Abstract
Purpose of review
Despite maximum medical and mechanical support therapy, heart failure remains a relentlessly progressive disorder with substantial morbidity and mortality. Autophagy, an evolutionarily conserved process of cellular cannibalization, has been implicated in virtually all forms of cardiovascular disease. Indeed, its role is context dependent, antagonizing or promoting disease depending on the circumstance. Here, we review current understanding of the role of autophagy in the pathogenesis of heart failure and explore this pathway as a target of therapeutic intervention.
Recent findings
In preclinical models of heart disease, cardiomyocyte autophagic flux is activated; indeed, its role in disease pathogenesis is the subject of intense investigation to define mechanism. Similarly, in failing human heart of a variety of etiologies, cardiomyocyte autophagic activity is upregulated, and therapy, such as with mechanical support systems, elicits declines in autophagy activity. However, when suppression of autophagy is complete, rapid and catastrophic cell death occurs, consistent with a model in which basal autophagic flux is required for proteostasis. Thus, a narrow zone of ‘optimal’ autophagy seems to exist. The challenge moving forward is to tune the stress-triggered autophagic response within that ‘sweet spot’ range for therapeutic benefit.
Summary
Whereas we have known for some years of the participation of lysosomal mechanisms in heart disease, it is only recently that upstream mechanisms (autophagy) are being explored. The challenge for the future is to dissect the underlying circuitry and titrate the response into an optimal, proteostasis-promoting range in hopes of mitigating the ever-expanding epidemic of heart failure.
Keywords: autophagy, cancer chemotherapy cardiotoxicity, cardiac hypertrophy, glycogen storage cardiomyopathy, heart failure, ischemic heart disease
Introduction
Our understanding of molecular mechanisms governing pathophysiology in the disease-stressed heart has expanded rapidly in recent decades. However, despite these important advances, emergence of new, clinically meaningful therapeutic strategies, apart from the rapid expansion of device-based therapies, has disappointed. Thus, most patients with heart failure experience unrelenting disease progression, and heart failure-associated morbidity and mortality remain high worldwide [1]. Further, the prevalence of this syndrome is increasing, impacted simultaneously by deteriorations in the western lifestyle and by success in taming the acutely lethal manifestations of other diseases. Given all this, there is great urgency to identify and exploit novel therapeutic targets of heart failure pathogenesis.
Recently, studies from laboratories around the world have shown that autophagy [from the Greek auto (self) and phagein (eating)], a nearly ubiquitous process of cellular cannibalization in which intracellular components are delivered to lysosomes for bulk degradation, is a key element of stress-triggered cardiac remodeling [2]. Autophagy is a dynamic process that ensures cellular homeostasis by eliminating damaged organelles and toxic protein aggregates, as well as by recycling nutrients during starvation and stress [3]. Three types of autophagy have been recognized: microautophagy, chaperone-mediated autophagy, and macroautophagy [4]. Here, we focus on macroautophagy (hereafter termed autophagy), the most highly characterized type.
Autophagy: a ubiquitous catabolic process
Molecular mechanisms governing mammalian autophagy are increasingly defined and will be discussed here only in broad overview. Readers are referred to several outstanding reviews for additional details [5-7].
Governing signaling pathways
In the presence of nutrient abundance and circulating insulin, class I phosphatidylinositol-3-kinases (PI3K-I) are activated, phosphorylating and activating AKT, a nexus of metabolic signaling in many cell types. AKT, in turn, triggers the protein mammalian Target of Rapamycin (mTOR), which blocks autophagy by inhibiting nucleation of a macromolecular complex composed of class III PI3K and vacuolar protein sorting 34 (Vps34). Activated class III PI3K (PI3K-III) and Vps34 then localize to the phagophore, in which they interact with Beclin 1/ATG6 and ATG14 to promote autophagic flux [3,8].
Under conditions of nutrient inadequacy, in contrast, mTOR is inhibited, and the autophagic flux pathway is released from repression [8]. Consistently with this, the class III PI3K inhibitor 3-methyladenine (3-MA) blocks autophagy [4]. In addition, AMP-activated protein kinase (AMPK) is activated by depletion of intracellular nutrient stores, inhibits mTOR, and upregulates autophagy [9]. Alternatively, in the setting of pathologic stress (e.g., starvation, ischemia, hypoxia, oxidative stress, mitochondrial damage, chemotherapeutic drugs), a Beclin 1–PI3K-III complex is formed, promoting initiation of autophagy [6]. Disrupting the interaction of Beclin 1 with the Vps34 complex, or depleting Beclin 1 by genetic knockdown, significantly diminishes progression of the autophagic cascade [3].
