Abstract
Somatic cell nuclear transfer, a technique used to generate clone embryos by transferring the nucleus of a somatic cell into an enucleated oocyte, is an excellent approach to study the reprogramming of the nuclei of differentiated cells. Here, we conducted a transcriptomic study by performing microarray analysis on single Sertoli cell nuclear transfer (SeCNT) embryos throughout preimplantation development. The extensive data collected from the oocyte to the blastocyst stage helped to identify specific genes that were incorrectly reprogrammed at each stage, thereby providing a novel perspective for understanding reprogramming progression in SeCNT embryos. This attempt provided an opportunity to discuss the possibility that ectopic gene expression could be involved in the developmental failure of SeCNT embryos. Network analysis at each stage suggested that in total, 127 networks were involved in developmental and functional disorders in SeCNT embryos. Furthermore, chromosome mapping using our time-lapse expression data highlighted temporal–spatial changes of the abnormal expression, showing the characteristic distribution of the genes on each chromosome. Thus, the present study revealed that the preimplantation development of SeCNT embryos appears normal; however, the progression of incorrect reprogramming is concealed throughout development.
Introduction
With progression of mammalian development, which starts at fertilization, the totipotency of embryonic cells is rapidly lost and cell fate is progressively determined. Somatic cell nuclear transfer (SCNT) is an excellent approach for understanding the nuclear reprogramming of somatic cells, because it is the only way that somatic cells are turned into a totipotent stage, from which all types of cells, including cells of extra-embryonic tissues, can be differentiated (Wilmut et al. 1997, Wakayama et al. 1998, Gurdon & Wilmut 2011). In many species, embryo-cloning studies have demonstrated that the differentiated cell status is not irreversible and can be switched by various regulatory molecules (Thuan et al. 2010). However, only a limited proportion of SCNT embryos are allowed to develop into normal individuals and the majority die at different stages of development (Ono et al. 2001, Ogonuki et al. 2002).
To date, a number of studies have been conducted to understand how donor nuclei acquire totipotency and/or why the majority of SCNT fails to develop to term (Rideout et al. 2001, Hochedlinger & Jaenisch 2006, Peat & Reik 2012). Reprogramming of donor cells begins shortly after the transplantation of a donor cell nucleus into an enucleated oocyte (Gurdon et al. 1976), and Q-PCR studies have revealed that many genes were abnormally expressed in SCNT embryos (Bortvin et al. 2003, Inoue et al. 2006, Jincho et al. 2008). Transcriptome studies using microarray analysis are often an effective approach in studying the reprogramming, which occurs in SCNT embryos (Inoue et al. 2002, 2010, Pfister-Genskow et al. 2005, Smith et al. 2005, Somers et al. 2006, Wakayama et al. 2006, Beyhan et al. 2007, Vassena et al. 2007, Fukuda et al. 2010). Such studies have shown that a number of genes avoid being reprogrammed during the first and second cell cycles (Vassena et al. 2007), and a considerable number of genes were abnormally expressed at blastocysts (Inoue et al. 2002, Wakayama et al. 2006, Fukuda et al. 2010). Meantime, these results suggested that some part of the genes were corrected during development to the blastocyst stage. Furthermore, Inoue et al. (2010) obtained very interesting observations that inappropriate reprogramming of X chromosome-related genes could be responsible for developmental failure in SCNTs. Their gene expression profiles showed that 80% of the downregulated genes in Sertoli cell nuclear transfer (SeCNT) embryos recovered when Xist-knockout donor cells were used. In a previous study, we conducted further comprehensive transcriptome analysis of mouse SCNT embryos at the blastocyst stage by using a microarray designed for single embryos so as to gain better understanding on the rapid embryo loss around implantation (Fukuda et al. 2010). The results revealed that each type of SCNT embryos had a unique gene expression profile that strictly depended on three donor cells: Sertoli cells, ES cells, and cumulus cells. Furthermore, the analysis identified 35 genes that were inappropriately reprogrammed in most of the SeCNT blastocysts. Transcriptome studies in bovine have also shown global gene expression profiles in nuclear-transferred embryos at the blastocyst stage, and suggested that restrictive reprogramming errors might be magnified in the subsequent developmental stages in SCNT embryos (Pfister-Genskow et al. 2005, Smith et al. 2005, Somers et al. 2006, Beyhan et al. 2007).
Studies on reprogramming need to show the changing aspect of the gene expression profile of SCNT embryos throughout the preimplantation stages. For example, Tong et al. (2006) reported aberrant gene expression in cumulus-cloned embryos detected at the 2-cell, 4-cell, and blastocyst stages and found candidate genes related to cell structure, protein modification and cell signaling, and transcription factors (TFs). Since time-lapse gene expression profiles certainly provide beneficial information for understanding reprogramming in SCNT embryos, further precise and comprehensive transcriptome analysis should be performed in mice. In the present study, we used a transcriptomic approach to analyze the progression of reprogramming in SeCNT embryos throughout preimplantation development by using microarray analysis of single embryos. This novel research angle provided a comprehensive data set, which provided a whole picture of the reprogramming process in SeCNT embryos.
Results and discussion
Gene expression profile during preimplantation development
To gain a better understanding of the molecular mechanisms that underlie the reprogramming of SeCNT embryos, we performed transcriptomic profiling using single preimplantation-stage embryos from all preimplantation stages. Here, we analyzed SeCNTs because of their relatively high and stable development to term compared with other SCNTs (Ogura et al. 2000, Meissner & Jaenisch 2006). Of the activated SeCNT embryos, more than 60% developed to the blastocyst stage, and nine pups (10.1%) were obtained after the embryos were transferred to recipients in the present study (Supplementary Table 1, see section on supplementary data given at the end of this article). However, the majority of the embryos were lethal. At present, we are unable to select viable embryos for further analysis, but this might be possible in the near future by using a real-time imaging system (Mizutani et al. 2012).
