Abstract
Before the advent of genome-wide association studies (GWASs), hundreds of candidate genes for obesity-susceptibility had been identified through a variety of approaches. We examined whether those obesity candidate genes are enriched for associations with body mass index (BMI) compared with non-candidate genes by using data from a large-scale GWAS. A thorough literature search identified 547 candidate genes for obesity-susceptibility based on evidence from animal studies, Mendelian syndromes, linkage studies, genetic association studies and expression studies. Genomic regions were defined to include the genes ±10 kb of flanking sequence around candidate and non-candidate genes. We used summary statistics publicly available from the discovery stage of the genome-wide meta-analysis for BMI performed by the genetic investigation of anthropometric traits consortium in 123 564 individuals. Hypergeometric, rank tail-strength and gene-set enrichment analysis tests were used to test for the enrichment of association in candidate compared with non-candidate genes. The hypergeometric test of enrichment was not significant at the 5% P-value quantile (P = 0.35), but was nominally significant at the 25% quantile (P = 0.015). The rank tail-strength and gene-set enrichment tests were nominally significant for the full set of genes and borderline significant for the subset without SNPs at P < 10−7. Taken together, the observed evidence for enrichment suggests that the candidate gene approach retains some value. However, the degree of enrichment is small despite the extensive number of candidate genes and the large sample size. Studies that focus on candidate genes have only slightly increased chances of detecting associations, and are likely to miss many true effects in non-candidate genes, at least for obesity-related traits.
INTRODUCTION
The genetic contribution to inter-individual variation in body mass index (BMI), as a measure of obesity risk, has been well-established with heritability estimates ranging between 40 and 70%, while the remaining variation is due to lifestyle factors (1). Since the mid-1990s, candidate gene studies have aimed to identify obesity-susceptibility genes. Candidate gene studies are hypothesis-driven, and hundreds of genes for which there is some evidence for a role in the regulation of energy balance in animal models or in extreme/monogenic forms of obesity have been tested for association with obesity-related traits (2). However, consistent associations have been reported for only a handful of these candidate genes (3). The main reasons for the limited success of the candidate gene approach include small sample sizes and thus low statistical power, low numbers of genetic variants tested per gene and thus incomplete coverage of the common variation and limited biological insights providing the basis for gene candidacy.
The advent of the genome-wide association approach in 2005 has changed the way and the speed by which genetic loci are being discovered. Among the strongest arguments in favour of performing genome-wide association studies (GWASs) is the fact that such studies are hypothesis-free; i.e. the whole genome is screened for association to a complex disease or trait, without prior hypotheses about which genes or regions are likely to be associated. The results of numerous GWASs performed in recent years have justified this approach, as many previously unsuspected regions have been reproducibly associated to numerous complex traits (4).
Nevertheless, interest remains in the analysis of candidate genes, for a number of reasons. First, although GWASs have identified many unsuspected regions, they have also detected association to many genes that are regarded as good (though sometimes a posteriori) candidates (5). Considerable recent progress has been made in delineating biological pathways and interactions between gene networks, providing richer information on the identification of candidate genes. Several groups have shown that functional elements, genes and relevant pathways are all enriched among the results of GWASs (4,6,7). Finally, the failure of GWASs results to explain the heritability of many traits can be partly attributed to stringent thresholds of statistical significance, due to a high multiple testing burden, and there is considerable interest in applying methods that use prior information to improve the power of tests and reduce multiple testing (8,9). With GWAS data now available on numerous large cohorts, it has become possible to embed candidate gene studies within GWASs, testing for association on a much larger number of candidate genes than previously possible.
It is still unclear, however, that candidate genes as defined by expert knowledge are more likely to be associated than random genes or genomic regions. Enrichment of association in genes compared with non-coding regions has been demonstrated in psychiatric disorders (6), and overlap between some candidate genes and GWAS-identified loci have been observed for some common traits and diseases (10–12), including BMI (5). However, this does not necessarily extend to candidate genes in general and the extent of overlap may vary across traits and diseases. If there is indeed an enrichment of association for candidate genes, then this could be exploited to improve statistical power, but if not, then the value of embedded candidate gene studies is open to question.
