Abstract
The packaging of eukaryotic DNA into nucleosomes, the fundamental unit of chromatin, creates a barrier to nuclear processes, such as transcription, DNA replication, recombination, and repair(1). This obstructive nature of chromatin can be overcome by the enzymatic activity of chromatin remodeling complexes which creates a more favorable environment for the association of essential factors and regulators to sequences within target genes. Here we describe a detailed approach for analyzing chromatin architecture and remodeling by restriction endonuclease hypersensitivity assay. This procedure uses restriction endonucleases to characterize changes in chromatin that accompany nucleosome remodeling. The specific experimental example described in this article is the BRG1 complex dependent chromatin remodeling of the steroid hormone-responsive mouse mammary tumor virus promoter. Through the use of these methodologies one is able to quantify changes at specific nucleosomes in response to regulatory signals.
1. Introduction
In the eukaryotic nucleus, DNA is packaged with histone and non-histone proteins to form a highly organized chromatin structure(2). This DNA-protein assembly can represents a significant barrier to a number of nuclear processes such as DNA repair, recombination, replication and transcription(3). Often the structural changes in chromatin that accompany transcriptional activation require multi-protein enzymatic complexes that manipulate the nucleosomal architecture(4). Alteration in the chromatin structure by ATP-dependent remodeling complexes is considered a significant initial step in transcriptional regulation(5). Chromatin remodeling complexes to use the energy of ATP-hydrolysis to alter local chromatin structure to facilitate a more “open/active” conformation conducive for the recruitment and binding of essential nuclear factors. A number of chromatin remodeling complexes have been identified that modulate the arrangement and stability of nucleosomes in a non-covalent manner (5). Generally, these ATP-dependent remodeling machines are divided into four major subfamilies, characterized by the identity of their central catalytic subunit, which include BRG1 (or hBrm), ISWI, Mi-2 and Ino80 of their respective complexes SWI/SNF, ISWI, NuRD and INO80 (6, 7)
The SWI/SNF remodeling complex was originally identified in yeast, by mutations in mating type switching (SWI) and sucrose fermentation (SNF), and its function is highly conserved in eukaryotes with homologous found in Drosophila and mammals. Human SWI/SNF is a large multi-protein complex that possesses either BRG1 or hBrm, two ATPases related to yeast SWI2/SNF2 and STH1, and are comprised of about 10 BRG1-associated factors (BAFs) subunits, most of which are orthologous to those found in yeast SWI/SNF and RSC (5).
This article provides a detailed approach for analyzing chromatin architecture and remodeling by the BRG1-SWI/SNF complex upon nuclear receptor signaling (Figure 1). Using the human SW-13 cell line, containing stably integrated mouse mammary tumor virus (MMTV) promoter, as a model system we are able to evaluate chromatin changes upon hormone signaling (8). This procedure allows the use restriction endonucleases to characterize alterations in the chromatin structure that accompany nucleosome remodeling and transcriptional activation (9). The methodology presented will focus on analyzing nucleosomal organization using isolated nuclei, restriction endonucleases, and reiteractive primer extension. All methods described here have been employed extensively to study the activity of BRG1 on chromatin structure and transcriptional regulation from the MMTV promoter (10). Although the focus of this article is on the MMTV promoter in SW-13 cells, these techniques have been applied to a variety of inducible and developmentally responsive gene promoters in higher eukaryotes to analyze changes within chromatin (11-15).
