black circles, green line), THF (black squares, red line), and DMSO (black triangles, blue line). (b) FTIR of the rilpivirine complex to the WT-RT in a single crystal (black squares), and its fit (redline, dash blue and dash green), (c) FTIR of the rilpivirine complex to the WT-RT in solution (black squares), (d) FTIR of the rilpivirine complex to the mutant M2-RT (Y181C/K103N) in solution (black squares), (e) FTIR of the rilpivirine complex to the mutant M1-RT (L100I/K103N) in solution (black squares), and their corresponding fits (red, dash blue, and dashed green lines).
The 2D-IR spectra of M1-RT/rilpivirine and M2-RT/rilpivirine complexes are shown in Figure 2 as plots of coherence, ωτ, versus detection, ωt, frequency for particular population times (T). They show a positive band along the diagonal (ωτ = ωt), which corresponds to the v=0→1 transitions, and its negative band corresponding to the v=1→2 transitions which is anharmonically shifted along ωt. In both mutants, the positive and negative bands are composed of two distinct diagonal transitions located at ~2215 cm−1 and ~2225 cm−1 in ωτ. The negative band is anharmonically shifted from the positive band by 24.0 cm−1 which is consistent with previous reports on nitriles.7 The nitrile T1 vibrational lifetimes estimated from the peak 2D IR signal decay were 3.8±0.5 ps for M1-RT and 4.1±0.5 for M2-RT benzonitriles in agreement with the value reported for WT-RT7 and 3.2±0.5 ps for M1-RT and 4.0±0.5 ps for M2-RT cinnamonitriles. These lifetimes are not considered to be significantly different and they are all much longer than the frequency relaxation. To estimate and compare the changes, without considering the vibrational lifetime, occurring in the 2D-IR spectra for the different enzyme/drug complexes, the traces of their signals along the diagonal were least square fitted with two Gaussians (see Table S2 of Supplementary Information). The 2D-IR modeling showed that the 2215 cm−1 transition always has the higher peak intensity, the 2225 cm−1 transition is always the broader, and the peak height of the 2215 cm−1 transition is twice that of 2225 cm−1 for any mutant or waiting time (T). However, statistically significant differences in the FWHM of the high frequency peak and peak area ratio are observed for the different mutants. Their FWHMs are 13.4 ± 2.3 cm−1, 14.8 ± 2.2 cm−1, and 10.5 ± 2.3 cm−1 for the M2-RT, M1-RT, and WT-RT complexes, respectively. These differences are also apparent in the ratio of the integrated areas of the low to high frequency peaks, which are 1.4 for M2-RT, 1.1 for M1-RT, and 2.1 for WT-RT. In contrast, the widths of the low frequency peak do not differ statistically (6.9 ± 0.9 cm−1 for M2-RT, 7.6 ± 0.8 cm−1 for M1-RT, and 8.8 ± 1.3 cm−1 for WT-RT). Furthermore, neither the spectral line shape nor the peak position nor the FWHM, for any of the enzyme/drug complexes, shows any waiting time evolution. Finally, the FFCF decay time of ~1 ps, determined from the waiting time dynamics of the integrated echo peak signal14,15, is observed for either the WT and mutant complexes (Figure 3). Although the dynamics of a water grating signal16 will interfere with the nitrile signal in the integrated echo thereby making it zero at long waiting times, the peak shift dynamics is only slightly influenced (Figure 3b,d). Moreover, the noise in the 2D IR spectra has negligible influence on the dynamics of the integrated echo (Figure 6).
Figure 2.
Absorptive 2D IR spectra of the two investigated mutant-rilvipirine complexes at different waiting times. The left and right columns correspond to the spectra of M1-RT (L100I/K103N)/rilpivirine and M2-RT (Y181C/K103N)/rilpivirine, respectively. The waiting time of each spectrum is indicated in the figure.
Figure 3.
Experimental peak shift decays extracted from 2D-IR data as a function of waiting time. Left and right columns correspond to the M1-RT (L100I/K103N)/rilpivirine and to the M2-RT (Y181C/K103N)/rilpivirine complexes, respectively. Panels (a) and (c) show the peak shift of the total signal and panels (b) and (d) to the peak shift with water signal subtracted. Red lines correspond to their fit as mentioned in the text.