Molecular anatomy of the autophagy cascade
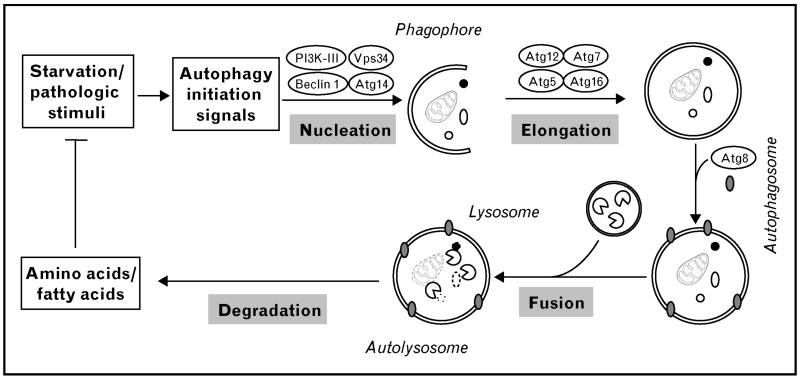
The first step of the autophagic pathway is termed nucleation, a class III PI3K-promoted process in which the membrane compartment phagophore begins to develop by utilizing intracellular membrane sources or by de-novo membrane synthesis (Fig. 1). Next, the elongation step is accomplished by activation of two parallel pathways: in one, the E1-like enzyme ATG7 mediates the conjugation of ATG12 to ATG5, which then couples with ATG16. This complex is required for autophagosome formation and dissociates from the vacuole upon maturation. In the other, microtubule-associated protein 1 light chain 3 (LC3/ATG8) is cleaved by ATG4, producing the so-called LC3-I form. LC3-I is subsequently activated by conjugation to phosphatidyl-ethanolamine by means of its interaction with ATG7 and ATG3 (E2-like enzymes), resulting in LC3-II [6,7]. LC3-II levels are a frequently tracked marker of autophagy, as the molecule remains attached to the autophagosome membrane until degradation is completed [4]. The final step is accomplished by the fusion of the autophagosome with a lysosome, forming an autolysosome. Degradation of the autolysosomal cargo is accomplished by several lysosomal hydrolases, and the catabolized products are released into the cytosol and recycled for nutrient and/or structural needs [7].
Figure 1. Schematic of autophagy pathways.
A variety of stimuli, including both physiological (e.g., starvation) and pathologic stresses, trigger cardiac autophagy. The first step in the process is nucleation. Classically, a complex composed of class III PI3K and Beclin 1 complex is involved. This complex initiates formation of an isolated double membrane and subsequent membrane elongation. The membrane then fuses upon itself, forming the distinctive double-membrane autophagosome. During the following fusion step, autophagosomes ultimately fuse with lysosomes, culminating in cargo degradation. The end-products of autolysosome cargo degradation provide both fuel and elemental building blocks to preserve vital cellular functions and remove toxic cellular elements. ATG, autophagy-related genes; PI3K-III, class III phosphatidylinositide-3-kinase; Vps34, vacuolar protein sorting 34.
Autophagy in human cardiovascular disease
Despite the rapid emergence of mechanistic insights into the role(s) of autophagy in preclinical models of heart disease (see below), much less is known about cardiomyocyte autophagy in human disease (Table 1 [10-12, 13••,14•,15,16•,17•]). Most evidence to date has been obtained in myocardial samples obtained from patients with heart failure, Danon disease, ischemic heart disease, and cancer chemotherapy cardiomyopathy. Genetic studies are largely lacking.
Table 1.