IVF B6D2F2 (B6D2F1/B6D2F1) embryos were used as controls, because 95% or more of the IVF eggs develop to the blastocyst stage in our laboratory. Moreover, inappropriate gene expression caused by in vitro culturing can be eliminated among differentially expressed genes in SeCNTs. We analyzed IVF embryos (B6D2F1/B6D2F1) (n=3 at each stage), SeCNT embryos (n=5 at each stage), and MII oocytes (B6D2F1, n=3). The genetic background of donor Sertoli cells (B6CBF1) differed from that of the IVF controls (B6D2F2). Therefore, the genetic background might have affected the differences in gene expression profiles. However, the differences were perhaps not significant because a comparison of the gene expression profiles between the B6D2F2 and B6CBF1 embryos revealed that the correlation coefficient was rather high (0.97). The CEL file data that were obtained from Mouse GeneChip 430 2.0 were entered into GeneSpring GX 12.0 for the analysis. A dendrogram was constructed by performing a hierarchical clustering analysis (Fig. 1A), and the principal component analysis (PCA) results are shown in a 2D display (Fig. 1B).
Figure 1.
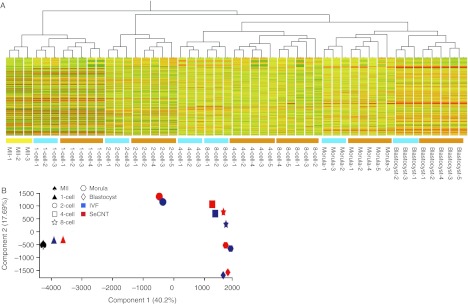
Hierarchical clustering dendrogram and principal component analysis (PCA). (A) Hierarchical clustering dendrogram of 51 samples. In total, 17 822 probe sets passed the cut-off RAW value of <100 and were used for hierarchical clustering. Yellow, metaphase II oocytes; blue, IVF samples; orange, SeCNT samples. (B) PCA results in a 2D display. Blue symbols, IVF embryos; red symbols, SeCNT embryos.
In the dendrogram (Fig. 1A), the samples were clearly divided into five groups according to the embryonic development stages in which SeCNT embryos were distinguished from IVF embryos at each stage. The gene expression profiles changed depending on the developmental stage rather than the difference between the IVF and SeCNT embryos. However, in the IVF groups, the gene expression profiles of 4-cell- and 8-cell-stage embryos were very similar and formed one cluster, and were definitely different from the SeCNT embryos. These results indicate that SeCNT embryos expressed transcriptomes that resembled that of IVF embryos. However, particular gene clusters were differentially expressed between the two groups (Kohda et al. 2005). The results from the PCA (Fig. 1B) supported the results of the dendrogram and showed three significant changes in the profiles between 1- and 2-cell stages, 2- and 4-cell stages, and the morula and blastocyst stages. These three changing phases reflected major events : firstly, the initial phase of reprogramming; secondly, zygotic gene activation; and finally, first cell differentiation into inner-cell-mass cells and trophoblastic cells (Chastant et al. 1996, Kawasumi et al. 2007, Sawai 2009, Egli et al. 2011).
Genes were differentially expressed in SeCNT embryos at each stage
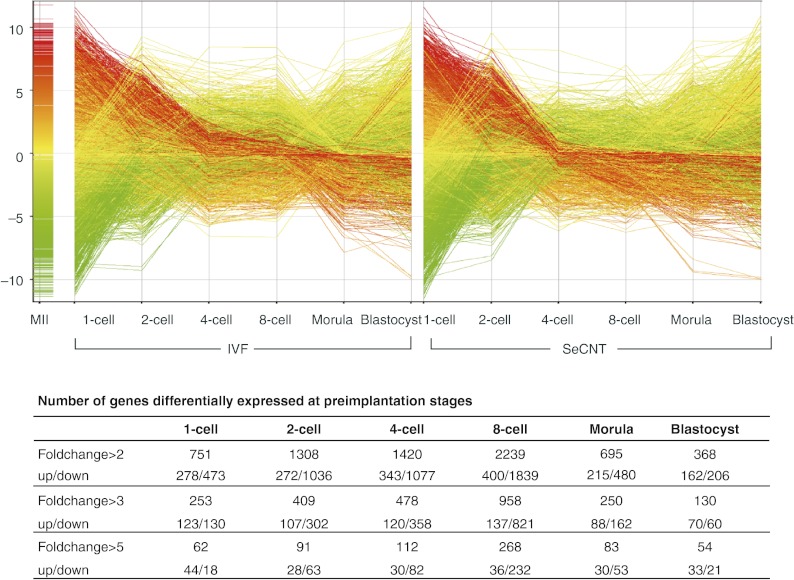
In an attempt to identify particular genes that needed to be reprogrammed correctly, we compared the gene expression profiles of SeCNT and control IVF embryos at each stage. The probe sets passed the filter of cut-off RAW values of <100 and subjected to a two-way ANOVA (P<0.001, 4793 probe sets). The number of differentially expressed genes is shown in Fig. 2, and genes that showed twofold or greater changes are listed in Supplementary Table 2, see section on supplementary data given at the end of this article.
Figure 2.
Time-lapse gene expression profile of the differentially expressed genes in SeCNT embryos during the preimplantation stages. In total, 4793 probe sets passed the two-way ANOVA and were used for fold-change selection at each stage. The number of genes differentially expressed at each stage is shown in the table by 2-, 3-, and 5-fold changes. Up/down means the probe sets were up/downregulated in SeCNT embryos. Each line represents one probe set: the color of the line represents the gene expression in MII; the line in red represents a higher expression in MII; and the line in green represents a lower expression.
Generally, the transcriptomic profiles of SeCNT embryos resembled those of IVF embryos (Fig. 2). A considerably large number of genes were differentially expressed at threefold or greater levels at each stage: a minimum of 130 genes at the blastocyst stage and a maximum of 958 genes at the 8-cell stage. However, genes showing fivefold or greater changes were relatively limited. Interestingly, of the differentially expressed genes, the number of downregulated genes in SeCNT embryos was higher than that of the upregulated genes from the 2-cell (107/302) to the morula (88/162) stage. The number was also six times higher at the 8-cell stage (137/821) but not at the blastocyst stage. Key genes, which would have affected the development of SeCNT embryos, were incorrectly reprogrammed throughout preimplantation development. Thus, the cloned embryos feigned seemingly proper embryonic development so that the progression of incorrect reprogramming was concealed throughout development. Therefore, SeCNT embryos rapidly lost their developmental capability from the 8-cell stage (Latham 2005), when the number of genes that were inappropriately expressed was greatest. Our previous study analyzed SCNT embryos that were constructed from three types of donor cells and showed that the abnormal gene expression profiles of SeCNT embryos were influenced by donor cell-specific gene expression (Fukuda et al. 2010). Furthermore, a gene ontology analysis suggested that the level of gene expression in the donor cells was responsible for disorders pertaining to specific biological functions in SeCNT embryos.