Here, we addressed this issue for obesity-susceptibility. We performed a thorough literature search to derive a comprehensive list of 547 candidate genes for obesity, based on studies of model organisms, monogenic syndromes of obesity, linkage studies, candidate gene association studies and other sources of evidence, including gene expression studies. We then referenced these genes in the results of a large meta-analysis of BMI conducted by the genetic investigation of anthropometric traits (GIANT) consortium (5). We compared their ranking within the full list of genes with the ranking of non-candidate genes, with the aim of observing more significant association statistics, on average, for candidate genes compared with non-candidate genes. We considered the regions around genes with no prior evidence for candidacy as a baseline for comparison, rather than intergenic regions, because we were interested in whether, given a focus on testing genes, there is value in specifically considering candidate genes.
RESULTS
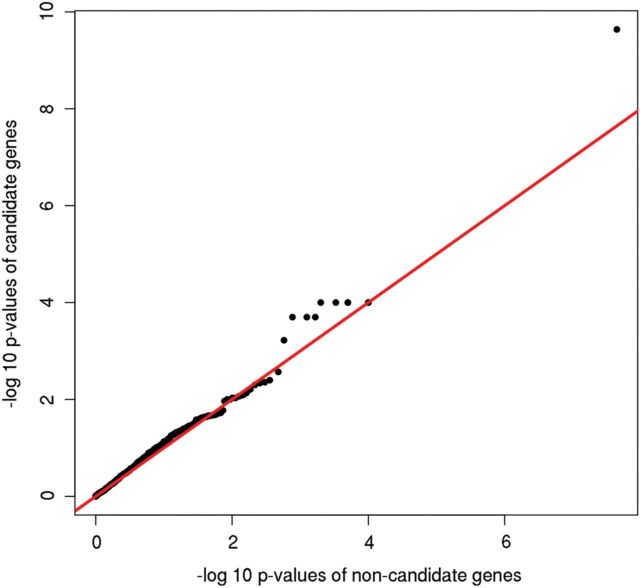
A quantile–quantile plot comparing gene-level P-values between candidate (ngenes = 547) and non-candidate genes (ngenes = 12 722) shows a possible enrichment of candidate genes (Fig. 1). The hypergeometric test of enrichment was not significant at the 5% P-value quantile (P = 0.35) but was nominally significant at the 25% quantile (P = 0.015) (Table 1). Similar results were obtained for the subset of genes (ncandidate = 544; nnon-candidate = 12 682) after excluding any SNPs with associations at P < 10−7 (P = 0.55 and P = 0.016, for the 5 and 25% threshold, respectively). The rank tail-strength and gene-set enrichments tests were nominally significant for the full set of genes and borderline significant for the subset without SNPs at P < 10−7 (Table 1). The Kolmogorov–Smirnov (KS) and Wilcoxon tests did not reach nominal significance, though the latter was borderline. Taken together, these results reflect greater power of the tests that are sensitive to the tail distribution of P-values. Although we applied multiple tests to the same data, the results are clearly correlated and it is reasonable to conclude that there is nominally significant evidence for enrichment in the tests best designed to detect such enrichment.
Figure 1.
Quantile–quantile plot comparing the observed adjusted minimum P-values of candidate gene SNPs with the observed adjusted minimum P-values of non-candidate gene SNPs.
Table 1.
P-values for enrichment of evidence for association in candidate genes compared with non-candidate genes
| GSEA | Hyper 5% | Hyper 25% | Rank TS | KS | Wilcoxon | |
|---|---|---|---|---|---|---|
| Genes | 0.0352 | 0.3452 | 0.0153 | 0.0424 | 0.1089 | 0.0562 |
| Genes (exclusion of associations with P < 10−7) | 0.0627 | 0.5506 | 0.0163 | 0.0684 | 0.1273 | 0.0665 |
| Sources of evidence | ||||||
| Animal studies | 0.2257 | 0.5324 | 0.0996 | 0.1412 | 0.3831 | 0.1418 |
| Monogenic obesity | 0.0320 | 0.0228 | 0.1597 | 0.0664 | 0.2552 | 0.1842 |
| Linkage | 0.2303 | 0.5642 | 0.0679 | 0.0474 | 0.0451 | 0.0295 |
| Association | 0.2975 | 0.3699 | 0.0766 | 0.0170 | 0.0124 | 0.0019 |
| Other including expression studies | 0.0596 | 0.1761 | 0.0248 | 0.0343 | 0.0534 | 0.0309 |
| Number of evidence sources | ||||||
| 4 | 0.2236 | 0.3036 | 0.0706 | 0.0434 | 0.0088 | 0.0043 |
| 3 | 0.1096 | 0.08703 | 0.2722 | 0.0465 | 0.0794 | 0.0358 |
| 2 | 0.4171 | 0.4703 | 0.1479 | 0.1273 | 0.1688 | 0.0584 |
| 1 | 0.0962 | 0.7082 | 0.0638 | 0.3466 | 0.2830 | 0.4845 |
| ≥3 | 0.0665 | 0.05169 | 0.1061 | 0.0115 | 0.0134 | 0.0043 |
| ≥2 | 0.1318 | 0.1365 | 0.0528 | 0.0113 | 0.0136 | 0.0036 |
GSEA, gene set enrichment analysis; Hyper x%, hypergeometric test with threshold at x percentile over P-value distribution; TS, tail strength; KS, Kolmogorov–Smirnov. Nominally significant (P < 0.05) results shown in bold, and all tests were one-sided.
