Abstract
The C. elegans epidermis forms one of the principal barrier epithelia of the animal. Differentiation of the epidermis begins in mid embryogenesis and involves apical-basal polarization of the cytoskeletal and secretory systems as well as cellular junction formation. Secretion of the external cuticle layers is one of the major developmental and physiological specializations of the epidermal epithelium. The four post-embryonic larval stages are separated by periodic moults, in which the epidermis generates a new cuticle with stage-specific characteristics. The differentiated epidermis also plays key roles in endocrine signaling, fat storage, and ionic homeostasis. The epidermis is intimately associated with the development and function of the nervous system, and may have glial-like roles in modulating neuronal function. The epidermis provides passive and active defenses against skin-penetrating pathogens and can repair small wounds. Finally, age-dependent deterioration of the epidermis is a prominent feature of aging and may affect organismal aging and lifespan.
Keywords: EPITHELIA, MORPHOGENESIS, PAR PROTEINS, barrier epithelia, wound healing
INTRODUCTION TO PART II
The first of these reviews addressed the anatomy and development of the C. elegans epidermis. Here we focus on the differentiation of the epidermis as a polarized barrier epithelium, its intimate relationship with the external cuticle, and its many roles in the physiology of the developing and mature worm.
THE EPIDERMIS AS A POLARIZED EPITHELIUM
The epidermis is one of the principal epithelia of the C. elegans body, and has provided important insights into epithelial differentiation. The epidermal cytoskeleton, junctional complexes, and apical basal polarity have been extensively reviewed 1–3 and are summarized here (FIGURE 1).
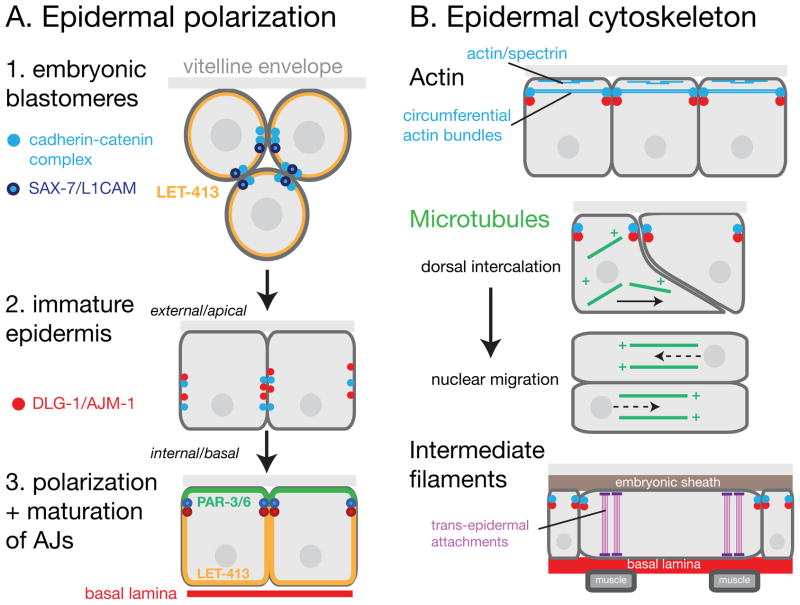
FIGURE 1. Development of epidermal polarity and cytoskeletal systems.
(A) Overview of development of polarity and epidermal features, based on 1. With a few exceptions, the order and dependency of the various polarization processes is not clearly established in the epidermis. All early embryonic blastomeres express classical cadherin/HMP-1 and LET-413/Scribble; outer blastomeres are contacted externally by the vitelline envelope. Epithelial differentiation begins with the onset of DLG-1 and AJM-1 expression in the premorphogenetic epidermis. In the early epidermis DLG-1 and AJM-1 are punctate and localized over the lateral surface. As the epidermis matures, DLG-1 and AJM-1 become confined to the subapical region to form adherens junctions (AJs), and LET-413 becomes basolaterally localized. PAR-3 and PAR-6 become apically localized in the epidermis, but are not essential for all aspects of apical-basal polarity. PAR-6 functions independently of PAR-3 to promote the assembly (but not localization) of the AJs.
(B) Epidermal cytoskeletal systems. Actin filaments (light blue lines) are found in the cortical spectrin cytoskeleton and in large circumferential bundles linking cadherin-catenin junctions. Microtubules (green lines) become polarized circumferentially in the dorsal epidermis during dorsal intercalation. Subsequent nuclear migrations are directed towards microtubule plus ends. Intermediate filaments (pink) such as IFB-1 are expressed in dorsal and ventral epidermal cells after enclosure and link apical and basal hemidesmosomes in muscle-adjacent regions of the epidermis.
Junctional complexes and polarity
Epidermal cells have provided an excellent model for understanding the development and function of junctions in epithelialization. Two major subapical junctional complexes play partly redundant functions in epidermal adhesion, the apical-most cadherin-catenin complex (CCC) and the more subapical DLG-1/AJM-1 complex (DAC) 1; together these complexes make up the so-called C. elegans apical junction (CeAJ) in mature epidermal cells. In early embryos the CCC is widely expressed and localized at all sites of cell-cell contact (FIGURE 1A); the CCC functions redundantly with the adhesion molecule SAX-7/L1CAM in early blastomere adhesion 4. The first signs of epithelial differentiation in epidermal precursors are the punctate expression of DAC components AJM-1 and DLG-1 in the dorsal epidermal cells (FIGURE 4A in Part I). Both the CCC and DAC are initially distributed along the lateral sides of epidermal cells. Polarization begins with the redistribution of the CCC and DAC components to subapical locations.
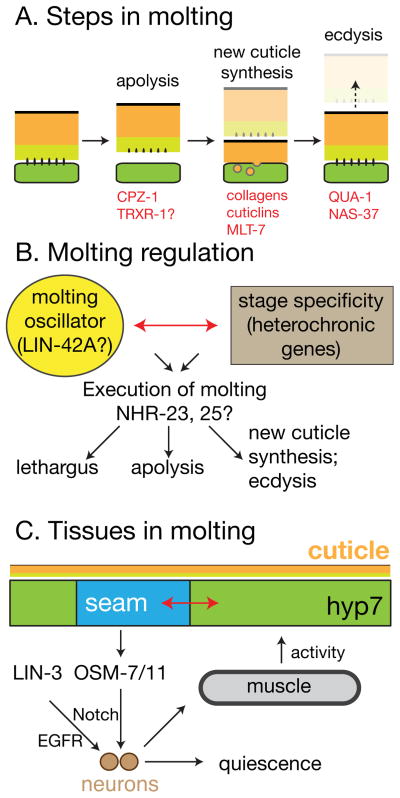
FIGURE 4. Stages, pathways, and tissues involved in molting.
(A) Molts have been divided into three main stages: apolysis, cuticle synthesis, and ecdysis. Some genes may be involved in more than one stage. Proteases such as CPZ-1 are involved in apolysis. Ecdysis (separation of old cuticle) also involves a complex set of motor behaviors not depicted here. (B) Genetic pathways regulating molting. Highly speculative outline of possible regulatory relationships, partly based on 126. Not all regulatory interactions may apply to all molts. (C) Tissue coordination during molts. The focus of LIN-42A function in molting appears to be in the lateral seam. LIN-42A might directly or indirectly regulate epidermal quiescence signals such as LIN-3 and OSM-7/11, which act on neurons. Quiescence involves decreased muscle contractions, which in turn may affect expression of epidermal molting genes 135.
Epithelial polarization involves several pathways, including the basolateral protein LET-413/Scribble 5 and the apical PAR-3/6 complex. LET-413 is uniformly expressed in early embryos; in differentiating epidermal cells it becomes basolaterally localized and serves to confine the AJ components to subapical domains. The PAR-3/6 complex is critical for polarization in many cell types, including the early embryo, complicating efforts to examine its role in the epidermis. Recent elegant experiments have revealed that neither PAR-6 nor PAR-3 are essential for epidermal polarity 6, 7. PAR-6 is required for maturation of apical junctions, independent of PAR-3; PAR-3 has a more restricted role in epidermal development 7. As epidermal cells are generated in an intrinsically asymmetric context, in that they contact other cells only on their basal and lateral sides, they may be less dependent on polarity mechanisms such as the PAR genes. In late gastrulation laminin deposition on the basal surface of the epidermis begins to establish the epidermal basal lamina 8; the role of the BL in epidermal polarization has yet to be precisely established.
The CCC and DAC can assemble independently of one another 9, 10, suggesting neither complex is individually essential for initial epidermal polarity. Embryos lacking the CCC generate a motile epidermal epithelium but fail to establish stable adhesion in late epidermal enclosure 11. Conversely, the DAC does not become essential until the 2-fold stage. Embryos lacking LET-413/Scribble display highly disorganized junctions yet develop to the 1.7-fold stage 5. Embryos lacking components of both the CCC and DAC show synergistic effects on adhesion, suggesting partial redundancy between the functions of the two complexes. It is unclear whether in such embryos the epidermis fails to polarize, or whether it initiates but fails to maintain polarization.
Apical junctions play critical roles later in embryonic morphogenesis. For example, the CCC links the circumferential actin bundles, allowing force transmission during epidermal elongation 12. Other modulatory CCC components such as JAC-1 are not individually essential for elongation, but their loss can exacerbate defects due to partial loss of CCC function 13. The CCC also localizes further junctional molecules such as the claudin VAB-9, itself required for normal epidermal elongation 14. Junctions are also highly dynamic: during ventral enclosure new junctions rapidly form at newly established cell contacts at the ventral midline, and then in the case of the posterior leading cells appear to be almost immediately disassembled as the two cells fuse. Indeed, epidermal cells that fuse with syncytia tend to be connected by apical junctions, with a few exceptions 15.
The epidermal cytoskeleton
The epidermis is richly endowed with cytoskeletal structures, many of which are critical for cell shape changes in the embryo. Indeed, the epidermis is one of the few tissues in which the interplay between actin microfilaments, microtubules, and cytoplasmic intermediate filaments (cIFs) can be studied in depth; the cIF cytoskeleton is considered in the next section.
The differentiated epidermal actin cytoskeleton alternates between a disordered meshwork of filaments and a set of highly organized circumferential bundles that form hoop-like structures along the body 16. Actomyosin-based contraction of circumferential actin bundles within seam cells is essential for the cell shape changes of epidermal elongation (see below). Within the dorsal and ventral epidermis, although not in the seam, actin bundles influence the position of secretory vesicles and correlate with the eventual formation of circumferential cuticle furrows 17. The cortical actin cytoskeleton is also critical for epidermal cell shape changes; association of cortical actin with the apical membrane is mediated by α-spectrin SPC-1 18 and βH spectrin SMA-1 19, 20. Anchoring of nuclei to stable positions within epidermal syncytia is mediated by the giant protein ANC-1, which tethers nuclei to the actin cytoskeleton 21.
