Abstract
Stenotrophomonas maltophilia strain C6, capable of utilizing phenanthrene as a sole source of carbon and energy, was isolated from creosote-contaminated sites at Hilo, Hawaii. Twenty-two metabolites of phenanthrene, covering from dihydrodiol to protocatechuic acid, were isolated and characterized. Phenanthrene was degraded via an initial dioxygenation on 1,2-, 3,4-, and 9,10-C, where the 3,4-dioxygenation and subsequent metabolisms were most dominant. The metabolic pathways were further branched by ortho- and meta-cleavage of phenanthrenediols to produce 1-hydroxy-2-naphthoic acid, 2-hydroxy-1-naphthoic acid, and naphthalene-1,2-dicarboxylic acid. These intermediates were then transformed to naphthalene-1,2-diol. 1-Hydroxy-2-naphthoic acid was also degraded via a direct ring cleavage. Naphthalene-1,2-diol underwent primarily ortho-cleavage to produce trans-2-carboxycinnamic acid and then to form phthalic acid, 4,5-dihydroxyphthalic acid and protocatechuic acid. Accumulation of salicylic acid in prolonged incubation indicated that a limited extent of meta-cleavage of naphthalene-1, 2-diol also occurred. This is the first study of detailed phenanthrene metabolic pathways by Stenotrophomonas maltophilia.
Keywords: phenanthrene, Stenotrophomonas maltophilia, dioxygenaton, ortho-cleavage, meta-cleavage
1. Introduction
Polycyclic aromatic hydrocarbons (PAHs) are aromatic hydrocarbons with two or more fused benzene rings from natural and anthropogenic sources (Haritash and Kaushik, 2009). PAHs have attracted particular research attentions for decades because of their prolonged persistence, recalcitrance, potential mutagenic and carcinogenic properties (Arulazhagan and Vasudevan, 2011; Feng et al. 2012;). On the basis of their abundance, toxicity and intrinsic chemical stability to various transformation processes, 16 PAHs have been identified by the U.S. Environmental Protection Agency as priority pollutants (Keith and Telliard, 1979). Phenanthrene, a simple PAH with three-fused rings in an angular fashion, is commonly used as a model compound for PAH biodegradation studies (Seo et al. 2009; Roy et al. 2012).
Three bacteria strains were first isolated and reported in 1928 for their degradation of phenanthrene (Evans et al. 1965), which initiated studies of bacterial degradation of PAHs. Evans et al. (1965) proposed metabolic pathways of phenanthrene by pseudomonas aeruginosa through 3,4-dioxygenation to form 1,2-dihydroxynaphthalene which then enters into the naphthalene pathway. Since then, a wide range of bacteria have been reported for their biodegradation functions. For example, the gram-negative bacteria include Alcaligenes sp. (Deveryshetty and Phale 2010; John et al. 2012), Anthrobacter sp. (Kallimanis et al. 2007; Kallimanis et al. 2009), Burkholderia sp. (Dandie et al 2004; Seo et al. 2007b), Martelella sp. (Feng et al. 2012), Micrococcus sp. (John 2012), Ochrobacterum sp. (Ghosal et al. 2010), Pseudomonas sp. (Rodrigues et al. 2005; Zhao et al. 2009), Sphingomonas sp. (Pinyakong et al. 2000; Roy et al.2012), Staphylococcus sp. (Mallick et al. 2007) and Stenotrophomonas sp. (Juhasz et al. 2000). The gram-positive bacteria include Brevibacterium sp. (Samanta et al. 1999 ), Mycobacterium sp. (Moody 2001; Hennessee et al. 2009), and Rhodococcus sp. (Song et al. 2011). Metabolic pathways of phenanthrene and other PAHs by different bacterial species have been documented (Meckenstock 2004; Seo et al. 2009). The initial dioxygenation often takes place at 1,2-, 3,4- and 9,10-carbon positions of phenanthrene. Subsequent meta-cleavage of the resultant diols from 1,2- and 3,4-dioxygenations led to the formation of 2-hydroxy-1-naphthoic acid and 1-hydroxy-2-naphthoic acid, respectively (Seo et al. 2009).
Stenotrophomonas maltophilia (formally known as pseudomonas maltophilia) can degrade PAHs. S. maltophilia strain VUN 10,003 is highly capable of degrading not only low molecular weight PAHs (i.e., 2–3 rings), but also high molecular weight PAHs (≥ 4 rings) such as benzo[a]pyrene, dibenz[a,h]anthracene, and coronene (Juhasz et al. 1996, 2000, 2002). Some other species in the genus of Stenotrophomonas can also degrade acenaphthylene, phenanthrene, chloroanilines and chlorocatechol (Andreoni et al. 2004; Radianingtyas et al. 2003; Nayak et al. 2009). To our knowledge, bacterial degradation of 6–7 rings PAHs was reported only with S. maltophilia (Juhasz et al. 2000). Stenotrophomonas sp. can degrade acenaphthylene via 1,2-dihydroxy-naphthalene, salicylate and catechol (Nayak et al. 2009). Recent studies suggested that S. maltophila may have at least two different dioxygenases (NCBI access number : AY588578.1, and AY588576.1), which are highly homologous with naphthalene dioxygenases from other gram-negative bacteria. However, detailed metabolism of phenanthrene is still uncertain in Stenotrophomonas spp.
Our previous preliminary study with S. maltophilia C6 indicated that this strain could utilize phenanthrene (Seo et al. 2007a). In the present study, we have focused the isolation and identification of metabolites of phenanthrene and described a detailed metabolic map. In addition to the previously reported pathways, strain C6 can metabolize phenanthrene via multiple pathways. To our knowledge, this is the first study of detailed metabolic pathway of phenanthrene by S. maltophilia. Understanding of metabolic pathways may lead to development of better bioremediation technologies for cleaning up PAH contaminated sites.