Figure 6.
Simulated absorptive 2D-IR spectra and peak shift dynamics of WT-RT/rilpivirine complex with (right column) and without (left column) Gaussian noise for different waiting times as indicated in the Figure. Top left panel peak shift dynamic calculated directly from the response functions. Top right panel peak shift dynamics calculated from a window in the simulated 2D IR spectrum. Squares and red line corresponds to simulations with and without noise, respectively. Parameters are presented in the Supporting Information (Table S5). No 2D-IR cross peak between the two nitrile transitions is located in the spectra for any of the mutant/drug complexes either in theory or experiments.
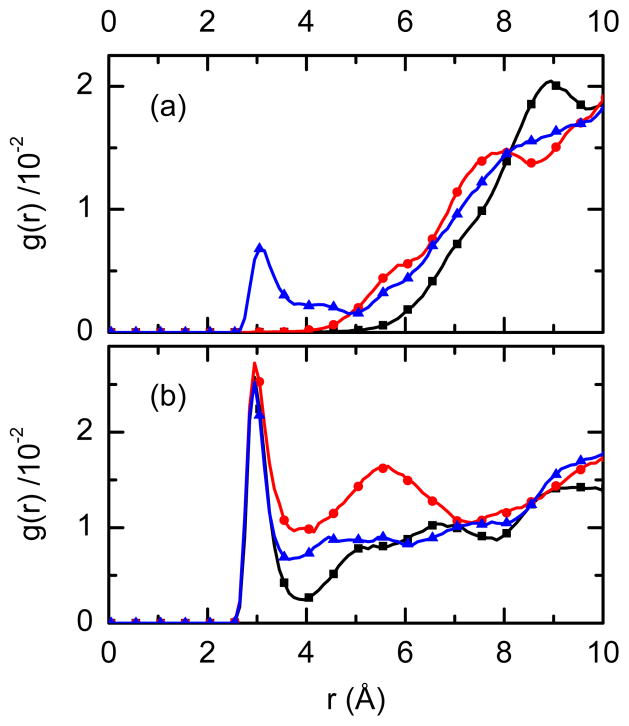
The interactions and spectral signature of the nitrile groups of rilpivirine were computed from classical MD simulations of the drug/RT complexes immersed in a classical bath of water. The similarity of the FTIR spectra of WT-RT/rilpivirine complex in solution and in single crystal provides confidence in basing the simulation on the crystal coordinates. Figure 4 shows calculated radial distribution functions gNCNOW(r) between the nitrogen atom of nitrile (NCN) in the cinnamonitrile and benzonitrile arms and oxygen atoms of water (OW) from our MD simulations. For all three studied drug/enzyme complexes, the g(r) functions show that there is water at H-bond distance to the nitrile of the cinnamonitrile arm (Figure 4b). Also, the water accessibility to the cinnamonitrile end of the pocket, as seen by the gNCNOW(r) in the region of 4–6 Å, is very different for the different mutants and follows the trend: WT-RT<M2-RT<M1-RT (where < means less accessible). In contrast, the benzonitrile arm does not have water at conventional H-bond distances (Figure 4a).
Figure 4.
Calculated radial distribution function g(r) highlighting differences between the two nitrile groups of rilpivirine. The radial distribution functions between the nitrogen of the nitrile group and the oxygen of water for: (a) benzonitrile arm and (b) cinnamonitrile arm of the inhibitor. Black, blue, and red lines correspond to the WT, M1-RT, and M2-RT enzymes, respectively.