The epidermal microtubule cytoskeleton is important for dorsal intercalation movements 22 and for the contralateral migrations of nuclei in the dorsal epidermis 23. Imaging of MT dynamics in the dorsal epidermis has established that during dorsal intercalation the epidermal MT arrays reorganize such that their plus ends grow to the new tip of the intercalated cell (FIGURE 1B). The plus-end directed motor kinesin-1 is required for nuclear migrations and is recruited to the nuclear envelope by the UNC-83 and UNC-84 protein complex 23. Pharmacological inhibition of MT assembly also disrupts epidermal elongation, suggesting a later role for MTs in distributing the tension generated by actomyosin contraction 16.
Trans-epidermal attachments: the epidermis as tendon
A key function of the epidermis is to provide a mechanically strong connection between body wall muscles and the external cuticle 24, 25. This tendon-like role is mediated by specific structures in the cIF cytoskeleton known as trans-epidermal attachments (TEAs) or fibrous organelles 26. Each TEA consists of two hemidesmosome-like junctional plaques connected by an array of intermediate filaments (pink lines in FIGURE 1B). The junctional plaques contain the spectraplakin VAB-10A 27. The VAB-10 spectraplakins also provide cross-links between the actin, microtubule and intermediate filament cytoskeletons. C. elegans encodes a large family of cytoskeletal intermediate filaments (cIFs) 28, several of which are required in the epidermis and appear functionally analogous to vertebrate skin keratins 29, 30. cIF assembly and function is tightly regulated by several mechanisms, including post-translational modification by SUMO 31. Intracellular components of TEAs also include the conserved ankyrin domain protein VAB-19 32, its interacting partner, the PTB domain protein EPS-8 33, and the nematode-specific protein PAT-12 34. All these components are essential for later epidermal elongation and muscle attachment.
The junctional plaques of TEAs are themselves linked to transmembrane proteins at the apical and basal plasma membranes: at the basal plaque, Myotactin/LET-805 functions to link epidermal cells to the muscle-adjacent basal lamina 35; at the apical plaque, the filaggrin-related transmembrane proteins MUA-3 36 and MUP-4 37 link the epidermis to the cuticle. The recruitment and maturation of TEAs has been a tractable genetic model for hemidesmosome biogenesis 38.
Epidermal TEAs are essential for muscle attachment and function, and are also essential for epidermal elongation beyond the two-fold stage. Epidermal elongation beyond the two-fold stage has long been known to require the contractile activity of the body wall muscles that attach to TEAs 39. The mechanism by which muscle contraction promotes epidermal cell shape changes has recently been elucidated and involves a tension-sensing process mediated by a p21-activated kinase pathway acting at TEAs 40. TEAs thus not only function as tendon-like attachments but also play mechanosensory roles.
The epidermal basal lamina and interactions with internal tissues
The basal (inner) surface of the epidermis is lined by a basal lamina (BL) that maintains epithelial integrity, allows adhesion between epidermis and muscles, and provides a permissive and instructive substrate for cell and axon migrations (FIGURE 2A,B). In the absence of BL components such as laminins the epidermis differentiates as a polarized epithelium but intermingles and adheres to other internal epithelia 8.
FIGURE 2. The epidermal basal lamina, and neuronal development.
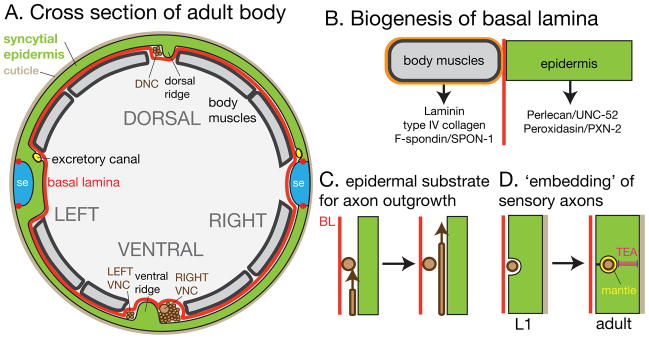
(A) Cross-section of adult body showing epidermis and adjacent tissues and structures. The external surface is covered by the cuticle; the internal face is covered by basal lamina (BL, red). Neurons (brown) reside on the epidermal side of the BL; the positions of the major nerve tracts relative to the dorsal and ventral epidermal ridges are shown. (B) Biogenesis of BL. The majority of known BL components are synthesized in muscles; the composition of the muscle-adjacent BL is likely distinct from that of non-muscle adjacent BL. (C) Developing axons appear to use the epidermis rather than the BL as substrate, as they are seen to pass on the epidermal side of other axons (R. Durbin, Ph.D thesis). (D). Sensory axons have a close relationship with the epidermis. Mechanosensory processes become progressively confined within the epidermis and secrete a specialized ECM, the mantle.
Biogenesis of the basal lamina
The epidermal BL separates ectodermal tissues (epidermis and neurons) from mesoderm, endoderm and the pseudocoelomic body cavity. As in other animals, laminins are the earliest components of the BL, and begin to be deposited during gastrulation. Interestingly, several BL components, including laminins, type IV collagens 41, and F-spondin/SPON-1 42, are expressed by muscles and not by the epidermis, and are required for later epidermal elongation. Recruitment of such proteins to the epidermal BL may involve epidermal cell-matrix receptors such as dystroglycan/DGN-1 43. However the mechanisms that target nascent BL to specific cell surfaces are not yet well understood in C. elegans. The BL that forms the tight mechanical linkage between body muscles and epidermis appears to be a fusion of the muscle and epidermal BL.
The epidermis itself makes a subset of BL components. The extracellular matrix peroxidasins PXN-1 and PXN-2 are expressed in the epidermis, and PXN-2 at least is likely to cross-link BL target proteins 44. Fibulin/FBL-1 is expressed both by muscles and the epidermis 45. UNC-52/Perlecan is expressed by the epidermis; multiple UNC-52 isoforms are generated by MEC-8-dependent alternative splicing of unc-52 46. unc-52 mutants with stop codons in alternatively spliced exons display normal embryonic development but are synthetic-lethal with mec-8. mec-8 itself is not essential for epidermal development but displays synthetic lethality with several sym genes 47, some of which are expressed and function in the epidermis 48. MEC-8-dependent alternative splicing appears to have an important yet cryptic role in epidermal differentiation.
The dynamics of the BL during embryogenesis remain incompletely understood. During epidermal elongation the BL is likely highly dynamic, accommodating the overall change in shape of the animal. Little is known of how the BL remodels during elongation while retaining tight mechanical coupling between muscles and epidermis.
The epidermis as a source of cues for patterning other tissues
The epidermis and development of the nervous system
The development of the epidermis and the nervous system are intimately connected. As discussed above, epidermal enclosure involves migration of epidermal epithelial cells over a largely neuronal substrate. Later, the epidermis provides a substrate for outgrowth and guidance of axons in the developing nervous system 49, 50. Almost all neurons extend axons in the compartment between the epidermis and the basal lamina (BL). The excretory cell also develops in this compartment (FIGURE 2A).
The dorsal and ventral midline regions of the epidermis form ‘ridges’ adjacent to the dorsal and ventral nerve cords 51, 52. Ridges are subcellular epidermal protrusions between regions of muscle attachment, and appear to express distinct molecular determinants 53. The dorsal epidermal ridge develops during embryogenesis whereas the ventral ridge only becomes prominent in larval development. Although initially symmetrical, both ridges become left-right asymmetric: the ventral ridge confines axons into a large right-hand bundle and a much smaller left-hand bundle; a few axons normally cross the epidermal ridge at defined positions. Conversely the dorsal ridge constrains axons to the left-hand side. The asymmetry of the ventral nerve cord appears to arise from initially asymmetric outgrowth of pioneer axons, and is only later reinforced by the epidermal ridge. It is unknown if the left-right asymmetry of the ridges is intrinsic to the epidermis, which otherwise appears bilaterally symmetrical 54. In larval development axons maintain their positions on either side of the ventral epidermal ridge; screens for mutants with ectopic axonal crossovers have identified many factors involved in maintenance of axon position at the midline. Among these, heparan sulfated proteoglycans and the FGF receptor EGL-15 are expressed in the epidermis 55, 56. The epidermal function of EGL-15 is kinase-dependent and involves the Ras pathway.
The epidermal BL appears to be permissive for the migrations of axonal growth cones and other migratory cells in the developing animal, and forms a matrix for instructive guidance cues 57. For example, ventral epidermal (P) cells are sources of netrin/UNC-6 that repels or attracts growth cones in the dorsoventral axis 58. Other proteins such as the ADAM UNC-71 may be secreted by the epidermis and act as short-range permissive factors for axon guidance 59. Although the BL is important for guidance, in many cases the growth substrate for developing axons appears to be the epidermis itself rather than its BL (FIGURE 2C; R. Durbin, Ph.D. thesis, WormAtlas). Motor neuron growth cones insert ‘fingers’ into the dorsal epidermal ridge (Durbin thesis, p. 69), suggesting that commissural growth cones stop at the dorsal midline not simply because they have reached the bottom of a concentration gradient but also because some attractive factor keeps them there.
Epidermal cells also provide localized cues for axonal branching and synaptogenesis, most intensively analyzed in the developing vulva. Vulval epidermal cells express the neurexin-like protein BAM-2, which locally represses branching of VC neurons 60. Another VC branch-inhibiting factor, the N-glycanase PNG-1 may also have an epidermal focus 61. Vulval epidermal cells express the nephrin SYG-2, a local ‘guidepost’ cue that enhances branching and synaptogenesis by the HSN neuron 62.
The epidermis and internal tissues
The epidermal BL provides substrate and guidance cues for migrations of a variety of mesodermal cells and tissues. In larval stages the somatic gonad and ventral epidermis undergo multiple reciprocal interactions during vulval development. Somatic gonadal cells first induce vulval differentiation in the ventral epidermis. Later the attachment of the uterine and vulval epithelia requires removal of the intervening BL 63. Epidermally expressed cues trigger the invasion of the BL by the gonadal anchor cell, and actively constrain the extent by which the BL is remodeled 64.
