Abstract
Circulating androgen levels are often used as indicators of physiological or pathological conditions. More than half of the variance for circulating androgen levels is thought to be genetically influenced. A genome-wide association study (GWAS) has identified two loci, SHBG at 17p13 and FAM9B at Xp22, for serum testosterone (T) levels; however, these explain only a small fraction of inter-individual variability. To identify additional genetic determinants of androgen levels, a GWAS of baseline serum T and dihydrotestosterone (DHT) levels was conducted in 3225 men of European ancestry from the REduction by DUtasteride of Prostate Cancer Events (REDUCE) study. Cross-validation was used to confirm the observed associations between the drug (n = 1581) and placebo (n = 1644) groups of REDUCE. In addition to confirming the associations of two known loci with serum T levels (rs727428 in SHBG: P = 1.26 × 10−12; rs5934505 in FAM9B: P = 1.61 × 10−8), we identified a new locus, JMJD1C at 10q21 that was associated with serum T levels at a genome-wide significance level (rs10822184: P = 1.12 × 10−8). We also observed that the SHBG locus was associated with serum DHT levels (rs727428: P = 1.47 × 10−11). Moreover, two additional variants in SHBG [rs72829446, in strong linkage equilibrium with the missense variant D356N (rs6259), and rs1799941] were also independently associated with circulating androgen levels in a statistical scale. These three loci (JMJD1C, SHBG and FAM9B) were estimated to account for ∼5.3 and 4.1% of the variance of serum T and DHT levels. Our findings may provide new insights into the regulation of circulating androgens and potential targets for androgen-based therapy.
INTRODUCTION
Androgens are male sex steroids derived from cholesterol and are essential for gender determination and maturation in men (1). Circulating levels of androgens are important physiological or pathological markers for several human diseases (such as benign prostatic hyperplasia and prostate cancer) and disorders (such as hypogonadism and congenital adrenal hyperplasia) as well as responses to androgen-based therapies (2). Testosterone (T) and dihydrotestosterone (DHT) are the two major androgens in men. T is the major male androgen in circulation, while DHT is the principal androgen in the prostate and hair follicle. About 90% of circulating T is secreted by Leydig cells in the testis (3). In contrast, only 25% of circulating DHT is produced in the testis, with most of the DHT (∼70%) arising from conversion of T in peripheral tissues (such as that of the prostate and skin) via 5α-reductase (4).
Evidence from twin studies has showed that more than half of the variance for circulating androgens levels is accounted by genetic factors (5,6). Recently, Ohlsson et al. (7) performed a meta-analysis of genome-wide association studies (GWASs) of serum T concentration in men and identified two loci of SHBG (sex hormone-binding globulin) at 17p13 [lead single nucleotide polymorphisms (SNPs): rs12150660 and rs6258] and FAM9B (family with sequence similarity 9, member B) at Xp22 (lead SNP: rs5934505) that were significantly associated with T levels. However, each variant conferred a modest effect and explained only a small fraction (0.6–2.3%) of inter-individual variability in serum T levels. This suggests that additional sequence variants influencing T levels remain to be discovered. In addition, no studies have reported genetic determinants for serum DHT levels to date.
To further understand the heritability of the circulating androgen levels, we conducted GWAS of baseline serum T and DHT levels in 3225 men of European descent within the REduction by DUtasteride of Prostate Cancer Events (REDUCE) study. The loci identified as associated with circulating androgen levels were also tested for association with serum prostate-specific antigen (PSA) levels and prostate volume at the baseline, and prostate cancer risk during a 4-year follow-up.
RESULTS
A total of 642 461 genotyped SNPs in 3225 individuals (Supplementary Material, Table S1) were analyzed for associations between SNPs and serum levels of the androgens T and DHT. The –log10 P-values by chromosome location for T and DHT are shown in Figure 1A and B, respectively. Q–Q plots for T and DHT are presented in Supplementary Material, Figure S1; the estimated inflation factors were modest (λ = 1.05 for T and 1.02 for DHT) and thus the reported P-values are not corrected for genomic inflation. As shown in Supplementary Material, Table S2, a total of 13 SNPs reached the genome-wide significance level (<5 × 10−8) for associations with T. Of these 13 SNPs, 4 were at 10q21 (lowest P-value of 1.20 × 10−8seen for rs10822186), 8 at 17p13 (lowest P-value of 1.26 × 10−12 seen for rs727428) and 1 at Xp22 (rs5934505: P = 1.61 × 10−8). For associations with DHT, there were nine significant SNPs, all of which are located at 17p13, with the most significant SNP being rs727428 (P = 1.47 × 10−11). SNP rs727428 was the most significant locus associated with both T and DHT. Significant association results were consistently observed among the two treatment groups (drug group: 1581 subjects and placebo group: 1644 subjects) for 10q21, 17p13 and Xp22 (Supplementary Material, Results; Supplementary Material, Table S3).
Figure 1.
