Abstract
The extracellular matrix (ECM) not only provides physical support for tissues, but it is also critical for tissue development, homeostasis and disease. Over 300 ECM molecules have been defined as comprising the “core matrisome” in mammals through the analysis of whole genome sequences. During tooth development, the structure and functions of the ECM dynamically change. In the early stages, basement membranes (BMs) separate two cell layers of the dental epithelium and the mesenchyme. Later in the differentiation stages, the BM layer is replaced with the enamel matrix and the dentin matrix, which are secreted by ameloblasts and odontoblasts, respectively. The enamel matrix genes and the dentin matrix genes are each clustered in two closed regions located on human chromosome 4 (mouse chromosome 5), except for the gene coded for amelogenin, the major enamel matrix protein, which is located on the sex chromosomes. These genes for enamel and dentin matrix proteins are derived from a common ancestral gene, but as a result of evolution, they diverged in terms of their specific functions. These matrix proteins play important roles in cell adhesion, polarity, and differentiation and mineralization of enamel and dentin matrices. Mutations of these genes cause diseases such as odontogenesis imperfect (OI) and amelogenesis imperfect (AI). In this review, we discuss the recently defined terms matrisome and matrixome for ECMs, as well as focus on genes and functions of enamel and dentin matrix proteins.
Keywords: Tooth extracellular matrix, evolution and function, development and diseases
1. Introduction
The extracellular matrix (ECM) regulates many aspects of cell processes, organogenesis, tissue homeostasis and functions, regeneration, and diseases. The ECM is constantly being remodeled and reshaped through degrading followed by reassembly. The remodeling is particularly prevalent during development and wound repair. The ECM can be remodeled in response to signals transmitted by ECM receptors such as integrins and syndecans, and by ECM modifying proteins such as matrix metalloproteinases.
Many ECM proteins are comprised of repeats of domains such as EGF-like, fibronectin type I, collagen triple helix, laminin G, fibulin, and link protein. There are three major classes of ECM proteins based on their structural characteristics: collagens, proteoglycans, and glycoproteins. Collagens form extracellular matrices and provide tissue strength. Among the collagen family, collagen I is the most abundant ECM in the body. It not only serves as a tissue scaffold, but also as a critical ECM for bone and dentin mineralization. Proteoglycans are expressed in many tissues, and their core proteins are substituted with glycosaminoglycans (GAG) such as heparan sulfate (HS) and chondroitin sulfate (CS) chains. Aggrecan, which used to be referred to as the proteoglycan, contains about 100 CS chains in its core protein and serves to resist compression in joint cartilage. Another example of proteoglycans is perlecan, the major basement membrane proteoglycan, which has many biological functions such as angiogenesis, skeletal formation, and cancer. Glycoproteins include many ECMs such as fibronectin, laminin, agrin, ameloblastin, amelogenin, and fibulin and have many biological activities including cell adhesion, migration, and differentiation via cell receptors and growth factor interactions.
In tooth development, three types of ECM structures play critical roles in morphogenesis. In early development, basement membranes (BMs), which consist of a unique set of ECM proteins, serve as a physical barrier and a regulatory structure for reciprocal signaling between the dental epithelium and mesenchyme. The dental epithelium and mesenchyme differentiate to enamel matrix-secreting ameloblasts and dentin matrix-secreting odontoblasts. Both enamel and dentin matrices are made up of a distinct set of ECM proteins. In dentin and bone, collagen I forms collagen fibrils and provides a scaffold for mineralization. Many non-collagenous dentin and bone matrix proteins are also required for mineralization in these tissues. In contrast, mineralization in enamel matrix is formed primary through activities of non-collagenous proteins. Some enamel and dentin matrix proteins show multiple cellular functions such as cell adhesion and polarity.
2. How many extracellular matrix molecules are coded by the whole genome in mammals? What are the definitions of “matrisome” and “matrixome”?
The completion of the entire genome sequencing allows all ECM proteins to be listed and categorized based primarily on the presence of characteristic domains. Hynes and Noba [1] defined the “core matrisome” as a list of all ECM proteins, excluding ECM-modifying proteins such as matrix metalloproteinases, which includes approximately 300 proteins. It is comprised of approximately 1.5% of proteasomes, a list of all proteins expressed by the genome. It includes 53 collagens, ~35 proteoglycans, and more than 200 glycoproteins. Sekiguchi and his group also proposed the term “matrixome” as a subset of the proteome to define whole collections of ECM molecules. In their matrixome project, a “mouse basement membrane body map” has been created in which 90% of about 50 of all basement membrane proteins and its associated proteins were complied and shown in high resolution digital images by immunohistochemistry in mouse embryos of varying stages of development [2]. In addition to the core matrisome, there is a large number of ECM-modifying enzymes such as those involved in proteolysis, sulfation, and glycosylation, ECM-binding growth factors such as TGFβs and BMPs, and other ECM-associated proteins. They cooperate to assemble and remodel ECMs and bind to cells through ECM receptors. Together with receptors, they provide multiple signals to cells that control the survival, proliferation, differentiation, shape, polarity, and motility of cells [1].
ECM proteins are widely expressed in almost all tissues in the human body and play a pivotal role in development and tissue functions. The total number of ECM molecules is surprisingly small. For example, the G-protein Coupled Receptor (GPCR) family and the zinc finger protein family consist of ~800 and ~750 members, respectively. The complexity of diverse developmental and tissue-specific functions of the ECM are the result of post-translational modifications, protein-protein interactions, availability of ECM cell surface receptors, and cell type-specific ECM expression, as well as variants of RNA splicing and alternative promoters. Alterations in the expression of ECM molecules are also often associated with many diseases such as diabetes, inflammation, and autoimmune diseases. There are several databases that are useful for finding information regarding the expression and distribution of various ECM genes/proteins (e.g., http://web.mit.edu/hyneslab/matrisome/; http://www.matrixome.com/bm/Home/home/home.asp; http://www.proteinatlas.org/;http://bite-it.helsinki.fi for teeth).
3. How have extracellular matrix genes evolved?
ECMs are widespread in metazoan; they underlay and surround many cells and comprise distinct morphological arrangements. ECM proteins are typically made of repeated domains, which are encoded in the genome as separate exonic units. Such domain repeats are evoked by exon shuffling, which is the earliest recognized and most elaborate examples in ECM evolution [3, 4]. These repeat arrangements are highly characteristic of ECMs. Analyses of the genomes in different organisms provide new insight into the evolution of the matrisome. Some ECMs in mammals appear to retain very ancient forms, which are found in all metazoans including sponges, and some are derived from newer proteins. A set of ECM proteins, which are components of the basic cores of basement membrane, is present in genomes of protostomes and deuterostomes and therefore must be common ancestors. These conserved ECM proteins are collagen IV (α1, α2 chains), laminin (2 α, β1, and γ1 chains), nidogen, and perlecan. Gene duplications have resulted in increases in the number of the genes for collagen IV chains (6), laminin chains (11), and nidogens (2), but not perlecan (one). The increase in the number of other family genes is also due to gene duplication and exon shuffling of ancestral genes that gave rise to diversity of the structure and functions of ECMs. During the evolution of the vertebrate subphylum, large numbers of ECM proteins have been created. Some of these ECM proteins, especially those in hard tissues such as teeth, cartilage, and bones, neural crest, vasculature, and advanced nervous and immune systems, must be newly acquired by domain shuffling and duplication events as well as the activation of intergenic and intronic sequences.