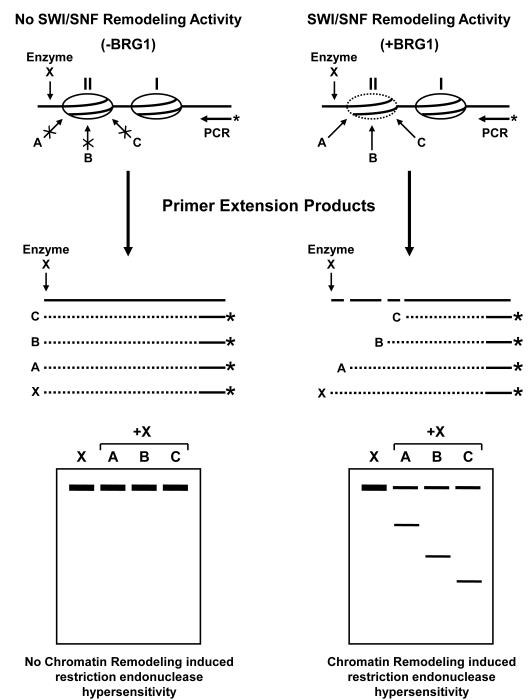
Figure 1.
Schematic of the restriction enzyme hypersensitivity assay used to evaluate chromatin structure and detect SWI/SNF-mediated chromatin remodeling activity. Nuclei are isolated from naïve or hormone-treated cells with (+BRG1) or without (-BRG1) expression of a functional SWI/SNF chromatin remodeling complex. The chromatin is partially digested with various restriction endonucleases (A, B, or C) that target the predicted hypersensitive region (II). After purification, the genomic DNA is digested to competition with a second restriction enzyme which cut outside the hypersensitive region (enzyme X). The resulting DNA is purified and analyzed by reiterative primer extension using Taq polymerase and a 32P-labeled oligonucleotide specific for the target of interest. The amplified products are resolved on a denaturing polyacrylamide gel and exposed to a PhosphorImager screen to detect the presence of bands that correspond, in length, to the predicted in vivo restriction endonuclease cleavage pattern. Re-digestion with the second restriction enzyme (in vitro digest) serves as an internal standard to ensure equal amounts of template DNA was loaded in each reaction. The prediction in this model would suggest that the SWI/SNF complex targets regions within nucleosome II where it remodeling the chromatin structure to allow access to transcription factors or, in this case, restriction endonucleases to the genomic DNA. In the absences of BRG1, SWI/SNF is unable to remodel nucleosome II where it act to block restriction enzyme access to the target DNA.
2. Materials (see Note 1)
2.1. Cell Culture
Dulbecco’s Modified Eagle’s Medium (DMEM) high glucose (Gibco/Invitrogen, Carlsbad, CA) supplemented with 10 % normal fetal bovine serum, 2 mM L-glutamine, 10 mM HEPS, 100 μg/ml penicillin, and 100 μg/ml streptomycin.
Cultured cells were grown in a humidified incubator at 37°C under 5% CO2 on 150 mm tissue culture plates.
Transient transfection of BRG1 expression plasmid using LipofectAMINE 2000 (Invitrogen, Carlsbad, CA) according to manufacturer’s recommendations. (see Note 2)
For the purpose of this procedure, cells were treated with 100 nM dexamethasone (Dex) or with equal volume vehicle (100% ethanol) for 1 h prior to nuclei isolation.
2.2. Nuclei Isolation (see Note 3)
Phosphate buffered saline (sterile-filtered).
Homogenization Buffer: 10 mM Tris-HCl, pH 7.4, 15 mM NaCl, 60 mM KCl, 1 mM EDTA, 0.1 mM EGTA, 0.1% Nonidet P-40 (NP-40), 5% sucrose, 0.15 mM spermine, and 0.5 mM spermidine.
Sucrose Pad: 10 mM Tris-HCl, pH 7.4, 15 mM NaCl, 60 mM KCl, 1 mM EDTA, 0.1 mM EGTA, 10% sucrose, 0.15 mM spermine, and 0.5 mM spermidine.
Wash Buffer: 10 mM Tris-HCl, pH 7.4, 15 mM NaCl, 60 mM KCl, 1 mM EDTA, 0.1 mM EGTA, 0.15 mM spermine, and 0.5 mM spermidine.
7.5-ml Dounce tissue grinder/homogenizer (Wheaton 357542) designed for fine particle size reduction without damage to cell nuclei.