For candidate genes within each category of evidence, there were nominally significant tests for all categories, except for genes identified by animal studies (Table 1). For genes associated with monogenic obesity, only the GSEA and hypergeometric (5%) tests were significant, in contrast to the other categories (linkage, association, and other) for which the rank tail-strength and Wilcoxon tests reached nominal significance. As we had no prior preference for which categories would show enrichment, a multiple test correction should apply, under which no categories show significant enrichment. Therefore our results on enrichment within categories of candidate genes should only be regarded as suggestive.
After grouping genes according to the number of categories providing evidence, there was nominally significant enrichment of candidate genes with at least two categories of evidence, of genes with at least three categories of evidence and of genes with exactly four categories of evidence. Again, significance was nominal and was attained only with the rank tail-strength and one-sided KS and Wilcoxon tests (Table 1).
Of the SNPs in or near candidate genes, SNPs in BDNF (brain-derived neurotrophic factor) showed the most significant associations with BMI (P = 5.55 × 10−13 for rs10767664), followed by associations of non-synonymous SNPs in SH2B1 (SH2B adaptor protein 1), MC4R (melanocortin 4 receptor) and GIPR (gastric inhibitory polypeptide receptor) (Table 2). These four genes had previously been highlighted by Speliotes et al. (5), as they reached genome-wide significance at the follow-up. SNPs in other four genes that ranked among the top 2% located in or near PMS2L3 (postmeiotic segregation increased 2-like 3), SCG3 (secretogranin III), VPS13B/COH1 (vacuolar protein sorting 13 homolog B/Cohen syndrome) and CFB (complement factor B) (Table 2).
Table 2.
The most significant SNPs in candidate genes that were rank in the top 2% in the GIANT consortium (5)
| Gene symbol | Gene name | Number of evidence sources | SNP | Location in/near gene | Chr. location | MAF | Betaa | SEa | P-value | Rankb |
|---|---|---|---|---|---|---|---|---|---|---|
| BDNF | Brain-derived neurotrophic factor | 3 | rs10767664 | Intronic (in LD (r2 = 0.77) with rs6265, V66M) | 11p13 | 0.21 | 0.04 | 0.005 | 5.55 × 10−13 | 158 |
| SH2B1 | SH2B adaptor protein 1 | 2 | rs7498665 | T484A | 16p11.2 | 0.40 | −0.03 | 0.004 | 1.76 × −10 | 215 |
| MC4R | Melanocortin 4 receptor | 4 | rs2229616 | V103I | 18q22 | 0.02 | −0.09 | 0.02 | 8.38 × −7 | 922 |
| GIPR | Gastric inhibitory polypeptide receptor | 1 | rs1800437 | E354Q | 19q13.3 | 0.22 | −0.03 | 0.006 | 8.56 × −7 | 925 |
| PMS2L3 | Postmeiotic segregation increased 2-like 3 | 1 | rs1167827 | 5′ | 7q11-q22 | 0.44 | −0.02 | 0.005 | 2.10 × −6 | 1091 |
| SCG3 | Secretogranin III | 1 | rs2652594 | Intronic | 15q21 | 0.44 | 0.02 | 0.004 | 3.82 × −6 | 1192 |
| VPS13B/COH1 | Vacuolar protein sorting 13 homolog B/Cohen syndrome | 1 | rs12550139 | Intronic | 8q22.2 | 0.12 | −0.03 | 0.006 | 1.61 × −5 | 1626 |
| CFB | Complement factor B | 2 | rs440454 | 3′ | 6p21.3 | 0.29 | 0.02 | 0.005 | 2.47 × −5 | 1866 |
aBeta (SE) represents the effect of the minor allele on BMI, which was inverse normally transformed. All summary statistics were derived from Speliotes et al. (5).
bRank represents the rank of the given SNP among all SNPs tested, when sorted from most significant to least significant.