Because TFs always have a significant influence on the regulation of gene expression, their reprogramming failure directly affects wide-scale gene expression and the development of SeCNT embryos. We found that TFs, which represented 10–25% of the differentially expressed genes, were abnormally expressed at several stages of development, and, in particular, a large number of genes were inappropriately expressed with threefold or greater changes at the 2-cell (up: 18, down: 89), 4-cell (up: 19, down: 97), and 8-cell (up: 33, down: 154) stages (Supplementary Table 3, see section on supplementary data given at the end of this article). Therefore, the reprogramming failure of TFs might have been partly responsible for the significant abnormality in the transcriptome in SeCNT embryos. Notably, TFs such as Yy1 (Donohoe et al. 1999) and Tial1 (Beck et al. 1998), which have been reported via knockout studies to be involved in embryonic lethality, were completely repressed in SeCNT embryos. Furthermore, as we discuss below, many TFs function upstream to regulate the network of differentially expressed genes.
To gain a better understanding of the functions of the differentially expressed genes, we carried out a gene annotation analysis by IPA using genes that were differentially expressed at levels threefold or greater. The top ten functions of the genes at each stage are listed in Table 1. Several gene functions were frequently listed in the top ten list throughout the preimplantation stages, such as post-translational modification, gene expression, DNA replication/recombination/repair, and cell cycling. The results show that characteristic disorders occurred at each stage. Interestingly, ‘cell death’ was listed as the top function in the morula stage and was also included in the list of the top ten functions at the 8-cell stage. This coincides well with SeCNT embryo development, because these embryos were significantly retarded and arrested around these stages (Tveden-Nyborg et al. 2005). It is thought that differentiation of the embryonic cells into cell lineages of trophoblasts or inner cell mass is determined after the morula stage, but in the cloned embryos, some specific errors such as those detected here might lead to functional disorders in cell death and cell cycling (Park et al. 2004). Furthermore, genes that are regarded as having ‘free radical scavenging’ functions are listed in the top ten list at the 2-cell stage. This suggests that SeCNT embryos are exposed to oxidative stress, by which the embryos may suffer short- and long-term disorders (Ohashi et al. 2006). Furthermore, these results suggest that genes whose functions are related to metabolic activity (e.g. lipid metabolism, vitamin and mineral metabolism, and carbohydrate metabolism) were insufficiently expressed, especially at the blastocyst stage.
Table 1.
Top ten functions of differentially expressed genes at each embryonic stage by IPA.
| Cell stage (number of genes included) | P value |
|---|---|
| 1-Cell stage | |
| Post-translational modification (16) | 6.36×10−6 |
| DNA replication, recombination, and repair (15) | 3.91×10−5 |
| Cell-to-cell signaling and interaction (14) | 2.92×10−4 |
| Cellular development (16) | 5.81×10−4 |
| Gene expression (38) | 8.29×10−4 |
| Cell cycle (11) | 9.62×10−4 |
| Cell morphology (18) | 9.62×10−4 |
| Cellular function and maintenance (19) | 9.62×10−4 |
| Cellular assembly and organization (18) | 1.74×10−3 |
| Cellular compromise (8) | 5.09×10−3 |
| 2-Cell stage | |
| Free radical scavenging (5) | 2.81×10−4 |
| Cellular growth and proliferation (88) | 3.01×10−4 |
| Nucleic acid metabolism (4) | 3.01×10−4 |
| Small-molecule biochemistry (29) | 3.01×10−4 |
| Protein synthesis (33) | 5.18×10−4 |
| Post-translational modification (21) | 5.93×10−4 |
| Cellular compromise (12) | 8.92×10−4 |
| Cellular movement (19) | 1.02×10−3 |
| Cell cycle (59) | 1.50×10−3 |
| Cellular morphology (23) | 1.64×10−3 |
| 4-Cell stage | |
| DNA replication, recombination, and repair (41) | 4.69×10−7 |
| Gene expression (72) | 4.69×10−7 |
| Cellular assembly and organization (29) | 8.04×10−5 |
| Cellular development (35) | 2.04×10−4 |
| Cell cycle (53) | 3.24×10−4 |
| Cellular growth and proliferation (91) | 3.38×10−4 |
| Amino acid metabolism (5) | 9.15×10−4 |
| Small-molecule biochemistry (26) | 9.15×10−4 |
| Post-translational modification (18) | 2.08×10−3 |
| Lipid metabolism (9) | 3.03×10−3 |
| 8-Cell stage | |
| Gene expression (146) | 2.14×10−6 |
| Cell cycle (108) | 4.60×10−6 |
| Cellular assembly and organization (103) | 4.60×10−6 |
| DNA replication, recombination, and repair (71) | 4.60×10−6 |
| Post-translational modification (36) | 1.33×10−5 |
| Cell-to-cell signaling and interaction (15) | 1.45×10−4 |
| Drug metabolism (9) | 1.45×10−4 |
| Small-molecule biochemistry (72) | 1.45×10−4 |
| Cell death (184) | 5.19×10−4 |
| Amino acid metabolism (22) | 6.20×10−4 |
| Morula stage | |
| Cell death (60) | 1.58×10−4 |
| Cellular movement (19) | 1.95×10−4 |
| Post-translational modification (6) | 7.01×10−4 |
| Protein folding (5) | 7.01×10−4 |
| Cellular growth and proliferation (31) | 1.47×10−3 |
| Cellular assembly and organization (19) | 1.57×10−3 |
| Cellular compromise (5) | 2.18×10−3 |
| DNA replication, recombination, and repair (7) | 2.22×10−3 |
| Gene expression (40) | 2.22×10−3 |
| Cell-to-cell signaling and interaction (21) | 2.89×10−3 |
| Blastocyst stage | |
| Lipid metabolism (19) | 3.23×10−6 |
| Small-molecule biochemistry (28) | 3.23×10−6 |
| Vitamin and mineral metabolism (15) | 3.23×10−6 |
| Cellular assembly and organization (10) | 5.88×10−5 |
| Cellular growth and proliferation (40) | 9.10×10−5 |
| DNA replication, recombination, and repair (5) | 2.17×10−4 |
| Gene expression (21) | 2.17×10−4 |
| Carbohydrate metabolism (12) | 2.76×10−4 |
| Cellular movement (21) | 3.08×10−4 |
| Cell-to-cell signaling and interaction (16) | 9.54×10−4 |
Zygotic gene activation during embryo development
The zygotic activation of genes involves de novo transcription from the embryonic genome, which mainly occurs from the late 1-cell stage after fertilization (Bouniol et al. 1995), and this transition is an essential program for normal development (Minami et al. 2007). First, by using the transcriptome data of IVF embryos, we attempted to identify the genes with greater than threefold expression changes during specific stages (Table 2 and Supplementary Table 4, see section on supplementary data given at the end of this article) and the number of genes at each stage: 1-cell 659, 2-cell 1301, 4-cell 331, 8-cell 31, morula 201, and blastocyst 281. Here, for convenience, these genes are named zygotic activated genes. A minority of these genes was transiently expressed only at specific stages. In addition, more than 50% of the zygotic activated genes expressed from the 2-cell stage corresponded to those that were identified by an α-amanitin inhibition test (Zeng & Schultz 2005). As well, these genes were activated mainly in the 2-cell stage; their levels decreased once by the 8-cell stage and then increased slightly at the morula and blastocyst stages. This finding suggests that dynamic changes in gene expression profiles occurred at the morula and blastocyst stages.