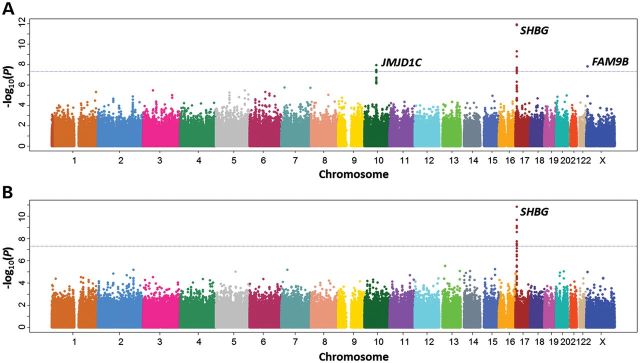
Manhattan plots of the strength of associations (–log10 P values; Y-axis) between SNPs (X-axis by chromosome and chromosomal position) and serum levels of testosterone (A) and DHT (B).
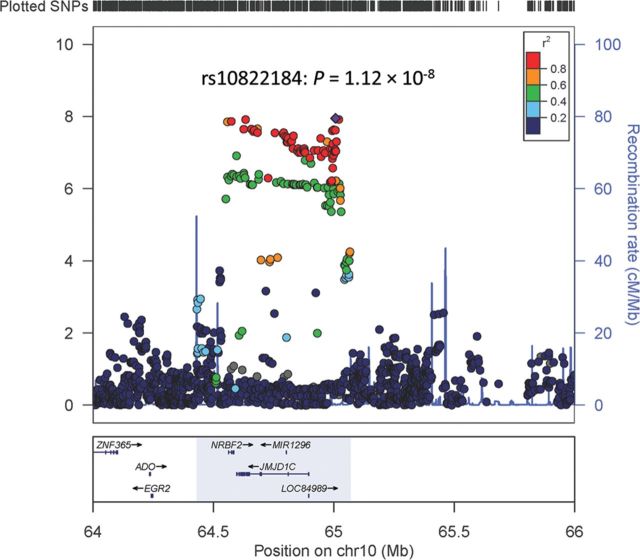
We identified a novel locus at 10q21 (rs10822186: P = 1.20 × 10−8; Table 1) that was associated with serum T levels at a genome-wide level of significance. To fine map the 10q21 region, we imputed additional SNPs at the region of chr10:64430000–65070000. As shown in Figure 2 and Supplementary Material, Table S4, 661 SNPs (including 69 genotyped and 592 imputed) were evaluated for association with T level. The most significant result was for rs10822184, an imputed SNP (P = 1.12 × 10−8), which was in strong linkage disequilibrium (LD) (r2 = 0.99) with the genotyped SNP rs10822186 (Table 1). After adjusting for rs10822184, none of the other SNPs at 10q21 remained significantly associated with T at P < 7.56 × 10−5 (0.05/661, the number of SNPs tested at this locus), suggesting no additional independent loci exist in this region. In addition, we found both rs10822184 and rs10822186 to be suggestively associated with DHT levels (P = 2.51 × 10−4 and 2.27 × 10−4, respectively) (Supplementary Material, Table S4). The SNP rs10822184 was estimated to account for 1.1 and 0.4% of the variance in serum T and DHT levels, respectively.
Table 1.
Summary result for SNPs at 10q21, 17p13 and Xp22 associated with serum testosterone (T) or DHT levels
| Chr. | SNP | Position | Gene (location) | Major/minor alleles (MAF) | T |
DHT |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| R2a | Beta (SE)a | Pa | R2a | Beta (SE)a | Pa | |||||
| 10q21 | rs10822184b | 65007159 | JMJD1C (∼111 kb at 5′-flank) | C/T (0.490) | 0.011 | −0.058 (0.010) | 1.12 × 10−8 | 0.004 | −0.055 (0.015) | 2.51 × 10−4 |
| rs10822186b | 65020389 | JMJD1C (∼125 kb at 5′-flank) | G/A (0.491) | 0.011 | −0.058 (0.010) | 1.20 × 10−8 | 0.004 | −0.055 (0.015) | 2.27 × 10−4 | |
| 17p13 | rs727428 | 7478517 | SHBG (1122 bp at 3′-flank) | C/T (0.414) | 0.014 | −0.073 (0.010) | 1.26 × 10−12 | 0.013 | −0.103 (0.015) | 1.47 × 10−11 |
| 17p13 | rs72829446b | 7492848 | SHBG (∼15 kb at 3′-flank) | C/T (0.101) | 0.010 | 0.099 (0.018) | 5.77 × 10−8 | 0.012 | 0.164 (0.027) | 9.46 × 10−10 |
| rs6259b | 7477252 | SHBG (exon 8 D356N) | G/A (0.107) | 0.008 | 0.091 (0.018) | 3.25 × 10−7 | 0.012 | 0.154 (0.026) | 4.04 × 10−9 | |
| 17p13 | rs1799941 | 7474148 | SHBG (8 bp at 5′-flank) | G/A (0.251) | 0.006 | 0.057 (0.013) | 1.53 × 10−5 | 0.005 | 0.076 (0.019) | 1.06 × 10−4 |
| Xp22 | rs5934505 | 8873826 | FAM9B (∼79 kb at 3′-flank)) | A/G (0.278) | 0.010 | 0.091 (0.016) | 1.61 × 10−8 | 0.006 | 0.104 (0.024) | 1.10 × 10−5 |
MAF, minor allele frequency.
aPartial R2 from linear regression models adjusted for age and Eigenvector. Models for rs72829446 and rs6259 were additionally adjusted for rs727428 while the model for rs1799941 was additionally adjusted for both rs727428 and rs72829446.
bThe imputed SNPs rs10822184 and rs72829446 are in strong LD with the genotyped SNPs rs10822186 (r2: 0.99) and rs6259 (r2: 0.88), respectively.