2. Materials and methods
2.1 Chemicals
Phenanthrene (>98% purity) and metabolites were purchased from Sigma-Aldrich (Milwaukee, WI), Fisher Scientific (Morris Plains, NJ), or TCI America (Portland, OR). All chemicals used for media were of at least reagent grade. Ethyl acetate and other solvents were highest grade commercially available.
2.2 Bacterial strain and growth conditions
Stenotrophomonas maltophilia strain C6 was isolated from a soil sample from a former oil gasification company site in Hilo, Hawaii (19° 49′ 20″ N latitude, 155° 05′ 01″ W longitude) via an enrichment cultivation in mineral medium (MM) supplemented with phenanthrene (50 mg/50 ml of MM) (Seo et al. 2007a). Strain C6 was cultured in LB medium to an exponential growth period. An aliquot of 3 ml of the C6 LB culture was centrifuged at 10000 rpm and the pellet was then washed 6 times with MM (Bastiaens et al. 2000). The washed strain C6 was re-suspended in 1 ml of MM. For degradation studies and cell growth detection, 100 μl of the strain C6 suspension was added into 5 ml MM supplemented with phenanthrene (50 μg/ml), a mixture of phenanthrene (50 μg/ml) and glucose (50 μg/ml), or glucose (50 μg/ml).
For metabolite isolation and characterization studies, an aliquot of 10 ml of the C6 LB culture was centrifuged. The C6 cell pellet was washed, re-suspended in 500 ml mineral medium supplemented with phenanthrene (500 mg/1.5 l) as a sole source of carbon, and then cultured at 28 °C and 150 rpm (C4 Rotary shaker, New Brunswick Scientific, NJ). All degradation studies were performed in triplicate. The dead C6 cells, boiled for 5 min at 100 °C, was used as the control.
2.3 Extraction and analysis of phenanthrene
Cell growth was spectrophotometrically determined at 600 nm (OD600) after periods of incubation. Phenanthrene in the cultures after varying periods of incubation was extracted with an equal volume of ethyl acetate for three times. The extracts were combined, dried over through anhydrous sodium sulfate, and concentrated to near dryness, followed by reconstitution of the residue in 1 ml of acetone. Phenanthrene was analyzed on a Shimadzu LC-10AS high performance liquid chromatograph connected to a fixed wavelength UV detector at 254 nm and a Hypersil Green PAH column (5 μm 100 × 4.6 mm) (Thermo Scientific, West Palm Beach, FL). Phenanthrene was eluted by linearly increasing 40% (v/v) aqueous acetonitrile to 100% (v/v) acetonitrile in 25 min at a flow rate of 1.6 ml/min.
A pseudo first-order rate model was applied to calculate the biodegradation rate constant (k) and half life (t1/2), C = C0 exp (−kt), t1/2 = ln 2/k, where C0 is the initial phenanthrene concentration, and C is the phenanthrene concentration at the time t. All experiments were done in triplicate unless stated otherwise.
2.4 Extraction and identification of metabolites
After incubation for 3, 7, and 14 d, the cultures were filtered through glass wool and centrifuged (6,000 × g, 10 min). Supernatant was acidified to pH 2–3 with 6 N HCl and extracted with ethyl acetate (3 × 500 ml). The combined organic layer was extracted with aqueous sodium hydroxide (10 mM, 3 × 500 ml,). The remaining organic phase was dried over anhydrous sodium sulfate and concentrated to 5 ml of ethyl acetate (neutral fraction). The aqueous layer was acidified to pH 2–3 and extracted with ethyl acetate (3 × 500 ml, acidic fraction) (Seo et al. 2007b).
Metabolites in the neutral fraction after derivatization or no derivatization were characterized with gas chromatography-mass spectrometry (GC-MS) (Seo et al. 2007b). For the detection of diol and cis-dihydrodiols, after removal of ethyl acetate the residue was dissolved in acetone (10 ml) containing n-butylboronic acid (50 mg). After refluxing for 30 min, the mixture was concentrated to 1 ml and analyzed by GC-MS. The residue was further derivatized with methyl iodide and re-analyzed with GC-MS. Metabolites in the acidic fraction were derivatized with diazomethane or methyl iodide (Seo et al. 2007b).
GC-MS analysis was performed on a Varian 3800 gas chromatograph (GC) - Saturn-2000 ion trap mass spectrometer system (ITMS) (Varian Inc., Walnut Creek, CA), equipped with a ZB-1 column (60 m, 0.25 μm, Phenomenex, Torrance, CA). An aliquot of 2.0 μL of sample was injected in splitless mode with an AS8400 autosampler. The purge valve was activated 3 min after the sample injection. Helium was the carrier gas as a flow rate of 2 ml/min. The column temperature was started at 120 °C for 2 min, programmed to 280 °C at a rate of 2 °C/min, and held 280 °C for 10 min. Injector and analyzer temperatures were set to 270 and 280 °C, respectively. The mass spectrometer was operated in electron impact mode(70 eV) (Seo et al., 2007b).
3. Results
3.1 Degradation kinetics of phenanthrene by S. maltophilia strain C6
S. maltophilia strain C6 could completely degrade phenanthrene in 14 days. No trace of phenanthrene was detected after 14 days (Fig. 1A). The degradation followed a pseudo first-order kinetic reaction. The degradation constant and half-life of phenanthrene were 0.177 day−1 and 2.42 days, respectively under the cultured conditions. Strain C6 can utilize phenanthrene for growth, while the boiled C6 could not grow (Fig. 1B). Strain C6 grew well in the culture supplemented with glucose and a mixture of phenanthrene and glucose.