Table 2.
Number of zygotic activated genes differentially expressed in SeCNT embryos.
| 1-cell | 2-cell | 4-cell | 8-cell | Morula | Blastocyst | |
|---|---|---|---|---|---|---|
| Zygotic activated in IVF | 659 (76)a | 1301 (120) | 331 (4) | 31 (6) | 201 (14) | 281 (–)b |
| Downregulated in SeCNTs | 76 (7) | 156 (19) | 37 (0) | 4 (1) | 6 (3) | 4 (–) |
| Repressed in SeCNTs | 11 | 30 | 16 | 0 | 0 | 0 |
| KO datac | 20 | 53 | 19 | 2 | 2 | 0 |
| Related to lethality | 14 | 35 | 13 | 0 | 1 | 0 |
Values in parentheses indicate the number of stage-specific expressed genes.
Stage-specific genes were identified as only having gene expression at a particular stage. The blastocyst stage was lacking at a later stage for comparison. (–) indicates no data.
Knockout data obtained from MGI (http://www.informatics.jax.org).
Notably, some of the zygotic activated genes were incompletely activated and showed reduced expression in the 1- to 4-cell stages of SeCNT embryos: 1-cell 76, 2-cell 168, and 4-cell 47. Interestingly, after the 8-cell stage, only some of the genes were incorrectly expressed, although the number of zygotic activated genes was significantly increased at the morula and blastocyst stages. Furthermore, we detected that some of the zygotic activated genes were completely repressed (<100 of the RAW value) at the 1-cell (n=11), 2-cell (n=30), and 4-cell (n=16) stages of SeCNT embryos. This observation showed that a number of zygotic activated genes could not be reprogrammed correctly in SeCNT embryos, which suggests that these genes are involved in prenatal and postnatal developmental disorders.
Gene network analysis using differentially expressed genes
To understand the interactions of the differentially expressed genes, we performed a gene network analysis at each stage by using IPA: in total, 127 networks were formed (Table 3 and Supplementary Table 5, see section on supplementary data given at the end of this article). The frequency of network descriptions is shown in Supplementary Table 6: the top ten were hereditary disorder (n=19), cancer (n=18), embryonic development (n=15), cell cycle (n=13), cellular assembly and organization (n=13), molecular transport (n=13), developmental disorder (n=12), neurological disease (n=11), and small-molecule biochemistry (n=11). Thus, the IPA network analysis strongly suggested that many genes that are critical to growth and survival were dysfunctional in SeCNT embryos, which resulted in embryo death.
Table 3.
Number of networks constructed by the differentially expressed genes (fold change >3).
| 1-cell | 2-cell | 4-cell | 8-cell | Morula | Blastocyst | |
|---|---|---|---|---|---|---|
| No. of networks | 12 | 21 | 24 | 45 | 15 | 10 |
| No. of overlapped networks | 1 | 2 | 1 | 1 | 1 | 2 |
| No. of networks in overlapped networks | 9 | 13, 2 | 15 | 40 | 7 | 8, 2 |
The four networks with extremely high score values of >50 are shown in Fig. 3 and these functions are as follows: 1-cell stage: post-translational modification, organismal injury and abnormalities, and digestive system development and function; 2-cell stage: free radical scavenging, cellular compromise, DNA replication, DNA recombination, and DNA repair; 4-cell stage: post-translational modification, protein degradation, and protein synthesis; and 8-cell stage: cell-to-cell signaling and interaction, tissue development, and cardiovascular system development and function.
Figure 3.
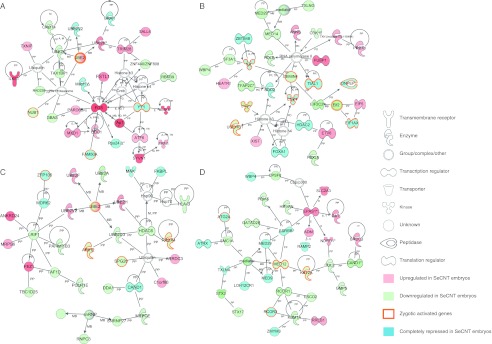
Top gene networks of the differentially expressed genes in SeCNT embryos. The networks were constructed by IPA using the differentially expressed gene list in Supplementary Table 2, fold change >3. Top network of the 1-cell stage (A), 2-cell stage (B), 4-cell stage (C), and 8-cell stage (D) that had a score >50. The score was given by IPA, which is based on the hypergeometric distribution and calculated with right-tailed Fisher's exact test. Green, genes downregulated in SeCNT embryos; blue, genes completely repressed in SeCNT embryos; red, genes upregulated in SeCNT embryos. Orange frames represent the zygotic activated genes.
Notably, several genes that were integrated into the networks were completely repressed. For example, eight genes, including zygotic activated genes and genes that encode TFs, were completely repressed in the top network at the 2-cell stage (Fig. 3B). Interestingly, it has been reported that the deletion of some of these repressed genes, Tial1 (Beck et al. 1998), Ilk (Lange et al. 2009), Hdac2 (Trivedi et al. 2007), Sod2 (Li et al. 1995), and Foxa1 (Kaestner et al. 1999), results in prenatal or postnatal death, which suggests that this network might be involved in developmental disorders of SeCNT embryos at the 2-cell stage. Furthermore, these observations provide an opportunity to discuss the possibility that these ectopic gene expressions could be involved in later embryo development. In addition, in total, eight overlapped networks were constructed (Supplementary Figure 1, see section on supplementary data given at the end of this article) and a hugely overlapped network was obtained for the 8-cell stage that consisted of 40 gene networks. This suggests that various functional disorders occur at the 8-cell stage in SeCNT embryos, which leads to a significant proportion of developmental arrest at this stage.