We report a very high-resolution (1.51 Å) X-ray crystal structure of WT-RT/rilpivirine complex achieved by developing a new sample preparation (see Supplementary Information). The refinement converged with an R and Rfree of 0.154 and 0.193, respectively (see Table S3 of Supplementary Information). In this structure, the conformation of rilpivirine is similar to the previously reported 1.80 Å structure of the complex.1 Remarkably, this new structure shows a crystallographic water molecule at a H-bonding distance (2.7 Å) to the nitrile of the cinnamonitrile arm, whereas the benzonitrile arm is in a water-free environment, in complete agreement with the 2D-IR data and MD simulations. Note, that the water H-bonded to the cinnamonitrile is apparent in the very high-resolution structure whereas it was not evident in the previous structures of the drug complexes with WT-RT, M1-RT, and M2-RT at lower resolutions.1
Discussion
Rilpivirine shows two completely different IR spectra when dissolved in bulk solvents and when complexed with the HIV-1 RT enzyme. While the benzonitrile and cinnamonitrile (Figure 1) vibrational bands of rilpivirine cannot be distinguished in bulk solvents (Figure 1a), two very distinct bands are observed in the solution of the enzyme/drug complex. However, the single absorption band in bulk solvents blue-shifts and widens with increasing solvent polarity, like many other reported CN stretch bands4–6,12,13. Moreover, the CN stretch band of rilpivirine is always broader than the typical CN stretch in protic and aprotic solvents.17 This is consistent with the single IR band being composed of two overlapping transitions with similar transition dipole moments where the environment sensed by each nitrile group is so similar that it fails to spectrally separate the two CN stretch transitions (see Supplementary Information). DFT calculations also agree with this result and show that the two drug CN stretches have similar transition dipole moments and are only separated by 4 cm−1 (see Table S4 of Supplementary Information).
Interestingly, the drug bound to the RT mutants shows two resolved transitions where the frequency separation between bands is caused exclusively by the drug being positioned inside the enzyme.7 Moreover, mutations in the pocket significantly modify the IR bandwidths without altering the transition dipoles. In the mutant RT/drug complex, the broader, higher frequency band resembles the CN stretch in an aqueous environment and the narrower, low frequency, transition has parameters similar to model compounds in polar aprotic solvents. Therefore, the spectral parameters of the two nitrile CN stretches in the mutant complexes indicate that one arm of the drug is in a water-free cavity while the other occupies a water-containing environment. These spectral changes in the CN bands of the drug for the two mutant complexes reinforce the idea that each nitrile group of the inhibitor senses the differences in the surrounding environment caused by different mutations in the pocket. However, an assignment of the two transitions to the benzonitrile and cinnamonitrile ends of the drug is not certain from the analysis of the linear spectral data alone. Thus, the FFCF of the vibrations is used to achieve this assignment. A correlation time of ~1 ps suggest the CN group is undergoing hydrogen bond rearrangements from water or charged side chains (0.8–1.5 ps18–21). This contrasts with the slower correlation times found in aprotic solvents (5–10 ps20,22), which was previously measured in the low frequency CN stretch transition of the WT-RT/drug complex7. Although the echo peak shift does not distinguish the two CN groups, simulations show that it will be dominated by the component that decays fastest and hence has the widest transition. Therefore, the fast correlation time decays of the two mutant RT/rilpivirine complexes are attributed to the relaxation of the higher frequency transition at ~2225 cm−1, corresponding to the frequency dynamics of the cinnamonitrile end of the drug as inferred from MD simulations and X-ray structure. Although in a given solvent, the CN stretch of model cinnamonitriles always have lower transition frequencies than benzonitriles, in a protein environment constituted from a specific sequence of amino acid side chains, all bearing different type of interactions with the drug, the ordering could be easily reversed. Moreover, it has been observed that through different side chain interactions the frequency of the benzonitrile CN stretch can vary within 18 cm−1.23 Given that the cinnamonitrile group is interacting with water (Figure 5), it is expected that its CN stretch