2.3. In vivo Digestion by Restriction Endonuclease (see Note 4)
Restriction endonuclease(s) which cleavages the target genomic DNA sequence within close proximity of a transcription factor binding site or region of chromatin to be assayed for remodeling.
Restriction enzyme digestion buffer: 10 mM Tris-HCl, pH 7.4, 15 mM NaCl, 60 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 5% glycerol, and 1 mM dithiothreitol (DTT).
Proteinase K buffer: 10 mM Tris-HCl, pH 7.6, 10 mM EDTA, 0.5% sodium dodecyl sulfate (SDS), and 0.2 mg/ml proteinase K (Invitrogen, Carlsbad, CA).
Phenol:Chloroform:Isoamyl alcohol (25:24:1) saturated with 10 mM Tris, pH 8.0, 1 mM EDTA.
Chloroform.
2.4. In vitro Re-digestion and Purification of DNA
Restriction endonuclease which recognizes a cleavage site 200-300 bp upstream of the in vivo restriction enzyme site.
Reaction buffer supplied with enzyme.
Phenol:Chloroform:Isoamyl alcohol (25:24:1) saturated with 10 mM Tris, pH 8.0, 1 mM EDTA.
Chloroform.
2.5. End Labeling of Oligonucleotide (see Note 5)
5 pmol gene-specific oligonucleotide
ATP, [γ-32P]-6000Ci/mmol 10mCi/ml EasyTide, (PerkinElmer, Covina, CA)
PNK - T4 polynucleotide kinase (NEB, Ipswich, MA)
T4 polynucleotide kinase buffer: 70 mM Tris-HCl, 10 mM MgCl2, 5 mM DTT, pH 7.6 (supplied with enzyme).
MicroSpin Sephadex G-25 columns (GE Healthcare Life Sciences, Piscataway, NJ).
Liquid scintillation counter.
2.6. Reiteractive Primer Extension
PCR reaction mix: 1X PCR buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl, 2-4 mM MgCl2, 0.5% Tween 20), 200 μM deoxynucleotides, 2.5 U Taq DNA polymerase (Invitrogen, Carlsbad, CA).
PCR stop buffer: 10 mM Tris-HCl, pH 7.4, 200 mM Sodium acetate, pH 7.0, 5 mM EDTA, 0.1 μg/μl yeast tRNA.
2.7. Analysis of Primer Extension products (see Note 6)
Acrylamide stock solution: 38% acrylamide and 2% bis-acrylamide (w/v)
For 8% denaturing polyacrylamide gel: 25.2 g urea, 6 ml 10X TBE, 112 ml acrylamide stock solution, water to 60 ml, 10 μl TEMED and 200 μl ammonium persulfate (10%). Pour gel immediately.
Sample loading buffer: 80% formamide, 0.01 M NaOH, 1 mM EDTA, 0.04% bromophenol blue, 0.04% xylene cyanol.
Sequi-Gen GT nucleic acid sequencing system; 38 × 30 cm gel (Bio-Rad, Hercules, CA).
PhosphorImager screen, scanner and ImageQuant software (GE Healthcare Life Sciences, Piscataway, NJ).
3. Methods
The methodology presented is suitable for the analysis of chromatin structure in vivo and can assist in the mapping of DNA-protein interactions and to elucidate their mechanistic implications regarding various nuclear processes including transcriptional regulation, DNA repair, and replication. The requirements for this procedure include 1) the genomic DNA of interest contains sequence-specific binding sites for and responds to a specific transcription factors and 2) has a restriction enzyme cleavage site located proximal to the binding region and is contained within the confines of a nucleosome or organized as chromatin.
3.1. Cell Culture
The experimental procedure outlined below is based on studies in human carcinoma cells that were stably transformed to contain multiple copies of the nuclear receptor-dependent MMTV promoter. For the purpose of this protocol, we have selected the human adrenal carcinoma cell line, SW-13, which do not express BRG1 protein (10). The use of this multi-copy promoter cell lines provides a very strong signal-to-noise ratio for the MMTV sequence, as well as, to enhance the ability to define the chromatin structure of the promoter. The use of this cell line has greatly enhanced our ability to study nuclear receptor-initiated cascades that lead to structural changes within chromatin upon receptor binding to specific hormone response elements within target promoters (16, 17). Therefore, these general growth conditions have been optimized for our established SW-13/MMTV cell line. Specific growth requirements may vary from this description depending on cell type used.