Chromosome maps of differentially expressed genes
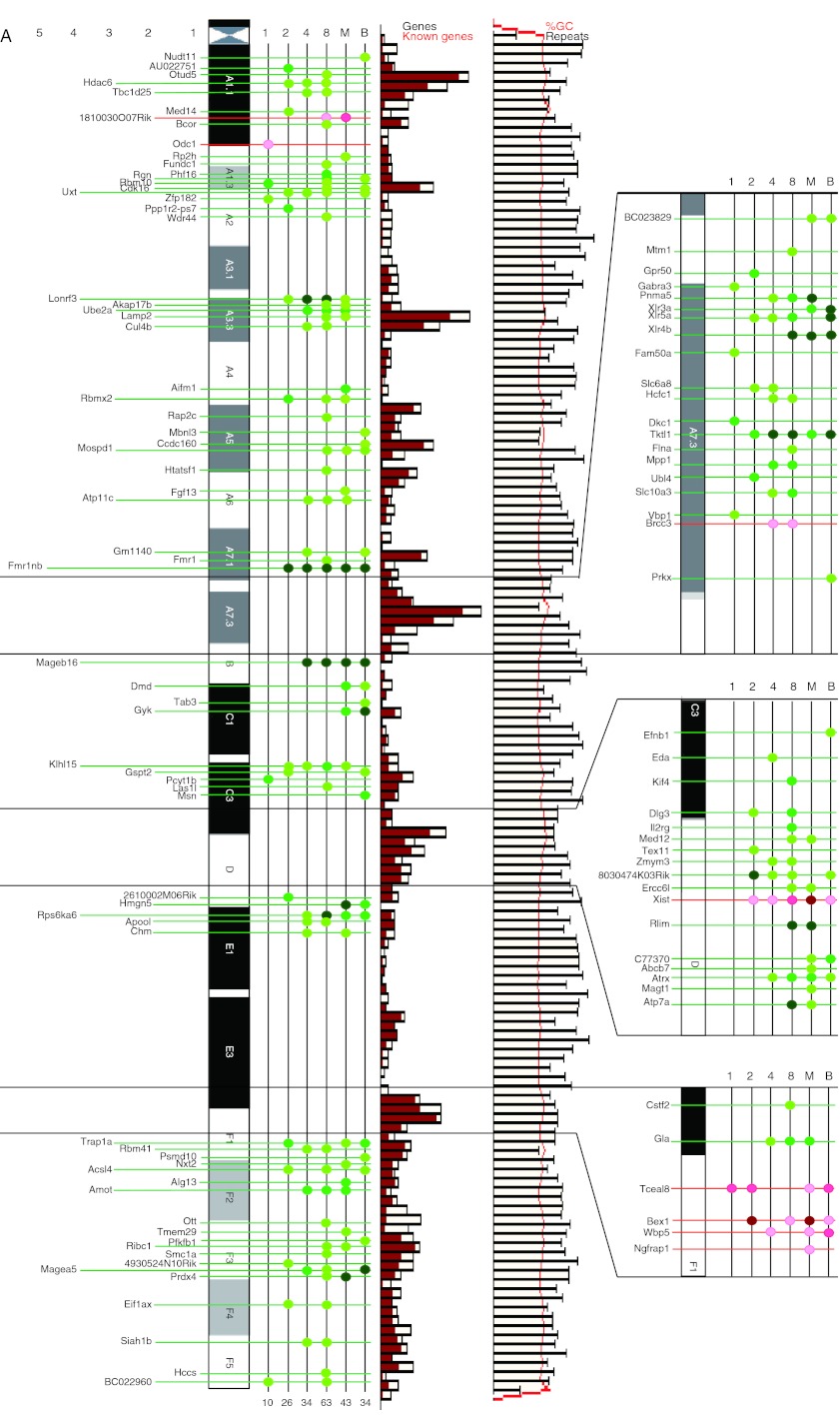
To obtain an overview of the global transcriptomic changes in SeCNT embryos throughout preimplantation development, we constructed a map of each chromosome by using the data on the differentially expressed genes (Fig. 4A and B). This analysis showed temporal–spatial changes in the abnormal expressions of the X-linked genes after nuclear transfer. The mapping data of the X chromosome showed that the abnormally reprogrammed genes were concentrated in specific regions, such as the A7.3 and D bands. However, the reason for this is still unclear. Interestingly, Tceal8, Bex1, Wbp5, and Ngfrap1, which are located on the pseudo-autosomal region in the F1 region of chromosome (Chr) X (Jeppesen & Turner 1993), were upregulated. Matoba et al. (2011) reported that the abnormal repression of X-linked genes could be corrected by either gene knockout or siRNA inhibition of Xist, which leads to a significant improvement in the survival rate of SeCNT embryos. However, we detected neither a positive nor a negative correlation between these genes and Xist. This finding implies that the expression errors that occurred in Chr X genes were not under the control of Xist alone and that other factors could have been involved.
Figure 4.
Chromosome map views of the differentially expressed genes on Chr X and Chr 7. (A) Temporal–spatial changing aspects of the differentially expressed genes on Chr X. (B) Temporal–spatial changing aspects of the differentially expressed genes on Chr 7. Genes differentially expressed in SeCNT embryos with a greater than threefold change were matching to the chromosome map. Genes that were upregulated in SeCNT embryos are shown as red dots and lines; genes downregulated are shown as green dots and lines. All the location data were mapped by using the database Ensembl (http://asia.ensembl.org). The gradient color from light to dark means a fold change >3, 5, and 10. The bar graph in the middle panel shows the gene density of the chromosome region. Each bar represents ∼800 kb long. Black frame represents the number of genes in that region; the red bar represent the number of known genes. The line graph in the right panel shows the GC% and the number of repeat sequences. The red line represents GC% in the region and the black line represents the number of repeat sequences. The magnified panels were five times enlarged from the original chromosome region.