4. Is there a common ancestral gene for enamel and dentin matrix proteins?
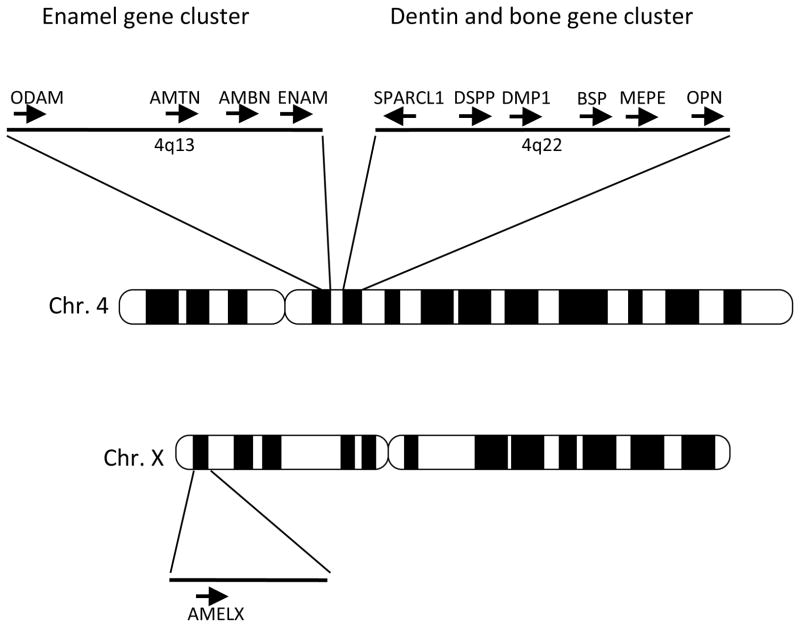
Mineralization is one of the hallmark processes during vertebrate evolution that incorporates minerals in soft matrices. Three major mineralized tissues are enamel/enameloids (enamel-like tissues), dentin, and bone. These tissues have evolved over 500 million years to gain species- and tissue-specific multiple functions such as body armor, tissue support, locomotion, and teeth [5]. Although the molecular mechanism of mineralization is not fully understood, the mineralization process is likely evolved from a common ancestral process [5]. The vertebrate tooth consists of three principal mineralized tissues: enamel, dentin, and the surrounding bone. Development of these tissues requires ECMs that contain distinct sets of proteins [6, 7]. Most dentin matrix proteins are also present in the bone matrix, while enamel matrix proteins are specifically expressed by ameloblasts. Genes encoding non-collagenous dentin matrix proteins expressed by odontoblasts are DSPP (dentin sialophosphoprotein), BSP (bone sialoprotein/IBSP, integrin-binding sialoprotein), DMP1 (dentin matrix acidic phosphoprotein), MEPE (matrix extracellular phosphoglycoprotein), and OPN (osteopontin/SPP1, secreted phosphoprotein 1) (Table 1). These dentin matrix genes are also expressed in bone, although their expression levels are different. The genes for enamel matrix proteins expressed by ameloblasts include AMEL (amelogenin), ENAM (enamelin), AMBN (ameloblastin/sheathlin/amelin), and recently identified AMTN (amelotin) and ODAM (odontogenic, ameloblast-associated, APIN). These genes evolved from a common ancestral gene encoding a secretory calcium-binding phosphoprotein (SCPP) by gene duplications [8, 9]. Some of the SCPP family proteins are also found in milk and saliva. SCPP family genes are mapped on human chromosome 4 in two clustered regions separated by 15 Mb (Fig. 1). Enamel matrix genes and milk/saliva SCPP genes are located on 4q13 (about 770 kb) and dentin matrix genes on 4q22 (about 375 kb). However, AMEL is located on sex chromosomes; AMELX on X chromosome, and AMELY on Y chromosome. About 90% of the AMEL transcripts are expressed on the X locus, where they are located on intron 1 of the ARHGAP6 gene in an opposite orientation. Comparative analysis of the enamel matrix genes in different species revealed that ENEM is the most ancient gene, and the other four enamel genes are derived from ENEM. AMEL is derived from AMBN as a result of a duplication and was later translocated to sex chromosomes [9]. The chromosomal synteny of these genes including milk/saliva SCPP is conserved in primates and rodents. In birds, who lost teeth about 100 million years ago, the chromosomal synteny of the dentin matrix genes is conserved, while the enamel and milk/saliva gene cluster is lacking [10].
Table 1.
Extracellular matrix proteins in dentin and enamel
| Non-collagenous Dentin Matrix Proteins
| |
|---|---|
| Gene Symbol | Protein Name |
| DSPP | Dentin sialophosphoprotein |
| DMP1 | Dentin matrix acidic phosphoprotein 1 |
| BSP (IBSP) | Bone sialoprotein (integrin-binding sialoprotein) |
| MEPE | Matrix extracellular phosphoglycoprotein |
| OPN (SPP1) | Osteopontin (Secreted phosphoprotein 1) |
| FBLN7a | Fibulin-7 (TM14) |
| Enamel Matrix Proteins
| |
|---|---|
| Gene Symbol | Protein Name |
| AMEL | Amelogenin |
| ENAM | Enamelin |
| AMBN | Ameloblastin (sheathlin, amelin) |
| AMTN | Amelotin |
| ODAM(APIN) | Odontogenic, ameloblast-associated (Apin protein) |
| TUFT1b | Tuftelin |
Non-dentin matrix proteins and enamel matrix proteins are shown.
Fibulin-7 and
Tuftelin are also expressed in soft tissues.
Figure 1. The tooth and bone gene cluster on human chromosome 4.
ENAM (enamelin) AMBN (ameloblastin), AMTN (amelotin), and ODAM (odontogenic, ameloblast-associated), which are enamel matrix genes, are located on 4q13 (about 770 kb) and SPARCL1, DSPP (dentin sialophosphoprotein), DMP1 (dentin matrix acidic phosphoprotein 1), BSP (bone sialoprotein/IBSP, integrin-binding sialoprotein), MEPE (matrix extracellular phosphoglycoprotein), and OPN (osteopontin/SPP1, secreted phosphoprotein 1), which are dentin and bone matrix genes are on 4q22 (about 375 kb). AMEL is located sex chromosomes. The X chromosome gene, AMELX, is shown. Each arrow illustrates a gene and the transcriptional direction.
Dentin and enamel matrix gene mutations are associated with several tooth diseases. For example, DSPP mutations are associated with dentin dysplasia (DP) and dentinogenesis imperfect (DGI–III). [11–14]. AMEL and ENAM mutations are associated with amelogenesis imperfect (AI) [15, 16].