Fig. 1.
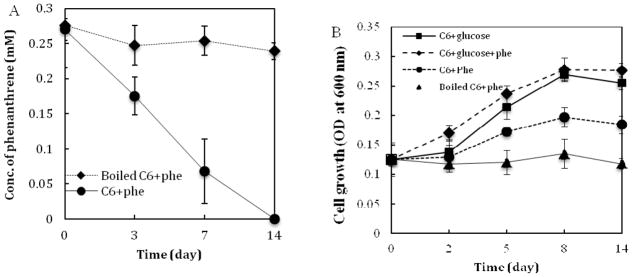
Degradation kinetics of phenanthrene (A) by and growth curve (B) of S. maltophilia strain C6. Error bars represent the standard deviations of triplicate samples. Phe, phenanthrene.
3.2 Metabolite identification and metabolic map of phenanthrene
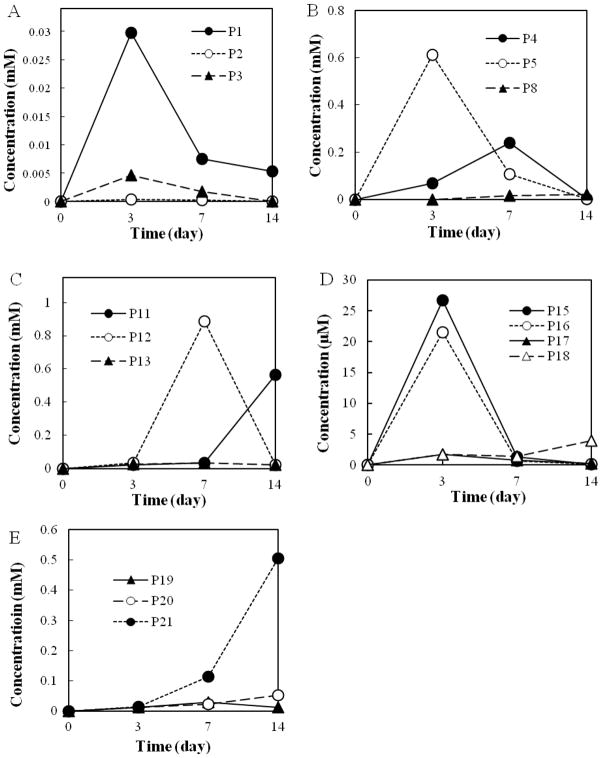
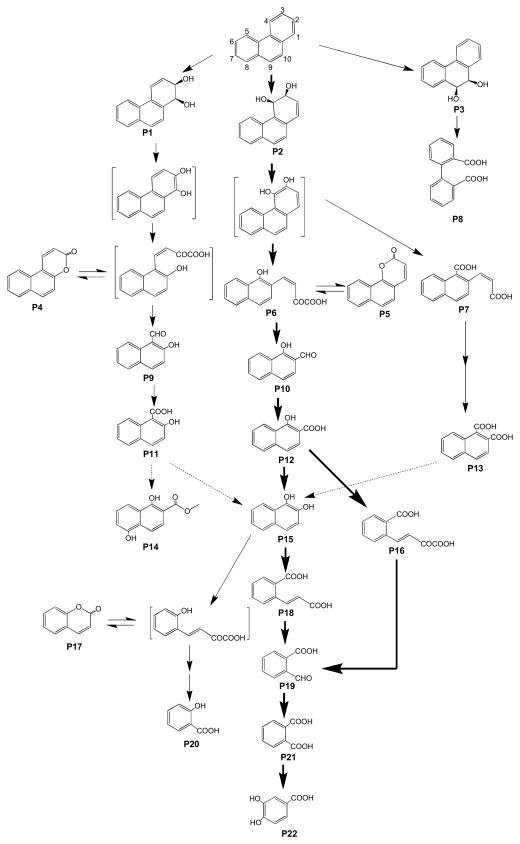
Twenty-two metabolites (P1–P22) were identified in acidic and neutral fractions (Figs. 2 and 3). Three dihydrodiols [P1, P2, and P3; GC retention time (tR) = 53.41, 50.78, and 51.34 min, respectively] were detected in the neutral fraction after an incubation of 3 to 14 days, while the highest concentrations of P1 and P3 occurred at day 3 (Fig. 2A). Their mass spectra [n-butylboromate ester; m/z 278 (M+), 221, 194, 178, 165] were consistent with those published in the literature (Krivobok et al. 2003) and authentic standard (n-butylboromate ester of cis-phenanthrene-9, 10-dihydrodiol). Although phenanthrene diols were not found, several ring cleavage products were detected as dominant metabolites in the first 3-d incubation period. Two benzocoumarins [P4 and P5; Rt = 41.02 and 39.81 min, respectively; m/z 196 (M+), 168, 139] were found in the acidic fraction. P4 and P5 were 5,6- and 7,8-benzocoumarin, respectively. The concentration of P5 was approximately 7-fold of P4 at day 3, whereas it decreased to an half of P4 at day 7 (Fig. 2B). Benzocoumarins are considered as rearrangement products of o-hydroxynaphthyl-α-oxobutenoate (Pinyakong et al. 2000).
Fig. 2.