Figure 2.
Regional plot of the associations between SNPs at 10q21 and serum testosterone levels. Association of individual SNP is plotted as −log10 P against the chromosomal position. Results of both genotyped and imputed SNPs are shown. Colors indicate the LD strength between the most significant SNP rs10822184 and the other SNPs assessed. The right Y-axis shows the recombination rate estimated from the 1000 Genomes CEU population.
We also confirmed the locus 17p13 associated with T in the study of Ohlsson et al. (7). The two previously reported SNPs, rs12150660 and rs6258, were not directly genotyped in our study, thus an imputation analysis was performed to infer known untyped SNPs at 17p13 (chr17:7255000–7540000) based on combined data from the 1000 Genomes and HapMap3 projects. The most significant SNP for both T and DHT remained rs727428 (Supplementary Material, Table S5). The known SNP rs12150660 was confirmed as associated with both T and DHT, exceeding the level of genome-wide significance (P = 7.47 × 10−11 and 9.09 × 10−9, respectively). The other reported SNP, rs6258, was in a low frequency [minor allele frequency (MAF) = 0.004] and was also confirmed as associated with T and DHT (P = 1.53 × 10−4 and 5.53 × 10−3, respectively).
We performed a stepwise conditional analysis to assess independent association with androgen levels for the most significant SNP in our study (rs727428), two previously reported SNPs (rs12150660 and rs6258) and other SNPs in the region. After adjusting for rs727428 (named as region A), two additional SNPs (rs72829446 and rs1799941) remained significantly associated with T (P = 5.77 × 10−8 and 1.53 × 10−5, respectively) and DHT (P = 9.46 × 10−10 and 1.06 × 10−4, respectively) (Table 1), and were named as regions B and C, respectively. No other SNPs at 17p13 were associated with T or DHT at P < 1.54 × 10−4 (0.05/325, the number of SNPs tested at this locus). As shown in Supplementary Material, Figure S2, the SNP rs727428 at region A is in modest LD with rs1799941 at region B (r2 = 0.23), and with rs72829446 at region C (r2 = 0.14). Low LD is found between SNPs rs1799941 at region B and rs72829446 at region C (r2 = 0.03). The SNP rs1799941 (region C) is in strong LD with the previously reported SNP rs12150660 (r2 = 0.94). None of the above three independent SNPs is in LD with another reported SNP rs6258 (r2 = 0). In total, the combination of these three SNPs is estimated to account for ∼3% of the variance in serum T or DHT levels (Table 1). Notably for region C, the imputed SNP rs72829446 is in strong LD with the genotyped SNP rs6259 (r2 = 0.88), a non-synonymous variant at codon 356 of the SHBG gene, which was also significantly associated with T (P = 3.25 × 10−7) and DHT (P = 4.04 × 10−9) after conditioning on rs727428 (Table 1).
Our results confirmed the associations of Xp22 with T identified previously by Ohlsson et al. (7). In the current study, rs5934505 was the most significant SNP at Xp22 (P = 1.61 × 10−8) associated with serum T levels (Table 1), which is same as reported by Ohlsson et al. (7). This variant was estimated to account for 1.0% of the variance in serum T levels. We also observed that this SNP was associated with serum DHT levels, having a P-value of 1.10 × 10−5. None of the other SNPs at Xp22 was associated with T or DHT at P < 0.01 after conditioning on rs5934505.
When the independent SNPs at 10q21 (rs10822184), 17p13 (rs727428, rs72829446 and rs1799941) and Xp22 (rs5934505) were included in a multivariate linear regression model, these five SNPs were estimated to account for a total of 5.31 and 4.12% of the observed variance in serum T and DHT levels, respectively (Supplementary Material, Table S6). We further examined the association between each of these five SNPs with serum PSA levels, prostate volume and prostate cancer risk in the participants from REDUCE. Only marginal associations were observed between rs10822184 at 10q21 and baseline serum PSA (P = 0.017), prostate volume (P = 0.065) and risk of aggressive prostate cancer [relative risk (RR) = 1.33, 95% confidence interval (CI): 1.01–1.69; P = 0.022] (Supplementary Material, Results; Supplementary Material, Tables S7 and S8). The pleiotropy effects of rs10822184 with the above prostate-related traits were not statistically significant if multiple testing corrections were applied.
DISCUSSION
In this study, we identified a new locus at 10q21that were associated with serum androgen levels using a genome-wide association approach in 3225 men of European descent, and confirmed two loci at 17p13 and Xp22 that were reported by Ohlsson et al. (7). Our results further indicate that three genetic markers (rs727428, rs72829446 and rs179941) at 17p13 were independently associated with androgen levels in a statistical scale. Results of this study may provide important insight into the regulation of endogenous androgen concentrations, and may also have some clinical relevance for other diseases and disorders that are influenced by androgens, such as type 2 diabetes (8), vascular disease (9) and metabolic syndrome (10).