5. What roles do extracellular matrix proteins play in tooth development and diseases?
Tooth morphogenesis is a result of reciprocal interactions between oral epithelium and ectomesenchyme, culminating in the formation of mineralized tissues, enamel, and dentin [17–19]. The earliest morphogenetic event of mouse tooth development occurs when the oral ectoderm invaginates into the underlying neural crest-derived mesenchyme. Continuation of this invagination results in the formation of epithelial tooth buds. Mesenchymal cells surrounding the bud form the dental papillae, which later develop into dentin-secreting odontoblasts and the tooth pulp. After the bud stage, the tooth germ progresses to the cap and bell stages, and the epithelium differentiates into enamel-secreting ameloblasts. During this transition stage, BMs disappear and are replaced with enamel and dentin matrices.
5.1. Basement membrane proteins
Basement membranes (BMs) are thin extracellular matrices that separate epithelial and mesenchymal cells and surround cells, such as endothelial, muscular, and neural cells. Basement membranes are the first extracellular matrices to appear in development and are critical for organ development and tissue repair [20, 21]. They not only provide the scaffold for cells and cell layers, but they also play an essential role in morphogenesis that affects cell adhesion, migration, proliferation, and differentiation. BMs consist of collagen IV, laminin, perlecan, nidogen/entactin, and other molecules that interact with each other to form the supramolecular structure [20, 22]. The structure and components of BMs differ between tissues, resulting in tissue-specific structures and functions. Some BM components, such as perlecan, are present in tissues like cartilage that have no basement membrane, suggesting their distinct role in these tissues. BM components also play an important role in tooth and cartilage development.
These BM matrices control proliferation, polarity, and attachment; they also determine tooth germ size and morphology [17, 23, 24]. For example, lamininα5 (Lama5), a component of laminin-10/11 (laminin-512 consisting of α5β1γ1 chains/laminin-521 consisting of α5β2γ1 chains[25])is the major lamininα chain in tooth basement membranes. Lama5 knockout (KO) mice die between embryonic days 14 (E14) and 19 (E19) with multiple developmental abnormalities such as a vascular glomeruli, impaired branching morphogenesis [26]. Lama5 KO mice develop a small tooth germ with no cusps, in which the inner dental epithelium is not polarized and enamel knot formation is defective [24]. Integrin α6/β4, a ligand of Lama5, is not localized at the basal layer of the epithelium but diffusely expressed around the cell surface. Laminins-10 and -11 promote spreading and filopodia-like micro-spike formation of dental epithelium, and the interaction of Lama5 and Integrin α6/β4 mediates these cellular changes through PI3 kinase-Cdc42/Rac signaling [23, 24]. These studies demonstrate that Lama5 regulates the polarity and formation of the monolayer of the inner dental epithelium, and these cellular processes are essential for tooth growth and morphogenesis. Another example is the involvement of lamininα2 (Lama2) in odontoblast differentiation. In the maturation stage of tooth development, BMs reappear. Lama2 is a component of laminin-2 (laminin-211) and laminin-4 (laminin-221), which consist of α2β1γ1 chains and α2β2γ1 chains, respectively [25]. Lama2 is a major component of BMs in skeletal muscle and the peripheral nervous system. Lama2 is also expressed in odontoblasts during the late stage of tooth germ development [27, 28]. Lama2-deficent mice display thin dentin and defective dentinal tube structure [28]. These phenotypes are similar to dentinogenesis imperfect (DI) in humans. In mutant mice, dentin sialoprotein expression is reduced in odontoblasts. In cell cultures, laminin-2, not collagen I and fibronectin, increases the expression of dentin sialoprotein in odontoblasts [28]. Thus Lama2 is required for odontoblast differentiation. Perlecan (HSPG2) is a major heparan sulfate proteoglycan in BMs and also present in some other tissues such as cartilage. In developing teeth, the perlecan protein is expressed in BMs, the intercellular spaces of the enamel organ, and the dental papilla/pulp. In situ hybridization showed that perlecan mRNA is expressed in stellate reticulum cells and dental papilla/pulp cells, including odontoblasts and fibroblastic cells in the dental follicle [29]. Overexpression of perlecan under the control of a keratin 5 promoter in the enamel organ of transgenic mice shows abnormal tooth morphology and dysregulation of growth factors such as bFGF and TGF-β1 [30].
5.2. Enamel matrix proteins
Dental epithelium differentiates into presecretory, secretory, and maturation stage ameloblasts [31]. At the secretory stage, ameloblasts synthesize and secrete specific proteins in the enamel matrix that are replaced by calcium and phosphate during the maturation stage for enamel formation. The major secretory proteins synthesized by ameloblasts can be categorized as amelogenin (AMEL) and nonamelogenin proteins. Amelogenin, the major component of the mineralizing enamel matrix, is critical for enamel formation [31–33]. Ameloblastin (AMBN) and enamelin (ENAM) are also members of the nonamelogenin protein category. Ameloblastin is a tooth-specific glycoprotein that is rich in proline, glycine, and leucine [34–36]. Enamelin contains hydrophobic, acidic, and basic domains in different regions of its molecule [37–39]. More recently, two more enamel matrix proteins, amelotin [40, 41]and odontogenic ameloblast-associated (ODAM/APIN) [42, 43], were identified. Tuftelin was originally identified from a bovine ameloblast-enriched cDNA library. Later, tuftelin cDNA was also cloned in mice and humans [44, 45]. Tuftelin is synthesized and secreted by ameloblasts into the enamel matrix and concentrates at the dentin–enamel junction (DEJ) region [46]. However, unlike amelogenin and other enamel matrix proteins, tuftelin lacks a signal peptide sequence and is also expressed in many other tissues such as kidney, liver, testis and lung [44]. The human tuftelin gene (TUFT1) maps on chromosome 1 at 21–31 [47].
Immunohistochemistry demonstrates that amelogenin is expressed in the ECM prior to ameloblastin [48]. At the presecretory stage, when the basement membrane still divides ameloblasts and odontoblasts, only amelogenin is detectable in secretory granules. Ameloblastin is not detectable extracellularly at this early stage, but it appears later. At the secretory stage, amelogenin and ameloblastin are produced by secretory ameloblasts. Amelogenin localizes in the enamel layer, whereas ameloblastin localizes at the apical region of the cells and serves as the adhesion molecule for ameloblasts [49]. In a gene-targeted mutant ameloblastin, in which exons 4 and 5 of the AMBN gene in mice are deleted, ameloblasts are detached from the enamel matrix, continue to proliferate and form multiple cell layers, and odontogenic tumors often develop in the maxilla with age. Using recombinant ameloblastin proteins, the heparin-binding domains at the C-terminal half of ameloblastin are shown to be critical for ameloblast binding to dental epithelial cells in cultures [50]. Overexpression of full-length ameloblastin protein inhibits the proliferation of human ameloblastoma AM-1 cells, but overexpression of ameloblastin protein deficient in heparin-binding domains shows no inhibitory effect. In full-length ameloblastin overexpressing AM-1 cells, the expression of Msx2, which regulates the dental epithelial progenitor phenotype, is decreased. In contrast, the expression of cell proliferation inhibitors p21 and p27 is increased. In addition, the expression of enamelin, a marker of differentiated ameloblasts, is induced, suggesting that ameloblastin promotes odontogenic tumor differentiation. These results suggest that ameloblasts promote cell binding through the heparin binding sites, and play an important role in preventing odontogenic tumor development by suppressing cell proliferation and maintaining differentiation phenotypes through Msx2, p21, and p27 [50].