Autophagy in human cardiovascular disease
| Cardiovascular disease | Sample source | No. of patients |
Methods to detect autophagy | Autophagic activity |
Reference |
|---|---|---|---|---|---|
| Heart failure (DCM) | Ventriculectomy | 27 | EM | Increased | [10] |
| Heart failure (unclassified cardiomyopathy) |
Endomyocardial biopsy | 1 | EM | Increased | [1] |
| Heart failure (idiopathic DCM) | Explanted hearts | 19 | EM, ubiquitinated proteins | Increased | [12] |
| Heart failure (idiopathic DCM, LVAD) | Explanted hearts | 9 | Beclin 1, ATG5, LC3II mRNA and protein level |
Decreased after LVAD |
[13••] |
| Glycogen storage disease | Animal model of human disease |
N/A | EM | Decreased | [14•] |
| Ischemic heart disease (ischemia–reperfusion) |
Animal model of human disease |
N/A | EM, Beclin 1, LC3, cathepsin D | Increased | [15] |
| Anticancer drug-induced cardiomyopathy (doxorubicin) |
Animal model of human disease |
N/A | EM, Beclin 1 | Increased | [16•] |
| Anticancer drug-induced cardiomyopathy (bortezomib) |
Animal model of human disease |
N/A | EM, LC3-GFP | Increased | [17•] |
DCM, dilated cardiomyopathy; EM, electron microscopy; LVAD, left ventricular assist device; N/A, not available.
Techniques used to detect autophagy in human samples are largely limited to electron microscopic ultrastructural and immunohistochemical analyses. And, not surprisingly, human cardiac tissue samples are obtained with limitations, such as being from either endomyocardial biopsies harvested during right heart catheterization, explanted hearts obtained at the time of cardiac transplantation, or ventricular tissue cores extracted at the time of assist device implantation. Further, a vast spectrum of disease type, severity, duration, and therapies further limits the mechanistic insights which can be derived from their analysis.
Heart failure
Evidence for autophagy in human heart disease emerged first from tissue samples of dilated cardiomyopathy [10]. Twenty-seven hearts explanted from end-stage heart failure patients undergoing partial ventriculectomy were examined. Ultrastructural analyses revealed numerous autophagic vacuoles containing cytoplasmic material and organelles that were localized within degenerated cardiomyocytes. In dilated cardiomyopathic hearts, autophagy appeared to be associated not only with degradation of damaged intracellular organelles but also with progressive destruction of cardiomyocytes [10]. Interestingly, in 19 explanted hearts from end-stage heart failure patients who underwent transplantation, cardiomyocyte death occurring by multiple mechanisms was inferred, with autophagic mechanisms a prominent example; the estimated prevalence of apoptotic, necrotic, and autophagic cells was 0.002, 0.06 and 0.08%, respectively [12].
Autophagic cell death, or programmed cell death type II, may be a significant contributor to the pathogenesis of heart failure [11,12]. In patients with isolated aortic valvular stenosis and varying degrees of left ventricular systolic dysfunction, cell loss, mainly by autophagy and oncosis (necrotic cellular morphology with swelling of cytoplasmic organelles), was associated with the progression of left ventricular systolic dysfunction [8]. More recently, biopsy samples of left ventricular myocardium from nine patients with idiopathic dilated cardiomyopathy were obtained at the time of implantation and explantation of a left ventricular assist device (LVAD) [13••]. Molecular studies of these samples showed that mechanical unloading of the failing human heart was associated with decreased markers of autophagy. The authors went on to suggest that autophagy may be an adaptive mechanism in the failing heart, a phenomenon which is attenuated by LVAD support [13••]. It is not known whether mechanical unloading of the failing heart leads to normalization of cardiomyocyte autophagic activity back to basal levels.
Glycogen storage disease-related cardiomyopathy
Glycogen storage disease can present as hypertrophic cardiomyopathy [14•,18,19•]. This is particularly the case for Danon disease, a condition characterized by defective autophagosome–lysosome fusion owing to a mutation in the lysosomal membrane receptor LAMP-2. Consequent accretion of unprocessed autophagosomes provokes cardiomyopathy [4]. In a mouse model of Pompe disease, a disorder marked by defective metabolism of glycogen due to insufficiency of lysosomal acid alpha-glucosidase, suppression of the initiation steps of autophagy by inactivating ATG7 facilitates successful enzyme replacement therapy [20•]. A novel LAMP-2-positive dilated cardiomyopathy has also been reported [21]. This late-onset cardiomyopathy is characterized by increased autophagic vacuoles along with clinical features suggestive of Danon disease, yet LAMP-2 gene mutations are lacking [21].
Ischemic heart disease
Cardiomyocyte autophagy is a prominent feature in ischemic disease. That said, analysis of tissue samples from patients with ischemic heart disease is typically confounded by co-existing heart failure [10]. Activation of cardiomyocyte autophagy has been reported in a porcine model of chronic ischemia–reperfusion (I/R) [15], and rodent I/R models have been employed extensively in studies focusing on mechanism.