Although the association with serum androgen levels at 10q21 was not described in the original published meta-analysis of Ohlsson et al (7), supporting evidence for the association existed in the meta-analysis (personal communication with Dr Ohlsson). The lead SNP at 10q21 (rs10822186) was significantly associated with serum androgen level in their meta-analysis of 8938 men from seven cohorts (P = 3.09 × 10−3, Supplementary Material, Table S9). This independent confirmation increases the confidence that the new locus likely represents a true association. Of interest, SNPs within the 10q21 locus have been associated with blood lipoprotein levels (rs7923609, rs12768534 and rs10761731) (11,12) and plasma alkaline phosphatase levels (rs12355784 and rs10761779) (13) in previous GWAS. These SNPs are in strong LD with rs10822184 (r2 ≥ 0.7) and also significantly associated with T and DHT levels (Supplementary Material, Table S10). Four genes (JMJD1C, NRBF2, MIR1296 and LOC84989) are located in the identified 10q21 region. Of these genes, JMJD1C looks to be the most possible candidate because a previous study shows that JMJD1C may have transcriptional regulatory functions in the development of mouse testis and may regulate the metabolism of cholesterol to testosterone through coordinated regulation of histone methylation and demethylation (14). Thus, it is biologically plausible that genetic variants in JMJD1C may modify circulating androgen levels, given that 90% of circulating T is secreted by the testis in humans (4). Interestingly, one of the SNPs (rs7910927) located in JMJD1C was also recently reported to be significantly associated with circulating SHBG concentrations through a GWAS meta-analysis (40). It is unknown which of the identified 10q21 SNPs, if any, may be functional, because the 146 SNPs that remained significant even after Bonferroni correction (Supplementary Material, Table S4), including rs10822184 and rs10822186, are all located in intronic or intergenic regions; it is also possible that the functional SNP has yet to be identified.
Although multiple genes localize to 17p13, one likely candidate gene is the well-known SHBG. The primary function of SHBG is to bind circulating sex hormones (such as androgens and estrogens) thereby influencing the hormones' bioavailable fractions and sequestering the circulating hormones from biologic action (15). There is some evidence suggesting that SHBG may also play a role in steroid signaling, through binding to a cell membrane receptor (RSHBG) and inducing cAMP synthesis (16). In the circulation, 50–60% of T is bound to SHBG, ∼40–50% is bound loosely to albumin and 1–2% is in a free state (4), whereas serum DHT levels are only 10% of the serum levels of T (17).
In the circulation, T and DHT levels are positively correlated to SHBG levels (18). Previous studies have shown that some genetic variants of SHBG are associated with circulating levels of SHBG and sex hormones. For rs6259, a non-synonymous variant that results in the substitution of asparagines for aspartic acid at codon 356 (D356N, also known as D327N) in exon 8 of SHBG, the variant allele has been associated with decreased SHBG's metabolic clearance rate in animal models (19). Consistently, the variant allele has been associated with increased SHBG levels in most (20–25) but not all (26,27) studies. This variant has also been associated with blood T levels in a previous study (25). In the current study, we found this non-synonymous variant, in strong LD with rs72829446 (r2 = 0.88), was independently associated with both T and DHT levels, which may be explained as a result of SHBG levels regulated by rs6259.
In the GWAS conducted by Ohlsson et al. (7), rs12150660 was identified as an independent variant associated with serum T levels. In the current study, we found this variant was also significantly associated with T and DHT levels and in strong LD with the independent SNP rs1799941 (r2 = 0.94). For rs1799941, located eight nucleotides upstream of the transcriptional start point of SHBG, its variant allele has been shown to be strongly associated with circulating levels of SHBG (20,23,26,28–31), T (29,30) and DHT (28). This variant is in strong LD with a (TAAAA)n pentanucleotide repeat polymorphism (rs35785886, r2 > 0.8) located within an alu sequence at the 5′ boundary of the SHBG promoter (23,30). This repeat polymorphism has been found to influence the transcription of SHBG through its interactions with downstream elements, including an SP1 binding site, and may contribute to differences in plasma SHBG levels between individuals (32).
The SNP rs727428 was the most significant variant at 17p13 associated with T and DHT levels. The SNP rs727428 has also been previously associated with blood SHBG levels (23,26). This variant is 1.1 kb downstream of the 3′-end of SHBG and is thought to influence transcription factor binding directly or indirectly due to its strong LD with a promoter variant (rs858518) in SHBG (23). Taken together with our findings, the mechanism by which altered levels of circulating androgens are influenced by rs724828 at 17p13 warrant further functional studies.
Of note, the above three independent SNPs at 17p13 were identified in terms of statistical scale. No obvious recombination hotspots or separated LD blocks were observed between these three SNPs. In addition, the non-synonymous SNP rs6258 (P156L) in exon 4 of SHBG, which was shown to affect SHBG's affinity for binding T and the measured free T fraction, and identified as an independent locus for serum T levels by Ohlsson et al. (7), was not identified as independent SNP in the current study according to the predefined criteria (P < 1.54 × 10−4 after stepwise regression analysis). Though the effect of this SNP could not be explained by the above three SNPs because no LDs are observed (Supplementary Material, Fig. S2), its relative low frequency (MAF = 0.004) might reduce the statistical power in this study.
We confirmed the genome-wide significant association of SNP rs5934505 at Xp22 with serum androgen levels that were originally reported by Ohlsson et al. (7). This variant is located 79 kb downstream of FAM9B and 145 kb upstream of FAM9A (family with sequence similarity 9, member A). FAM9B and FAM9A share 46% amino acid identity and are expressed exclusively in the testis (33). The region consisting of FAM9B and FAM9A at Xp22 can be involved in a translocation with chromosome 10q24 (33). However, very little is known about the biological functions of these two genes.