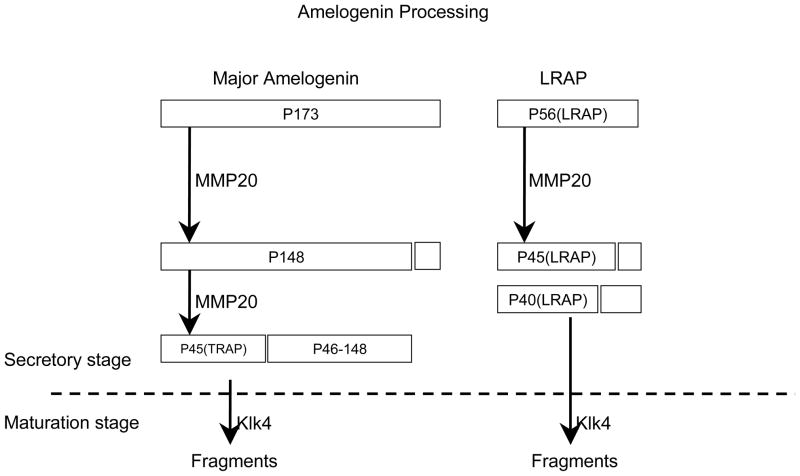
Amelogenin is the major secretory product of ameloblasts and is critical for proper tooth enamel formation. Amelogenin comprises over 80% of total secretory stage enamel proteins. Four secreted amelogenin isoforms have been isolated from developing porcine enamel [51]. The major porcine amelogenin (P173) has 173 amino acids (Fig. 2). The second most abundant amelogenin isoform is the leucine-rich amelogenin polypeptide (LRAP) having 56 amino acids [52]. These amelogenin isoforms undergo characteristic patterns of digestion by two extracellular proteases: Mmp20 and Klk4. Both enzymes are critical for proper dental enamel formation. Mmp20 is expressed throughout the secretory stage [53], while Klk4 is expressed during the maturation stage, including the short transitional phase. Therefore, Mmp20 has the only significant proteolytic activity in the enamel extracellular matrix during the secretory stage. The study of Mmp20 and Klk4 digestion of P173 and LRAP showed a unique processing pathway [51]. Mmp20 cleaves P173 and generates the most abundant amelogenin cleavage products, P148 (tyrosine rich amelogenin polypeptide: TRAP) and the P46-148 amelogenins (Fig 2). Mmp20 shows little or no activity against TRAP and LRAP, whereas Klk4 degrades both of them [54]. These results suggest that TRAP and LRAP amelogenin peptides accumulate and are slowly degraded by Mmp20 during the secretory stage. In the maturation stage, Klk4 is expressed in the enamel matrix space and degrades TRAP and LRAP (Fig 2). Digested enamel matrices are replaced by calcium and phosphate and then create highly calcified enamel. Amelogenin (AMLX) KO mice show severe enamel hypoplasia similar to human X-linked amelogenesis imperfect [32]. Analysis of the AMLX-deficient teeth of the mutant mice reveals that amelogenin is not required to initiate mineral crystal formation, but rather for the organization of crystal patterning and the regulation of enamel thickness. The AMLX KO tooth phenotypes can be rescued with two amelogenin transgenes, M180 (the most abundant amelogenin protein in murine) and LRAP [55]. When the KO mice were mated with mice that expressed the transgene M180, a partial rescue of the phenotype occurred in terms of enamel thickness and volume. However, the transgene LRAP alone fails to rescue the amelogenin null phenotype [56]. Adding LRAP transgene to M180-KO mice leads to an added improvement in both the amount of enamel and enamel structure. These results suggest that M180 contributes to enamel thickness and volume. On the other hand, LRAP tends to involve calcification. Double-knockout mice for ameloblastin (AMBN) and amelogenin (AMLX) show enamel defects and an irregular ameloblast layer, as well as adetachment from the enamel surface, similar to AMBN KO teeth [57]. However, the enamel width was significantly reduced in the double-KO mice when compared with single KO mice. These results suggest the synergistic role of ameloblastin and amelogenin.
Figure 2. Porcine amelogenin isoforms and cleavage fragments.
The major amelogenin (P173) and the second most abundant isoform LRAP are digested by two extracellular proteases, Mmp20 and Klk4. Mmp20 is expressed the secretory stage, while Klk4 is expressed during the maturation stage.
Amelotin and ODAM are induced during a transition stage of ameloblast differentiation, and their expression continues to the maturation stage [40, 43, 58, 59]. These expression patterns are distinct from those of the three other enamel matrix proteins, ameloblastin, amelogenin and enamelin. Transgenic mice that produce amelotin under the control of the amelogenin gene promoter show thinner enamel and a highly irregular enamel surface structure. The overexpression of amelotin disrupts the formation of Tomes’ process and causes defects in the orderly growth of enamel prisms [60].
Transgenic mice that produce amelogenin signal peptide-fusion tuftelin under the control of the amelogenin promoter display a loss of restricted growth of enamel crystallites along their a-axis and b-axis [61].
5.3. Dentin matrix proteins
Dentin and bone consist of similar ECM proteins including collagen and non-collagen matrix proteins [62]. These ECM proteins are secreted from odontoblasts, and osteoblasts, both which are derived from the neural crest. Collagen I fibrils serve as the major scaffold for mineralization in dentin and bone. However, non-collagenous matrix proteins are also required for mineralization. Although many non-collagenous dentin matrix proteins are also present in the bone matrix, relative ratios of their compositions vary significantly between dentin and bone. For example, the dentin sialophosphoprotein (DSPP) expression level is more than 100 times in the dentin matrix, compared with bone matrix. Human dentin diseases have been classified into two major groups: dentin dysplasia (DD) and dentinogenesis imperfecta (DGI–III) [11]. Mutations of collagen I cause osteogenesis imperfect (OI), which is characterized by brittle bone. DGI is often associated with OI. Mutations of DSPP have been associated with DD and DGI–III [11, 63].