DISCUSSION
Based on an extensive search of the literature, we developed a comprehensive list of candidate genes for obesity. Using genome-wide association data from a large consortium meta-analysis, we show nominally significant evidence of enrichment for association among candidate genes compared with non-candidate genes. This enrichment is particularly evident among genes identified from human linkage and association studies, and for genes proposed from at least three separate lines of evidence.
These results suggest that, to a mild extent, candidate genes are more likely to be truly associated than other non-candidate genes, at least for obesity-susceptibility. However, the degree of enrichment is small and the statistical significance in our data is marginal despite the extensive length of the candidate gene list and the large sample size. The fact that candidate genes will have made the list based on evidence of small studies and false-positive observations may have contributed to the limited enrichment, in particular if there was only one source of evidence. Analysis methods can be devised that give more weight to candidate than non-candidate genes, but the relative weighting cannot be high and any improvement in power would be slight. Similarly, focusing exclusively on candidate genes is unlikely to lead to significant gains in power. Since most genes in the genome are not candidate genes, it is likely that much of the unexplained heritability for BMI can be attributed to genes that are not currently considered as candidates.
We considered the regions around genes with no prior evidence for candidacy as a baseline for comparison, rather than intergenic regions. Other authors have presented evidence that SNPs in genes are more strongly associated than those in non-coding regions (6). We were interested in whether, given a focus on testing genes, there is value in specifically considering candidate genes. Our results suggest that although there is evidence for the enrichment of association in candidate genes for BMI, the minor extent of that enrichment implies limited practical utility for improving the power of association studies.
As a side issue, we considered several possible tests of enrichment and introduced a novel test, the rank tail-strength, which appears to yield the most significant results across a range of tests. It was not the most significant test for any of the analyses we performed, but was nominally or borderline significant in all the cases in which other tests were significant. Given the potentially confusing array of enrichment tests available, the rank tail-strength shows promise as a test with consistent power. Its analytic distribution is known when the number of genes is large, as is the case here, ensuring the type-1 error rate is correct. To more thoroughly characterize its properties, its power should be assessed under a range of simulated conditions. This is beyond the scope of the present work, but our results suggest that further study of this method is warranted.
For the three candidate genes with the strongest associations in GIANT (BDNF, SH2B1, MC4R), there was evidence for their candidacy from multiple sources, whereas the other candidate genes with the next strongest associations only had evidence from one category each, with the exception of CFB. Of interest is that the SNPs in the top four candidate genes were either non-synonymous or in moderately high LD with a non-synonymous SNP.
BMI and related obesity-susceptibility traits are common phenotypes with a modest heritability (h2: ∼40–70%) and a non-Mendelian pattern of inheritance. It remains to be determined whether the degree of the enrichment of association in candidate genes can be generalized to similar traits and conditions, and which characteristics (e.g. heritability, prevalence, inheritance pattern) influence the degree of enrichment most. We should also note that we only considered genes for overall obesity-susceptibility, as assessed by BMI, rather than body fat distribution or body shape.
Taken together, we observed evidence for the enrichment of association in candidate genes, suggesting that the candidate gene approach retains some value. However, the degree of enrichment is small despite the extensive number of candidate genes and the large sample size. Studies that focus exclusively on candidate genes have only slightly increased chances of detecting associations, and are likely to miss many true effects in non-candidate genes, at least for obesity-related traits.
MATERIAL AND METHODS
Candidate genes
We defined ‘candidate genes’ as genes for which there was at least some a priori evidence for a role in obesity-susceptibility. As such, genetic loci identified by GWASs for obesity-related traits were not per se considered as candidate genes, but only when they overlapped with our a priori defined list.