Limitations of our study should be noted. First, the variants at 10q21, 17p13 and Xp22 only account for a small proportion of the overall observed variance in serum T and DHT levels. This suggests there may be other genetic determinants of serum androgen levels that remain unknown. Secondly, the statistical power of our study may be relatively limited, especially for assessing the association between SNPs and risk of prostate cancer, given that there were only 410 prostate cancer cases, and 124 with aggressive disease, among 1644 individuals in the placebo group. Thus, future studies with larger sample size may be needed to confirm and extend our findings in other populations.
In conclusion, we confirmed two loci, SHBG at 17p13and FAM9B at Xp22, and identified a new locus, JMJD1C at 10q21, as associated with circulating androgen levels, which are estimated to account for ∼5.3 and 4.1% of the variances observed for serum T and DHT levels, respectively. These findings provide new insights into the regulation of circulating androgen levels and may be of clinical relevance for androgen-related diseases.
MATERIALS AND METHODS
Study subjects
Subjects included in this study came from the REDUCE study, a multicenter, randomized, double-blind, placebo-controlled clinical trial, which was designed to evaluate the clinical value of dutasteride, a dual-5α-reductase inhibitor, in reducing the risk of incident prostate cancer. Details of the REDUCE study design and implementation have been described elsewhere (34,35). Briefly, men were enrolled if they were 50–75 years old; had a serum PSA level of 2.5–10.0 ng per milliliter; and had undergone a single prostate biopsy within 6 months before enrollment. Men were excluded if they had any of the following items: (i) undergone more than one biopsy; (ii) had prostate cancer of any grade, high-grade intraepithelial neoplasia, atypical small acinar proliferation, a history of prostate cancer or a prostate volume >80 ml; (iii) had undergone previous prostate surgery; or (iv) had an International Prostate Symptom Score of 25 or higher, or 20 or higher in the case of men taking alpha-blockers. For the current study, we included 3239 men of European descent who consented for genetic studies in REDUCE (drug group: 1585 subjects and placebo group: 1654 subjects). The characteristics of these 3239 participants were similar to the 2918 participants who did not provide consent for genetic studies (Supplementary Material, Table S1). For examining the associations of identified SNPs with prostate cancer risk, we restricted our study subjects to only the placebo group, using case- and non-case status after 4 years of follow-up (410 of 1654 men in the placebo group developed prostate cancer within this follow-up period). We further examined the identified SNPs for association with aggressive prostate cancer, defined as men (n = 124) that developed prostate cancer with a Gleason score of 7 or higher, stage T3b or higher, and/or were lymph node or metastasis positive (N+ or M+, respectively).
Measurement of serum levels of PSA, T and DHT and prostate volume
Serum levels of PSA at baseline were determined with an enzyme immunoassay at Quest Diagnostics (Van Nuys, CA, USA and Heston, Middlesex, UK). Serum testosterone levels were determined with a radioimmunoassay at Quest Diagnostics (San Juan Capistrano, CA, USA). Serum DHT levels were determined with a radioimmunoassay at Quest Diagnostics, Nichols Institute (San Juan Capistrano). Transrectal ultrasonography measurements by individual REDUCE investigators were used to calculate prostate volume in the formula: π/6 (anteroposterior width × cephalocaudal width ×transverse width).
Genotyping and imputation
DNA samples were genotyped using the Illumina HumanOmniExpress BeadChip, consisting of 729 755 SNPs, at the Center for Cancer Genomics, Wake Forest University. All 3225 samples genotyped (14 samples were not genotyped due to poor DNA quality) had a genome-wide call rate of ≥95% with an overall call rate of 99.7%; no individuals were excluded from GWAS analysis. The following quality control (QC) criteria were used to exclude SNPs from further analysis: MAF < 0.01 (n = 75 244), genotype call rate <95% (n = 6953) and P < 0.001 (n = 8666) for the Hardy–Weinberg equilibrium test. After exclusions, 642 461 SNPs remained for final genome-wide association analysis. To infer genotypes of SNPs that were not genotyped, we applied the IMPUTE computer program (36) using the combined data of the 1000 Genomes low-coverage pilot project and HapMap3 data as reference haplotype maps. A posterior probability of >0.90 was applied to call genotypes that were imputed from IMPUTE and the same QC procedure for excluding genotyped SNPs was applied to imputed SNPs.
Statistical analysis
A linear regression model was used to analyze the association of each SNP with quantitative traits, assuming an additive genetic model, as implemented in the PLINK software package (37). To limit potential bias due to deviation from normality of T and DHT, we tested associations after log transformation. Population stratification was estimated by a principal component approach, as implemented by the EIGENSTRAT software (38). The top Eigenvector as well as age was adjusted for as covariates in the linear regression analysis. A P of 5 × 10−8 was used as cutoff for genome-wide significance.