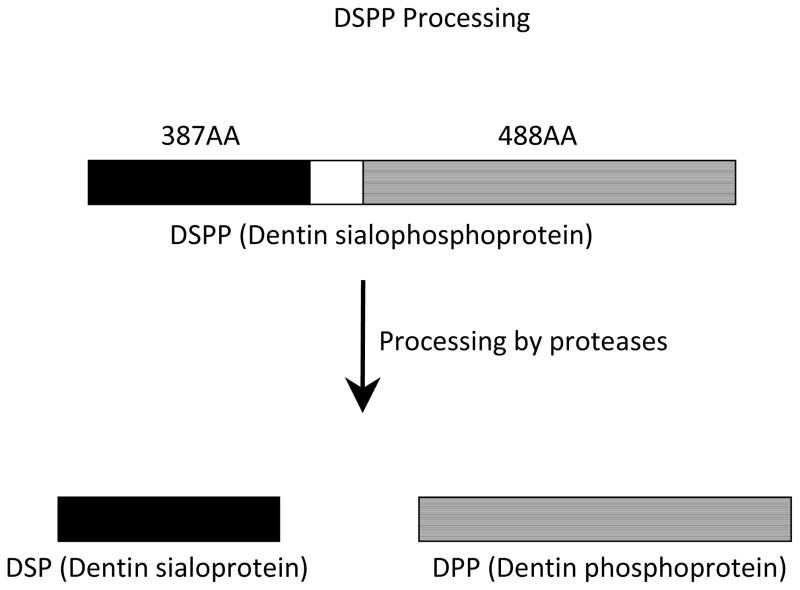
The DSPP gene has been mapped to 4q21, which includes the overlapping regions for DGI–II, DGI–III, and DD type II loci. Within a 375 kb region on human chromosome 4q21, there is a cluster of genes (DSPP, DMP1, BSP, MEPE, and OPN)coding for the non-collagenous dentin matrix proteins (Fig. 1). Dentinogenesis is a continuous process of matrix deposition throughout the life of a tooth. The odontoblast secretes dentin matrix and the cell leaves its process embedded in the dentin matrix. This process involves the initial odontoblastic synthesis of a collagen-rich ECM and predentin that is converted to dentin when the collagen becomes mineralized. Similarly, osteogenesis involves an initial unmineralized osteoid that is mineralized and converted to bone [6]. Dentin sialoprotein (DSP) and dentin phosphoprotein (DPP) are major non-collagenous proteins and play a critical role in dentinogenesis. These two proteins are derived from the cleavage of a 940 amino acid polypeptide, DSPP (Fig. 3) [64–66]. DSPP is expressed in differentiating and mature odontoblasts, with expression by mature ameloblasts. The function of DSPP in ameloblasts is unknown. Low levels of DSPP have been observed in the ears and bones [67, 68]. DSP, the amino terminal domain of DSPP, is a sialic acid-rich and glycosylated protein that shares similarities with the other sialoproteins, such as BSP, DMP-1, and OPN. DPP is a highly phosphorylated protein with repeats of aspartic acid and phosphoserine. DPP plays an important role in the nucleation of hydroxyapatite formation during dentin calcification [69–71].
Figure 3. Processing of DSPP.
The DSPP (dentin sialophosphoprotein) protein is coded by the DSPP gene and is the precursor of the DSP (dentin sialoprotein) and DPP (dentin phosphoprotein) proteins. The DSPP protein is cleaved by proteases into N-terminal DSP, and C-terminal DPP proteins.
DSSP KO mice display enlarged pulp chambers, increased predentin width, and hypomineralization, similar to human dentinogenesis imperfecta III [72]. In order to investigate the in vivo roles of DSP and DPP as individual matrix proteins in dentin mineralization, transgenic mice that express only the DSP part but not DPP part were created using the DSP transgene expressed by odontoblasts under the control of the DSPP promoter in a DSPP KO genomic background [73]. The mutant mice showed a partial rescue of the DSPP KO phenotype except for the dentin mineral density. These results indicate the distinct roles of DSP and DPP in dentin mineralization, with DSP regulating the initiation of dentin mineralization, and DPP being involved in the maturation of mineralized dentin.
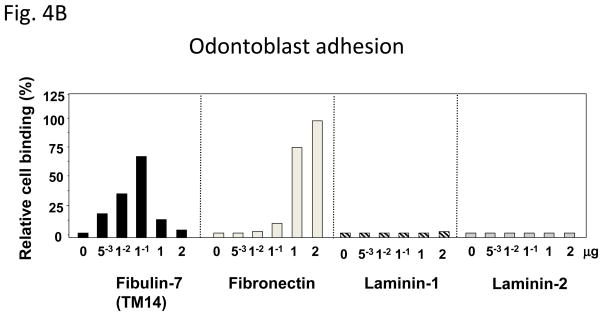
Recently, a new dentin matrix protein, TM14 (Fibulin-7), was identified by differential hybridization using mouse tooth germ cDNA microarrays [74]. TM14 contains three EGF modules at the center, a C-terminal domain homologous to the fibulin module, and a unique Sushi domain at the N-terminus (Fig. 4A). Because of the similarity of its domain structure, TM14 protein can be classified as a new member of the extracellular protein fibulin family: fibulin-7 (FBLN7) [75]. Fibulin-7 mRNA is expressed by preodontoblasts and odontoblasts in developing teeth. Immunostaining revealed that fibulin-7 is localized at the apical pericellular regions of preodontoblasts. When the dentin matrix is fully formed and dentin mineralization has occurred, fibulin-7 is localized in the predentin matrix and along the dentinal tubules [74]. The recombinant fibulin-7 protein interacts with heparin, fibronectin, fibulin-1, and dentin sialophosphoprotein [74]. Odontoblasts bind to fibulin-7 and fibronectin but not to laminin-1 and laminin-2 using substrate-coated wells in culture (Fig. 4B) [74]. Cell binding is reduced when fibulin-7 concentrations are increased to 1 and 2 μg/well. This may be because fibulin-7 may form multimeric complexes and mask an active site for cell binding. Fibulin-7 also binds to dental mesenchyme cells but not dental epithelial cells, or non tooth cells. such as NIH3T3, HeLa, and COS7 cells [74]. Heparin, EDTA, and anti-integrin β1 antibodies inhibit fibulin-7 binding to dental mesenchyme cells, suggesting that both a heparin sulfate-containing cell surface receptor and an integrin are involved in fibulin-7 cell binding [74]. These data suggest that fibulin-7 plays an important role in both the differentiation and maintenance of odontoblasts as well as in dentin formation. Fibulin-7 is also expressed in other tissues such cartilage, capillary walls, placenta, and the eye [74, 75]. The human fibulin-7 gene (FBLN7) maps on chromosome 2, at 2q13. The expression patterns and chromosome location of the fibulin-7 gene is distinct from those of other enamel and dentin matrix genes.
Figure 4. Fibulin-7 structure and cell binding activity.
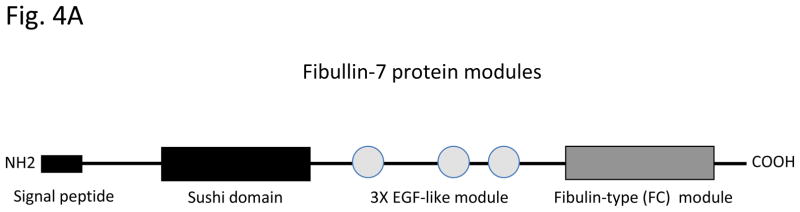
(A) Fibulin-7 protein modules. Fibulin-7 (TM14) protein consists of 3 domains, I, II, III. Profibulin-7 contains a typical signal peptide sequence. At the N-terminus, the sushi domain represents unique in fibulin-7 among other fibulin family proteins. Domain II contains 3 EGF-like calcium-binding modules. At the C-terminus, the FC globular domain is characteristic to the fibulin family and shared by all fibulin proteins. Fibulin-7 shows similar modular arrangement as the other fibulin members. (B) Fibulin-7 binding to odontoblasts. Odontoblasts attach to recombinant fibulin-7 and fibronectin but not to laminins-1 and laminins-2. The data are from Fig. 8B in J Biol Chem 2007, 282: 30878–30888.
6. Conclusion
Analysis of genome sequences reveals that there are approximately 300 genes encoding ECM proteins in mammals. Tooth ECM proteins consist of collagenous and non-collagenous proteins that are important for tooth morphogenesis. Genes for many non-collagenous proteins expressed in the enamel and dentin matrices are clustered in two close regions on the same chromosome. Evolutional genome analysis suggests that these genes arose from a common ancestral gene that encoded a secretory calcium-binding protein. During evolution, these genes gained unique functions in tooth mineralization tissues. Studies of ECM structure/function and interactions, as well as genetic analyses, will provide further insight into understanding the mechanisms of tooth morphogenesis and mineralization processes.