Concentration profiles of metabolites. (A) cis-phenanthrene dihydrodiols (P1, P2, P3); (B) benzocoumarins (P4, P5) and diphenic acid (P8); (C) o-hydroxynaphthoic acids (P11, P12) and naphthalene-1,2-dicarboxylic acid (P13); (D) naphthalene-1,2-diol (P15), 2-carboxybenzalpyruvate (P16), coumarin (P17) and 2-carboxycinnamic acid (P18); (E) 2-formylbenzoic acid (P19), salicylic acid (P20) and phthalic acid (P21). Results were average values of two separate experiments. The metabolites P6, P7, P9, P10 and P14 were not shown in Fig. 2.
Fig. 3.
Proposed metabolic pathways of phenanthrene by S. maltophilia strain C6. Carbon atoms in phenanthrene was numbered. The compounds are 1,2-dihydroxyphenanthrene (P1), 3,4-dihydroxyphenanthrene (P2), cis-9,10-hidroxyphenanthrene (P3), 5,6-benzocoumarin (P4), 7,8-benzocoumarin (P5), methyl trans-4-(1-methoxy-2-naphthyl)-2-oxobut-3-enoate (P6), 2-carboxyvinyl-1-naphthoate (P7), 2-2′-diphenic acid (P8), 2-hydroxy-1-naphthaldehyde (P9); 1-hydroxy-2-naphthaldehyde (P10); 2-hydroxy-1-naphthoic acid (P11); 1-hydroxy-2-naphthoic acid (P12); naphthalene-1,2-dicarboxylic acid (P13), 1,4-dihydroxy-2-naphthoic acid methyl ester (P14), naphthalene-1,2-diol (P15), 2-carboxybenzalpyruvate (P16), coumarin (P17), 2-carboxycinnamic acid (P18), 2-formylbenzoic acid (P19), salicylic acid (P20), phthalic acid (P21), and protocatechuic acid (P22). Bold and thin arrows: major and minor pathways, respectively; dotted line arrow, tentative pathway. Metabolites in brackets, not detected.
Among the metabolites, derivatized with diazomethane, P6 showed a similar GC retention time and mass spectrum [tR = 54.34 min, m/z 270 (M+, 23), 239 (100), 211 (53), 196 (57), 168 (34), 152 (8), 139 (19)] with the standard methyl trans-4-(1-methoxy-2-naphthyl)-2-oxobut-3-enoate. P7 was found in the cultures on day 4 and 7, but not in day 14 (data not shown). The mass spectrum of P7 [tR = 50.12 min, m/z 270 (M+, 9), 239 (10), 211 (100), 196 (7), 179 (10), 168 (15), 151 (18), 139 (10)] was very similar with that of the synthetic 1-carboxyvinyl-2-naphthoate dimethyl ester [m/z 270 (M+, 10), 239 (7), 211 (100), 196 (7), 179 (15), 168 (12), 151 (13), 139 (7)], but with a different retention time (tR = 49.57 min). Although the mass spectrum resembled with those of fully methylated o-hydroxynaphthyl-α-oxobutenoates (HNOBA), the base peak of HNOBA was m/z 239, which corresponds to the loss of methoxy group. According to these results, P7 was tentatively identified as 2-carboxyvinyl-1-naphthoate (CVNA). P8 from 7- and 14-days acidic fraction (tR = 36.78 min) was confirmed as diphenic acid. A trace amount of two o-hydroxynaphthaldehydes (P9 and P10, tR = 31.64 and 25.80 min, respectively) were found in 7-days samples (data not shown). P9 and P10 were 2-hydroy-1-naphthaldehyde and 1-hydroxy-2-naphthaldehyde, respectively. P9 and P10 are presumably oxidized to 2-hydroxy-1-naphthoic acid (P11) and 1-hydroxy-2-naphthoic acid (P12), respectively.
Both P11 and P12 were found in the acidic fraction with different abundance (Fig. 2C). The concentration of P12 was 20-fold higher than P11 at day 7. P12, however, was rapidly degraded, while P11 was relatively persistent to degradation (Fig. 2C). At day 14, the concentration of P12 was very low, while the concentration of P11 was quite high. Metabolites P13 [as dimethyl ester, tR = 39.54 min, m/z 244 (M+, 47), 229 (1), 213 (100), 198 (2), 170 (8), 154 (7)] was confirmed with the standard naphthalene-1,2-dicarboxylic acid. Although the concentrations were much lower than o-hydroxynaphthoic acids (approximately 10%), P13 was continuously found during the entire experiment.
The mass spectrum of P14 [tR = 35.45 min, m/z 218 (M+, 22), 186 (100), 158 (53), 130 (29), 102 ( 53) was very similar with that of 1,5-dihydroxy-2-naphthoic acid methyl ester (Pinyakong et al. 2000). Naphthalene 1,2-diol (P15, tR = 33.40 min) was detected during the entire experiment in the neutral fraction. P16 (tR = 38.25 min) was found to be 2-carboxybenzalpyruvate [dimethyl ester, m/z 248 (M+ 1), 217 (2), 189 (100), 161 (38), 145 (68), 130 (22)]. P17 (tR = 15.34 min) was coumarin. The concentrations of P15 and P16 reached maximum (approximately 29 and 22 μM) after 3 days of incubation and P15 and P16 were much more dominant than P17 and P18 (Fig. 2D). The metabolite P18 (tR = 28.48 min) was confirmed as 2-carboxycinnamic acid, while P19 (tR = 12.64 min) was 2-formylbenzoic acid. Both P18 and P19 were found in a trace level. P18, however, was continuously accumulated till the end of experiment to 4 μM at day 14. The concentrations of salicylic acid (P20, tR = 8.23 min) were below the limit of detection at day 3. It was, however, accumulated exponentially and comprised 0.1% and 1% of total identified metabolites at day 7 and day 14, respectively (Fig. 2E). No trace of gentisic acid and catechol were found during the entire experiment. Phthalic acid (P21, tR = 15.63 min) detected in the acidic fraction was rapidly accumulated and comprised approximately 42% of the total metabolites at day 14. Concentrations of protocatechuic acid [(P22), tR = 21.83 min] detected in the acidic fraction also increased over the incubation.