frequency will be blue-shifted by the H-bonding as is usually observed in the CN stretch modes of nitrile model compounds,5 including rilpivirine. Even when the CN group is in contact with only one water molecule, its CN stretch frequency can shift significantly depending on the angle and distance between the CN and the OH groups.17 On the contrary, the CN stretch transition of the benzonitrile arm inside the enzyme is down shifted by 3–4 cm−1 from its typical frequencies for aprotic solvents such as tetrahydrofuran (THF). The assignments made here do not follow the direct comparison with model compounds in bulk solvent as it was assumed in Ref. 7. The vibrational frequencies of CN in structurally defined pockets of a protein may not conform to those in bulk solvents because of spatial variations in the dielectric constants or specific interactions arising from the unique pocket structures. Thus the ordering of benzo- and cinnamonitrile CN frequencies need not be those of the model compounds. The signatures presented by nitrile groups of their local environments in proteins are also evident from the peak heights observed in the FTIR and 2D-IR diagonal traces. An oscillator immersed in water will have comparatively large frequency fluctuations that do not remain correlated for more than ~1 ps6,19,20,24 because of the fast dynamics of H-bond making and breaking25,26. Thus, the water induced frequency fluctuations translate into a broadening of the IR lineshape, which decreases the peak intensity to maintain its transition dipole magnitude. This hypothesis is supported by theoretically modeling the frequency fluctuations of the nitrile groups of rilpivirine in various environments. In bulk solvents, the distributions of frequency fluctuations for both CN stretches have very similar standard deviations (σ) consistent with each group sensing the same type of environment, but both are strongly solvent dependent: σ = 9 cm−1 for water and σ = 3.5 cm−1 for THF (see Figure S1 of Supplementary Information). Inside the protein, the frequency fluctuations are distinctly different for each nitrile group: σ = 6 cm−1 and for cinnamonitrile and σ = 3 cm−1 for benzonitrile. Also, the simulation indicates that the main contribution to the vibrational frequency fluctuations in the enzyme is from the water interacting with the nitrile group (see Figure S2 of Supplementary Information). The through bond effects on the CN mode caused by other groups, such as -NH2 H-bonding to water, have not been considered but the through-space effects would be expected to be negligible. Future experiments on other vibrations of the drug such as the 15N-H stretch would also add understanding to the dynamics of the pockets. The experimental probes of the pocket dynamics are limited to the spectral densities of the nitrile vibrations which we assume are not influenced by other parts of the drug undergoing hydrogen bonding although additional experiments are needed to prove this statement.
Figure 5.

Snapshot of rilpivirine in the NNRTI-binding pocket as observed in our MD simulations and X-ray crystal structure. Left and middle panels correspond to the different views of the pocket from the MD simulation where cyan, blue, red, and white correspond to carbon, nitrogen, oxygen, and hydrogen atoms, respectively, and the grey surface to the protein. Right panel corresponds to the thermal ellipsoid representation of rilpivirine in the NNRTI-binding pocket; the ellipsoids represent anisotropic thermal parameters of individual atoms that were refined using 1.51 Å resolution X-ray diffraction data. Two water molecules that interact with rilpivirine are represented as red ellipsoids.
From the analysis of the linear and nonlinear experimental results, we propose a spectral model for the bound rilpivirine which is tested by simulations of the FTIR and 2D-IR using response functions27 and the parameters observed in these and previous experiments7 (see Table S5 of Supplementary Information), where the FFCF is represented by an exponential decay of the form: . The simulation leads to a 2D-IR spectrum of WT-RT/rilpivirine at different waiting times (Figure 6) which reproduces well the observed elongated spectral shape of positive and negative peaks with the two distinctive peak maxima and their diagonal peak height ratio of ~ 2:1. However, these spectral features have reduced discernibility because of the noise contribution (Figure 6). They also correctly predict the frequency dynamics giving rise to the peak shift (Figure 6). These same parameters reproduce the linear IR spectrum of the WT-RT/rilpivirine complex (see Figure S3 of Supplementary Information).