Anticancer drug-induced cardiomyopathy
Cancer chemotherapy, particularly with anthracyclines, has long been associated with significant cardiotoxicity, cardiomyopathy, and heart failure [22]. However, the fact that cancer patients are typically treated with multiple drugs in combination has made it difficult to pinpoint a unique culprit. Of course, availability of human tissues in this context is rare. However, in a rat model of doxorubicin-induced cardiomyopathy, cardiomyocyte autophagy was implicated as a catabolic pathway important in the development of heart failure [16•]. Furthermore, in cancer patients treated with the reversible proteasome inhibitor bortezomib, drug-related cardiotoxicity has been suspected [17•]. Rats exposed to bortezomib developed heart failure, and endoplasmic reticulum stress and upregulated autophagy have been described [17•].
Mechanistic studies in animal models of cardiovascular disease
Increases in autophagic flux have been documented in virtually all forms of human cardiovascular disease, including ventricular hypertrophy, heart failure, ischemic disease, and glycogen storage disorders. However, whether this catabolic process is adaptive or maladaptive remains unknown. To address this knowledge gap, a number of studies have been performed in preclinical animal models seeking to decipher the role of this process in disease pathogenesis, tease out its mechanistic underpinnings, and ultimately discover molecular targets for potential therapeutic intervention.
Autophagy in the transition from hypertrophy to heart failure
The initial response of the heart to increases in afterload is hypertrophic growth [23]. If the afterload stress persists, the heart will eventually become dilated, contractile function will decline, and heart failure ensues [24]. Indeed, this progressive course of disease occurs commonly in patients with hypertension or ischemic heart disease [25]. Now, recent work has demonstrated that autophagic flux in cardiomyocytes is activated in this context. For example, in a model of pressure overload induced surgically by transverse aortic constriction (TAC), we have reported that autophagic activity increases rapidly after TAC, peaks at 72 h, and is maintained at elevated levels for at least 3–4 weeks [26]. The degree of autophagic activity correlates with the magnitude of hypertrophic growth and with the rate of transition to heart failure [26], and steady-state levels of autophagic flux correlate with heart mass [27••]. Consistently with these findings, transgenic mice with cardiomyocyte-restricted overexpression of Beclin 1, a rate-limiting protein in the autophagic cascade, manifest increased autophagic activity in the setting of elevated afterload and a correspondingly amplified pathological remodeling response, including ventricular dilation, systolic dysfunction, and early mortality [26,27••]. Conversely, suppression of autophagy in the context of Beclin 1 haploinsufficiency halved the stress-induced autophagic response and partially rescued the phenotype [26].
Collectively, these data suggest that autophagy is mal-adaptive under conditions of pressure overload, a common clinical scenario. One potential mechanism for this disease-promoting behavior is that autophagy may facilitate hypertrophic growth and allow the sustenance of greater degrees of hypertrophy; this, in turn, promotes the emergence of systolic dysfunction and heart failure. In this context, afterload-induced cardiomyocyte autophagy is a potential target for therapeutic intervention. To test this, we recently employed small molecule inhibitors of histone deacetylases (HDACs). We, and others, have shown previously that HDAC inhibitors are efficacious in the suppression of pathological cardiac remodeling [28]. Given this, we hypothesized that HDAC-dependent pathological autophagy may contribute to the disease process and that suppression of pathological autophagy by HDAC inhibition (HDACi) may underlie their beneficial effects. Consistently with this model, we found that HDACi was, in fact, capable of profoundly suppressing load-induced cardiomyocyte autophagy, and this autophagic response is required for much of the pathological growth response [27••].