To confirm the significant loci from the analysis using the entire data set and to identify additional SNPs that were associated with baseline T or DHT but may not have reached genome-wide significance level, separate analysis was further performed to test consistent associations by partitioning based on the REDUCE treatment group (drug and placebo). Associations were considered true if P-values were <1 × 10−3 and had the same direction of effect in both the groups. For the regions that were genome-wide associated with T or DHT levels, known ungenotyped SNPs were imputed and conditional analysis was applied to test the independence of SNPs in stepwise manner, using the originally significant SNPs as covariates for subsequent analyses. The significance level for the independence test was defined as the Bonferroni-corrected significance level (0.05/n) according to the number of SNPs (n) tested in a given region.
T- or DHT-related SNPs were tested for association with baseline PSA levels and prostate volume using linear regression models adjusted for age and the top Eigenvector. The effects of identified SNPs on prostate cancer risk were further evaluated in the REDUCE placebo group. Cumulative incidence of prostate cancer among different genotype groups was used to estimate the RRs and 95% CIs for prostate cancer risk using log-binomial regression models (39). A similar approach was also applied to evaluate the relationship of SNPs with aggressive prostate cancer risk.
SUPPLEMENTARY MATERIAL
FUNDING
The study is partially supported by a national cancer institute RC2 grant (ca148463) to J.X.
ACKNOWLEDGEMENTS
We thank the patients enrolled in REDUCE who provided consent and genetic samples which enabled this study, and the clinicians who contributed their expertise in recruiting study patients for the REDUCE clinical study. Dave Pulford, Jennifer Aponte, Jon Charnecki and Mary Ellyn Volk participated in consent reconciliation and sample management to enable genetic sample selection for inclusion and genotype determination. Karen King provided data management support for this project. We appreciate the assistance of Lauren Marmor in coordinating the support of the AVODART collaborative research team. We also thank Claes Ohlsson, Henri Wallaschofski, Claudia Schurmann and their consortium of testosterone GWAS meta-analysis for sharing the results with us.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Mooradian A.D., Morley J.E., Korenman S.G. Biological actions of androgens. Endocr. Rev. 1987;8:1–28. doi: 10.1210/edrv-8-1-1. doi:10.1210/edrv-8-1-1. [DOI] [PubMed] [Google Scholar]
- 2.Penning T.M. New frontiers in androgen biosynthesis and metabolism. Curr. Opin. Endocrinol. Diabetes Obes. 2010;17:233–239. doi: 10.1097/MED.0b013e3283381a31. doi:10.1097/MED.0b013e3283381a31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Labrie F. Adrenal androgens and intracrinology. Semin. Reprod. Med. 2004;22:299–309. doi: 10.1055/s-2004-861547. doi:10.1055/s-2004-861547. [DOI] [PubMed] [Google Scholar]
- 4.Imamoto T., Suzuki H., Yano M., Kawamura K., Kamiya N., Araki K., Komiya A., Nihei N., Naya Y., Ichikawa T. The role of testosterone in the pathogenesis of prostate cancer. Int. J. Urol. 2008;15:472–480. doi: 10.1111/j.1442-2042.2008.02074.x. doi:10.1111/j.1442-2042.2008.02074.x. [DOI] [PubMed] [Google Scholar]
- 5.Ring H.Z., Lessov C.N., Reed T., Marcus R., Holloway L., Swan G.E., Carmelli D. Heritability of plasma sex hormones and hormone binding globulin in adult male twins. J. Clin. Endocrinol. Metab. 2005;90:3653–3658. doi: 10.1210/jc.2004-1025. doi:10.1210/jc.2004-1025. [DOI] [PubMed] [Google Scholar]
- 6.Kuijper E.A., Lambalk C.B., Boomsma D.I., van der Sluis S., Blankenstein M.A., de Geus E.J., Posthuma D. Heritability of reproductive hormones in adult male twins. Hum. Reprod. 2007;22:2153–2159. doi: 10.1093/humrep/dem145. doi:10.1093/humrep/dem145. [DOI] [PubMed] [Google Scholar]
- 7.Ohlsson C., Wallaschofski H., Lunetta K.L., Stolk L., Perry J.R., Koster A., Petersen A.K., Eriksson J., Lehtimaki T., Huhtaniemi I.T., et al. Genetic determinants of serum testosterone concentrations in men. PLoS Genet. 2011;7:e1002313. doi: 10.1371/journal.pgen.1002313. doi:10.1371/journal.pgen.1002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding E.L., Song Y., Malik V.S., Liu S. Sex differences of endogenous sex hormones and risk of type 2 diabetes: a systematic review and meta-analysis. JAMA. 2006;295:1288–1299. doi: 10.1001/jama.295.11.1288. doi:10.1001/jama.295.11.1288. [DOI] [PubMed] [Google Scholar]
- 9.Ruige J.B., Mahmoud A.M., De Bacquer D., Kaufman J.M. Endogenous testosterone and cardiovascular disease in healthy men: a meta-analysis. Heart. 2011;97:870–875. doi: 10.1136/hrt.2010.210757. doi:10.1136/hrt.2010.210757. [DOI] [PubMed] [Google Scholar]