3.2. Isolation of nuclei
This protocol will outline the standard procedure for nuclei isolation from tissue cultured cells. All steps are performed on ice with pre-chilled equipment and solutions at 4°C. Cells were treated with hormone or vehicle prior to nuclei isolation to stimulate glucocorticoid receptor signaling.
Treat cells with 100 nM Dex or vehicle for 1 h prior to nuclei isolation.
Rinse cells with cold PBS and detach from 150mm plates by scraping cells into 10 ml cold PBS. Transfer cells to a pre-chilled 15-ml conical centrifuge tube.
Pellet cells by centrifugation at 500 × g for 5 min at 4°C. Remove PBS from cell pellet.
Add cold homogenization buffer (5 ml) and gently to dislodge cell pellet by pipetting. Transfer cells and buffer to a pre-chilled 7-ml Dounce tissue grinder/homogenizer and incubate on ice for 2 min.
Lyse cells by gently using four complete strokes of the Dounce pestle (tight pestle). Transfer lysate to a pre-chilled 15-ml conical tube. (see Note 7)
Gently add 1 ml sucrose pad directly to the bottom of the tube using a micropipette.
Sediment nuclei through sucrose pad by centrifugation at 1400 × g for 20 min at 4°C. Delicately remove supernatant from nuclei pellet.
Add 1-ml wash buffer to nuclei and gently resuspend pellet using a micropipette. Once fully in solution, add an additional 4-ml wash buffer and centrifuge at 750 × g for 5 min at 4°C.
Carefully remove all traces of wash buffer and store nuclei on ice.
3.3. In vivo Digestion by Restriction Endonuclease
The choice of restriction endonuclease to use for this accessibility assay is dependent upon the availability of cleavage sites within the genomic region of interest. Most analysis will require the testing of numerous restriction enzymes found to cleave within the target sequence until an optimal enzyme(s) is identified. The cleavage buffer selected for this assay was chosen because it maintains the structural integrity of the nuclei and is compatible with a broad range of restriction enzymes (see Note 8). The quantity of enzyme to use should be derived empirically and will depend on the efficiency at which the enzyme cleaves DNA when using the buffer recommended for hypersensitivity assays.
Gently resuspend nuclei in cold restriction enzyme digestion buffer (use a 3:1 ration of buffer to nuclei pellet; i.e. resuspend a 50 μl compact nuclei pellet in 150 μl buffer). Transfer aliquots of 100 μl nuclei to pre-chilled 5-ml polypropylene tubes (Falcon 2063).
Digest nuclei with appropriate restriction endonuclease (100-1000U/ml) at 30°C for 15 min (in vivo digest). Use 100μl resuspended nuclei for each digest.
Stop reactions by adding 1-ml Proteinase K buffer. Mix each sample by inverting five times and incubate overnight at 37°C.
Purify total DNA by four extractions with phenol/chloroform/isoamyl alcohol (PCI-25:24:1, v/v) and two extractions with chloroform. The first two extractions should be carried out with twice the volume PCI (2.0 ml) and each subsequent extraction with one volume (1.0 ml). Mix samples by vigorous shaking and centrifuge at 10,000 × g for 5 min at room temperature.
Precipitate the DNA by addition of 1/10th volume of 1 m NaCl and 3 volumes ice-cold 95% ethanol. Incubate samples at −20°C for 1 h then pellet the DNA by centrifugation at 12,000 × g for 30 min at 4°C.
Wash DNA pellet with 1-ml cold 70% (v/v) ethanol and centrifuge at 12,000g for 10 min at 4°C.