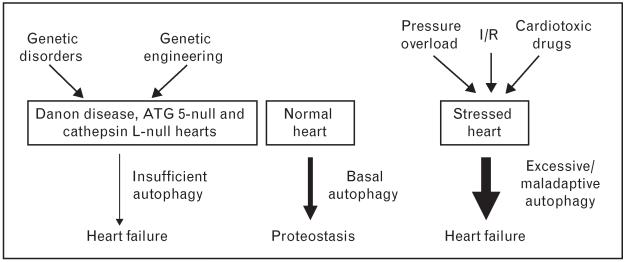
Even as overexuberant cardiomyocyte autophagy can be maladaptive, complete abrogation of the catabolic response is similarly maladaptive. For example, inactivation of the gene coding for ATG5 in the heart triggers rapid-onset heart failure [29••]. More recently, in a model of systemic inactivation of the gene coding for the lysosomal enzyme cysteine endopeptidase cathepsin L (Ctsl), emergence of large dysmorphic vesicles in the cytoplasm and dilated cardiomyopathy were documented [30]. Nutrient deprivation and mTOR suppression with rapamycin, two robust triggers of autophagy, were each incapable of activating autophagic flux. From this, the authors concluded that impaired degradation of autolysosomal content in the absence of Ctsl was a major mechanism underlying the cardiomyopathic phenotype [30]. These observations, then, are consistent with the notion that basal levels of cardiomyocyte autophagy are critically required for cellular proteostasis. Given this, we favor a model in which titration of cardiomyocyte autophagy within an optimal, adaptive zone is an approach of therapeutic interest [2] (Fig. 2). Importantly, HDACi suppresses, but does not eliminate, the autophagy response to stress and hence is an attractive strategy worthy of additional investigation [27••].
Figure 2. Adaptive and maladaptive autophagy in the heart.
Under normal physiological conditions, basal autophagy is critically required for protein quality control, removal of damaged organelles, and recycling of intracellular elements to maintain cardiac homeostasis. Excessive, or abrogated, autophagy is maladaptive. In the setting(s) of maladaptive, pathological stress, overactivated autophagy fuels hypertrophic growth, leading to autophagic cell death and ultimately heart failure. Conversely, complete abrogation of cellular autophagy eliminates critical housekeeping functions of autophagy, triggering rapid and catastrophic cardiac dysfunction.
Autophagy in ischemic heart disease
Multiple studies have demonstrated that cardiomyocyte autophagy is activated during ischemia, and suppression of that autophagic response can be detrimental [31,32]. Underlying mechanisms may relate to autophagy-dependent replenishment of cellular metabolic needs in the setting of their inadequacy and elimination of dysfunctional mitochondria, which would otherwise release reactive oxygen species (ROS) and pro-apoptotic mediators [32]. However, most patients with ischemic heart disease recanalize their coronary vessels spontaneously, or this is effected mechanically by percutaneous intervention. As such, in most clinical scenarios, myocardial ischemia is coupled with restoration of blood flow and myriad associated events, including robust release of ROS [33]. Here, reperfusion following coronary artery occlusion triggers robust increases in autophagy [33]. Multiple studies, conducted in tissue culture [34], rodents [26], and large animal models [15], have revealed marked activation of autophagic flux during the reperfusion phase. Oxidative stress is thought to be a major underlying mechanism [33]. However, whether this upregulated autophagy is adaptive or maladaptive is the subject of debate. In heterozygous Beclin 1-null mice, I/R-induced autophagy was significantly attenuated compared with wild-type controls. Following experimental I/R, these mice developed less infarction and less apoptosis [31]. Similar findings have been reported in cultured neonatal cardiomyocytes exposed to simulated I/R, in which chemical suppression of autophagy with 3-MA improved cell viability [35]. In contrast, it has been reported that simulated I/R-induced autophagy in cultured cell lines can be protective [34,36]. Recently, it was reported that sulfaphenazole, an inhibitor of cytochrome P450-2C9, activates autophagy and is protective against I/R injury, both in tissue culture and in isolated perfused rat hearts [37]. At present, the extent to which these discrepancies derive from differing cell types, model systems, or experimental paradigms is unclear.
Anticancer drug-induced cardiac autophagy
Doxorubicin-induced cardiomyopathy has been studied extensively [16•,38]. However, precise understanding of underlying mechanisms is lacking. Cardiomyocyte death by apoptosis and necrosis is thought to be the primary mechanism of doxorubicin-induced cardiomyopathy. However, in a rat model of doxorubicin-induced cardiomyopathy, pharmacological suppression of autophagy was associated with significant rescue of cardiac function [16•]. Furthermore, doxorubicin-induced autophagic vacuoles were blocked by 3-MA in failing rat heart [16•]. A recent report showed that the transcription factor GATA4 inhibits doxorubicin-induced autophagy and cardiomyocyte death through upregulating the survival factor Bcl2 and downregulating autophagy-related genes [38]. Current management of doxorubicin-induced cardiotoxicity includes regular monitoring of cardiac function and limiting the maximum dose of exposure. One day, inhibition of maladaptive autophagy may be a therapeutic option for this common condition.