Acknowledgments
This work was supported by grants from the Intramural Program of National Institute of Dental and Craniofacial Research and the National Institutes of Health to Y.Y. K.Y. was supported in part by the Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hynes RO, Naba A. Overview of the matrisome--an inventory of extracellular matrix constituents and functions. Cold Spring Harb Perspect Biol. 2012;4:a004903. doi: 10.1101/cshperspect.a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manabe R, Tsutsui K, Yamada T, Kimura M, Nakano I, Shimono C, et al. Transcriptome-based systematic identification of extracellular matrix proteins. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12849–54. doi: 10.1073/pnas.0803640105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engel J. Domain organizations of modular extracellular matrix proteins and their evolution. Matrix biology: journal of the International Society for Matrix Biology. 1996;15:295–9. doi: 10.1016/s0945-053x(96)90130-4. [DOI] [PubMed] [Google Scholar]
- 4.Patthy L. Genome evolution and the evolution of exon-shuffling--a review. Gene. 1999;238:103–14. doi: 10.1016/s0378-1119(99)00228-0. [DOI] [PubMed] [Google Scholar]
- 5.Kawasaki K, Suzuki T, Weiss KM. Genetic basis for the evolution of vertebrate mineralized tissue. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11356–61. doi: 10.1073/pnas.0404279101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler WT, Brunn JC, Qin C. Dentin extracellular matrix (ECM) proteins: comparison to bone ECM and contribution to dynamics of dentinogenesis. Connective Tissue Research. 2003;44 (Suppl 1):171–8. [PubMed] [Google Scholar]
- 7.Bartlett JD, Ganss B, Goldberg M, Moradian-Oldak J, Paine ML, Snead ML, et al. 3. Protein-protein interactions of the developing enamel matrix. Curr Top Dev Biol. 2006;74:57–115. doi: 10.1016/S0070-2153(06)74003-0. [DOI] [PubMed] [Google Scholar]
- 8.Kawasaki K, Weiss KM. Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4060–5. doi: 10.1073/pnas.0638023100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sire JY, Davit-Beal T, Delgado S, Gu X. The origin and evolution of enamel mineralization genes. Cells Tissues Organs. 2007;186:25–48. doi: 10.1159/000102679. [DOI] [PubMed] [Google Scholar]
- 10.Kawasaki K, Weiss KM. Evolutionary genetics of vertebrate tissue mineralization: the origin and evolution of the secretory calcium-binding phosphoprotein family. Journal of experimental zoology Part B, Molecular and developmental evolution. 2006;306:295–316. doi: 10.1002/jez.b.21088. [DOI] [PubMed] [Google Scholar]
- 11.MacDougall M, Dong J, Acevedo AC. Molecular basis of human dentin diseases. Am J Med Genet A. 2006;140:2536–46. doi: 10.1002/ajmg.a.31359. [DOI] [PubMed] [Google Scholar]
- 12.Hart PS, Hart TC. Disorders of human dentin. Cells Tissues Organs. 2007;186:70–7. doi: 10.1159/000102682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JW, Simmer JP. Hereditary dentin defects. Journal of dental research. 2007;86:392–9. doi: 10.1177/154405910708600502. [DOI] [PubMed] [Google Scholar]
- 14.McKnight DA, Simmer JP, Hart PS, Hart TC, Fisher LW. Overlapping DSPP mutations cause dentin dysplasia and dentinogenesis imperfecta. Journal of dental research. 2008;87:1108–11. doi: 10.1177/154405910808701217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu JC, Chun YH, Al Hazzazzi T, Simmer JP. Enamel formation and amelogenesis imperfecta. Cells Tissues Organs. 2007;186:78–85. doi: 10.1159/000102683. [DOI] [PubMed] [Google Scholar]
- 16.Wright JT, Torain M, Long K, Seow K, Crawford P, Aldred MJ, et al. Amelogenesis imperfecta: genotype-phenotype studies in 71 families. Cells Tissues Organs. 2011;194:279–83. doi: 10.1159/000324339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thesleff I, Barrach HJ, Foidart JM, Vaheri A, Pratt RM, Martin GR. Changes in the distribution of type IV collagen, laminin, proteoglycan, and fibronectin during mouse tooth development. Developmental biology. 1981;81:182–92. doi: 10.1016/0012-1606(81)90361-4. [DOI] [PubMed] [Google Scholar]
- 18.Aberg T, Wozney J, Thesleff I. Expression patterns of bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and cell differentiation. Developmental dynamics: an official publication of the American Association of Anatomists. 1997;210:383–96. doi: 10.1002/(SICI)1097-0177(199712)210:4<383::AID-AJA3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 19.Jernvall J, Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mechanisms of development. 2000;92:19–29. doi: 10.1016/s0925-4773(99)00322-6. [DOI] [PubMed] [Google Scholar]
- 20.Martin GR, Timpl R. Laminin and other basement membrane components. Annual review of cell biology. 1987;3:57–85. doi: 10.1146/annurev.cb.03.110187.000421. [DOI] [PubMed] [Google Scholar]
- 21.Kleinman HK, McGarvey ML, Hassell JR, Star VL, Cannon FB, Laurie GW, et al. Basement membrane complexes with biological activity. Biochemistry. 1986;25:312–8. doi: 10.1021/bi00350a005. [DOI] [PubMed] [Google Scholar]
- 22.Timpl R. Macromolecular organization of basement membranes. Curr Opin Cell Biol. 1996;8:618–24. doi: 10.1016/s0955-0674(96)80102-5. [DOI] [PubMed] [Google Scholar]
- 23.Fukumoto S, Yamada Y. Review: Extracellular matrix regulates tooth morphogenesis. Connective Tissue Research. 2005;46:220–6. doi: 10.1080/03008200500344017. [DOI] [PubMed] [Google Scholar]
- 24.Fukumoto S, Miner JH, Ida H, Fukumoto E, Yuasa K, Miyazaki H, et al. Laminin alpha5 is required for dental epithelium growth and polarity and the development of tooth bud and shape. The Journal of biological chemistry. 2006;281:5008–16. doi: 10.1074/jbc.M509295200. [DOI] [PubMed] [Google Scholar]
- 25.Aumailley M, Bruckner-Tuderman L, Carter WG, Deutzmann R, Edgar D, Ekblom P, et al. A simplified laminin nomenclature. Matrix biology: journal of the International Society for Matrix Biology. 2005;24:326–32. doi: 10.1016/j.matbio.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Miner JH, Li C. Defective glomerulogenesis in the absence of laminin alpha5 demonstrates a developmental role for the kidney glomerular basement membrane. Developmental biology. 2000;217:278–89. doi: 10.1006/dbio.1999.9546. [DOI] [PubMed] [Google Scholar]