Allow DNA pellet to air-dry for 1 h at room temperature, resuspend in 100 μl sterile water, and transfer to 1.5-ml microfuge tube. (see Note 9).
3.4. In vitro Re-digestion and Purification of DNA
The purified in vivo digested genomic DNA is cut to completion with a second restriction endonuclease which recognizes a cleavage site upstream or downstream of the in vivo restriction enzyme site depending on which template strand is to be extended. This in vitro digestion is performed overnight usually with 100U of endonuclease to ensure complete digestion of the target sequence. The in vitro digest serves as an internal control to ensure equal amounts of DNA template was used in the reiterative primer extension reaction.
Digest DNA to completion with a second restriction enzyme recognizing a site upstream of the in vivo restriction enzyme site. Use 100 Units of enzyme with manufacture’s provided buffer according to manufacturer’s instructions (in vitro digest). Incubate reaction overnight at 37°C. (see Note 10)
Purify digested DNA by two extractions with phenol/chloroform/isoamyl alcohol (PCI-25:24:1, v/v) and one extractions with chloroform. Mix samples by vigorous shaking and centrifuge at high speed (20,800 × g) for 5 min at room temperature.
Precipitate the DNA by addition of 625 μl ice-cold 95% ethanol and 5 μl 5 M NaCl. Incubate samples at −20°C for 30 min and pellet the DNA by centrifuge at high speed (20,800 × g) for 30 min at 4°C.
Allow DNA pellet to air-dry for 1 h at room temperature, resuspend in 100 μl sterile water, and determine DNA concentration by A260/280 absorbance reading.
3.5. End labeling of Sequence-specific oligonucleotides
End-labeling is a rapid and sensitive method for radioactively labeling DNA fragments such as oligonucleotide and is useful for visualizing small amounts of DNA. All of the enzymes employed are specific to either the 3′ or 5′ termini of DNA and will, consequently, only incorporate one radio-phosphate per oligonucleotide. The most common method of radio-labeling oligonucleotides uses polynucleotide kinase (PNK) to transfer a single radioactive phosphate group from γ-32P-ATP to the 5′-end of the oligonucleotide.
In a 1.5-ml microfuge tube add the following in the order indicated; combine PCR-grade water for a total volume of 20 μl, 2 μl 10X T4 polynucleotide kinase buffer (supplied with enzyme), 2 μl target-specific oligonucleotide (5 μM), 50 μCi ATP, [γ-32P]-6000 Ci/mmol, and 20U T4 polynucleotide kinase.
Mix the reaction by vortex and briefly centrifuge.
Incubate the reaction mixture for 10 min at 37°C.
Prepare MicroSpin G-25 columns according to manufacturer’s guidelines.
Pass labeling reaction over readied G-25 column as directed by manufacturer’s protocol.
Determine incorporation of radiolabel (cpm/μl) by liquid scintillation counter measurement.
3.6. Reiterative Primer Extension
The extent of restriction endonuclease hypersensitivity for a given target sequence is determined using reiterative primer extension with Taq polymerase and a template-specific 32P-labeled oligonucleotide. The primer selected to detect restriction endonuclease hypersensitivity regions should be located either upstream or downstream of the transcription factor binding site, ideally 200-300bp from the in vivo cleavage site.
The concentration of Mg and oligonucleotide selected, for primer extension, greatly influences the specificity and yield of the reaction; therefore, the amounts of both should be titrated to achieve optimal results. The primer selected should anneal downstream of the in vivo restriction endonuclease cleavage site and be greater than 18 bases in length with melting temperatures ranging between 45 and 70°C. The reiterative primer extension/PCR reaction, described above, has been optimized for studies involving the MMTV promoter (10). These conditions may be employed for other steroid-responsive promoters although optimization should be performed. (see Note 11)
Amplify 10-20 μg purified in vivo/in vitro digested genomic DNA in 30 μl PCR reaction mix using 1-5 × 106 cpm 32P-labled sequence-specific oligonucleotide. (see Note 12)
Thermocycler program should include an initial cycle of denaturation at 94°C for 4 min, annealing for 60°C for 2 min, and primer extension at 72°C for 2 min. Follow initial cycle with 29 cycles of 2 min denaturation at 94°C, 2 min annealing at 60°C, and 2 min primer extension at 72°C with final extension for 10 min.