Therapeutic targets of maladaptive autophagy
Clearly, cardiomyocyte autophagy participates, in one form or another, in virtually all forms of heart disease. As such, titration of autophagic flux is an objective of potential therapeutic interest: suppression of excessive autophagy without eliminating basal fluxes required for cell survival may be the key.
Currently, there are no active clinical trials testing anti-autophagic interventions in cardiac disease. However, several possible targets in the autophagy pathways have been identified. In the UM-X7.1 hamster model of dilated cardiomyopathy, heart failure develops progressively, culminating in 50% mortality by 30 weeks of age [39]. Treatment with granulocyte colony-stimulating factor (G-CSF) significantly improves both survival and cardiac function, and suppression of autophagy, as opposed to suppression of apoptosis or promotion of regeneration, has been implicated [39]. In a porcine I/R model, the antibiotic chloramphenicol succinate is protective in association with increased levels of the autophagy proteins Beclin 1 and LC3 [40]. AMPK regulates autophagy and has been implicated in pro-cell survival effects of autophagy in cardiac ischemia [31]. In a canine model of pacing-induced heart failure, AMPK activation by metformin, a commonly used antidiabetic medication, prevents apoptosis and promotes relative functional preservation of cardiac performance [41]. Although autophagy was not examined in this model, it is tempting to speculate that its effects on autophagy contribute to the salubrious actions. Finally, urocortin, an endogenous cardiac peptide, blocked autophagy induced by I/R, and downregulated Beclin 1 was implicated [35].
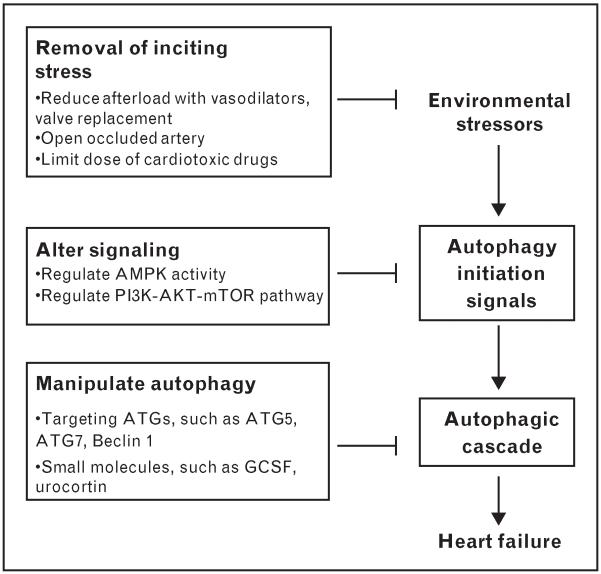
Owing to the dearth of small-molecule agents regulating autophagy, new therapeutic options must be developed, such as siRNA targeting autophagy genes. However, given the critical housekeeping function of basal autophagy in all cell types, additional molecular detail regarding the autophagic circuitry – and how it interfaces with metabolic, anabolic, and cell death-inducing events – is required (Fig. 3).
Figure 3. Therapeutic targets for maladaptive autophagy.
Therapies targeting cardiomyocyte autophagy can be envisioned at three levels: removal of inciting stressors, regulation of autophagy-initiating signals, and direct targeting of the autophagic molecular machinery. Potential molecular targets are indicated. AMPK, AMP-activated protein kinase; ATG, autophagy-related genes; I/R, ischemia–reperfusion; GCSF, granulocyte colony-stimulating factor; mTOR, mammalian target of rapamycin; PI3K, phosphatidylinositide-3-kinase.
Conclusion
Whereas involvement of autophagosomal or lysosomal mechanisms in heart disease is long-established, their context-dependent role in disease promotion and disease antagonism is just now emerging. A great deal of data from preclinical models demonstrate that excessive autophagy elicited by pathological stimuli, such as pressure overload and ischemia–reperfusion, is maladaptive and promotes cell death. Conversely, basal levels of constitutive autophagy are essential to maintain proteostasis, and elimination of this means of protein quality control triggers rapid cell death. Our vision for the future includes elucidation of the autophagic circuitry in the heart such that precise tuning of its actions – promoting proteostasis and inhibiting cell death – can be accomplished for therapeutic gain.
Key points.
Autophagy is an evolutionarily conserved pathway of protein and organelle catabolism, active in virtually all forms of cardiovascular pathology.