Structural changes of the binding pocket are clearly seen in the M1-RT and M2-RT mutants as reported from X-ray studies1 and by vibrational spectroscopy. The crystal structure data indicate that the drug undergoes significant conformational and positional rearrangements inside the M1-RT mutant when compared to the WT-RT complex.1 Vibrational spectroscopy supports, through changes in their spectral features, these structural modifications and shows that even when the pocket is mutated the cinnamonitrile end of the drug maintains its H-bond to water. The MD simulations also predict an enlargement of the binding pocket that increases the access of water to the pocket and the mobility of the drug. The pocket widening is clearly seen in the RMSD of drug coordinates in the computed trajectories for the complex with RT (see Table S4 of Supplementary Information). It will be interesting to determine whether the highly mobile waters found so readily by the 2D IR method in the mutants will be discernible in the X-ray diffraction electron density maps when they are obtained at high resolution. Moreover, MD simulations predict that the benzonitrile group of the drug bound to the M1-RT also interacts with a water molecule although with a significantly smaller probability than with cinnamonitrile (see Figure 4). All experimental spectral effects are reproduced by the simulation. Although the structural changes within the binding pocket in the mutants are significant, the drug maintains its position through a network of hydrogen bonds of which the newly discovered interaction between water and the cinnamonitrile group is essential. This interaction prevents the drug from being ejected from the binding pocket even when confronted with significant structural modifications (mutations). Thus, this interaction of the cinnamonitrile with water explains not only why the cinnamonitrile containing drugs are less susceptible to common mutations of the binding pocket, but also their strong potency.
In summary, 2D-IR experiments and MD simulations of two double mutant RT/rilpivirine complexes show that the spectral signature of the drug CN vibrations reveals essential, functional, and structural changes within the binding pocket which are not readily seen by other methodologies. In combination with MD simulations, 2D-IR experiments show that mutations producing M1-RT and M2-RT variants significantly affect the water accessibility to the binding pocket in the presence of rilpivirine. Because of increased flexibility, the DAPY class of NNRTIs have better properties overall than the first generation NNRTIs. However, the presence of the cinnamonitrile group has also enhanced the potency of rilpivirine by ~3-fold compared to its predecessor, dapivirine/TMC1201. The current study using IR, MD, and crystallography shows that the conserved interaction of the cinnamonitrile with a water molecule contributes to the enhanced binding of rilpivirine, and the interaction may be exploited in designing new NNRTIs.
Methods
Sample preparation
RT constructs were expressed and purified as previously described.28 Complexes of rilpivirine with L100I/K103N RT (RT51A) and Y181C/K103N RT (RT55A) in 10 mM Tris buffer, 75 mM NaCl at pH 8 were originally prepared in 2.5% DMSO and 1% β-octyl glucoside whose molar ratios were reduced to less than 0.002% by buffer exchange. The final concentrations of rilpivirine bound to RT were determined to be 1.29 mM (L100I/K103N) and 1.55 mM (Y181C/K103N). The engineered forms of the RT used for the solution phase studies were identical to those in the X-ray studies both for wild-type and double mutant samples. Sources of chemicals are given in SI.
Protein crystals were grown28 from drops (see Supplementary Information) on CaF2 then placed into a cell with two CaF2 windows separated by a Teflon spacer of 56 μm thickness. The integrity of crystals grown on CaF2 windows was verified by X-ray diffraction analysis. The dimensions of each single crystal in the plane of the windows were less than or equal to 200 × 300 μm with a solvent content of approximately 56%. The concentration of rilpivirine in the crystal is ~3.5 mM based on the Matthews coefficient.
2D-IR and linear IR spectroscopy
The FTIR spectra were recorded on a Nicolet 6700 spectrometer. The 2D-IR signals were collected and processed by interferometry as previously described.29 An IR pulsed source of 70 fs duration centered at 4.5 μm with a spectral band width of ~210 cm−1 was generated from a home made OPA pumped with a home made ultrafast Ti:Sapphire amplifier. The IR source was divided into three beams of approximately 300 nJ energy and a fourth beam of ~30 nJ. The three 300 nJ pulses (labeled as k1, k2, and k3) were focused onto the sample and the IR photon echo field in the −k1+k2+k3 phase matching direction was collinearly overlapped with the local oscillator pulse (kLO) preceding k3 by a time delay of ~1 ps. The interferometric signal was dispersed off of a 100 grooves/mm grating and detected using an amplified 64 element MercuryCadmiumTelluride detector. After Fourier transformation the signals along the ωτ and ωt (detection frequency) intervals of the two-dimensional spectrum were obtained. A detailed description of the two-dimensional analysis can be found elsewhere.29 The 2D-IR spectra are plots of coherence, ωτ, versus detection, ωt, frequency.