3.3 Comparison among 1,2-, 3,4- and 9,10-dioxygenations of phenanthrene
S. maltophilia strain C6 can degrade phenanthrene via 1,2-, 3,4-, and 9,10-dioxygenations (Fig. 2). Fig. 4 shows the concentration profiles of metabolites in the upper metabolic pathways. P1, P4, P9, and P11 are products of 1,2-dioxygenation, while P2, P5, P6, P7, P12 and P13 are 3,4-dioxygenation metabolites. Because of a trace amount of P9 and tentative identification of P6 and P7, the amounts of P9, P6 and P7 were not included in the calculation. P3 and P8 represent 9,10-dioxygenation. The concentration of 3,4-dioxygenation metabolites was approximately 6 times of 1,2-dioxygenation metabolites and 146 times of 9,10-C metabolites after 3 days of incubation. Up to 7 days, 3,4-C metabolites reached the highest concentration. After 14 days of incubation, the concentration of 3,4-C metabolites P2, P5, P12, P13 decreased to 4% of the concentration on the 7th day. The concentrations of the 1,2-C metabolites (P1, P4, and P11) and 9,10-C metabolites (P3 and P8) were gradually accumulated over the incubation period, although the concentrations of 9,10-C metabolites were low. The results suggest that the 3,4-dioxygenation of phenanthrene is significantly more dominant than 1,2- and 9,10-dioxygenations in S. maltophilia strain C6.
Fig. 4.
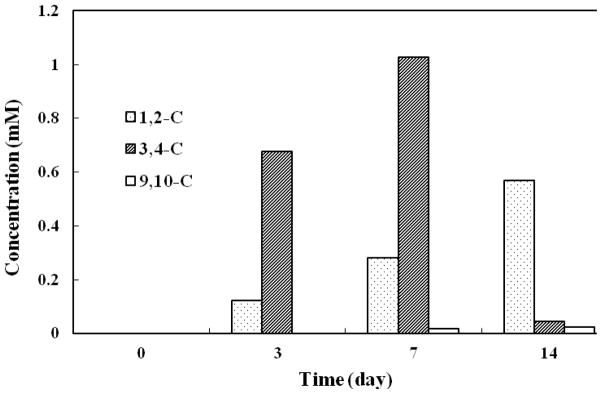
Concentration profiles of metabolites in upper metabolic pathways. 1,2-C, 3,4-C, and 9,10-C denote the metabolites from the dioxygenations on corresponding positions and subsequent metabolism. Metabolites included for the concentration calculation were P1, P4, P9 and P11 for 1,2-dioxygenation; P2, P5, P6, P7, P12 and P13 for 3,4-dioxygenation; and P3 and P8 for 9,10-dioxygenation.
4. Discussion
Strain C6 showed to utilize phenanthrene for slow growth (Fig. 1). It grew in a similar rate utilizing glucose as a mixture of phenanthrene and glucose (Fig. 1B), which suggests that phenanthrene is not toxic to strain C6.
Strain C6 can decompose phenanthrene through very complex metabolic pathways (Fig. 3). Initial dioxygenations occurred at 1,2-, 3,4- and 9,10-C positions of phenanthrene. Dioxygenations in multiple positions are common in PAH metabolism by Mycobacterium spp. and Sphingomonas spp. (Kim et al. 2005; Pinyakong et al. 2000; Seo et al. 2012). Although either dihydrodiols or diols were found, 3,4-dioxygenation was the major pathway since the metabolites of consecutive reactions were dominant during the initial incubation period (3–7 days, Fig. 4). In comparison with metabolites from 3,4-dioxygenation and subsequent reactions, the amount of 1,2-dioxygenation related products gradually increased over the incubation period. 5,6-Benzocoumarin (P4) was accumulated till day 7, but rapidly decreased to a trace level at day 14. 2-Hydroxy-1-naphthaldehyde (P9) was detectable at day 7, but not at day 14. These results suggest that at least, enzymes in upper PAH metabolism, from dihydrodiols (P1, P2) to o-hydroxynaphthaldehydes (P9, P10), can decompose metabolites both from 1,2- and 3,4-dioxygenations. Rapid accumulation of 2-hydroxy-1-naphthoic acid (P11) in a prolonged experiment period indicates that there may be no catabolic enzymes for P11 or the specificity of enzymes may be limited. A common bacterial metabolite of 9,10-dioxygenation of phenanthrene is diphenic acid (P8), which is usually the most dominant metabolites from the culture of Mycobacterium sp. (Vila et al. 2001; Kim et al. 2005). Although strain C6 could produce P8, the amount was very limited in comparison with other metabolites and no possible degradation product of P8 was detected (e.g., biphenyl-2-carboxylic acid, dihydroxybiphenyl-2-carboxylic acid). These results suggest that this pathway has a minor role in phenanthrene degradation. Among the well-established initial metabolic pathways of phenanthrene diols, meta-cleavage (also called extradiol cleavage) is known as the most dominant degradation pathways of 3,4- (or 1,2-) diol, which produce o-hydroxynaphthyl-α-oxobut-3-enoate (e.g., P6). In this experiment, high concentrations of benzocoumarins (P4, P5) also revealed the same ring opening (i.e., meta-cleavage). Ring opening of phenanthrene-9,10-diol, however, predominantly occurred via ortho-cleavage.