Generally, the male Chr X is protected from inactivation and remains active (Barakat & Gribnau 2012). Recent studies have shown that abnormal reprogramming of Chr X inactivation, which is regulated by a non-coding RNA encoded on Chr X called Xist, is the main cause of the developmental failure of SCNT embryos (Inoue et al. 2010). Interestingly, Xist expression was first detectable at the 2-cell stage and lasted until the blastocyst stage in SeCNT embryos (Supplementary Figure 2, see section on supplementary data given at the end of this article). However, its expression was not detected in IVF male embryos throughout development. Because of this abnormal activation of Xist in SeCNT embryos, a number of X-linked genes were abnormally downregulated.
The chromosome maps of Chr 7 and others are shown in Fig. 4B and Supplementary Figure 3 respectively. In addition to Chr X, the differentially expressed genes formed many clusters on every autosome. Some chromosomal regions contained large regions that did not show any differentially expressed genes, for example the particularly large regions in Chr 7 (B4–B5, 35 Mb; Fig. 4B), Chr 1 (E1.1–E3.1, 36 Mb), and Chr 14 (D3–E4, 38 Mb) (Supplementary Figure 3). On further examination, we found that some of these regions showed low gene density and a high repeat sequence frequency. Furthermore, other regions, such as the B1–B2.1 region of Chr 6, did not contain any differentially expressed genes despite their high gene density. Therefore, some factors other than gene density were possibly involved in the distribution of the differentially expressed genes. Figure 4B highlights the interesting fact that Ilk and Tial1 on Chr 7, whose knockouts are lethal (Beck et al. 1998, Lange et al. 2009), were repressed from the 2-cell stage to the blastocyst stage. This suggests that these genes are involved in developmental disorder in SeCNT embryos during the preimplantation stage.
Finally, we focused on imprinted genes, which are expressed from either the paternal or the maternal allele, because aberrant expression and DNA methylation are considered to be the causes of abnormal development in SCNT embryos. To date, the number of imprinted genes that have been identified is over 100, excluding small transcripts. However, only five imprinted genes, Phf17 (Chr 3), H19 and Cd81 (Chr 7), Impact (Chr 18), and Xlr4b (Chr X), are abnormally expressed during the preimplantation stage; the expression of all the other imprinted genes is established to be after implantation (Dean et al. 2001, Kang et al. 2001). This result implies that the abnormal expression of many imprinted genes in SCNT embryos is caused by the failure to reverse the methylation of the imprinted genes in DNA methylation regions after implantation. Further comparison between methylome (Kobayashi et al. 2012) and transcriptome data may provide insight into the molecular mechanism that underlies reprogramming in SCNT embryos.
Conclusion
Our study is the first to provide a comprehensive gene expression profile of single SeCNT embryos from the 1-cell to the blastocyst stage, and it provides a temporal–spatial view of the reprogramming mechanisms in preimplantation SeCNT embryos (Supplementary Figure 4, see section on supplementary data given at the end of this article). The present results show that the structural features of a chromosome, which are due to the presence of specific epigenetic modifications, could be responsible for the distribution of the differentially expressed genes, i.e. the formation of high- and low-density domains on each chromosome. Furthermore, the differentially expressed genes may control the changes in epigenetic modifications. The data obtained here suggest that the genes expressed abnormally during the zygotic activation period (around the 1-cell and 2-cell stages) are responsible for early embryo loss both during and after implantation. Stage-specific gene expression regulates the activation of several zygotic genes, and incorrect reprogramming may be potentially responsible for embryo loss during the post-implantation period. Thus, the present study revealed that the SeCNT feign proper preimplantation development; however, the progression of incorrect reprogramming is concealed throughout development.
Materials and Methods
Embryos
Recipient MII oocytes were collected from mature B6D2F1 (C57BL/6NJcl×DBA/2JJcl) female mice after superovulation was induced in these mice. The donor SR cells were obtained from 3-day-old male B6CBF1 mice and harvested, as described previously (Ogura et al. 2000). Oocytes IVF with male B6D2F1 sperm were used as control wild types. MII oocytes were collected 14 h after hCG injection. For the 1-cell, late 2-cell, 4-cell, 8-cell, morula, and blastocyst stages, the embryos were cultured for 16, 32, 48, 62, 72, and 84 h after sperm addition or activation.
Nuclear transfer and culture
Sertoli cell-cloned embryos were produced by the injection of a donor nucleus into enucleated oocytes with a piezo-driven system (Prime Tech Ltd, Ibaraki, Japan), by using a previously described method (Ogura et al. 2000). The activated cloned embryos were cultured in potassium simplex optimization medium (KSOM) at 37 °C, under an atmosphere of 5% CO2, 5% O2, and 90% N2 until use. Each embryo was lysed in 50 μl of buffer RLT containing 1% β-mercaptoethanol (Qiagen).
Microarray and data analysis
Samples of cRNA were amplified according to a previously described modified protocol (Fukuda et al. 2010). After the quality of the amplified product was verified by Experion capillary electrophoresis (Bio-Rad), 10 μg of fragmented cRNA were hybridized to a GeneChip Mouse Genome 430 2.0 Array (Affymetrix, Santa Clara, CA, USA), which contains 45 101 probe sets. Affymetrix data file CEL output by GeneChip Operating Software (GCOS) version 1.3 (Affymetrix) was then loaded into GeneSpring GX 12.0 (Agilent Technologies, Santa Clara, CA, USA) with default normalization: summarization algorithm with RMA (robust M-algorithm) and baseline to the median of all samples. The box plots of all signal values for each sample are shown in Supplementary Figure 5, see section on supplementary data given at the end of this article.
To determine which genes were differentially expressed during each stage, we compared the two sets (IVF vs SeCNT) of data at each stage separately. PCA was employed to analyze the gene expression patterns of all the embryos. Hierarchical clustering was performed with Pearson's correlation analysis for the measurement of similarity and clustering algorithm with average linkage. The genes were investigated by gene ontology analysis using Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Inc., Red Wood, CA, USA). All microarray data are compliant with the Minimum Information About a Microarray Experiment (MIAME). The raw CEL file data have been deposited in a MIAME-compliant database (GEO; http://www.ncbi.nlm.nih.gov/geoprofiles, GSE41358).
The chromosome map views of the differentially expressed genes were constructed by mapping the genes showing threefold or greater changes onto the chromosome maps obtained from the database Ensembl (http://asia.ensembl.org). The chromosome map views constructed were compared with gene density and GC% summaries obtained from the chromosome summary view of Ensembl.