The epidermis is both source and recipient of Wnt signals throughout development. Wnt signals control polarity and patterning of seam cells and other dividing cells within the epidermal layer 65, 66. Epidermally expressed Wnts instruct migrations of neurons and neuroblasts over the epidermis 67, and the guidance and polarity of axons along the anteroposterior axis 68. Wnts themselves are expressed by subsets of epidermal cells: EGL-20 is expressed in posterior ventral and rectal epidermal cells 69 and LIN-44 is expressed in tail epidermis 70.
CUTICLE STRUCTURE, SECRETION, AND MOLTING
Structure of the cuticle
The true external surfaces of animals are generally not skin cells but the extracellular matrices secreted by skin cells. These matrices can consist of dead skin cells encased in a lipid-rich matrix (stratum corneum of mammals), or rigid extracellular chitinous cuticles (arthropods). In nematodes the extracellular layers consist of a flexible collagenous cuticle, a lipid rich epicuticle, and a glycoprotein-rich surface coat (FIGURE 3). All of these extracellular structures play important roles in the integrity of the animal and its interactions with the environment. The cuticle is not a rigid exoskeleton, but acts in concert with muscle tone and the hydrostatic skeleton to maintain the animal’s shape (see BOX). However the cuticle has limits to its flexibility, and is shed four times to accommodate larval growth.
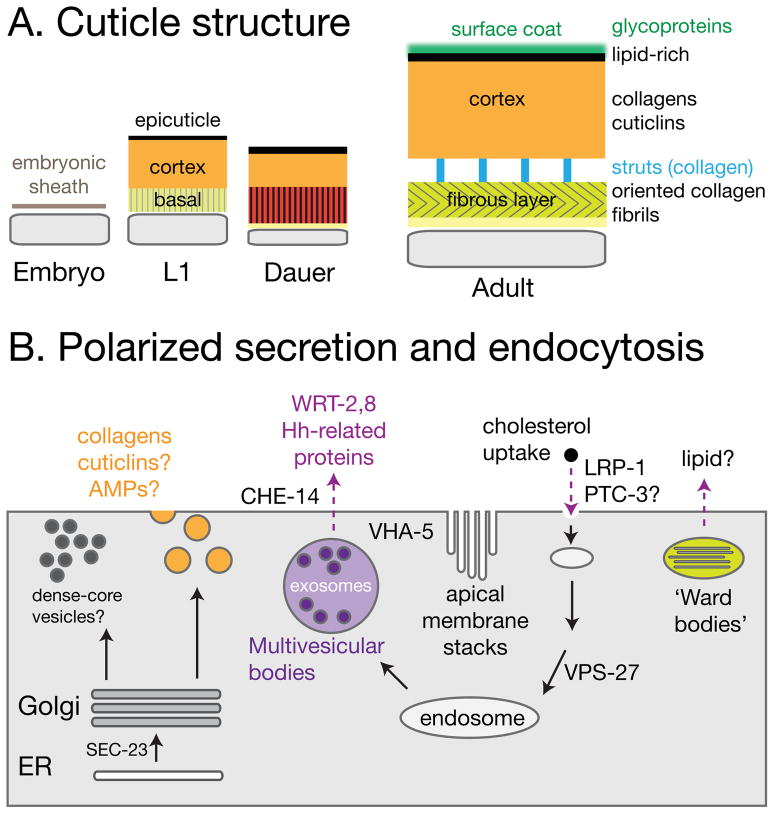
FIGURE 3. Cuticle structure and secretory apparatus.
(A) Schematic cross-sections of cuticle in different larval stages and Dauer; the surface coat is likely present in all stages but may differ in composition. (B) Aspects of polarized secretion and endocytic pathways in the epidermis. Cuticle collagens are thought to be secreted through classical apical secretory pathway. Large dense-cored vesicles have been reported in the epidermis prior to molting, but may differ from the dense-core vesicles characterized in neurons. The epidermis is abundant in MVBs, involved in exosome-based secretion of hedgehog family proteins such as the WRTs 101. The V0 ATPase subunit VHA-5 may function in MVB fusion with the apical membrane; VHA-5 is also localized to the prominent apical membrane stacks, of unknown function. Ellipsoidal organelles dubbed ‘Ward bodies’ contain membranous stacks; they have been observed in electron micrographs but are of unknown function.
BOX 1. The epidermis and the ‘hydrostatic skeleton’.
C. elegans has few rigid skeletal structures. It has long been thought that forces from muscle contraction can be transmitted because internal fluids and tissues are under hydrostatic pressure, the ‘hydrostatic skeleton’. The internal pressure of Ascaris was measured to be between 70–225 mm Hg 205 and C. elegans may be similar. Thus, a sufficiently drastic puncture wound can lead to flaccidity and death. However C. elegans can seal and repair small wounds such as those caused by microinjection needles 159. Needle wounds have been found to cause a relatively modest (20%) decrease in body stiffness 206, suggesting the shape of the nematode body mostly reflects cuticle stiffness and that hydrostatic pressure has a secondary role. C. elegans body stiffness thus results from a combination of cuticle mechanics, muscle tone, and internal pressure 207. The relative importance of these mechanisms likely varies between species; for example, in some nematodes the epidermis itself has been proposed to act as a compartmentalized hydrostatic skeleton 208.
The collagenous cuticle
The structure and composition of the cuticle have been reviewed extensively 71 and are briefly summarized here. Collagens make up the bulk of the soluble protein component of the cuticle. C. elegans cuticle collagens form a multigene family with ~170 members. A small subset of collagen genes were identified from their mutant phenotypes (Dumpy, Blistered, Roller, Squat, Long, Ray morphology). RNAi of most collagen genes does not cause overt defects; collagens may have highly redundant roles in development. A distinct protein family, the cuticlins (see Note), makes up the insoluble fraction of the cuticle 72–74. Cuticlins are important for formation of patterned structures within the cuticle such as the lateral ridges or alae. In contrast to the external cuticle produced by the epidermis, the more rigid cuticle lining the pharyngeal lumen contains chitin 75.
Collagens and cuticlin genes are expressed either in the syncytial epidermis or in the lateral seam cells or in both, and undergo extensive post-translational processing in the epidermal secretory pathway. Collagens and noncollagenous proteins are extensively crosslinked by peroxidases secreted by the epidermis 76, 77. The cuticle is built according to a common ‘ground plan’ that is modified in each stage by expression of stage-specific components. Some collagens are expressed in all larval stages, in cyclic patterns associated with cuticle synthesis at each molt, whereas others are highly stage-specific (e.g. col-19 is only expressed in the L4 and adult). Collagen genes have been classified as early, intermediate, or late with respect to their expression peak in each stage 78. Early and intermediate collagens form distinct substructures in the cuticle 79, suggesting that tightly regulated transcription allows components of a common structure to assemble most efficiently. Along these lines, it seems plausible that cuticle would be synthesized in an outside-in sequence, but this has not yet been established.
Some collagens such as dpy-7 may be directly regulated by the epidermal GATA factors 80 and by NHRs 81. Indeed, expression of NHR-23 and NHR-25 is also staggered within each intermolt 82, although it is unclear if this is sufficient to explain the sequence of collagen expression. Other transcription factors implicated in collagen transcription include ELL-1 83 and LIN-29, which directly activates the L4-specific col-17/19 loci 84, 85. However a systematic understanding of collagen transcription within the epidermis remains lacking.
The epicuticle and surface coat
The epicuticle and surface coat are the two outermost layers of the cuticle (FIGURE 3A). In EM sections the epicuticle has a trilaminar structure resembling a thickened plasma membrane. The membrane-like properties of the epicuticle suggest the cuticle should be seen as a dynamic, homeostatic acellular compartment rather than an inert exoskeleton 86. The epicuticle itself is covered by an outer layer known as the surface coat (or glycocalyx), sometimes visible as a thin amorphous layer in electron micrographs. The molecular biology of the epicuticle and surface coat remain very poorly understood. Freeze fracture studies of the C. elegans epicuticle are consistent with a lipid bilayer in which glycoproteins are embedded 87. The surface coat is also thought to be composed of glycoproteins such as lectins and mucins; its composition varies during development, perhaps to allow evasion of pathogens 88. Mutants defective in the surface coat have been identified via aberrant expression of surface antigens 89, altered lectin binding 90, or defects in bacterial adhesion 91, 92. Some of these genes (e.g. bah-1) 93 could encode components of the epicuticle or surface coat, whereas others encode carbohydrate biosynthetic enzymes likely required for post-translational modification of coat proteins 94. The surface coat and epicuticle pose a challenge to genetic analysis as individual components may be redundant or have subtle roles under lab conditions.
The epicuticle is synthesized by the epidermis, as the first step in formation of a new cuticle at a molt. The embryonic sheath resembles the epicuticle in ultrastructure and may be the precursor to the epicuticle of the L1. The cellular origin of the surface coat is less clear. Studies of other nematodes have suggested that excretory or pharyngeal gland cells secrete surface coat that spreads over the cuticle surface. On the other hand, several bus genes, likely involved in surface coat or epicuticle biosynthesis, are expressed in epidermal seam cells and not in the syncytial epidermis 94. Conversely many cuticular collagens are expressed by syncytial epidermis and not seam. These observations raise the conundrum of how the seam and non-seam epidermis coordinate the production of the various cuticle compartments. ‘Trans-cuticular’ transport mechanisms or channels for surface coat components have been reported in parasitic nematodes but not in C. elegans. Both mechanisms may apply: seam cells could be responsible for surface coat production during a molt whereas glands could maintain a flow of surface coat material in intermolt or adult periods.
Secretion of the embryonic sheath and first cuticle
Prior to formation of the skin the developing embryo is confined within a eggshell secreted by the zygote after fertilization. The developing epidermis generates an external extracellular matrix, the embryonic sheath, shortly after enclosure; this embryonic sheath is essential for epidermal elongation 16. In its earliest state the sheath is a thin sheet that contacts the epidermal membrane close to the circumferential actin bundles discussed above. The composition of the early sheath is unknown; it later develops into a trilaminar lipid-rich layer 17 that resembles the larval epicuticle. Lipid-binding proteins expressed by the embryonic epidermis could be important in formation of the sheath 95; a family of leucine-rich repeat proteins secreted by the embryonic epidermis may also play roles in epicuticle biogenesis 96.