Two metabolic pathways of 1-hydroxy-2-naphthoic acid (P12) have been proposed (Adachi et al. 1999; Balashova et al. 2001; Seo et al. 2009). Those are (1) dioxygenation and ortho-cleavage to 2-carboxybenzapyruvate (P16) and (2) decarboxylation and hydroxylation by salicylate monooxygenase to naphthalene-1,2-diol (P15). Concentration profiles of P15 and P16 show that strain C6 could use both pathways in a nearly similar extent (Fig. 2D). As how phenanthrene diols degraded, naphthalene-1,2-diol (P15) can also undergo ortho- (Annweiler et al. 2000; Vila et al. 2001) and meta-cleavage (Eaton and chapman 1992), from which the products are 2-carboxycinnamic acid (P18) and 2-hydroxybenzalyruvate. Although 2-hydroxybenzapyruvate was not detected, the presence of coumarin (P17) and salicylic acid (P20) revealed that strain C6 could use meta- and ortho-cleavage to break naphthalene-1,2-diol.
Based the metabolite profiles, 2-formylbenzoic acid (P19), a precursor of phthalic acid (P21), can be produced from naphthalene-1,2-diol (P15) and 2-carboxybenzalpyruvate (P16) and the contribution of P15 and P16 are approximately equal on the production of P19. The amount of phthalic acid (P21) increased up to 42% of the total identifiable metabolites by the end of experiment (14 d), which suggests a limited degradation. However, a concomitant increase of protocatechuic acid (P22) showed that phthalic acid is an intermediate of further metabolism. The concentration of salicylic acid (P20) also increased. No metabolites of salicylic acid (e.g., gentisic acid and catechol), however, were detected during the entire experiment. These results suggest that salicylic acid and related metabolites are not major metabolites and enzymes in strain C6 may have a limited degradation potential for salicylates.
5. Conclusion
Phenanthrene can be completely degraded by S. maltophilia strain C6 through 1,2-, 3,4-, and 9,10-dioxygenations. 3,4-Dioxygenation and subsequent metabolisms were most dominant. Phenanthrene diols were transformed to o-hydroxynaphthoates or naphthalene-1,2-dicarboxylic acid via ortho-, and meta-cleavage. Subsequent metabolism of these acids produced naphthalene-1,2-diol or 2-carboxybenzalpyruvate, which is further degraded to protocatechuic acid and salicylic acid. The metabolite profiles suggest involvement of many enzymes in PAH metabolism in this strain. Complex metabolic pathways indicate that S. maltophilia C6 has been well adapted to use phenanthrene as a substrate and it has application potentials for bioremediation of PAHs contamination.
Highlights.
Stenotrophomonas maltophilia C6 utilizes phenanthrene for growth.
C6 degrades phenanthrene via multi-site dioxygenation and ortho-/meta-cleavage.
Part of the branched pathways is merged into 1,2-dihydroxynaphthalene.
3,4-dioxygenation dominates.
Acknowledgments
This work was supported in part the U.S. ONR HEET award N00014-09-1-0709 and the National Institute on Minority Health and Health Disparities grant 8 G12 MD007601-26. SG was a recipient of scholarship from the Chinese Scholarship Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi K, Iwabuchi T, Sano H, Harayama S. Structure of the ring cleavage product of 1-hydroxy-2-naphthoate, and intermediate of the phenanthrene-degradative pathway of Nocardioides sp. strain KP7. Journal of Bacteriology. 1999;181:757–763. doi: 10.1128/jb.181.3.757-763.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arulazhagan P, Vasudevan N. Biodegradation of polycyclic aromatic hydrocarbons by a halotolerant bacterial strain Ochrobactrum sp. VA1. Marine Pollution Bulletin. 2011;62:388–394. doi: 10.1016/j.marpolbul.2010.09.020. [DOI] [PubMed] [Google Scholar]
- Andreoni V, Cavalca L, Rao MA, Nocerino G, Bernasconi S, DellÕAmico E, Colombo M, Gianfreda L. Bacterial communities and enzyme activities of PAHs polluted soils. Chemosphere. 2004;57:401–412. doi: 10.1016/j.chemosphere.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Annweilaer E, Richnow HH, Antranikian G, Hebenbrock S, Grams C, Franke S, Franke W, Michaelis W. Naphthalene degradation and incorporation of naphthalene-derived carbon into biomass by the thermophile Bacillus thermoleovorans. Applied and Environmental Microbiology. 2000;66:518–523. doi: 10.1128/aem.66.2.518-523.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashova NV, Stolz A, Knackmuss HJ, Kosheleva IA, Naumov AV, Boronin AM. Purification and characterization of a salicylate hydroxylase involved in 1-hydroxy-2-naphthoic acid hydroxylation from the naphthalene and phenanthrene-degrading bacterial strain Pseudomonas putida BS202-P1. Biodegradation. 2001;12:179–188. doi: 10.1023/a:1013126723719. [DOI] [PubMed] [Google Scholar]