Supplementary data
This is linked to the online version of the paper at http://dx.doi.org/10.1530/REP-12-0435.
Author contribution statement
F Cao and T Kono conceived the idea and designed the experimental strategy; A Fukuda and H Watanabe produced all embryos for microarray; F Cao and T Kono wrote the paper; and T Kono supervised the study.
Supplementary Material
Acknowledgements
The authors thank Drs Hidehiko Ogawa, Yayoi Obata, Hisato Kobayashi, and Takuya Wakai for their critical discussion on the manuscript and Mr Tomoaki Morisono for his kind help.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
T Kono was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (grant no. 20062009).
References
- Barakat TS, Gribnau J. X chromosome inactivation in the cycle of life. Development. 2012;139:2085–2089. doi: 10.1242/dev.069328. [DOI] [PubMed] [Google Scholar]
- Beck AR, Miller IJ, Anderson P, Streuli M. RNA-binding protein TIAR is essential for primordial germ cell development. PNAS. 1998;95:2331–2336. doi: 10.1073/pnas.95.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyhan Z, Ross PJ, Iager AE, Kocabas AM, Cunniff K, Rosa GJ, Cibelli JB. Transcriptional reprogramming of somatic cell nuclei during preimplantation development of cloned bovine embryos. Developmental Biology. 2007;305:637–649. doi: 10.1016/j.ydbio.2007.01.041. [DOI] [PubMed] [Google Scholar]
- Bortvin A, Eggan K, Skaletsky H, Akutsu H, Berry DL, Yanagimachi R, Page DC, Jaenisch R. Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development. 2003;130:1673–1680. doi: 10.1242/dev.00366. [DOI] [PubMed] [Google Scholar]
- Bouniol C, Nguyen E, Debey P. Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Experimental Cell Research. 1995;218:57–62. doi: 10.1006/excr.1995.1130. [DOI] [PubMed] [Google Scholar]
- Chastant S, Christians E, Campion E, Renard JP. Quantitative control of gene expression by nucleocytoplasmic interactions in early mouse embryos: consequence for reprogrammation by nuclear transfer. Molecular Reproduction and Development. 1996;44:423–432. doi: 10.1002/(SICI)1098-2795(199608)44:4%3c423::AID-MRD1%3e3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. PNAS. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Molecular and Cellular Biology. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D, Chen AE, Saphier G, Ichida J, Fitzgerald C, Go KJ, Acevedo N, Patel J, Baetscher M, Kearns WG, et al. Reprogramming within hours following nuclear transfer into mouse but not human zygotes. Nature Communications. 2011;2:488. doi: 10.1038/ncomms1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A, Cao F, Morita S, Yamada K, Jincho Y, Tane S, Sotomaru Y, Kono T. Identification of inappropriately reprogrammed genes by large-scale transcriptome analysis of individual cloned mouse blastocysts. PLoS ONE. 2010;5:e11274. doi: 10.1371/journal.pone.0011274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, Wilmut I. Nuclear transfer to eggs and oocytes. Cold Spring Harbor Perspectives in Biology. 2011;3:a002659. doi: 10.1101/cshperspect.a002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, De Robertis EM, Partington G. Injected nuclei in frog oocytes provide a living cell system for the study of transcriptional control. Nature. 1976;260:116–120. doi: 10.1038/260116a0. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061–1067. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- Inoue K, Kohda T, Lee J, Ogonuki N, Mochida K, Noguchi Y, Tanemura K, Kaneko-Ishino T, Ishino F, Ogura A. Faithful expression of imprinted genes in cloned mice. Science. 2002;295:297. doi: 10.1126/science.295.5553.297. [DOI] [PubMed] [Google Scholar]
- Inoue K, Ogonuki N, Miki H, Hirose M, Noda S, Kim JM, Aoki F, Miyoshi H, Ogura A. Inefficient reprogramming of the hematopoietic stem cell genome following nuclear transfer. Journal of Cell Science. 2006;119:1985–1991. doi: 10.1242/jcs.02913. [DOI] [PubMed] [Google Scholar]
- Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, Matoba S, Shiura H, Ikeda R, Mochida K, Fujii T, et al. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science. 2010;330:496–499. doi: 10.1126/science.1194174. [DOI] [PubMed] [Google Scholar]
- Jeppesen P, Turner BM. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-Q. [DOI] [PubMed] [Google Scholar]
- Jincho Y, Sotomaru Y, Kawahara M, Ono Y, Ogawa H, Obata Y, Kono T. Identification of genes aberrantly expressed in mouse embryonic stem cell-cloned blastocysts. Biology of Reproduction. 2008;78:568–576. doi: 10.1095/biolreprod.107.064634. [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Katz J, Liu Y, Drucker DJ, Schutz G. Inactivation of the winged helix transcription factor HNF3alpha affects glucose homeostasis and islet glucagon gene expression in vivo. Genes and Development. 1999;13:495–504. doi: 10.1101/gad.13.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YK, Koo DB, Park JS, Choi YH, Chung AS, Lee KK, Han YM. Aberrant methylation of donor genome in cloned bovine embryos. Nature Genetics. 2001;28:173–177. doi: 10.1038/88903. [DOI] [PubMed] [Google Scholar]
- Kawasumi M, Anzai M, Takehara T, Mitani T, Kato H, Saeki K, Iritani A, Matsumoto K, Hosoi Y. Abnormal distribution of chromosomes in the first division of nuclear transferred mouse embryos. Journal of Reproduction and Development. 2007;53:615–622. doi: 10.1262/jrd.19007. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genetics. 2012;8:e1002440. doi: 10.1371/journal.pgen.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohda T, Inoue K, Ogonuki N, Miki H, Naruse M, Kaneko-Ishino T, Ogura A, Ishino F. Variation in gene expression and aberrantly regulated chromosome regions in cloned mice. Biology of Reproduction. 2005;73:1302–1311. doi: 10.1095/biolreprod.105.044958. [DOI] [PubMed] [Google Scholar]
- Lange A, Wickstrom SA, Jakobson M, Zent R, Sainio K, Fassler R. Integrin-linked kinase is an adaptor with essential functions during mouse development. Nature. 2009;461:1002–1006. doi: 10.1038/nature08468. [DOI] [PubMed] [Google Scholar]