Depending on the context, cardiomyocyte autophagy can be adaptive or maladaptive.
Autophagy is regulated by multiple signaling pathways, including class I PI3K–AKT–mTOR, class III PI3K and Beclin, insulin, and AMPK pathways.
Judicious titration (tuning) of autophagy within an optimal zone of activation may be a therapeutic strategy with clinical relevance.
Acknowledgements
This work was supported by grants from the NIH (HL-075173, J.A.H.; HL-080144, J.A.H.; HL-090842, J.A.H.; T32, M.X.), AHA (0640084N, J.A.H.), (7-08-MN-21-ADA, J.A.H.), the AHA-Jon Holden DeHaan Foundation (0970518N, J.A.H.), and the Fondo Nacional de Desarrollo (FONDECYT 1080436, S.L.; FONDAP 15010006, S.L.). S.L. is on sabbatical leave at the Department of Internal Medicine (Cardiology), University of Texas Southwestern Medical Center, Dallas, Texas, USA.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Additional references related to this topic can also be found in the Current World Literature section in this issue (p. 272).
- 1.Roger VL, Go AS, Lloyd-Jones DM, et al. Executive summary: Heart Disease and Stroke Statistics – 2011 update: a report from the American Heart Association. Circulation. 2011;123:459–463. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothermel BA, Hill JA. Autophagy in load-induced heart disease. Circ Res. 2008;103:1363–1369. doi: 10.1161/CIRCRESAHA.108.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. Mammalian autophagy, core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravikumar B, Sarkar S, Davies JE, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 8.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottlieb RA, Mentzer RM. Autophagy during cardiac stress: joys and frustrations of autophagy. Annu Rev Physiol. 2010;72:45–59. doi: 10.1146/annurev-physiol-021909-135757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimomura H, Terasaki F, Hayashi T, et al. Autophagic degeneration as a possible mechanism of myocardial cell death in dilated cardiomyopathy. Jpn Circ J. 2001;65:965–968. doi: 10.1253/jcj.65.965. [DOI] [PubMed] [Google Scholar]
- 11.Saijo M, Takemura G, Koda M, et al. Cardiomyopathy with prominent autophagic degeneration, accompanied by an elevated plasma brain natriuretic peptide level despite the lack of overt heart failure. Intern Med. 2004;43:700–703. doi: 10.2169/internalmedicine.43.700. [DOI] [PubMed] [Google Scholar]
- 12.Kostin S, Pool L, Elsasser A, et al. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92:715–724. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 13••.Kassiotis C, Ballal K, Wellnitz K, et al. Markers of autophagy are down-regulated in failing human heart after mechanical unloading. Circulation. 2009;120(Suppl 11):S191–S197. doi: 10.1161/CIRCULATIONAHA.108.842252. First study of human samples employing modern autophagic molecular markers.
- 14•.Tanaka Y, Guhde G, Suter A, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. First animal model of LAMP-2 mutation-induced cardiomyopathy.
- 15.Yan L, Vatner DE, Kim SJ, et al. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci U S A. 2005;102:13807–13812. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Lu L, Wu W, Yan J, et al. Adriamycin-induced autophagic cardiomyocyte death plays a pathogenic role in a rat model of heart failure. Int J Cardiol. 2009;134:82–90. doi: 10.1016/j.ijcard.2008.01.043. This study implicated autophagy in doxorubicin-induced cardiomyopathy.
- 17•.Nowis D, Maczewski M, Mackiewicz U, et al. Cardiotoxicity of the anticancer therapeutic agent bortezomib. Am J Pathol. 2010;176:2658–2668. doi: 10.2353/ajpath.2010.090690. This study implicated autophagy in bortezomib-induced cardiomyopathy.
- 18.Arad M, Maron BJ, Gorham JM, et al. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med. 2005;352:362–372. doi: 10.1056/NEJMoa033349. [DOI] [PubMed] [Google Scholar]
- 19•.Nishino I, Fu J, Tanji K, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. First paper to report disruption of the autophagic pathway in human glycogen storage disease.
- 20•.Raben N, Schreiner C, Baum R, et al. Suppression of autophagy permits successful enzyme replacement therapy in a lysosomal storage disorder–murine Pompe disease. Autophagy. 2010;6:1078–1089. doi: 10.4161/auto.6.8.13378. This study suggested that ATG7 may be a therapeutic target in diseases with disordered autophagy.