Stop each PCR reaction with addition of 150 μl PCR stop buffer.
Purify extended products by extraction with two rounds of phenol/chloroform/isoamyl alcohol (500 μl) and one round chloroform (500 μl). Mix samples by vortexing for 5 sec and centrifuge at high speed (20,800 × g) for 5 min at room temperature.
Precipitate extension products by addition of 625 μl cold 95% ethanol and 5 μl 5 M NaCl followed by centrifugation at high speed (20,800 × g) for 30 min at 4°C. Precipitated products were washed with 70% cold ethanol, recovered by centrifugation (20,800 × g) for 10 min at 4°C and allow pellet to air-dry.
3.7. Analysis of Polymerase Extension Products
Primer extension products are resolved on denaturing polyacrylamide gels. Samples should be allowed to electrophorese to yield maximal separation between bands corresponding to the in vivo restriction endonuclease cleavage site and the in vitro extension terminal cleavage site. For the purposes of this procedure, samples were resolved on 38 × 30 cm gels using Sequi-Gen GT nucleic acid sequencing system (see Note 13).
1- Resuspend DNA pellets in 7 μl sample loading buffer, vortex at high speed for 10 sec, briefly centrifuge, heat for 5 min at 95°C, vortex again, and re-centrifuge to collect sample.
2- Pour denaturing polyacrylamide gel and pre-run according to manufacture’s specifications.
6- Load primer extension products and separate on denaturing polyacrylamide gel.
7- After electrophoresis, allow gel to cool then transfer to filter paper.
8- Dry gel under vacuum for 1 h at 80°C.
9- Expose to PhosphorImager Screen, scan and analyze suing ImageQuant software (see Note 14).
5. Concluding Remarks
The authors have described the use of restriction enzyme hypersensitivity/ reiterative primer extension for assaying nucleosome remodeling which is often a prerequisite for transcriptional activation (15). This method allows for a high-resolution analysis of changes in the chromatin architecture upon promoter stimulation. The individual example presented here demonstrates the use of restriction enzymes to determine steroid-responsive SWI/SNF-mediated promoter hypersensitivity (Figure 2). This method has also been employed to characterize chromatin remodeling and induction of hypersensitivity regions within endogenous nuclear receptor-dependent promoters. Restriction endonuclease hypersensitivity assays were used to identify differences in the chromatin structure of the IκBα and MMTV promoters upon induction by the glucocorticoid and progesterone receptors (18). Nuclease hypersensitivity has been used to demonstrate that estrogen induction of ERRα involves chromatin remodeling around the multiple hormone-response element (19).
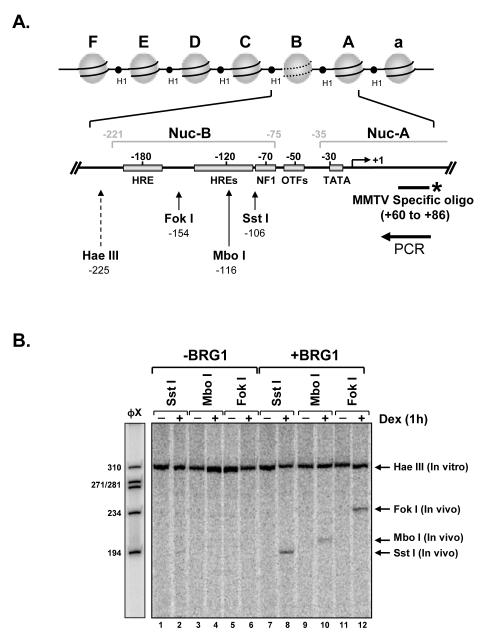
Figure 2.