- 27.Salmivirta K, Sorokin LM, Ekblom P. Differential expression of laminin alpha chains during murine tooth development. Developmental dynamics: an official publication of the American Association of Anatomists. 1997;210:206–15. doi: 10.1002/(SICI)1097-0177(199711)210:3<206::AID-AJA2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 28.Yuasa K, Fukumoto S, Kamasaki Y, Yamada A, Fukumoto E, Kanaoka K, et al. Laminin alpha2 is essential for odontoblast differentiation regulating dentin sialoprotein expression. The Journal of biological chemistry. 2004;279:10286–92. doi: 10.1074/jbc.M310013200. [DOI] [PubMed] [Google Scholar]
- 29.Ida-Yonemochi H, Ohshiro K, Swelam W, Metwaly H, Saku T. Perlecan, a basement membrane-type heparan sulfate proteoglycan, in the enamel organ: its intraepithelial localization in the stellate reticulum. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 2005;53:763–72. doi: 10.1369/jhc.4A6479.2005. [DOI] [PubMed] [Google Scholar]
- 30.Ida-Yonemochi H, Satokata I, Ohshima H, Sato T, Yokoyama M, Yamada Y, et al. Morphogenetic roles of perlecan in the tooth enamel organ: an analysis of overexpression using transgenic mice. Matrix biology: journal of the International Society for Matrix Biology. 2011;30:379–88. doi: 10.1016/j.matbio.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Fincham AG, Moradian-Oldak J, Simmer JP. The structural biology of the developing dental enamel matrix. J Struct Biol. 1999;126:270–99. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- 32.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, et al. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. The Journal of biological chemistry. 2001;276:31871–5. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 33.Moradian-Oldak J. Amelogenins: assembly, processing and control of crystal morphology. Matrix biology: journal of the International Society for Matrix Biology. 2001;20:293–305. doi: 10.1016/s0945-053x(01)00154-8. [DOI] [PubMed] [Google Scholar]
- 34.Fong CD, Slaby I, Hammarstrom L. Amelin: an enamel-related protein, transcribed in the cells of epithelial root sheath. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 1996;11:892–8. doi: 10.1002/jbmr.5650110704. [DOI] [PubMed] [Google Scholar]
- 35.Krebsbach PH, Lee SK, Matsuki Y, Kozak CA, Yamada KM, Yamada Y. Full-length sequence, localization, and chromosomal mapping of ameloblastin. A novel tooth-specific gene. Journal of Biological Chemistry. 1996;271:4431–5. doi: 10.1074/jbc.271.8.4431. [DOI] [PubMed] [Google Scholar]
- 36.Hu CC, Fukae M, Uchida T, Qian Q, Zhang CH, Ryu OH, et al. Sheathlin: cloning, cDNA/polypeptide sequences, and immunolocalization of porcine enamel sheath proteins. Journal of dental research. 1997;76:648–57. doi: 10.1177/00220345970760020501. [DOI] [PubMed] [Google Scholar]
- 37.Uchida T, Tanabe T, Fukae M, Shimizu M. Immunocytochemical and immunochemical detection of a 32 kDa nonamelogenin and related proteins in porcine tooth germs. Arch Histol Cytol. 1991;54:527–38. doi: 10.1679/aohc.54.527. [DOI] [PubMed] [Google Scholar]
- 38.Fukae M, Tanabe T, Murakami C, Dohi N, Uchida T, Shimizu M. Primary structure of the porcine 89-kDa enamelin. Adv Dent Res. 1996;10:111–8. doi: 10.1177/08959374960100020201. [DOI] [PubMed] [Google Scholar]
- 39.Hu CC, Simmer JP, Bartlett JD, Qian Q, Zhang C, Ryu OH, et al. Murine enamelin: cDNA and derived protein sequences. Connective Tissue Research. 1998;39:47–61. doi: 10.3109/03008209809023911. discussion 3–7. [DOI] [PubMed] [Google Scholar]
- 40.Iwasaki K, Bajenova E, Somogyi-Ganss E, Miller M, Nguyen V, Nourkeyhani H, et al. Amelotin--a Novel Secreted, Ameloblast-specific Protein. Journal of dental research. 2005;84:1127–32. doi: 10.1177/154405910508401207. [DOI] [PubMed] [Google Scholar]
- 41.Moffatt P, Smith CE, St-Arnaud R, Simmons D, Wright JT, Nanci A. Cloning of rat amelotin and localization of the protein to the basal lamina of maturation stage ameloblasts and junctional epithelium. The Biochemical journal. 2006;399:37–46. doi: 10.1042/BJ20060662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moffatt P, Smith CE, Sooknanan R, St-Arnaud R, Nanci A. Identification of secreted and membrane proteins in the rat incisor enamel organ using a signal-trap screening approach. European journal of oral sciences. 2006;114(Suppl 1):139–46. doi: 10.1111/j.1600-0722.2006.00318.x. discussion 64–5, 380–1. [DOI] [PubMed] [Google Scholar]
- 43.Moffatt P, Smith CE, St-Arnaud R, Nanci A. Characterization of Apin, a secreted protein highly expressed in tooth-associated epithelia. Journal of cellular biochemistry. 2008;103:941–56. doi: 10.1002/jcb.21465. [DOI] [PubMed] [Google Scholar]
- 44.MacDougall M, Simmons D, Dodds A, Knight C, Luan X, Zeichner-David M, et al. Cloning, characterization, and tissue expression pattern of mouse tuftelin cDNA. Journal of dental research. 1998;77:1970–8. doi: 10.1177/00220345980770120401. [DOI] [PubMed] [Google Scholar]
- 45.Mao Z, Shay B, Hekmati M, Fermon E, Taylor A, Dafni L, et al. The human tuftelin gene: cloning and characterization. Gene. 2001;279:181–96. doi: 10.1016/s0378-1119(01)00749-1. [DOI] [PubMed] [Google Scholar]
- 46.Deutsch D, Palmon A, Fisher LW, Kolodny N, Termine JD, Young MF. Sequencing of bovine enamelin (“tuftelin”) a novel acidic enamel protein. Journal of Biological Chemistry. 1991;266:16021–8. [PubMed] [Google Scholar]
- 47.Deutsch D, Palmon A, Young MF, Selig S, Kearns WG, Fisher LW. Mapping of the human tuftelin (TUFT1) gene to chromosome 1 by fluorescence in situ hybridization. Mammalian genome: official journal of the International Mammalian Genome Society. 1994;5:461–2. doi: 10.1007/BF00357011. [DOI] [PubMed] [Google Scholar]
- 48.Nanci A, Zalzal S, Lavoie P, Kunikata M, Chen W, Krebsbach PH, et al. Comparative immunochemical analyses of the developmental expression and distribution of ameloblastin and amelogenin in rat incisors. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 1998;46:911–34. doi: 10.1177/002215549804600806. [DOI] [PubMed] [Google Scholar]
- 49.Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, et al. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973–83. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonoda A, Iwamoto T, Nakamura T, Fukumoto E, Yoshizaki K, Yamada A, et al. Critical role of heparin binding domains of ameloblastin for dental epithelium cell adhesion and ameloblastoma proliferation. The Journal of biological chemistry. 2009;284:27176–84. doi: 10.1074/jbc.M109.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamakoshi Y, Richardson AS, Nunez SM, Yamakoshi F, Milkovich RN, Hu JC, et al. Enamel proteins and proteases in Mmp20 and Klk4 null and double-null mice. European journal of oral sciences. 2011;119 (Suppl 1):206–16. doi: 10.1111/j.1600-0722.2011.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fincham AG, Belcourt AB, Termine JD, Butler WT, Cothran WC. Dental enamel matrix: sequences of two amelogenin polypeptides. Bioscience reports. 1981;1:771–8. doi: 10.1007/BF01114799. [DOI] [PubMed] [Google Scholar]