- Bastiaens L, Springael D, Wattiau P, Harms H, DeWachter R, Verachtert H, Diels L. Isolation of adherent polycyclic aromatic hydrocarbon (PAH)-degrading bacteria using PAH-sorbing carriers. Applied and Environmental Microbiology. 2000;66:1834–1843. doi: 10.1128/aem.66.5.1834-1843.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandie CE, Thomas SM, Bentham RH, McClure NC. Physiological characterization of Mycobacterium sp. strain 1B isolated from a bacterial culture able to degrade high-molecular-weight polycyclic aromatic hydrocarbons. Journal of Applied Microbiology. 2004;97:246–255. doi: 10.1111/j.1365-2672.2004.02087.x. [DOI] [PubMed] [Google Scholar]
- Deveryshetty J, Phale PS. Biodegradation of phenanthrene by Alcaligenes sp. strain PPH: partial purification and characterization of 1-hydroxy-2-naphthoic acid hydroxylase. FEMS Microbiology Letters. 2010;311:93–101. doi: 10.1111/j.1574-6968.2010.02079.x. [DOI] [PubMed] [Google Scholar]
- Evans WC, Fernley HN, Griffiths E. Oxidative Metabolism of Phenanthrene and Anthracene by Soil Pseudomonads. Biochemical Journal. 1965;95:819–831. doi: 10.1042/bj0950819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton RW, Chapman PJ. Bacterial metabolism of naphthalene: construction and use of recombinant bacteria to study ring cleavage of 1,2-dihydroxynaphthalene and subsequent reaction. Journal of Bacteriology. 1992;174:7542–7554. doi: 10.1128/jb.174.23.7542-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng TC, Cui CZ, Dong F, Feng YY, Liu YD, Yang XM. Phenanthrene biodegradation by halophilic Martelella sp.AD-3. Journal of Applied Microbiology. 2012;113:779–789. doi: 10.1111/j.1365-2672.2012.05386.x. [DOI] [PubMed] [Google Scholar]
- Ghosal D, Chakraborty J, Khara P, Dutta TK. Degradation of phenanthrene via meta-cleavage of 2-hydroxy-1-naphthoic acid by Ochrobactrum sp. strain PWTJD. FEMS Microbiology Letters. 2010;313:103–110. doi: 10.1111/j.1574-6968.2010.02129.x. [DOI] [PubMed] [Google Scholar]
- Haritash AK, Kaushik CP. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): A review. Journal of Hazardous Materials. 2009;169:1–15. doi: 10.1016/j.jhazmat.2009.03.137. [DOI] [PubMed] [Google Scholar]
- Hennessee CT, Seo JS, Alvarez AM, Li QX. Isolation and characterization of five new polycyclic aromatic hydrocarbon-degrading species isolated from Hawaiian soils: Mycobacterium crocinum sp. nov., Mycobacterium pallens sp. nov., Mycobacterium rutilum sp. nov., Mycobacterium rufum sp. nov., and Mycobacterium aromaticivorans sp. nov. International Journal of Systematic and Evolutionary Microbiology. 2009;59:378–387. doi: 10.1099/ijs.0.65827-0. [DOI] [PubMed] [Google Scholar]
- John RC, Essien JP, Akpan SB, Okpokwasili GC. Polycyclic aromatic hydrocarbon-degrading bacteria from aviation fuel spill site at Ibeno, Nigeria. Bulletin Environmental Contamination Toxicology. 2012;88:1014–1019. doi: 10.1007/s00128-012-0598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz AL, Britz ML, Stanley GA. Degradation of high molecular weight polycyclic aromatic hydrocarbons by Pseudomonas cepacia. Biotechnology Letters. 1996;18:577–582. [Google Scholar]
- Juhasz AL, Stanley GA, Britz ML. Microbial degradation and detoxification of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophilia strain VUN 10,003. Letters in Applied Microbiology. 2000;30:396–401. doi: 10.1046/j.1472-765x.2000.00733.x. [DOI] [PubMed] [Google Scholar]
- Juhasz AL, Stanley GA, Britz ML. Metabolite repression inhibits degradation of benzo[a]pyrene and dibenz[a,h]anthracene by Stenotrophomonas maltophilia VUN 10,003. Journal of Industrial Microbiology and Biotechnology. 2002;28:88–96. doi: 10.1038/sj/jim/7000216. [DOI] [PubMed] [Google Scholar]
- Kallimanis A, Frillingos S, Drainas C, Koukkou AI. Taxonomic identification, phenanthrene uptake activity, and membrane lipid alterations of the PAH degrading Arthrobacter sp. strain Sphe3. Appllied Microbiology and Biotechnology. 2007;76:709–717. doi: 10.1007/s00253-007-1036-3. [DOI] [PubMed] [Google Scholar]
- Kallimanis A, Kavakiotis K, Perisynakis A, Sproêr C, Pukall R, Drainas C, Koukkou AI. Arthrobacter phenanthrenivorans sp. nov., to accommodate the phenanthrene-degrading bacterium Arthrobacter sp. strain Sphe3. International Journal of Systematic and Evolutionary Microbiology. 2009;59:275–279. doi: 10.1099/ijs.0.000984-0. [DOI] [PubMed] [Google Scholar]
- Keith L, Telliard W. Priority pollutants. I. A perspective view. Environmental Science & Technology. 1979;13:416–423. [Google Scholar]
- Krivobok S, Kuony S, Meyer C, Louwagie M, Willison JC, Jouanneau Y. Identification of pyrene-induced proteins in Mycobacterium sp. strain 6PY1: evidence for two ring-hydroxylating dioxygenases. Journal of Bacteriology. 2003;185:3828–41. doi: 10.1128/JB.185.13.3828-3841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Freeman JP, Moody JD, Engesser KH, Cerniglia CE. Effects of pH on the degradation of phenanthrene and pyrene by Mycobacterium vanbaalenii PYR-1. Applied Microbiology and Biotechnology. 2005;67:275–285. doi: 10.1007/s00253-004-1796-y. [DOI] [PubMed] [Google Scholar]