X-ray crystallography
Crystallization was performed using the hanging-drop method with EasyXtal DG-Tools (Qiagen, Valencia, CA) crystallization trays (see Supplementary Information). Data collection was performed at the National Synchrotron Light Source (NSLS) X29 beamline. The diffraction data were indexed, processed, scaled and merged using HKL2000I30. Structure refinement and model building were performed using PHENIX31 and Coot32, respectively. The data processing and refinement statistics are listed in Table S3 of Supplementary Information and the coordinates and structure factors have been deposited in the Protein Data Bank (PDB; accession code 4G1Q).
Molecular dynamics simulations and frequency calculations
Simulations of all three RT/inhibitor complexes were performed using AMBER 1133 with the AMBER ff9934 force field for the protein, the GAFF35 force field for the drug and THF, and the TIP3P36 model for water. Both the drug and THF parameters were built with the antechamber software from AMBER tools. The X-ray crystal structures of rilpivirine bound to the hydrophobic pocket of wild-type and two double mutant RTs (PDB: 2ZD1, 2ZE2, 3BGR), including crystallographic water, was solvated in a truncated octahedral periodic container, with a minimum distance of 8 Å between any protein or drug atom and the closest container edge. Chloride counterions established charge neutrality. The ~130,000 atom system was energy minimized using steepest decent and conjugate gradient methods to remove any steric conflicts produced during the solvation. In the first step of the minimization, the protein-drug complex was restrained with a 500 kcal/(mol.Å2) harmonic potential, while the water and counterions were allow to move freely during the 2000 steps of this minimization. After the restrained minimization, an unrestrained energy minimization was performed on another 2500 steps. In all of the minimization steps, the converge gradient was set to 10−4 kcal/(mol.Å). The system temperature was then raised from 0 K to 298 K during a constant volume MD simulation (NVT) of 20 ps with a Langevin thermostat and where the RT-TMC278 complex was restrained (10 kcal/(mol.Å2)). The NVT step was followed by a 100 ps fully unrestrained run at constant pressure (NPT) using a Berendsen barostat (taup = 2.0 ps) to maintain the constant pressure. During this NPT step, the system was checked to achieve constant density. The next run involved a 100 ps constant volume MD (NVT) trajectory after which the temperature control was switched off and a final run of 5 ns under NVE conditions was performed. In this NVE run the RMS of protein was checked to confirm sample stability. The production runs were obtained after the latest NVE run by running a 1ns trajectory at NVE conditions in which each snapshot was recorded every 20 fs. The following parameters were used during the MD simulations: a 2 fs step, a SHAKE algorithm to constrain all bonds involving hydrogen atoms, periodic boundary conditions, and a particle-mesh Ewald summation method with a cutoff of 8 Å to compute the long-range electrostatic interactions. Frequency calculations of the two nitrile groups were performed according to the ab initio methodology developed by Cho et al.17
DFT calculations
DFT calculations were performed with the Gaussian 09 software package. In the computation of frequencies and transition dipole magnitudes, the structure of rilvipirine and other compounds was first fully optimized (energy minimized) at the B3LYP/6-31++G(d,p) level. The frequencies were later computed from all the optimized structures with the same level of theory. The correction factor for the ab initio frequencies of the drug was computed from the experimental frequency of four other model compounds listed in the SI (Table S2) in THF.
Supplementary Material
Acknowledgments
We thank Adrian Roitberg for advising us with the molecular dynamics simulations. This research was supported by National Institutes of Health Grants GM12592 (to R.M.H.) and NIH MERIT Award R37 AI27690 (to E.A.) by using instrumentation developed at NIH P41RR001348/9P41GM104605. We acknowledge Brookhaven National Laboratory (BNL) X29 beam-line facility for X-ray data collection and R.S.K. Vijayan for helpful discussions.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
E.A. and R.M.H. conceived and designed the experiments. J.R.C. and T.T. performed the infrared experiments. D.G.K, J.R.C., and T.T. analyzed the data. D.G.K. performed theoretical calculations and simulations. J.D.B. expressed and purified the protein samples, and grew the crystals. J.D.B. and D.P. performed the X-ray data collection. J.D.B. and K.D. analyzed and refined the crystal structure. D.G.K. and R.M.H. co-wrote the paper. All authors discussed the results and commented on the manuscript.