- 10.Brand J.S., van der Tweel I., Grobbee D.E., Emmelot-Vonk M.H., van der Schouw Y.T. Testosterone, sex hormone-binding globulin and the metabolic syndrome: a systematic review and meta-analysis of observational studies. Int. J. Epidemiol. 2011;40:189–207. doi: 10.1093/ije/dyq158. doi:10.1093/ije/dyq158. [DOI] [PubMed] [Google Scholar]
- 11.Chasman D.I., Pare G., Mora S., Hopewell J.C., Peloso G., Clarke R., Cupples L.A., Hamsten A., Kathiresan S., Malarstig A., et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. doi:10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. doi:10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan X., Waterworth D., Perry J.R., Lim N., Song K., Chambers J.C., Zhang W., Vollenweider P., Stirnadel H., Johnson T., et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am. J. Hum. Genet. 2008;83:520–528. doi: 10.1016/j.ajhg.2008.09.012. doi:10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S.M., Kim J.Y., Choe N.W., Cho I.H., Kim J.R., Kim D.W., Seol J.E., Lee S.E., Kook H., Nam K.I., et al. Regulation of mouse steroidogenesis by WHISTLE and JMJD1C through histone methylation balance. Nucleic Acids Res. 2010;38:6389–6403. doi: 10.1093/nar/gkq491. doi:10.1093/nar/gkq491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siiteri P.K., Murai J.T., Hammond G.L., Nisker J.A., Raymoure W.J., Kuhn R.W. The serum transport of steroid hormones. Recent Prog. Horm. Res. 1982;38:457–510. doi: 10.1016/b978-0-12-571138-8.50016-0. [DOI] [PubMed] [Google Scholar]
- 16.Rosner W., Hryb D.J., Kahn S.M., Nakhla A.M., Romas N.A. Interactions of sex hormone-binding globulin with target cells. Mol. Cell Endocrinol. 2010;316:79–85. doi: 10.1016/j.mce.2009.08.009. doi:10.1016/j.mce.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Hsing A.W., Chu L.W., Stanczyk F.Z. Androgen and prostate cancer: is the hypothesis dead? Cancer Epidemiol. Biomarkers Prev. 2008;17:2525–2530. doi: 10.1158/1055-9965.EPI-08-0448. doi:10.1158/1055-9965.EPI-08-0448. [DOI] [PubMed] [Google Scholar]
- 18.Roddam A.W., Allen N.E., Appleby P., Key T.J. Endogenous sex hormones and prostate cancer: a collaborative analysis of 18 prospective studies. J. Natl Cancer Inst. 2008;100:170–183. doi: 10.1093/jnci/djm323. doi:10.1093/jnci/djm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cousin P., Dechaud H., Grenot C., Lejeune H., Pugeat M. Human variant sex hormone-binding globulin (SHBG) with an additional carbohydrate chain has a reduced clearance rate in rabbit. J. Clin. Endocrinol. Metab. 1998;83:235–240. doi: 10.1210/jcem.83.1.4515. doi:10.1210/jc.83.1.235. [DOI] [PubMed] [Google Scholar]
- 20.Dunning A.M., Dowsett M., Healey C.S., Tee L., Luben R.N., Folkerd E., Novik K.L., Kelemen L., Ogata S., Pharoah P.D., et al. Polymorphisms associated with circulating sex hormone levels in postmenopausal women. J. Natl Cancer Inst. 2004;96:936–945. doi: 10.1093/jnci/djh167. doi:10.1093/jnci/djh167. [DOI] [PubMed] [Google Scholar]
- 21.Cousin P., Calemard-Michel L., Lejeune H., Raverot G., Yessaad N., Emptoz-Bonneton A., Morel Y., Pugeat M. Influence of SHBG gene pentanucleotide TAAAA repeat and D327N polymorphism on serum sex hormone-binding globulin concentration in hirsute women. J. Clin. Endocrinol. Metab. 2004;89:917–924. doi: 10.1210/jc.2002-021553. doi:10.1210/jc.2002-021553. [DOI] [PubMed] [Google Scholar]
- 22.Haiman C.A., Riley S.E., Freedman M.L., Setiawan V.W., Conti D.V., Le Marchand L. Common genetic variation in the sex steroid hormone-binding globulin (SHBG) gene and circulating shbg levels among postmenopausal women: the Multiethnic Cohort. J. Clin. Endocrinol. Metab. 2005;90:2198–2204. doi: 10.1210/jc.2004-1417. doi:10.1210/jc.2004-1417. [DOI] [PubMed] [Google Scholar]
- 23.Thompson D.J., Healey C.S., Baynes C., Kalmyrzaev B., Ahmed S., Dowsett M., Folkerd E., Luben R.N., Cox D., Ballinger D., et al. Identification of common variants in the SHBG gene affecting sex hormone-binding globulin levels and breast cancer risk in postmenopausal women. Cancer Epidemiol. Biomarkers Prev. 2008;17:3490–3498. doi: 10.1158/1055-9965.EPI-08-0734. doi:10.1158/1055-9965.EPI-08-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding E.L., Song Y., Manson J.E., Hunter D.J., Lee C.C., Rifai N., Buring J.E., Gaziano J.M., Liu S. Sex hormone-binding globulin and risk of type 2 diabetes in women and men. N. Engl. J. Med. 2009;361:1152–1163. doi: 10.1056/NEJMoa0804381. doi:10.1056/NEJMoa0804381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanbillemont G., Bogaert V., De Bacquer D., Lapauw B., Goemaere S., Toye K., Van Steen K., Taes Y., Kaufman J.M. Polymorphisms of the SHBG gene contribute to the interindividual variation of sex steroid hormone blood levels in young, middle-aged and elderly men. Clin. Endocrinol. 2009;70:303–310. doi: 10.1111/j.1365-2265.2008.03365.x. doi:10.1111/j.1365-2265.2008.03365.x. [DOI] [PubMed] [Google Scholar]