The embryonic sheath, unlike the cuticle, must be able to accommodate rapid radial contraction and axial elongation. The circumferential actin bundles in the epidermis correlate with circumferential furrows in the sheath and in the later cuticle; secretion of the sheath and cuticle is somehow patterned by the epidermal cytoskeleton. The collagenous cuticle of the L1 stage is not deposited until late embryogenesis, after elongation, and is essential for the embryo to retain its fully elongated form. Mutants lacking cuticle collagens such as sqt-3 undergo normal elongation to the threefold stage then retract to an almost un-elongated length. Loss of function in the secretory pathway blocks embryogenesis late in elongation, likely reflecting a failure in cuticle secretion 97.
The epidermal secretory apparatus
In keeping with its massive secretory needs, the epidermis is richly endowed with specialized secretory structures and organelles at its apical surface (FIGURE 3B). Golgi bodies are extremely abundant in epidermal cells, especially during cuticle synthesis at molts. Secretion of collagen and other cuticle components seems likely to be via the classical secretory pathway (ER-Golgi-post-Golgi vesicles); for example loss of function in sec-23 (involved in ER-Golgi transport) blocks development due to failure of cuticle secretion. Collagen biosynthesis involves extensive post-translational modifications. By analogy with vertebrate fibrillar collagen biosynthesis, early modifications such as prolyl 4-hydroxylation and trimerization are likely to occur in the epidermal ER. Later steps in collagen processing and crosslinking presumably occur after secretion into the cuticle compartment 71.
Vesicles with electron-dense cores are seen in the epidermis prior to molting 98; the molecular identity of these vesicles remains elusive. Genes critical for biosynthesis of neuronal dense-core vesicles, such as unc-31/CAPS or unc-108/Rab2, do not appear to act in the epidermis, suggesting the epidermal vesicles could represent a distinct secretory pathway. In contrast, secretion of hedgehog-related proteins uses an exosome-based mechanism. The epidermis contains numerous multivesicular bodies (MVBs) 99, 100, required for apical secretion of proteins such as WRT-2 and WRT-8 101. The V0 ATPase component VHA-5 is involved in MVB secretion, possibly in fusion of MVBs with the apical membrane.
In addition to these better-known secretory structures, other epidermis-specific organelles have been observed, of uncertain function. The apical epidermal membrane is frequently infolded into stacked sheets up to 1 μm in diameter, associated with ribosome-like particles on their cytoplasmic faces. The resulting increased surface area might enhance apical secretion or uptake. Interestingly, the V0 ATPase subunit VHA-5, required for MVB secretion, is enriched on apical membrane stacks 99; perhaps membrane stacks boost recycling of secretory components that have fused with the apical membrane. Finally epidermal cells also contain membrane-bound ellipsoidal organelles containing stacked membrane-like sheets (‘Ward bodies’; see Figure 2A in 98). The function of Ward bodies is unknown; speculatively, they could play roles in cuticle lipid (epicuticle) secretion, analogous to the lamellar bodies of vertebrate keratinocytes.
Molting
Molting, the periodic shedding and resynthesis of the cuticle to accommodate growth, is a central feature of the nematode life cycle 102. Molecular analysis of C. elegans molting has begun to uncover mechanistic similarities with other kinds of molting such as those of arthropods.
Execution of molting: apolysis, cuticle synthesis, and ecdysis
Each molt involves three stages: separation of the old cuticle (apolysis), synthesis of new cuticle, and shedding of the old cuticle (ecdysis) (FIGURE 4A). The overall timing and coordination of molting are likely under the control of as-yet unknown endocrine steroid hormones acting via epidermal nuclear hormone receptors that include NHR-23 and NHR-25 103. C. elegans requires dietary cholesterol 104, most likely for synthesis of steroid hormones involved in molting; epidermal uptake of cholesterol by the cholesterol receptor LRP-1 is essential for viability 105. The roles of other cholesterol trafficking proteins in molting 106, 107 may be explained by the need for cholesterol in steroid hormone synthesis.
Within a given larval stage molting is tightly regulated to occur at the correct time relative to other developmental events. Insight into how the molting cycle is integrated with other developmental processes came from the finding that inappropriate activation of nicotinic acetylcholine receptors delays developmental events but does not affect onset of molting, leading to lethality 108. This uncoupling of developmental events and molting cycles requires activity of nAChRs, DAF-12 and the epidermally expressed cation transporter CATP-1 109.
The first phase in molting, apolysis, allows the new cuticle to be synthesised between the epidermis and the old cuticle. Separation of the old cuticle from the epidermis requires the elimination of linkages between the epidermis and the cuticle matrix. Interactions between epidermal receptors and the cuticle could be broken by several mechanisms, including downregulation of the receptors, alteration of cuticle composition, or degradation of the receptor-ligand complexes. Epidermally expressed thioredoxin and glutathione reductases are required for molting 110. These enzymes have been proposed to generate an intracellular pool of GSH that is released to the extracellular milieu at apolysis. The resulting reduction of disulfide bonds within the cuticle could disrupt its attachment to the epidermis. Apolysis likely involves large-scale proteolysis of cuticle-epidermal attachments, and a variety of peptidases and proteases have been implicated in molting 111, including the cathepsin-like protease CPZ-1 112. Activity of such proteases must be under stringent control so as not to interfere with the synthesis of new cuticle; at least two serine protease inhibitors (serpins), BLI-5 113 and MLT-11 102, are required for molting.
The final stage in molting is ecdysis, the shedding and escape from old cuticle. C. elegans ecdysis involves a complex set of behaviors that include rotation, body contraction and thrusting, and regurgitation of pharyngeal cuticle. Together these result in mechanical disruption of any remaining linkage between old and new cuticle. The astacin metalloprotease NAS-37 is localized to the old cuticle and may be specifically involved in its degradation at the tip of the head during ecdysis 114; the related protease NAS-36 is also involved in ecdysis 115. Nematode-specific proteins such as MLT-10 and its relatives are important for cuticle synthesis and ecdysis although their biochemical functions are not yet known 116.
Hedgehog-related signals appear to play multiple roles in epidermal development, including molting. The epidermis secretes many members of the Warthog, Groundhog families of Hedgehog-related secreted proteins 117, 118. Several of these, including the quahog gene qua-1, are expressed cyclically, peaking during molts 119. QUA-1 is a candidate for a secreted cue to initiate ecdysis and is localized to the detaching cuticle; it may have other roles in cuticle development, as qua-1 mutant L1 cuticles, which have not experienced a molt, are structurally abnormal 119. Hh protein distribution can be influenced by HS proteoglycans, and indeed HSPG synthetic genes such as PST-1 are expressed in the epidermis and required for cuticle integrity 120. The epidermis expresses several Patched and Patched-related receptors 121, raising the possibility of autocrine Hh signaling within the epidermis. Other signaling proteins such as the amyloid precursor protein APL-1 are expressed in the epidermis and required for molting 122, 123.
Genetic regulation of molting: the molting cycle and heterochronic genes
Molting has been postulated to involve two distinct yet coordinated sets of regulatory controls (FIGURE 4B). First, a ‘molting oscillator’ has been proposed to initiate molts in a periodic fashion. Second, stage-specificity could be imposed by heterochronic genes, which ensure that each larval stage has the appropriate cuticle type. Certain heterochronic mutants lose stage-specific characters but continue to molt, indicating that stage progression can be uncoupled from the molt cycle. These inputs together trigger the processes that execute molting.
Several genes are known whose expression oscillates in a periodic fashion, peaking during each molt, including the nuclear hormone receptors NHR-23, NHR-25, and the primary transcript of the miRNA let-7 124. It remains unknown how the oscillations of these transcripts are related; although let-7 has been shown to negatively regulate the NHRs at the larval to adult transition 125, mature let-7 transcripts do not accumulate in earlier molts, suggesting other mechanisms must be responsible for periodic NHR transcript accumulation. The PERIOD protein LIN-42A has recently been found to be a critical component of a molting oscillator 126. lin-42 mRNA levels were known to oscillate in larval cycles 127. The lin-42 locus encodes multiple isoforms; loss of function in some of these isoforms results in heterochronic (but not molting) defects 127. Recent findings indicate that complete loss of function in the lin-42 locus results in delayed or arrythmic molts; conversely overexpression of the LIN-42A isoform triggers anachronistic molts 126, suggesting that LIN-42A is necessary and sufficient to initiate the molting program. The relationship of LIN-42A to the other cycling genes remains to be defined; NHR-25 appears to act partly in parallel to LIN-42A.
LIN-42 is a member of the PERIOD family of transcription factors, originally known from their roles in circadian rhythms in Drosophila and mammals. Although the C. elegans molting cycle is ultradian (8–10 h period), these results suggest the possibility of mechanistic similarities between the molting cycle and other biological clocks. Many biological oscillators are based around negative feedback loops, and indeed let-7 and the DAF-12 steroid hormone receptor comprise a negative feedback loop 128, 129. The coupling of stage-specific phenotypes and the molting cycle could reflect coordinate regulation of molting genes by miRNAs also involved in heterochronic progression 125, although this remains to be shown. It seems likely that the mechanistic basis of the C. elegans molting oscillator and its interconnections with the heterochronic genes will be soon elucidated.
Cessation of molting
The L4-to-adult molt is the last in the normal life cycle. How is the molting oscillator switched off? A key upstream event in the larval-to-adult transition appears to be a massive upregulation of let-7 and related miRNAs, resulting in strong inhibition of the molting factors such as NHR-23 and NHR-25 125. Loss of function in let-7, or in other factors required for the larval-to-adult transition, results in supernumerary molts that require NHR-23 and -25. These supernumerary molts may differ from normal molts; for example, NHR-23 and -25 are required for lethargus in let-7 supernumerary molts but are not required for lethargus of regular molts 130. Other molting regulators such as pqn-47, have complex requirements: PQN-47 is required for ecdysis, but partial loss of pqn-47 function results in a supernumerary adult molt 131.
The systemic nature of molting
Where does the molting oscillator reside in the animal? LIN-42A expression in seam cells is sufficient to rescue lin-42 molting defects, suggesting the seam is critical for molting cues 126. However the seam must synchronize with hyp7 and other epidermal cells (FIGURE 4C). The seam is directly adjacent to hyp7, and is connected to it via gap junctions 132, affording a variety of possibilities for regulation. Seam-derived cuticular structures such as alae are overtly stage-specific; however hyp7 also displays stage specific characteristics. For example, stage-specific collagens are expressed throughout the epidermis. As hyp7 is post-mitotic it is interesting to imagine how it adopts these stage-specific features.