The candidate genes for obesity were selected based on five lines of evidence from literature. These evidences include reports on animal models, Mendelian forms of obesity, location near linkage peaks or quantitative trait loci, human genetic association studies and other sources of evidence including expression studies. All of the relevant studies were identified by extended computer-based searches of PubMed databases. The following search terms were used: ‘obesity in mice’ or ‘obesity in animals’; ‘monogenic obesity’ or ‘Mendelian obesity syndrome’; ‘genes for obesity’ or ‘obesity pathway genes’ or ‘obesity candidate genes’; ‘obesity linkage studies’, ‘obesity expression analysis’; and ‘obesity genetic association studies’ or ‘obesity polymorphism’ or ‘obesity variant’. The retrieved studies were then read in their entirety to assess their appropriateness for inclusion as a candidate gene. In total, we identified 547 candidate genes (Supplementary Material, Table S1). Genomic regions studied were defined to include the candidate genes ±10 kb of flanking sequence around each gene (defined according to NCBI Build 35). Next, we assigned a rank to each gene based on the number of evidences. A rank of 4, 3, 2 and 1 indicates the genes having 1, 2, 3 and 4 lines of evidence, respectively. There were seven candidate genes with a rank of 1, 47 genes with a rank of 2148 genes with a rank of 3 and 334 with a rank of 4 (Supplementary Material, Table S1). Of the 547 candidate genes, 277 had evidence from animal studies, 57 from monogenic forms of obesity, 58 from linkage studies, 210 from association studies and 141 from other sources of evidence. The complete list of candidate genes with their sources of evidence is provided in Supplementary Material, Table S1.
The total number of genes in the analyses, including the candidate genes, was 13 269 (547 candidate and 12 722 non-candidate genes.
Information on gene–SNP annotation was obtained according to the University of California Santa Cruz (UCSC) genome server, http://genome.ucsc.edu/cgi-bin/hgTables?command=start, through combining appropriate fields in ‘Variation and Repeats’ and ‘Genes and Gene Prediction Tracks’ groups.
Available genome-wide association data
To examine enrichment of candidate gene associations, we used summary statistics available from the discovery stage of the genome-wide meta-analysis for BMI performed by the GIANT consortium as described previously (5). Briefly, in this study, summary statistics of 46 GWASs, each of which had tested association between ∼2.8 m SNPs and BMI in sex-specific strata, were combined in a meta-analysis using the inverse variance method, including up to 123 865 individuals of white European descent. Quality control was applied to each GWAS prior to the meta-analysis, excluding poorly imputed SNPs and SNPs with a minor allele count less than six. Genomic control correction was applied to each individual GWAS and to the final meta-analysed results. The GIANT GWAS data are publically available at http://www.broadinstitute.org/collaboration/giant/.
Statistical analysis
We are primarily interested in whether associations observed for candidate genes are stronger than those for non-candidate genes. To allow for possible confounding by gene size, linkage disequilibrium and the number of SNPs per gene, we adjusted SNP P-values within each gene to obtain gene-level P-values. As we did not have individual-level genotype data, we made this adjustment by taking the corresponding CEU founder genotypes from HapMap2, assigning a random binary phenotype to each HapMap individual and then using permutation tests implemented in PRESTO (13) to estimate the null distribution for the minimum P-value within each gene. The gene-level P-value was estimated as (R + 1)/(N + 1), where R is the number of permutations in which the minimum SNP P-value in the gene was less than or equal to the value seen in the meta-analysis, and N is the total number of permutations, which was 10 000.
There are many methods to test for the enrichment of association, defined as a systematically lower distribution of P-values among candidate genes compared with non-candidate genes. A simple and commonly used method is a hypergeometric test, or its binomial approximation, comparing the proportion of candidate genes with P-values lower than a given threshold with the corresponding proportion of non-candidate genes. We applied a one-sided hypergeometric test using the 5 and 25% points of the empirical distribution of P-values as thresholds for the entire set of genes. These thresholds were intended to capture enrichment in the tail of the overall P-value distribution, allowing either a minority or majority of the candidate genes to contribute to the enrichment, noting that the candidate genes comprise about one-quarter of the full set.
Some methods have been proposed that consider the whole distribution of P-values rather than just the number exceeding a threshold. In principle, these tests could be more powerful than the hypergeometric test (14). We considered the tail-strength statistic (15), which is sensitive to the departure of P-values from uniformity in the tail of the distribution. Both candidate and non-candidate genes could have a departure from uniformity; therefore, to test enrichment of the candidate genes, we calculated the tail strength for the ranks of the candidate gene P-values among the whole list of genes, scaled to lie within (0.1). This test, which is novel, is termed the rank tail-strength test.
We also applied the gene-set enrichment analysis test (16), using the normalized enrichment score and the −log transform of the P-values. This is a modification of the KS test towards greater sensitivity in the tail, and for comparison we also applied the standard KS test and the Wilcoxon rank-sum test to compare the sets of P-values in candidate and non-candidate genes.