BRG1-dependent restriction endonuclease chromatin accessibility within the MMTV promoter. A) Nucleosomal organization of the MMTV promoter. When stably integrated into the host genome the MMTV promoter is organized into a phased array of six positioned nucleosomes (A-F). The hormone inducible hypersensitivity region is encompassed by nucleosome B (Nuc-B). The expanded schematic of the proximal portion of the MMTV promoter shows the location of hormone-response elements (HREs), nuclear factor 1 (NF1), octamer transcription factors (OTFs) and the TATA binding protein (TBP) and identifies cleavage sites for restriction enzymes to test for induced hypersensitivity and chromatin remodeling (21). B) SW-13/MMTV cells, expressing GR, were transiently transfected with empty vector (-BRG1) or BRG1 expression plasmid (+BRG1). Nuclei, from untreated (-) or dexamethasone treated (10−7M) cells, were isolated and digested with restriction endonucleases targeting sequences within nucleosome-B (in vivo: Sst I, Mbo I, or Fok I). Purified genomic DNA was digested to completion with Hae III (In vitro). Fifteen micrograms of each sample was analyzed using linear Taq polymerase reiterative primer extension with a 32P-labeled single-stranded specific for MMTV. Extension products were analyzed on a 6% polyacrylamide denaturing gel and exposed to PhosphorImage screen.
Genome-wide strategies can be used to in concert with restriction endonuclease hypersensitivity to identify regulatory domains and elucidate specificity and function. DNase I hypersensitivity assays can be used to map discrete sites in the genome where changes in chromatin conformation yields DNA hypersensitivity to digest by the endonuclease DNase I. These DNase I hypersensitivity sites have been found in the regulatory domains of promoters, enhancers, silencers, insulators and locus control regions (20). Taken together, the use of these in vivo chromatin analysis techniques can provide insight into the roles structured chromatin and remodeling complexes play in the regulation of nuclear processes such as transcription.
5. Notes
The authors do not endorse any products or manufactures listed protocol
Transfection of BRG1 expression plasmids carried out in order to determine the dependence of SWI/SNF chromatin remodeling on restriction enzyme accessibility. Depending on cell line being used transfection may not be necessary.
Generally, buffers used for nuclei isolation are not maintained at 4°C for more that two months. The following buffer stock solution may be used in preparation of these solutions. 10X Salt Buffer: 100 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 600 mM KCl (store at room temperature). Spermine (0.15 M) and Spermidine (0.5 M) stock solutions (1000X) should be prepared and added to each buffer just prior to isolation. Store 1000X stock solutions at −20°C.
Prepare DTT as a 1M (1000X) stock and add prior to use. Proteinase K should be prepared as a 20 mg/ml (100X) stock and added prior to use.
Sequence specific real-time primer pairs may substitute for 32P-labeled oligonucleotide for real-time PCR analysis of cleavage products.
Our laboratory uses the following premixed sequencing solutions: SequaGel Sequencing System (National Diagnostics, Atlanta, GA).
The number of Dounce homogenizer pestle stroke needed for efficient nuclei isolation is cell type specific. It is recommended, after using four pestle strokes, to evaluate extent of nuclei isolation under a light microscope.
The restriction enzyme buffer supplied by the manufacture may be used for in vivo digest.
If using a Speed-Vac, be careful not to over dry DNA pellet.
The recognition sequence for this second restriction enzyme should located upstream of the in vivo restriction enzyme site. This digest serves as an internal standard to ensure equal loading of DNA into the Taq polymerase reiterative primer extension assay. This second digest also makes the sample less viscous and the genomic DNA easier to pipette.
The PCR conditions described here have been optimized for analysis of stably integrated MMTV promoter. However, these conditions have been used to evaluate in vivo restriction enzyme hypersensitivity from other nuclear receptor-dependent promoters and should prove to be good starting point for analyzing other target sequences.