As well as cuticle synthesis and removal, molts involve highly stereotyped behaviors, implying the coordinated regulation of multiple tissues. During most of the molt animals enter a quiescent sleep-like phase known as lethargus, regulated by the EGF family protein LIN-3 133;. LIN-3 activity may be induced by LIN-42A 126, and appears to act on a single neuron, ALA, which may play a neuroendocrine role in inducing lethargus. ALA neuronal processes are directly adjacent to seam cells. A second pathway regulating lethargus during molting involves OSM-7 and OSM-11, which are secreted by epidermal seam cells and act on neuronally expressed Notch family receptors to promote quiescence 134. Thus, at least two signals may originate in the epidermis to induce behavioral quiescence during molting.
In addition, muscle activity itself has been implicated in molting control. Loss of function in components of the muscle dense body or the BL results in upregulation of molting-related genes in the epidermis 135. Muscle contractions are inhibited during lethargus, perhaps to avoid rupturing the epidermis while it is unattached to cuticle; during molts muscles also autonomously downregulate expression of attachment site components 136. Such reduction in muscle-generated force could be sufficient to affect expression of molting related genes, but mechanisms underlying this non-autonomous regulation of epidermal gene expression are not yet known.
The epidermis and cuticle of the dauer larva
The dauer larva is an alternate L3 stage formed when L1 or L2 animals are exposed to insufficient food or excessive crowding. The dauer larva is morphologically specialized to withstand stress or poor environmental conditions. The epidermis in particular undergoes major remodeling during the transition into the dauer state. The epidermal layer becomes thinner, leading to ‘radial shrinkage’ of the entire animal 137. Remodeling of the epidermis is dependent on autophagy pathways 138.
The cuticle of the dauer larva is highly impermeable; for example, dauer larvae are resistant to treatment in 1% SDS. The dauer cuticle is thicker than that of non-dauers, and has ultrastructurally distinct features 139, most striking of which is a highly ordered striated layer 80 nm thick that may act as an impermeable paracrystalline lattice. Two cuticlins, CUT-1 and CUT-6, play specific roles in the dauer cuticle 73, 140, but are not required for the striated layer itself. Little is otherwise known about the molecular composition of the dauer epidermis/cuticle.
The epidermis is important in generating and responding to the intercellular signals that coordinate dauer development. As the epidermis consists of gap-junction coupled syncytia that enclose the animal, it is well placed to act as an endocrine organ that regulates the entire organism. The nuclear hormone receptor DAF-12 and the cytochrome P450 DAF-9 141, 142 act within the epidermis to promote normal development; in the absence of these signals, animals enter the dauer state and do not exit. DAF-9 may be important for production of steroidal hormones such as the dafachronic acids by the epidermis, suggesting the epidermis is a site of autocrine feedback and signal amplification in the dauer decision. The large number of NHRs expressed by the epidermis suggests the epidermis integrates multiple steroid hormone signals 143. Insulin signaling is also an important regulator of the dauer decision, and may involve the epidermally expressed insulin-like ligand INS-33 144.
FUNCTIONS OF THE DIFFERENTIATED EPIDERMIS
The differentiated epidermis plays many roles in the life of C. elegans. Like all skin layers, a primary function of the epidermis is as a barrier epithelium. In addition to this ‘passive barrier’ function, the epidermis actively repairs wounds and activates cutaneous innate immune responses to skin-penetrating pathogens. The epidermis functions in ionic homeostasis, metabolic storage, and phagocytosis of cellular debris. Finally, the epidermis mediates communication of sensory neurons to the external environment.
Barrier function of the epidermis-cuticle complex
In its natural environment, either soil or rotting fruit, C. elegans is subject to repeated desiccation and rehydration. The epidermis-cuticle complex prevents leakage of internal solutes to the environment (the inside-outside barrier), and blocks entry to molecules in the environment (the outside-inside barrier). The low permeability of the C. elegans cuticle to small molecules is well known to researchers attempting pharmacological manipulations; drug concentrations effective on C. elegans are often several orders of magnitude higher than those used for cultured cells (Rand and Johnson 1995). Permeability barrier function can be assayed by incubation in Hoechst 33258, which does not cross the plasma membrane or cuticle 145, or Hoechst 33342, which crosses membrane but not cuticle 146.
Analysis of mutants with increased permeability suggests one or more layers of the cuticle confer barrier function. Several genes required for adhesion to the pathogenic bacterium M. nematophilum are also required for cuticle integrity, including the glycosyltransferases BUS-8 147 and BUS-17 91, 148. The precise location of the barrier in the cuticle remains to be determined: it may reside in the cuticle proper, the lipid-rich epicuticle, the surface coat, or a combination of all three. In addition, the epidermis itself contributes to barrier function. Tight junctions prevent solute leakage between adjacent epidermal cells. The tetraspanin TSP-15 is important for the epidermal contribution to barrier function 145.
Cuticle integrity and the permeability barrier are also compromised in mutants defective in tetrahydrobiopterin (BH4) synthesis such as the GTP cyclohydrolase cat-4 149, 150. Although BH4 is essential for cuticle integrity in Drosophila, the requirement for BH4 in C. elegans cuticle is mechanistically distinct. The rigidity of Drosophila cuticle is due to crosslinking of chitin by reactive quinones synthesized by Dopa decarboxylase (Ddc) or tyrosine hydroxylase (TH), both of which require BH4 as cofactor. However C. elegans Ddc BAS-1 and TH (CAT-2) are not expressed in the epidermis, nor are they required for cuticle integrity. In contrast, the phenylalanine hydroxylase PAH-1 is expressed in the epidermis and is required for synthesis of cuticle melanin; pah-1 mutants do not display overt cuticle defects but synergize with other cuticle mutants 150. At present the role of BH4 in cuticle integrity remains mysterious, but appears distinct from the Bus genes. Fatty acid synthesis is also important for the permeability barrier, for unknown reasons 146.
The epidermis in defense: skin-penetrating pathogens and skin wounding
The epidermis and cuticle form the worm’s first line of defense against pathogens or mechanical damage by environmental insults. A wide variety of pathogenic bacteria, fungi, or protozoa can attack C. elegans or related nematodes via the epidermis. Many fungal pathogens of free-living and plant-parasitic nematodes (‘nematophagous’ fungi) enter via the cuticle/epidermis and have evolved specialized structures that adhere to or penetrate nematode cuticles (FIGURE 5A). The response to cuticle-penetrating pathogens is closely related to the response to physical wounding (FIGURE 5B). Although wound healing has only recently been investigated in C. elegans, other nematodes such as Oncholaimus exhibit ‘traumatic insemination’ in which the male punctures the hermaphrodite cuticle in mating; observations of ‘wound plugs’ after mating support an endogenous wound healing capacity in nematodes 151. To survive in the wild, nematodes must actively and passively defend themselves against such kinds of potentially fatal damage. Other pathogens such as M. nematophilum or Yersinia do not penetrate the cuticle but establish a stable infection on the cuticle surface 92, 152; infection is somehow sensed by the nematode, triggering swelling of underlying epidermal cells (FIGURE 5C).
FIGURE 5. Epidermal responses to infection, wounding, and stress.
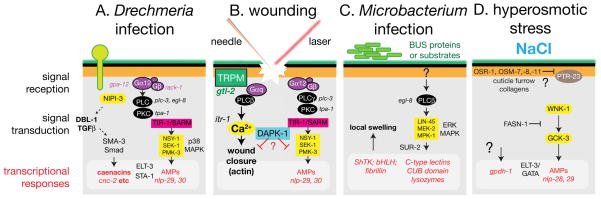
(A) Pathways implicated in responses to infection by Drechmeria coniospora. Drechmeria spores adhere to the cuticle and extend hyphae that penetrate the epidermis 209. A signal transduction pathway involving PKC, the SARM ortholog TIR-1, and the p38 MAPK cascade is involved in upregulation of transcription of the nlp-class AMPs 153, 155. A G-protein coupled receptor may be involved early in recognition of damage, as the same pathway is activated by sterile wounding; conversely, the Tribbles-like kinase is required for the response to infection but not to wounding. The GATA factor ELT-3 appears to be permissive for nlp upregulation 154. Drechmeria infection also triggers transcription of caenacin AMPs; cnc induction is partly independent of the p38 MAPK cascade and involves a non-cell-autonomous TGFβ signal, possibly from neurons 157. (B) Epidermal wounding by microinjection needle or laser irradiation triggers expression of the NLP-type AMPs, via a pathway similar to that involved in Drechmeria responses, but does not induce the cnc genes 159. A Gαq-PLCβ-Ca2+ signalling pαathway is involved in wound closure after puncture wounding 158; DAPK-1 can negatively regulate both the innate immune response pathway and the wound closure pathway. (C) Infection by the bacterium M. nematophilum induces swelling of rectal epidermal cells. Genes required for bacterial adhesion are likely involved in biogenesis of the surface coat and/or epicuticle. M. nematophilum infection triggers expression of a variety of genes, some of which are involved in the epidermal swelling response 210. Swelling requires phospholipase C β (EGL-8) 148 and the ERK MAPK cascade 211; it is not known if all transcriptional responses are ERK-dependent. (D) Osmotic stress may be sensed by structures at cuticle furrows coupled to epidermal cell surface receptors. Epidermal responses to hyperosmotic shock include elevated transcription of gpdh-1 (leading to glycerol accumulation) and expression of a subset of AMPs 165.
The response of the epidermis to infection by the cuticle-penetrating fungus Drechmeria coniospora has been extensively studied as a model for host-pathogen interaction and cutaneous innate immunity. Infection of C. elegans by Drechmeria triggers transcriptional upregulation in the epidermis of numerous small peptides, initially annotated as neuropeptide-like (nlp) but now known to have antimicrobial activity 153, 154. The pathways transducing the innate immune response to infection have been analyzed in detail. Signal transduction in the epidermis involves G protein signaling 155, the Toll-interleukin like adaptor TIR-1, a p38 MAPK cascade and the transcription factors ELT-3 and STA-1 156. A partly independent infection response involves the DBL-1 pathway, which triggers epidermal expression of caenacin family peptides 157. The fate of epidermally synthesized AMPs remains unclear; they may be delivered to the cuticle by one of the secretory mechanisms described above.