For the gene-set enrichment analysis, significance was calculated by a permutation test that randomly allocates each gene to the candidate or non-candidate list, keeping the total size of each list fixed. Each statistic is then calculated on the randomized data and an empirical P-value estimated as the proportion of random permutations in which the statistic exceeds the value observed in the original data. Results were based on 10 000 random permutations. For the rank tail-strength, significance was calculated from the asymptotic distribution for 547 candidate genes, given by N(0, 547−1) (15).
We compared the distribution of gene-level P-values for all candidate and non-candidate genes, and also for the reduced lists obtained by excluding all genes that included any SNP with P < 10−7. This was because the results could be skewed towards the enrichment of either set by a few extreme results, and also because we are interested in whether future studies would benefit by studying candidate genes that are not significant at the genome-wide level, and could therefore be found by standard GWASs. We also compared the distributions of P-values for candidate genes in each of the five evidence categories described above, and for sets of genes defined according to the number of categories providing evidence in favour of candidacy.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Medical Research Council through funding granted to the Epidemiology and Biostatistics Units.
ACKNOWLEDGEMENTS
The publicly available summary statistics of the genome-wide association meta-analyses of BMI (Speliotes et al. Nature Genetics 2010), published by the GIANT (Genetic Investigation of Anthropometric Traits) Consortium, was an important resource for the current study.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Maes H.H., Neale M.C., Eaves L.J. Genetic and environmental factors in relative body weight and human adiposity. Behav. Genet. 1997;27:325–351. doi: 10.1023/a:1025635913927. doi:10.1023/A:1025635913927. [DOI] [PubMed] [Google Scholar]
- 2.Rankinen T., Zuberi A., Chagnon Y.C., Weisnagel S.J., Argyropoulos G., Walts B., Perusse L., Bouchard C. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. doi:10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 3.Loos R.J. Genetic determinants of adiposity. In: Symonds M.E., editor. Adipose Tissue Biology. New York, NY: Springer Science; 2012. pp. 317–378. [Google Scholar]
- 4.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. doi:10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Allen H.L., Lindgren C.M., Luan J., Magi R., et al. Association analyses of 249796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. doi:10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moskvina V., Craddock N., Holmans P., Nikolov I., Pahwa J.S., Green E., Owen M.J., O'Donovan M.C. Gene-wide analyses of genome-wide association data sets: evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Mol. Psychiatry. 2009;14:252–260. doi: 10.1038/mp.2008.133. doi:10.1038/mp.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. doi:10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knight J., Barnes M.R., Breen G., Weale M.E. Using functional annotation for the empirical determination of Bayes factors for genome-wide association study analysis. PLoS One. 2011;6:e14808. doi: 10.1371/journal.pone.0014808. doi:10.1371/journal.pone.0014808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoo Y.J., Bull S.B., Paterson A.D., Waggott D., Sun L. Were genome-wide linkage studies a waste of time? Exploiting candidate regions within genome-wide association studies. Genet. Epidemiol. 2010;34:107–118. doi: 10.1002/gepi.20438. doi:10.1002/gepi.20438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S., et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. doi:10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. doi:10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho J.H., Gregersen P.K. Genomics and the multifactorial nature of human autoimmune disease. N. Engl. J. Med. 2011;365:1612–1623. doi: 10.1056/NEJMra1100030. doi:10.1056/NEJMra1100030. [DOI] [PubMed] [Google Scholar]
- 13.Browning B.L. PRESTO: rapid calculation of order statistic distributions and multiple-testing adjusted P-values via permutation for one and two-stage genetic association studies. BMC Bioinformatics. 2008;9:309. doi: 10.1186/1471-2105-9-309. doi:10.1186/1471-2105-9-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fridley B.L., Jenkins G.D., Biernacka J.M. Self-contained gene-set analysis of expression data: an evaluation of existing and novel methods. PLoS One. 2010;5:e12693. doi: 10.1371/journal.pone.0012693. doi:10.1371/journal.pone.0012693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor J., Tibshirani R. A tail strength measure for assessing the overall univariate significance in a dataset. Biostatistics. 2006;7:167–181. doi: 10.1093/biostatistics/kxj009. doi:10.1093/biostatistics/kxj009. [DOI] [PubMed] [Google Scholar]
- 16.Wang K., Li M., Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 2007;81:1278–1283. doi: 10.1086/522374. doi:10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]