Sequencing reactions can be performed to map predicted restriction enzyme cleavage sites using the PCR reactions mix and conditions described above with three deoxynucleotides at 200 μM and the fourth at 100 μM ddGTP/20 μM dGTP, 200 μM ddATP/5 μM dATP, 200 μM ddCTP/ 20 μM dCTP, or 200 μM ddTTP/20 μM dTTP.
Detection of multiple transcription factor-binding sites can be visualized more effectively suing longer denaturing gels such as 40-50 cm.
Kodak X-OMAT Blue film can be used for gel exposure if PhosphorImager screen is unavailable.
References
- 1.Wolffe AP. Transcription: in tune with the histones. Cell. 1994;77:13–16. doi: 10.1016/0092-8674(94)90229-1. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD, Lorch Y. Twenty-Five Years of the Nucleosome, Fundamental Particle of the Eukaryote Chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 3.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 4.Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 5.Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nucl Recept Signal. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sif S. ATP-dependent nucleosome remodeling complexes: enzymes tailored to deal with chromatin. J Cell Biochem. 2004;91:1087–1098. doi: 10.1002/jcb.20005. [DOI] [PubMed] [Google Scholar]
- 7.Eberharter A, Becker PB. ATP-dependent nucleosome remodelling: factors and functions. J Cell Sci. 2004;117:3707–3711. doi: 10.1242/jcs.01175. [DOI] [PubMed] [Google Scholar]
- 8.Trotter KW, Archer TK. Nuclear receptors and chromatin remodeling machinery. Molecular and cellular endocrinology. 2007:265–266. 162–167. doi: 10.1016/j.mce.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Archer TK, Lefebvre P, Wolford RG, Hager GL. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 10.Trotter KW, Archer TK. Reconstitution of glucocorticoid receptor-dependent transcription in vivo. Molecular and cellular biology. 2004;24:3347–3358. doi: 10.1128/MCB.24.8.3347-3358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Almer A, Horz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. The EMBO journal. 1986;5:2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyes J, Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. The EMBO journal. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- 13.Okino ST, Whitlock JP., Jr. Dioxin induces localized, graded changes in chromatin structure: implications for Cyp1A1 gene transcription. Molecular and cellular biology. 1995;15:3714–3721. doi: 10.1128/mcb.15.7.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verdin E, Paras P, Jr., Van Lint C. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. The EMBO journal. 1993;12:3249–3259. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Archer TK, Cordingley MG, Wolford RG, Hager GL. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Molecular and cellular biology. 1991;11:688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Archer TK, Deroo BJ, Fryer CJ. Chromatin modulation of glucocorticoid and progesterone receptor activity. Trends Endocrinol Metab. 1997;8:384–390. doi: 10.1016/s1043-2760(97)00159-8. [DOI] [PubMed] [Google Scholar]
- 17.Deroo BJ, Archer TK. Glucocorticoid receptor-mediated chromatin remodeling in vivo. Oncogene. 2001;20:3039–3046. doi: 10.1038/sj.onc.1204328. [DOI] [PubMed] [Google Scholar]
- 18.Deroo BJ, Archer TK. Glucocorticoid receptor activation of the I kappa B alpha promoter within chromatin. Mol Biol Cell. 2001;12:3365–3374. doi: 10.1091/mbc.12.11.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu P, Kinyamu HK, Wang L, Martin J, Archer TK, Teng C. Estrogen induces estrogen-related receptor alpha gene expression and chromatin structural changes in estrogen receptor (ER)-positive and ER-negative breast cancer cells. J Biol Chem. 2008;283:6752–6763. doi: 10.1074/jbc.M705937200. [DOI] [PubMed] [Google Scholar]
- 20.Pipkin ME, Lichtenheld MG. A reliable method to display authentic DNase I hypersensitive sites at long-ranges in single-copy genes from large genomes. Nucleic Acids Res. 2006;34:e34. doi: 10.1093/nar/gkl006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hebbar PB, Archer TK. Chromatin remodeling by nuclear receptors. Chromosoma. 2003;111:495–504. doi: 10.1007/s00412-003-0232-x. [DOI] [PubMed] [Google Scholar]