Wounding of the epidermis, either by a microinjection needle or by laser irradiation, triggers responses that collectively allow the animal to survive and heal damage. Immediate responses to wounding include rapid and sustained elevation in epidermal Ca2+ 158, membrane sealing, and formation of an autofluorescent scab 159. Wounding also induces transcription of AMPs such as nlp-29. In general the peptides induced by wounding are a subset of those induced by infection, suggesting the response to infection includes a core response to non-specific damage. Other responses are infection-specific and require the Tribbles-like kinase NIPI-3 159. The pathways mediating AMP induction after infection and wounding are similar but not identical, indicating that the epidermis somehow discriminates between sterile wounding and pathogen infection. Loss of function in the C. elegans ortholog of death associated protein kinase DAPK-1 results in constitutive upregulation of epidermal AMPs and cuticle thickening, suggesting DAPK-1 negatively regulates wound responses 160.
Epidermal innate immune responses to wounding appear to be independent of the Ca2+ response to wounding, which functions specifically in wound closure 158. Epidermal Ca2+ dynamics involve putative epidermal Ca2+ channels such as GTL-2, and ITR-1-dependent release from internal stores. A G protein pathway involving Gαq/EGL-30 and PLCβ/EGL-8 is required for the epidermal Ca2+ response and may act in a common pathway with GTL-2. Epidermal Ca2+ signals promote wound closure by triggering formation of an actin ring at wound sites; this actin ring closes over a period of 1–2 h in the wild type. Several actin regulators and polymerization factors are required for proper formation of the actin ring. However, loss of function in non-muscle myosins results in faster ring closure, suggesting that actomyosin contractility may retard closure. Instead, wound closure may be driven by local polymerization of filopodia-like structures. Loss of DAPK-1 also causes faster wound closure, suggesting that DAPK-1 acts as a coordinate negative regulator of wound responses. Aspects of the dapk-1 epidermal phenotype are suppressed by loss of function in the mRNA polyadenylation regulator sydn-1 161. It will be illuminating to explore how DAPK-1 is regulated and how it limits wound responses.
Physiological functions of the differentiated epidermis
Resistance to osmotic shock and other stresses
The mature epidermis plays a variety of roles in the animal, including osmoregulation and ionic homeostasis, metabolic storage, and mechanosensory function. Hyperosmotic shock activates epidermal transcription of GPDH-1, elevating glycerol levels (FIGURE 5D) 162. However high glycerol expression is not sufficient for osmotic resistance, suggesting the existence of additional pathways. The kinases GCK-3 and WNK-1 act in the epidermis to control fluid uptake in recovery from hypertonic stress 163. Loss of function in the epidermally expressed chloride channel clh-1 results in increased epidermal size, a phenotype reversed by incubation in hyperosmotic media 164. Epidermal responses to osmotic stress overlap with transcriptional responses to infection 165. However the responses to pathogens and osmotic stress are not identical, indicating that the epidermis can distinguish between stresses.
How does the epidermis sense osmotic stress? Initial genetic screens for mutants resistant to osmotic stress (Osr) identified the nematode-specific gene OSR-1 166. Subsequent screens identified an unrelated family of genes that includes OSM-7 and OSM-11 167. Both OSR-1 and OSM-7/11 are expressed in the epidermis and contain N-terminal secretion signals, hinting at a role for the cuticle in osmotic resistance. Interestingly, a subset of cuticle mutants display resistance to osmotic stress, including animals lacking the cuticle collagens DPY-2/7/10 166 and the mucin OSM-8 168. As DPY-2/-7/-10 are localized to cuticular furrows between annuli, osmotic stress might be sensed via decreased tension between cuticle furrows and the epidermal plasma membrane. The epidermal patched-related transmembrane protein PTR-23 is required for the upregulation of osmotic responses in osm-8 mutants and could be involved in transduction of a cuticle tension signal 168; another epidermal patched family member PTC-3 has also been implicated in osmosensation 169.
Most of the above Osr mutants display high basal levels of glycerol, contributing to their ability to withstand stress 167. The NSY-1/SEK-1/p38 cascade is not required for acute osmotic stress resistance, but is required for the chronic Osr phenotype of osr-1 mutants. In contrast, the NSY-1 cascade is not required for the chronic Osr phenotype of osm-7 mutants. Thus, the mechanisms of osmotic stress resistance may differ between these mutants. Although glycerol accumulation may be sufficient for chronic osmotic stress resistance, additional responses may be involved in acute stress resistance.
In addition to these roles in osmotic stress resistance, the epidermis has a critical role in resistance to environmental toxins. For example, the MEK-1 MAP kinase cascade acts in the epidermis to confer resistance to heavy metal stress 170. Resistance to the toxic effects of H2S or HCN involves epidermal expression of oxidizing enzymes such as SQRD-1, presumably limiting exposure of more internal tissues 171. Finally, heme deficiency upregulates the expression of the HRG-2 transporter required for epidermal heme uptake 172, indicating the existence of a regulated pathway dedicated to epidermal heme homeostasis.
Lipid storage
While the intestine is the major adipose tissue of the worm, the epidermis is also a significant site of fat accumulation. The fat storage role of the epidermis may have been underestimated by the use of Nile red to detect fat, as Nile red does not efficiently stain the epidermis 173. Quantitative microscopy indicates that the epidermis accounts for ~10% of fat stores in the wild type and ~50% of the fat stores of dauer larvae 174. Indeed, L2 ‘predauer’ animals committed to enter the dauer stage can be recognized by the accumulation of fat in the epidermis 175. The epidermis is also the site of action of the AMP kinase (AMPK) pathway, which promotes the long-term survival of dauers by restraining lipid hydrolysis.
The epidermis, cell death, and cell damage
C. elegans lacks dedicated ‘professional’ phagocytic cells. Instead, cellular debris from dead cells is typically phagocytosed by adjacent tissue, in most cases the epidermis. Phagocytosis of apoptotic cell corpses by the epidermis can be seen during epidermal enclosure (e.g. Movie 7 in Chisholm and Hardin 2005), indicating the epidermis expresses the phagocytic machinery early in its development. In post-embryonic development of the nervous system, dying apoptotic cells are engulfed by adjacent epidermis 176. The epidermis also phagocytoses necrotic or degenerating neurons 177, 178. Epidermal phagocytosis may be responsible for the degeneration and clearance of axonal fragments generated after laser axotomy 179 or in axon fragility mutants 180, analogous to the clearance function of glia in other organisms 181.
Only a few programmed cell deaths occur in epidermal lineages in C. elegans, the exceptions including the epidermal-like tail spike cells 182. In contrast, ventral epidermal blast cells in P. pacificus undergo apoptosis unless prevented from doing so by expression of LIN-39 183. Pathological cell death of C. elegans ventral epidermal blast cells occurs in lin-24 and lin-33 mutants 184; such ‘cytotoxic’ deaths do not resemble apoptotic or necrotic deaths. The cytotoxic deaths of Pn.p cells in these mutants are independent of the CED-3 caspase but partly dependent on the phagocytic machinery. pvl-5 mutants display a distinct kind of CED-3-dependent non-apoptotic cell death 185. Epidermal cells may not express components of the apoptotic machinery and therefore (as in the case of the tail spike cells) rely on transcriptional activation of CED-3 to induce death.
The epidermis is also a major site of autophagy. Autophagosomes are prominent in the epidermis and as mentioned above, autophagy is important for the remodeling of the epidermis during dauer development. Decreased autophagy has been reported to result in decreased epidermal cell size and decreased body size 186. Autophagy and apoptosis may have redundant roles in epidermal morphogenesis 187.
The epidermis and neuronal function
Most nematode sensory neurons are closely associated with epidermis or cuticle, in a ways that reflect their sensory modality. Neurons sensing water-soluble chemicals such as amphid and phasmid neurons are exposed to the external environment via openings in the cuticle. Other sensory endings are embedded in specialized cuticle structures. In the case of the deirid, a putative mechanosensory neuron, the modified cuticle (‘substructure’) appears to be generated by the socket cell, as ectopic socket cells are sufficient to make ectopic substructures 188. The mechanisms by which sensilla or socket cells modify the cuticle remain unknown.
Other mechanosensory neurons do not contact the cuticle directly but are embedded within the epidermis (FIGURE 2D). The processes of ALM and PLM gradually become enveloped by the epidermis during larval development, and secrete a specialized ECM (the mantle) that attaches them to epidermal cells 189. Mechanosensory neurons recruit the ECM protein HIM-4, which then induces trans-epidermal attachments in the adjacent epidermis 190. Although HIM-4-dependent recruitment of trans-epidermal attachments is not essential for mechanosensation it might enhance sensory acuity under certain conditions. The PVD and FLP neurons form highly branched processes in close proximity to and in some cases partly embedded in the epidermis 191, 192. PVD and FLP neurons have nociceptive and proprioceptive roles that may depend on mechanical coupling to epidermal cells, but these cells do not recruit trans-epidermal attachments.
The dorsal and ventral nerve cords of nematodes are associated to various degrees with the adjacent epidermis. In C. elegans these axon bundles run alongside but are not enclosed by the epidermal ridges. At the other extreme, exemplified by Ascaris, nerve cords run almost entirely inside a channel termed the ‘hypodermal chalice’ 193, 194. Nematodes such as Trichuris display intermediate levels of association, with nerve cords partly embedded in epidermis 194. In C. elegans the epidermis frequently extends finger-like projections into adjacent axon bundles [cf. Figure 18 in 195]. It is unknown if these hypodermal protrusions play any role in neuronal function.
There is increasing evidence that the epidermis can affect neuronal function, either by controlling local ionic homeostasis adjacent to neurons or by providing other signals or trophic factors. Increased motor neuron excitability can be suppressed by loss of function in the epidermal TRPM channel GTL-2 196. As mentioned above, OSM-11 is secreted by epidermal seam cells and signals to neurons to regulate behavioral quiescence 134. Conversely, synapse-like structures between neurons and the epidermis have been noted in serial section EM reconstructions, suggesting neuronal activity could feed back on the epidermis. Interactions between the epidermis and nervous system are ripe for future investigation.
The aging epidermis
Early investigations into nematode aging focused on the epidermis and cuticle. Older nematodes, of various species, display progressive deterioration in cuticle structure and in barrier function 197, and possibly decreased surface coat thickness 198. The epidermis adjacent to muscles expands and bulges, suggesting the cumulative effects of muscle contraction result in a local loss of epidermal integrity 199. More recent studies have revealed major structural changes in the aging C. elegans epidermis 200, including subcellular deterioration, accumulation of lipid inclusions, and cuticle thickening. Older animals displayed ‘random but extreme local crises’ involving disruption of the plasma membrane, suggesting that a proximate cause of death of aged nematodes may be a decline in epidermal integrity leading to a loss of barrier function.