- 21.Sugimoto S, Shiomi K, Yamamoto A, et al. LAMP-2 positive vacuolar myopathy with dilated cardiomyopathy. Intern Med. 2007;46:757–760. doi: 10.2169/internalmedicine.46.6265. [DOI] [PubMed] [Google Scholar]
- 22.Zbinden G, Bachmann E, Holderegger C. Model systems for cardiotoxic effects of anthracyclines. Antibiot Chemother. 1978;23:255–270. doi: 10.1159/000401489. [DOI] [PubMed] [Google Scholar]
- 23.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 24.Hein S, Arnon E, Kostin S, et al. Progression from compensated hypertrophy to failure in the pressure-overloaded human heart: structural deterioration and compensatory mechanisms. Circulation. 2003;107:984–991. doi: 10.1161/01.cir.0000051865.66123.b7. [DOI] [PubMed] [Google Scholar]
- 25.St John Sutton MG, Plappert T, Rahmouni H. Assessment of left ventricular-systolic function by echocardiography. Heart Fail Clin. 2009;5:177–190. doi: 10.1016/j.hfc.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 26.Zhu H, Tannous P, Johnstone JL, et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1790. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Cao DL, Wang ZV, Battiprolu PK, et al. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci U S A. 2011;108:4123–4128. doi: 10.1073/pnas.1015081108. This paper demonstrates that suppressing cardiomyocyte autophagy by HDAC inhibition not only blocks hypertrophic growth but also reverses existing cardiac hypertrophy.
- 28.Kong Y, Tannous P, Lu G, et al. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113:2579–2588. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29••.Nakai A, Yamaguchi O, Takeda T, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. This paper demonstrated that complete abrogation of cardiomyocyte autophagy is detrimental to cardiac homeostasis under basal conditions and alters the myocyte response to pressure-overload stress.
- 30.Dennemarker J, Lohmuller T, Muller S, et al. Impaired turnover of autophagolysosomes in cathepsin L deficiency. Biol Chem. 2010;391:913–922. doi: 10.1515/BC.2010.097. [DOI] [PubMed] [Google Scholar]
- 31.Matsui Y, Takagi H, Qu X, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, Bosch-Marce M, Shimoda LA, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Hariharan N, Zhai P, Sadoshima J. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. Antioxid Redox Signal. 2010 doi: 10.1089/ars.2010.3488. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gurusamy N, Lekli I, Gorbunov NV, et al. Cardioprotection by adaptation to ischaemia augments autophagy in association with BAG-1 protein. J Cell Mol Med. 2009;13:373–387. doi: 10.1111/j.1582-4934.2008.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valentim L, Laurence KM, Townsend PA, et al. Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/ reperfusion injury. J Mol Cell Cardiol. 2006;40:846–852. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- 36.Yitzhaki S, Huang C, Liu W, et al. Autophagy is required for preconditioning by the adenosine A1 receptor-selective agonist CCPA. Basic Res Cardiol. 2009;104:157–167. doi: 10.1007/s00395-009-0006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C, Liu W, Perry CN, et al. Autophagy and protein kinase C are required for cardioprotection by sulfaphenazole. Am J Physiol Heart Circ Physiol. 2010;298:H570–H579. doi: 10.1152/ajpheart.00716.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi S, Volden P, Timm D, et al. Transcription factor GATA4 inhibits doxorubicin-induced autophagy and cardiomyocyte death. J Biol Chem. 2010;285:793–804. doi: 10.1074/jbc.M109.070037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyata S, Takemura G, Kawase Y, et al. Autophagic cardiomyocyte death in cardiomyopathic hamsters and its prevention by granulocyte colony-stimulating factor. Am J Pathol. 2006;168:386–397. doi: 10.2353/ajpath.2006.050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sala-Mercado JA, Wider J, Undyala VV, et al. Profound cardioprotection with chloramphenicol succinate in the swine model of myocardial ischemia–reperfusion injury. Circulation. 2010;122(Suppl 11):S179–S184. doi: 10.1161/CIRCULATIONAHA.109.928242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sasaki H, Asanuma H, Fujita M, et al. Metformin prevents progression of heart failure in dogs: role of AMP-activated protein kinase. Circulation. 2009;119:2568–2577. doi: 10.1161/CIRCULATIONAHA.108.798561. [DOI] [PubMed] [Google Scholar]