- 53.Hu JC, Sun X, Zhang C, Liu S, Bartlett JD, Simmer JP. Enamelysin and kallikrein-4 mRNA expression in developing mouse molars. European journal of oral sciences. 2002;110:307–15. doi: 10.1034/j.1600-0722.2002.21301.x. [DOI] [PubMed] [Google Scholar]
- 54.Nagano T, Kakegawa A, Yamakoshi Y, Tsuchiya S, Hu JC, Gomi K, et al. Mmp-20 and Klk4 cleavage site preferences for amelogenin sequences. Journal of dental research. 2009;88:823–8. doi: 10.1177/0022034509342694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibson CW, Li Y, Suggs C, Kuehl MA, Pugach MK, Kulkarni AB, et al. Rescue of the murine amelogenin null phenotype with two amelogenin transgenes. European journal of oral sciences. 2011;119 (Suppl 1):70–4. doi: 10.1111/j.1600-0722.2011.00882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen E, Yuan ZA, Wright JT, Hong SP, Li Y, Collier PM, et al. The small bovine amelogenin LRAP fails to rescue the amelogenin null phenotype. Calcified tissue international. 2003;73:487–95. doi: 10.1007/s00223-002-0036-7. [DOI] [PubMed] [Google Scholar]
- 57.Hatakeyama J, Fukumoto S, Nakamura T, Haruyama N, Suzuki S, Hatakeyama Y, et al. Synergistic roles of amelogenin and ameloblastin. Journal of dental research. 2009;88:318–22. doi: 10.1177/0022034509334749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Somogyi-Ganss E, Nakayama Y, Iwasaki K, Nakano Y, Stolf D, McKee MD, et al. Comparative temporospatial expression profiling of murine amelotin protein during amelogenesis. Cells Tissues Organs. 2012;195:535–49. doi: 10.1159/000329255. [DOI] [PubMed] [Google Scholar]
- 59.Kestler DP, Foster JS, Macy SD, Murphy CL, Weiss DT, Solomon A. Expression of odontogenic ameloblast-associated protein (ODAM) in dental and other epithelial neoplasms. Mol Med. 2008;14:318–26. doi: 10.2119/2008-00010.Kestler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lacruz RS, Nakayama Y, Holcroft J, Nguyen V, Somogyi-Ganss E, Snead ML, et al. Targeted overexpression of amelotin disrupts the microstructure of dental enamel. PloS one. 2012;7:e35200. doi: 10.1371/journal.pone.0035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo W, Wen X, Wang HJ, MacDougall M, Snead ML, Paine ML. In vivo overexpression of tuftelin in the enamel organic matrix. Cells Tissues Organs. 2004;177:212–20. doi: 10.1159/000080134. [DOI] [PubMed] [Google Scholar]
- 62.Huang B, Sun Y, Maciejewska I, Qin D, Peng T, McIntyre B, et al. Distribution of SIBLING proteins in the organic and inorganic phases of rat dentin and bone. European journal of oral sciences. 2008;116:104–12. doi: 10.1111/j.1600-0722.2008.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McKnight DA, Suzanne Hart P, Hart TC, Hartsfield JK, Wilson A, Wright JT, et al. A comprehensive analysis of normal variation and disease-causing mutations in the human DSPP gene. Human mutation. 2008;29:1392–404. doi: 10.1002/humu.20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.MacDougall M, Simmons D, Luan X, Nydegger J, Feng J, Gu TT. Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4. Dentin phosphoprotein DNA sequence determination. Journal of Biological Chemistry. 1997;272:835–42. doi: 10.1074/jbc.272.2.835. [DOI] [PubMed] [Google Scholar]
- 65.Veis A, Perry A. The phosphoprotein of the dentin matrix. Biochemistry. 1967;6:2409–16. doi: 10.1021/bi00860a017. [DOI] [PubMed] [Google Scholar]
- 66.Butler WT, Bhown M, Dimuzio MT, Linde A. Nonocollagenous proteins of dentin. Isolation and partial characterization of rat dentin proteins and proteoglycans using a three-step preparative method. Coll Relat Res. 1981;1:187–99. doi: 10.1016/s0174-173x(81)80019-2. [DOI] [PubMed] [Google Scholar]
- 67.Xiao S, Yu C, Chou X, Yuan W, Wang Y, Bu L, et al. Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat Genet. 2001;27:201–4. doi: 10.1038/84848. [DOI] [PubMed] [Google Scholar]
- 68.Qin C, Brunn JC, Cadena E, Ridall A, Butler WT. Dentin sialoprotein in bone and dentin sialophosphoprotein gene expressed by osteoblasts. Connective Tissue Research. 2003;44 (Suppl 1):179–83. [PubMed] [Google Scholar]
- 69.Veis A. Mineral-matrix interactions in bone and dentin. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 1993;8 (Suppl 2):S493–7. doi: 10.1002/jbmr.5650081312. [DOI] [PubMed] [Google Scholar]
- 70.George A, Bannon L, Sabsay B, Dillon JW, Malone J, Veis A, et al. The carboxyl-terminal domain of phosphophoryn contains unique extended triplet amino acid repeat sequences forming ordered carboxyl-phosphate interaction ridges that may be essential in the biomineralization process. Journal of Biological Chemistry. 1996;271:32869–73. doi: 10.1074/jbc.271.51.32869. [DOI] [PubMed] [Google Scholar]
- 71.Butler WT. Dentin matrix proteins. European journal of oral sciences. 1998;106 (Suppl 1):204–10. doi: 10.1111/j.1600-0722.1998.tb02177.x. [DOI] [PubMed] [Google Scholar]
- 72.Sreenath T, Thyagarajan T, Hall B, Longenecker G, D’Souza R, Hong S, et al. Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. The Journal of biological chemistry. 2003;278:24874–80. doi: 10.1074/jbc.M303908200. [DOI] [PubMed] [Google Scholar]
- 73.Suzuki S, Sreenath T, Haruyama N, Honeycutt C, Terse A, Cho A, et al. Dentin sialoprotein and dentin phosphoprotein have distinct roles in dentin mineralization. Matrix biology: journal of the International Society for Matrix Biology. 2009;28:221–9. doi: 10.1016/j.matbio.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Vega S, Iwamoto T, Nakamura T, Hozumi K, McKnight DA, Fisher LW, et al. TM14 is a new member of the fibulin family (fibulin-7) that interacts with extracellular matrix molecules and is active for cell binding. The Journal of biological chemistry. 2007;282:30878–88. doi: 10.1074/jbc.M705847200. [DOI] [PubMed] [Google Scholar]
- 75.de Vega S, Iwamoto T, Yamada Y. Fibulins: multiple roles in matrix structures and tissue functions. Cellular and molecular life sciences: CMLS. 2009;66:1890–902. doi: 10.1007/s00018-009-8632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]