Genetic and genomic studies suggest the epidermis may actively regulate age-dependent changes. Age-related declines in oocyte quality in the germline are affected by an epidermally expressed TGFβ pathway 201, suggesting communication between the epidermis and the germline. Global age-dependent changes in gene expression are regulated by the epidermal GATA factors ELT-3,-5 and -6 202. ELT-5 and ELT-6 expression levels progressively increase with adult age, resulting in progressive repression of ELT-3. It is not yet known whether the increase in ELT-5/6 expression reflects ‘selective drift’ in post-reproductive stages, or whether accumulated external stresses to the epidermis (mechanical trauma, oxidative damage) might underlie this regulatory shift.
Conclusions and Future Directions
Analysis of the differentiated C. elegans epidermis has begun to yield insights into its physiological roles, particularly in innate immunity, wound healing, and aging. Still, the analysis of the biology of the differentiated C. elegans epidermis remains in its early stages. Although the epidermis is a large and accessible organ, many of its functions are essential, potentially complicating genetic analysis. However the wild-type epidermis is generally sensitive to RNAi, so large scale RNAi screens for epidermal defects should be feasible. Epidermal-specific RNAi provides added specificity 203 and has been used to address epidermal-specific roles of essential genes 158. Recent advances in transcriptional profiling of specific tissues 204 should stimulate more systems-level analyses of the molecular basis of epidermal biology.
The C. elegans cuticle retains an enduring fascination as an example of a self-assembling complex extracellular structure. The biomechanical properties of the cuticle may be of interest in bioengineering of impermeable layers. The epicuticle is a remarkable extracellular lipid bilayer, possibly analogous to extracellular lipid bilayers of the vertebrate stratum corneum, and deserves more attention from molecular genetics. Finally, the surface coat plays a critical role in a nematode’s interaction with its environment. A better understanding of these surface structures will inform efforts to control plant and animal parasitic nematodes.
Acknowledgments
We thank Yishi Jin and the anonymous reviewers for comments. Our work on C. elegans epidermal development and wound healing is supported by NIH R01 GM54657.
Footnotes
Cuticlins (non-collagenous proteins of the nematode cuticle) should not be confused with cuticulin, the outermost layer of the insect epicuticle. Nematode and insect epicuticle layers are both thought to be lipid-rich, are deposited early in synthesis of new cuticles, and likely have permeability barrier function. It is not known whether nematode and insect epicuticles share any other similarities in terms of biogenesis or molecular composition.
Contributor Information
Andrew D. Chisholm, Email: chisholm@ucsd.edu, Division of Biological Sciences, Section of Cell and Developmental Biology, University of California San Diego, La Jolla, CA 92093
Suhong Xu, Division of Biological Sciences, Section of Cell and Developmental Biology, University of California San Diego, La Jolla, CA 92093.
References
- 1.Labouesse M. Epithelial junctions and attachments. WormBook. 2006:1–21. doi: 10.1895/wormbook.1.56.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding M, Woo WM, Chisholm AD. The cytoskeleton and epidermal morphogenesis in C. elegans. Exp Cell Res. 2004;301:84–90. doi: 10.1016/j.yexcr.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Gally C, Labouesse M. Tissue morphogenesis: how multiple cells cooperate to generate a tissue. Curr Opin Cell Biol. 2010;22:575–582. doi: 10.1016/j.ceb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Grana TM, Cox EA, Lynch AM, Hardin J. SAX-7/L1CAM and HMR-1/cadherin function redundantly in blastomere compaction and non-muscle myosin accumulation during Caenorhabditis elegans gastrulation. Developmental biology. 2010;344:731–744. doi: 10.1016/j.ydbio.2010.05.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Legouis R, Gansmuller A, Sookhareea S, Bosher JM, Baillie DL, Labouesse M. LET-413 is a basolateral protein required for the assembly of adherens junctions in Caenorhabditis elegans. Nat Cell Biol. 2000;2:415–422. doi: 10.1038/35017046. [DOI] [PubMed] [Google Scholar]
- 6.Totong R, Achilleos A, Nance J. PAR-6 is required for junction formation but not apicobasal polarization in C. elegans embryonic epithelial cells. Development. 2007;134:1259–1268. doi: 10.1242/dev.02833. [DOI] [PubMed] [Google Scholar]
- 7.Achilleos A, Wehman AM, Nance J. PAR-3 mediates the initial clustering and apical localization of junction and polarity proteins during C. elegans intestinal epithelial cell polarization. Development. 2010;137:1833–1842. doi: 10.1242/dev.047647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang CC, Hall DH, Hedgecock EM, Kao G, Karantza V, Vogel BE, Hutter H, Chisholm AD, Yurchenco PD, Wadsworth WG. Laminin alpha subunits and their role in C. elegans development. Development. 2003;130:3343–3358. doi: 10.1242/dev.00481. [DOI] [PubMed] [Google Scholar]
- 9.Koppen M, Simske JS, Sims PA, Firestein BL, Hall DH, Radice AD, Rongo C, Hardin JD. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat Cell Biol. 2001;3:983–991. doi: 10.1038/ncb1101-983. [DOI] [PubMed] [Google Scholar]
- 10.Firestein BL, Rongo C. DLG-1 is a MAGUK similar to SAP97 and is required for adherens junction formation. Mol Biol Cell. 2001;12:3465–3475. doi: 10.1091/mbc.12.11.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raich WB, Agbunag C, Hardin J. Rapid epithelial-sheet sealing in the Caenorhabditis elegans embryo requires cadherin-dependent filopodial priming. Curr Biol. 1999;9:1139–1146. doi: 10.1016/S0960-9822(00)80015-9. [DOI] [PubMed] [Google Scholar]
- 12.Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pettitt J, Cox EA, Broadbent ID, Flett A, Hardin J. The Caenorhabditis elegans p120 catenin homologue, JAC-1, modulates cadherin-catenin function during epidermal morphogenesis. J Cell Biol. 2003;162:15–22. doi: 10.1083/jcb.200212136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simske JS, Koppen M, Sims P, Hodgkin J, Yonkof A, Hardin J. The cell junction protein VAB-9 regulates adhesion and epidermal morphology in C. elegans. Nat Cell Biol. 2003;5:619–625. doi: 10.1038/ncb1002. [DOI] [PubMed] [Google Scholar]
- 15.Gattegno T, Mittal A, Valansi C, Nguyen KC, Hall DH, Chernomordik LV, Podbilewicz B. Genetic control of fusion pore expansion in the epidermis of Caenorhabditis elegans. Mol Biol Cell. 2007;18:1153–1166. doi: 10.1091/mbc.E06-09-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Priess JR, Hirsh DI. Caenorhabditis elegans morphogenesis: the role of the cytoskeleton in elongation of the embryo. Dev Biol. 1986;117:156–173. doi: 10.1016/0012-1606(86)90358-1. [DOI] [PubMed] [Google Scholar]
- 17.Costa M, Draper BW, Priess JR. The role of actin filaments in patterning the Caenorhabditis elegans cuticle. Dev Biol. 1997;184:373–384. doi: 10.1006/dbio.1997.8530. [DOI] [PubMed] [Google Scholar]
- 18.Norman KR, Moerman DG. Alpha spectrin is essential for morphogenesis and body wall muscle formation in Caenorhabditis elegans. J Cell Biol. 2002;157:665–677. doi: 10.1083/jcb.200111051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKeown C, Praitis V, Austin J. sma-1 encodes a βH-spectrin homolog required for Caenorhabditis elegans morphogenesis. Development. 1998;125:2087–2098. doi: 10.1242/dev.125.11.2087. [DOI] [PubMed] [Google Scholar]
- 20.Praitis V, Ciccone E, Austin J. SMA-1 spectrin has essential roles in epithelial cell sheet morphogenesis in C. elegans. Dev Biol. 2005;283:157–170. doi: 10.1016/j.ydbio.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- 22.Williams-Masson EM, Heid PJ, Lavin CA, Hardin J. The cellular mechanism of epithelial rearrangement during morphogenesis of the Caenorhabditis elegans dorsal hypodermis. Dev Biol. 1998;204:263–276. doi: 10.1006/dbio.1998.9048. [DOI] [PubMed] [Google Scholar]
- 23.Fridolfsson HN, Starr DA. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol. 2010;191:115–128. doi: 10.1083/jcb.201004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francis R, Waterston RH. Muscle cell attachment in Caenorhabditis elegans. J Cell Biol. 1991;114:465–479. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plenefisch JD, Zhu X, Hedgecock EM. Fragile skeletal muscle attachments in dystrophic mutants of Caenorhabditis elegans: isolation and characterization of the mua genes. Development. 2000;127:1197–1207. doi: 10.1242/dev.127.6.1197. [DOI] [PubMed] [Google Scholar]
- 26.Hresko MC, Williams BD, Waterston RH. Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J Cell Biol. 1994;124:491–506. doi: 10.1083/jcb.124.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosher JM, Hahn BS, Legouis R, Sookhareea S, Weimer RM, Gansmuller A, Chisholm AD, Rose AM, Bessereau JL, Labouesse M. The Caenorhabditis elegans vab-10 spectraplakin isoforms protect the epidermis against internal and external forces. J Cell Biol. 2003;161:757–768. doi: 10.1083/jcb.200302151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karabinos A, Schmidt H, Harborth J, Schnabel R, Weber K. Essential roles for four cytoplasmic intermediate filament proteins in Caenorhabditis elegans development. Proc Natl Acad Sci U S A. 2001;98:7863–7868. doi: 10.1073/pnas.121169998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hapiak V, Hresko MC, Schriefer LA, Saiyasisongkhram K, Bercher M, Plenefisch J. mua-6, a gene required for tissue integrity in Caenorhabditis elegans, encodes a cytoplasmic intermediate filament. Dev Biol. 2003;263:330–342. doi: 10.1016/j.ydbio.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Woo WM, Goncharov A, Jin Y, Chisholm AD. Intermediate filaments are required for C. elegans epidermal elongation. Dev Biol. 2004;267:216–229. doi: 10.1016/j.ydbio.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Kaminsky R, Denison C, Bening-Abu-Shach U, Chisholm AD, Gygi SP, Broday L. SUMO regulates the assembly and function of a cytoplasmic intermediate filament protein in C. elegans. Dev Cell. 2009;17:724–735. doi: 10.1016/j.devcel.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding M, Goncharov A, Jin Y, Chisholm AD. C. elegans ankyrin repeat protein VAB-19 is a component of epidermal attachment structures and is essential for epidermal morphogenesis. Development. 2003;130:5791–5801. doi: 10.1242/dev.00791. [DOI] [PubMed] [Google Scholar]