- Mallick S, Chatterjee S, Dutta TK. A novel degradation pathway in the assimilation of phenanthrene by Staphylococcus sp. strain PN/Y via meta-cleavage of 2-hydroxy-1-naphthoic acid: formation of trans-2,3-dioxo-5-(29-hydroxyphenyl)-pent-4-enoic acid. Microbiology. 2007;153:2104–2115. doi: 10.1099/mic.0.2006/004218-0. [DOI] [PubMed] [Google Scholar]
- Meckenstock RU, Safinowski M, Griebler C. Anaerobic degradation of polycyclic aromatic hydrocarbons. FEMS Microbiology Ecology. 2004;49:27–36. doi: 10.1016/j.femsec.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Moody JD, Freeman JP, Doerge DR, Cerniglia CE. Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR-1. Applied and Environmental Microbiology. 2001;67:1476–1483. doi: 10.1128/AEM.67.4.1476-1483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak AS, Veeranagouda Y, Lee K, Karegoudar TB. Metabolism of acenaphthylene via 1,2-dihydroxynaphthalene and catechol by Stenotrophomonas sp. RMSK. Biodegradation. 2009;20:837–843. doi: 10.1007/s10532-009-9271-1. [DOI] [PubMed] [Google Scholar]
- Pinyakong O, Habe H, Supaka N, Pinpanichkarn P, Juntongjin K, Yoshida T, Furihata K, Nojiri H, Yamane H, Omori T. Identification of novel metabolites in the degradation of phenanthrene by Sphingomonas sp. strain P2. FEMS Microbiology Letters. 2000;191:115–121. doi: 10.1111/j.1574-6968.2000.tb09327.x. [DOI] [PubMed] [Google Scholar]
- Radianingtyas H, Robinson GK, Bull AT. Characterization of a soil-derived bacterial consortium degrading 4-chloroaniline. Microbiology. 2003;149:3279–3287. doi: 10.1099/mic.0.26303-0. [DOI] [PubMed] [Google Scholar]
- Rodrigues AC, Wuertz S, Brito AG, Melo LF. Fluorene and Phenanthrene Uptake by Pseudomonas putida ATCC 17514: Kinetics and Physiological Aspects. Biotechnology and Bioengineering. 2005;90:281–289. doi: 10.1002/bit.20377. [DOI] [PubMed] [Google Scholar]
- Roy M, Khara P, Dutta TK. meta-Cleavage of hydroxynaphthoic acids in the degradation of phenanthrene by Sphingobium sp. strain PNB. Microbiology. 2012;158:685–695. doi: 10.1099/mic.0.053363-0. [DOI] [PubMed] [Google Scholar]
- Samanta SK, Chakraborti AK, Jain PK. Degradation of phenanthrene by different bacteria: evidence for novel transformation sequences involving the formation of 1-naphthol. Applied Microbiology and Biotechnology. 1999;53:98–107. doi: 10.1007/s002530051621. [DOI] [PubMed] [Google Scholar]
- Seo JS, Keum YS, Harada RM, Li QX. Isolation and characterization of bacteria capable of degrading polycyclic aromatic hydrocarbons (PAHs) and organophosphorus pesticides from PAH-contaminated soil in Hilo, Hawaii. Journal of Agricultural and Food Chemistry. 2007a;55:5383–5389. doi: 10.1021/jf0637630. [DOI] [PubMed] [Google Scholar]
- Seo JS, Keum YS, Hu YT, Lee SE, Li QX. Degradation of phenanthrene by Burkholderia sp. C3: initial 1,2- and 3,4-dioxygenation and meta- and ortho-cleavage of naphthalene-1,2-diol. Biodegradation. 2007b;18:123–131. doi: 10.1007/s10532-006-9048-8. [DOI] [PubMed] [Google Scholar]
- Seo JS, Keum YS, Li QX. Bacterial degradation of aromatic compounds. International Journal of Environmental Research and Public Health. 2009;6:278–309. doi: 10.3390/ijerph6010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JS, Keum YS, Li QX. Mycobacterium aromativorans JS19b1(T) degrades phenanthrene through C-1,2, C-3,4 and C-9,10 dioxygenation pathways. International Biodeterioration and Biodegradation. 2012;70:96–103. doi: 10.1016/j.ibiod.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XH, Xu Y, Li GM, Zhang Y, Huang TW, Hu Z. Isolation, characterization of Rhodococcus sp. P14 capable of degrading high-molecular-weight polycyclic aromatic hydrocarbons and aliphatic hydrocarbons. Marine Pollution Bulletin. 2011;62:2122–2128. doi: 10.1016/j.marpolbul.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Sutherland JB, Freeman JP, Selby AL, Fu PP, Miller DW, Cerniglia CE. Stereoselective formation of a K-region dihydrodiol from phenanthrene by Streptomyces flavovirens. Archives of Microbiology. 1990;154:260–266. doi: 10.1007/BF00248965. [DOI] [PubMed] [Google Scholar]
- Vila J, López Z, Sabaté J, Minguilló C, Solanas AM, Grifoll M. Identification of a Novel Metabolite in the Degradation of Pyrene by Mycobacterium sp. Strain AP1: Actions of the isolate on two- and three-ring polycyclic aromatic hydrocarbons. Applied and Environmental Microbiology. 2001;67:5497–5505. doi: 10.1128/AEM.67.12.5497-5505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HP, Wu QS, Wang L, Zhao XT, Gao HW. Degradation of phenanthrene by bacterial strain isolated from soil in oil refinery fields in Shanghai China. Journal of Hazardous Materials. 2009;164:863–869. doi: 10.1016/j.jhazmat.2008.08.098. [DOI] [PubMed] [Google Scholar]