- Latham KE. Early and delayed aspects of nuclear reprogramming during cloning. Biology of the Cell. 2005;97:119–132. doi: 10.1042/BC20040068. [DOI] [PubMed] [Google Scholar]
- Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nature Genetics. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- Matoba S, Inoue K, Kohda T, Sugimoto M, Mizutani E, Ogonuki N, Nakamura T, Abe K, Nakano T, Ishino F, et al. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. PNAS. 2011;108:20621–20626. doi: 10.1073/pnas.1112664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Jaenisch R. Mammalian nuclear transfer. Developmental Dynamics. 2006;235:2460–2469. doi: 10.1002/dvdy.20915. [DOI] [PubMed] [Google Scholar]
- Minami N, Suzuki T, Tsukamoto S. Zygotic gene activation and maternal factors in mammals. Journal of Reproduction and Development. 2007;53:707–715. doi: 10.1262/jrd.19029. [DOI] [PubMed] [Google Scholar]
- Mizutani E, Yamagata K, Ono T, Akagi S, Geshi M, Wakayama T. Abnormal chromosome segregation at early cleavage is a major cause of the full-term developmental failure of mouse clones. Developmental Biology. 2012;364:56–65. doi: 10.1016/j.ydbio.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Ogonuki N, Inoue K, Yamamoto Y, Noguchi Y, Tanemura K, Suzuki O, Nakayama H, Doi K, Ohtomo Y, Satoh M, et al. Early death of mice cloned from somatic cells. Nature Genetics. 2002;30:253–254. doi: 10.1038/ng841. [DOI] [PubMed] [Google Scholar]
- Ogura A, Inoue K, Ogonuki N, Noguchi A, Takano K, Nagano R, Suzuki O, Lee J, Ishino F, Matsuda J. Production of male cloned mice from fresh, cultured, and cryopreserved immature Sertoli cells. Biology of Reproduction. 2000;62:1579–1584. doi: 10.1095/biolreprod62.6.1579. [DOI] [PubMed] [Google Scholar]
- Ohashi M, Runge MS, Faraci FM, Heistad DD. MnSOD deficiency increases endothelial dysfunction in ApoE-deficient mice. Arteriosclerosis, Thrombosis, and Vascular Biology. 2006;26:2331–2336. doi: 10.1161/01.ATV.0000238347.77590.c9. [DOI] [PubMed] [Google Scholar]
- Ono Y, Shimozawa N, Ito M, Kono T. Cloned mice from fetal fibroblast cells arrested at metaphase by a serial nuclear transfer. Biology of Reproduction. 2001;64:44–50. doi: 10.1095/biolreprod64.1.44. [DOI] [PubMed] [Google Scholar]
- Park ES, Hwang WS, Jang G, Cho JK, Kang SK, Lee BC, Han JY, Lim JM. Incidence of apoptosis in clone embryos and improved development by the treatment of donor somatic cells with putative apoptosis inhibitors. Molecular Reproduction and Development. 2004;68:65–71. doi: 10.1002/mrd.20046. [DOI] [PubMed] [Google Scholar]
- Peat JR, Reik W. Incomplete methylation reprogramming in SCNT embryos. Nature Genetics. 2012;44:965–966. doi: 10.1038/ng.2393. [DOI] [PubMed] [Google Scholar]
- Pfister-Genskow M, Myers C, Childs LA, Lacson JC, Patterson T, Betthauser JM, Goueleke PJ, Koppang RW, Lange G, Fisher P, et al. Identification of differentially expressed genes in individual bovine preimplantation embryos produced by nuclear transfer: improper reprogramming of genes required for development. Biology of Reproduction. 2005;72:546–555. doi: 10.1095/biolreprod.104.031799. [DOI] [PubMed] [Google Scholar]
- Rideout WM, III, Eggan K, Jaenisch R. Nuclear cloning and epigenetic reprogramming of the genome. Science. 2001;293:1093–1098. doi: 10.1126/science.1063206. [DOI] [PubMed] [Google Scholar]
- Sawai K. Studies on gene expression in bovine embryos derived from somatic cell nuclear transfer. Journal of Reproduction and Development. 2009;55:11–16. doi: 10.1262/jrd.20131. [DOI] [PubMed] [Google Scholar]
- Smith SL, Everts RE, Tian XC, Du F, Sung LY, Rodriguez-Zas SL, Jeong BS, Renard JP, Lewin HA, Yang X. Global gene expression profiles reveal significant nuclear reprogramming by the blastocyst stage after cloning. PNAS. 2005;102:17582–17587. doi: 10.1073/pnas.0508952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers J, Smith C, Donnison M, Wells DN, Henderson H, McLeay L, Pfeffer PL. Gene expression profiling of individual bovine nuclear transfer blastocysts. Reproduction. 2006;131:1073–1084. doi: 10.1530/rep.1.00967. [DOI] [PubMed] [Google Scholar]
- Thuan NV, Kishigami S, Wakayama T. How to improve the success rate of mouse cloning technology. Journal of Reproduction and Development. 2010;56:20–30. doi: 10.1262/jrd.09-221A. [DOI] [PubMed] [Google Scholar]
- Tong GQ, Heng BC, Tan LG, Ng SC. Aberrant profile of gene expression in cloned mouse embryos derived from donor cumulus nuclei. Cell and Tissue Research. 2006;325:231–243. doi: 10.1007/s00441-006-0189-1. [DOI] [PubMed] [Google Scholar]
- Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nature Medicine. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- Tveden-Nyborg P, Peura TT, Hartwich KM, Walker SK, Maddox-Hyttel P. Morphological characterization of pre- and peri-implantation in vitro cultured, somatic cell nuclear transfer and in vivo derived ovine embryos. Reproduction. 2005;130:681–694. doi: 10.1530/rep.1.00850. [DOI] [PubMed] [Google Scholar]
- Vassena R, Han Z, Gao S, Baldwin DA, Schultz RM, Latham KE. Tough beginnings: alterations in the transcriptome of cloned embryos during the first two cell cycles. Developmental Biology. 2007;304:75–89. doi: 10.1016/j.ydbio.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- Wakayama S, Jakt ML, Suzuki M, Araki R, Hikichi T, Kishigami S, Ohta H, Van Thuan N, Mizutani E, Sakaide Y, et al. Equivalency of nuclear transfer-derived embryonic stem cells to those derived from fertilized mouse blastocysts. Stem Cells. 2006;24:2023–2033. doi: 10.1634/stemcells.2005-0537. [DOI] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Zeng F, Schultz RM. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Developmental Biology. 2005;283:40–57. doi: 10.1016/j.ydbio.2005.03.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.