Atomic coordinates and structure factors for the WT-RT structure at 1.51 Å resolution has been deposited in the Protein Data Bank under accession codes 4G1Q.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Contributor Information
Daniel G. Kuroda, Ultrafast Optical Processes Laboratory, Department of Chemistry, University of Pennsylvania, PA 19067.
Joseph D. Bauman, Center for Advanced Biotechnology and Medicine, Piscataway, NJ 08854. Department of Chemistry and Chemical Biology, Rutgers University, Piscataway, NJ 08854.
J. Reddy Challa, Ultrafast Optical Processes Laboratory, Department of Chemistry, University of Pennsylvania, PA 19067.
Disha Patel, Center for Advanced Biotechnology and Medicine, Piscataway, NJ 08854. Department of Medicinal Chemistry, Rutgers University, Piscataway, NJ 08854.
Thomas Troxler, Ultrafast Optical Processes Laboratory, Department of Chemistry, University of Pennsylvania, PA 19067.
Kalyan Das, Center for Advanced Biotechnology and Medicine, Piscataway, NJ 08854. Department of Chemistry and Chemical Biology, Rutgers University, Piscataway, NJ 08854.
Eddy Arnold, Center for Advanced Biotechnology and Medicine, Piscataway, NJ 08854. Department of Chemistry and Chemical Biology, Rutgers University, Piscataway, NJ 08854. Department of Medicinal Chemistry, Rutgers University, Piscataway, NJ 08854.
Robin M. Hochstrasser, Ultrafast Optical Processes Laboratory, Department of Chemistry, University of Pennsylvania, PA 19067
References
- 1.Das K, et al. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci USA. 2008;105:1466–1471. doi: 10.1073/pnas.0711209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bollini M, et al. Computationally-Guided Optimization of a Docking Hit to Yield Catechol Diethers as Potent Anti-HIV Agents. J Med Chem. 2011;54:8582–8591. doi: 10.1021/jm201134m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mccoy S, Caughey WS. Infrared Studies of Azido, Cyano, and Other Derivatives of Metmyoglobin, Methemoglobin, and Hemins. Biochemistry. 1970;9:2387. doi: 10.1021/bi00814a001. [DOI] [PubMed] [Google Scholar]
- 4.Getahun Z, et al. Using nitrile-derivatized amino acids as infrared probes of local environment. J Am Chem Soc. 2003;125:405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh A, Remorino A, Tucker MJ, Hochstrasser RM. 2D IR photon echo spectroscopy reveals hydrogen bond dynamics of aromatic nitriles. Chem Phys Lett. 2009;469:325–330. doi: 10.1016/j.cplett.2008.12.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim YS, Hochstrasser RM. Chemical exchange 2D IR of hydrogen-bond making and breaking. Proc Natl Acad Sci USA. 2005;102:11185–11190. doi: 10.1073/pnas.0504865102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang C, et al. Two-dimensional infrared spectra reveal relaxation of the nonnucleoside inhibitor TMC278 complexed with HIV-1 reverse transcriptase. Proc Natl Acad Sci USA. 2008;105:1472–1477. doi: 10.1073/pnas.0709320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamm P, Lim MH, Hochstrasser RM. Structure of the amide I band of peptides measured by femtosecond nonlinear-infrared spectroscopy. J Phys Chem B. 1998;102:6123–6138. [Google Scholar]
- 9.Hochstrasser RM. Two-dimensional spectroscopy at infrared and optical frequencies. Proc Natl Acad Sci USA. 2007;104:14190–14196. doi: 10.1073/pnas.0704079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkelstein IJ, et al. Probing dynamics of complex molecular systems with ultrafast 2D IR vibrational echo spectroscopy. Phys Chem Chem Phys. 2007;9:1533–1549. doi: 10.1039/b618158a. [DOI] [PubMed] [Google Scholar]
- 11.Ganim Z, et al. Amide I two-dimensional infrared Spectroscopy of proteins. Acc Chem Res. 2008;41:432–441. doi: 10.1021/ar700188n. [DOI] [PubMed] [Google Scholar]