- 26.Wickham E.P., 3rd, Ewens K.G., Legro R.S., Dunaif A., Nestler J.E., Strauss J.F., 3rd Polymorphisms in the SHBG gene influence serum SHBG levels in women with polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 2011;96:E719–E727. doi: 10.1210/jc.2010-1842. doi:10.1210/jc.2010-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turk A., Kopp P., Colangelo L.A., Urbanek M., Wood K., Liu K., Skinner H.G., Gapstur S.M. Associations of serum sex hormone binding globulin (SHBG) levels with SHBG gene polymorphisms in the CARDIA Male Hormone Study. Am. J. Epidemiol. 2008;167:412–418. doi: 10.1093/aje/kwm332. doi:10.1093/aje/kwm332. [DOI] [PubMed] [Google Scholar]
- 28.Eriksson A.L., Lorentzon M., Mellstrom D., Vandenput L., Swanson C., Andersson N., Hammond G.L., Jakobsson J., Rane A., Orwoll E.S., et al. SHBG gene promoter polymorphisms in men are associated with serum sex hormone-binding globulin, androgen and androgen metabolite levels, and hip bone mineral density. J. Clin. Endocrinol. Metab. 2006;91:5029–5037. doi: 10.1210/jc.2006-0679. doi:10.1210/jc.2006-0679. [DOI] [PubMed] [Google Scholar]
- 29.Ahn J., Schumacher F.R., Berndt S.I., Pfeiffer R., Albanes D., Andriole G.L., Ardanaz E., Boeing H., Bueno-de-Mesquita B., Chanock S.J., et al. Quantitative trait loci predicting circulating sex steroid hormones in men from the NCI-Breast and Prostate Cancer Cohort Consortium (BPC3) Hum. Mol. Genet. 2009;18:3749–3757. doi: 10.1093/hmg/ddp302. doi:10.1093/hmg/ddp302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huhtaniemi I.T., Pye S.R., Holliday K.L., Thomson W., O'Neill T.W., Platt H., Payne D., John S.L., Jiang M., Bartfai G., et al. Effect of polymorphisms in selected genes involved in pituitary-testicular function on reproductive hormones and phenotype in aging men. J. Clin. Endocrinol. Metab. 2010;95:1898–1908. doi: 10.1210/jc.2009-2071. doi:10.1210/jc.2009-2071. [DOI] [PubMed] [Google Scholar]
- 31.Melzer D., Perry J.R., Hernandez D., Corsi A.M., Stevens K., Rafferty I., Lauretani F., Murray A., Gibbs J.R., Paolisso G., et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. doi:10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hogeveen K.N., Talikka M., Hammond G.L. Human sex hormone-binding globulin promoter activity is influenced by a (TAAAA)n repeat element within an Alu sequence. J. Biol. Chem. 2001;276:36383–36390. doi: 10.1074/jbc.M104681200. doi:10.1074/jbc.M104681200. [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Garay I., Jablonka S., Sutajova M., Steuernagel P., Gal A., Kutsche K. A new gene family (FAM9) of low-copy repeats in Xp22.3 expressed exclusively in testis: implications for recombinations in this region. Genomics. 2002;80:259–267. doi: 10.1006/geno.2002.6834. doi:10.1006/geno.2002.6834. [DOI] [PubMed] [Google Scholar]
- 34.Andriole G., Bostwick D., Brawley O., Gomella L., Marberger M., Tindall D., Breed S., Somerville M., Rittmaster R. Chemoprevention of prostate cancer in men at high risk: rationale and design of the REduction by DUtasteride of Prostate Cancer Events (REDUCE) trial. J. Urol. 2004;172:1314–1317. doi: 10.1097/01.ju.0000139320.78673.2a. doi:10.1097/01.ju.0000139320.78673.2a. [DOI] [PubMed] [Google Scholar]
- 35.Andriole G.L., Bostwick D.G., Brawley O.W., Gomella L.G., Marberger M., Montorsi F., Pettaway C.A., Tammela T.L., Teloken C., Tindall D.J., et al. Effect of dutasteride on the risk of prostate cancer. N. Engl. J. Med. 2010;362:1192–1202. doi: 10.1056/NEJMoa0908127. doi:10.1056/NEJMoa0908127. [DOI] [PubMed] [Google Scholar]
- 36.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. doi:10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 37.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. doi:10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 39.McNutt L.A., Wu C., Xue X., Hafner J.P. Estimating the relative risk in cohort studies and clinical trials of common outcomes. Am. J. Epidemiol. 2003;157:940–943. doi: 10.1093/aje/kwg074. doi:10.1093/aje/kwg074. [DOI] [PubMed] [Google Scholar]
- 40.Coviello A.D., Haring R., Wellons M., Vaidya D., Lehtimaki T., Keildson S., Lunetta K.L., HE C., Fornage M., Lagou V., et al. A genome-wide association meta-analysis of circulating sex hormone-binding globulin reveals multiple loci implicated in sex steroid hormone regulation. PLoS Genet. 2012;8:e1002805. doi: 10.1371/journal.pgen.1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]