- 12.Andrews SS, Boxer SG. Vibrational stark effects of nitriles I. Methods and experimental results. J Phys Chem A. 2000;104:11853–11863. [Google Scholar]
- 13.Andrews SS, Boxer SG. Vibrational Stark effects of nitriles II. Physical origins of stark effects from experiment and perturbation models. J Phys Chem A. 2002;106:469–477. [Google Scholar]
- 14.Cho MH, et al. The integrated photon echo and solvation dynamics. J Phys Chem. 1996;100:11944–11953. [Google Scholar]
- 15.deBoeij WP, Pshenichnikov MS, Wiersma DA. On the relation between the echo-peak shift and Brownian-oscillator correlation function. Chem Phys Lett. 1996;253:53–60. [Google Scholar]
- 16.Kuo CH, Hochstrasser RM. Two dimensional infrared spectroscopy and relaxation of aqueous cyanide. Chem Phys. 2007;341:21–28. [Google Scholar]
- 17.Choi JH, et al. Nitrile and thiocyanate IR probes: Quantum chemistry calculation studies and multivariate least-square fitting analysis. J Chem Phys. 2008;128:134506. doi: 10.1063/1.2844787. [DOI] [PubMed] [Google Scholar]
- 18.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. 2D IR provides evidence for mobile water molecules in beta-amyloid fibrils. Proc Natl Acad Sci USA. 2009;106:17751–17756. doi: 10.1073/pnas.0909888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh A, Qiu J, DeGrado WF, Hochstrasser RM. Tidal surge in the M2 proton channel, sensed by 2D IR spectroscopy. Proc Natl Acad Sci USA. 2011;108:6115–6120. doi: 10.1073/pnas.1103027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tucker MJ, et al. 2D IR photon echo of azido-probes for biomolecular dynamics. Phys Chem Chem Phys. 2011;13:2237–2241. doi: 10.1039/c0cp01625j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuroda DG, Vorobyev DY, Hochstrasser RM. Ultrafast relaxation and 2D IR of the aqueous trifluorocarboxylate ion. J Chem Phys. 2010;132 doi: 10.1063/1.3285265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonner GM, et al. Probing the effect of the solution environment on the vibrational dynamics of an enzyme model system with ultrafast 2D-IR spectroscopy. Faraday Discuss. 2010;145:429–442. [Google Scholar]
- 23.Schultz KC, et al. A genetically encoded infrared probe. J Am Chem Soc. 2006;128:13984–13985. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- 24.Kuroda DG, Hochstrasser RM. Dynamic structures of aqueous oxalate and the effects of counterions seen by 2D IR. Phys Chem Chem Phys. 2012;14:6219–6224. doi: 10.1039/c2cp23892f. [DOI] [PubMed] [Google Scholar]
- 25.Corcelli SA, Lawrence CP, Skinner JL. Combined electronic structure/molecular dynamics approach for ultrafast infrared spectroscopy of dilute HOD in liquid H2O and D2O. J Chem Phys. 2004;120:8107–8117. doi: 10.1063/1.1683072. [DOI] [PubMed] [Google Scholar]
- 26.Moller KB, Rey R, Hynes JT. Hydrogen bond dynamics in water and ultrafast infrared spectroscopy: A theoretical study. J Phys Chem A. 2004;108:1275–1289. [Google Scholar]
- 27.Ge NH, Zanni MT, Hochstrasser RM. Effects of vibrational frequency correlations on two-dimensional infrared spectra. J Phys Chem A. 2002;106:962–972. [Google Scholar]
- 28.Bauman JD, et al. Crystal engineering of HIV-1 reverse transcriptase for structure-based drug design. Nucleic Acids Res. 2008;36:5083–5092. doi: 10.1093/nar/gkn464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YS, Wang JP, Hochstrasser RM. Two-dimensional infrared spectroscopy of the alanine dipeptide in aqueous solution. J Phys Chem B. 2005;109:7511–7521. doi: 10.1021/jp044989d. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Case DA, et al. AMBER 11. University of California; San Francisco: 2010. [Google Scholar]
- 34.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 35.Wang JM, et al. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen WL, et al. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.