- 33.Ding M, King RS, Berry EC, Wang Y, Hardin J, Chisholm AD. The cell signaling adaptor protein EPS-8 is essential for C. elegans epidermal elongation and interacts with the ankyrin repeat protein VAB-19. PLoS One. 2008;3:e3346. doi: 10.1371/journal.pone.0003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hetherington S, Gally C, Fritz JA, Polanowska J, Reboul J, Schwab Y, Zahreddine H, Behm C, Labouesse M. PAT-12, a potential anti-nematode target, is a new spectraplakin partner essential for Caenorhabditis elegans hemidesmosome integrity and embryonic morphogenesis. Dev Biol. 2011;350:267–278. doi: 10.1016/j.ydbio.2010.11.025. [DOI] [PubMed] [Google Scholar]
- 35.Hresko MC, Schriefer LA, Shrimankar P, Waterston RH. Myotactin, a novel hypodermal protein involved in muscle-cell adhesion in Caenorhabditis elegans. J Cell Biol. 1999;146:659–672. doi: 10.1083/jcb.146.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bercher M, Wahl J, Vogel BE, Lu C, Hedgecock EM, Hall DH, Plenefisch JD. mua-3, a gene required for mechanical tissue integrity in Caenorhabditis elegans, encodes a novel transmembrane protein of epithelial attachment complexes. J Cell Biol. 2001;154:415–426. doi: 10.1083/jcb.200103035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong L, Elbl T, Ward J, Franzini-Armstrong C, Rybicka KK, Gatewood BK, Baillie DL, Bucher EA. MUP-4 is a novel transmembrane protein with functions in epithelial cell adhesion in Caenorhabditis elegans. J Cell Biol. 2001;154:403–414. doi: 10.1083/jcb.200007075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zahreddine H, Zhang H, Diogon M, Nagamatsu Y, Labouesse M. CRT-1/calreticulin and the E3 ligase EEL-1/HUWE1 control hemidesmosome maturation in C. elegans development. Curr Biol. 2010;20:322–327. doi: 10.1016/j.cub.2009.12.061. [DOI] [PubMed] [Google Scholar]
- 39.Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M. A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature. 2011;471:99–103. doi: 10.1038/nature09765. [DOI] [PubMed] [Google Scholar]
- 41.Graham PL, Johnson JJ, Wang S, Sibley MH, Gupta MC, Kramer JM. Type IV collagen is detectable in most, but not all, basement membranes of Caenorhabditis elegans and assembles on tissues that do not express it. J Cell Biol. 1997;137:1171–1183. doi: 10.1083/jcb.137.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woo WM, Berry EC, Hudson ML, Swale RE, Goncharov A, Chisholm AD. The C. elegans F-spondin family protein SPON-1 maintains cell adhesion in neural and non-neural tissues. Development. 2008;135:2747–2756. doi: 10.1242/dev.015289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson RP, Kang SH, Kramer JM. C. elegans dystroglycan DGN-1 functions in epithelia and neurons, but not muscle, and independently of dystrophin. Development. 2006;133:1911–1921. doi: 10.1242/dev.02363. [DOI] [PubMed] [Google Scholar]
- 44.Gotenstein JR, Swale RE, Fukuda T, Wu Z, Giurumescu CA, Goncharov A, Jin Y, Chisholm AD. The C. elegans peroxidasin PXN-2 is essential for embryonic morphogenesis and inhibits adult axon regeneration. Development. 2010;137:3603–3613. doi: 10.1242/dev.049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muriel JM, Dong C, Hutter H, Vogel BE. Fibulin-1C and Fibulin-1D splice variants have distinct functions and assemble in a hemicentin-dependent manner. Development. 2005;132:4223–4234. doi: 10.1242/dev.02007. [DOI] [PubMed] [Google Scholar]
- 46.Spike CA, Davies AG, Shaw JE, Herman RK. MEC-8 regulates alternative splicing of unc-52 transcripts in C. elegans hypodermal cells. Development. 2002;129:4999–5008. doi: 10.1242/dev.129.21.4999. [DOI] [PubMed] [Google Scholar]
- 47.Davies AG, Spike CA, Shaw JE, Herman RK. Functional overlap between the mec-8 gene and five sym genes in Caenorhabditis elegans. Genetics. 1999;153:117–134. doi: 10.1093/genetics/153.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yochem J, Bell LR, Herman RK. The identities of sym-2, sym-3 and sym-4, three genes that are synthetically lethal with mec-8 in Caenorhabditis elegans. Genetics. 2004;168:1293–1306. doi: 10.1534/genetics.104.029827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hedgecock EM, Culotti JG, Hall DH, Stern BD. Genetics of cell and axon migrations in Caenorhabditis elegans. Development. 1987;100:365–382. doi: 10.1242/dev.100.3.365. [DOI] [PubMed] [Google Scholar]
- 50.Knobel KM, Jorgensen EM, Bastiani MJ. Growth cones stall and collapse during axon outgrowth in Caenorhabditis elegans. Development. 1999;126:4489–4498. doi: 10.1242/dev.126.20.4489. [DOI] [PubMed] [Google Scholar]
- 51.White JG, Southgate E, Thomson JN, Brenner S. The structure of the ventral nerve cord of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:327–348. doi: 10.1098/rstb.1976.0086. [DOI] [PubMed] [Google Scholar]
- 52.Hobert O, Bulow H. Development and maintenance of neuronal architecture at the ventral midline of C. elegans. Curr Opin Neurobiol. 2003;13:70–78. doi: 10.1016/s0959-4388(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 53.Shibata Y, Fujii T, Dent JA, Fujisawa H, Takagi S. EAT-20, a novel transmembrane protein with EGF motifs, is required for efficient feeding in Caenorhabditis elegans. Genetics. 2000;154:635–646. doi: 10.1093/genetics/154.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bergmann DC, Crew JR, Kramer JM, Wood WB. Cuticle chirality and body handedness in Caenorhabditis elegans. Dev Genet. 1998;23:164–174. doi: 10.1002/(SICI)1520-6408(1998)23:3<164::AID-DVG2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 55.Bulow HE, Boulin T, Hobert O. Differential functions of the C. elegans FGF receptor in axon outgrowth and maintenance of axon position. Neuron. 2004;42:367–374. doi: 10.1016/s0896-6273(04)00246-6. [DOI] [PubMed] [Google Scholar]
- 56.Bulow HE, Hobert O. Differential sulfations and epimerization define heparan sulfate specificity in nervous system development. Neuron. 2004;41:723–736. doi: 10.1016/s0896-6273(04)00084-4. [DOI] [PubMed] [Google Scholar]
- 57.Hedgecock EM, Culotti JG, Hall DH. The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron. 1990;4:61–85. doi: 10.1016/0896-6273(90)90444-k. [DOI] [PubMed] [Google Scholar]
- 58.Wadsworth WG, Bhatt H, Hedgecock EM. Neuroglia and pioneer neurons express UNC-6 to provide global and local netrin cues for guiding migrations in C. elegans. Neuron. 1996;16:35–46. doi: 10.1016/s0896-6273(00)80021-5. [DOI] [PubMed] [Google Scholar]
- 59.Huang X, Huang P, Robinson MK, Stern MJ, Jin Y. UNC-71, a disintegrin and metalloprotease (ADAM) protein, regulates motor axon guidance and sex myoblast migration in C. elegans. Development. 2003;130:3147–3161. doi: 10.1242/dev.00518. [DOI] [PubMed] [Google Scholar]
- 60.Colavita A, Tessier-Lavigne M. A Neurexin-related protein, BAM-2, terminates axonal branches in C. elegans. Science. 2003;302:293–296. doi: 10.1126/science.1089163. [DOI] [PubMed] [Google Scholar]
- 61.Habibi-Babadi N, Su A, de Carvalho CE, Colavita A. The N-glycanase png-1 acts to limit axon branching during organ formation in Caenorhabditis elegans. J Neurosci. 2010;30:1766–1776. doi: 10.1523/JNEUROSCI.4962-08.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen K, Fetter RD, Bargmann CI. Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG-1. Cell. 2004;116:869–881. doi: 10.1016/s0092-8674(04)00251-x. [DOI] [PubMed] [Google Scholar]
- 63.Hagedorn EJ, Sherwood DR. Cell invasion through basement membrane: the anchor cell breaches the barrier. Curr Opin Cell Biol. 2011 doi: 10.1016/j.ceb.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ihara S, Hagedorn EJ, Morrissey MA, Chi Q, Motegi F, Kramer JM, Sherwood DR. Basement membrane sliding and targeted adhesion remodels tissue boundaries during uterine-vulval attachment in Caenorhabditis elegans. Nat Cell Biol. 2011;13:641–651. doi: 10.1038/ncb2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whangbo J, Harris J, Kenyon C. Multiple levels of regulation specify the polarity of an asymmetric cell division in C. elegans. Development. 2000;127:4587–4598. doi: 10.1242/dev.127.21.4587. [DOI] [PubMed] [Google Scholar]
- 66.Gleason JE, Eisenmann DM. Wnt signaling controls the stem cell-like asymmetric division of the epithelial seam cells during C. elegans larval development. Dev Biol. 2010;348:58–66. doi: 10.1016/j.ydbio.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pan CL, Howell JE, Clark SG, Hilliard M, Cordes S, Bargmann CI, Garriga G. Multiple Wnts and frizzled receptors regulate anteriorly directed cell and growth cone migrations in Caenorhabditis elegans. Dev Cell. 2006;10:367–377. doi: 10.1016/j.devcel.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 68.Hilliard MA, Bargmann CI. Wnt signals and frizzled activity orient anterior-posterior axon outgrowth in C. elegans. Dev Cell. 2006;10:379–390. doi: 10.1016/j.devcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 69.Whangbo J, Kenyon C. A Wnt signaling system that specifies two patterns of cell migration in C. elegans. Mol Cell. 1999;4:851–858. doi: 10.1016/s1097-2765(00)80394-9. [DOI] [PubMed] [Google Scholar]
- 70.Herman MA, Vassilieva LL, Horvitz HR, Shaw JE, Herman RK. The C. elegans gene lin-44, which controls the polarity of certain asymmetric cell divisions, encodes a Wnt protein and acts cell nonautonomously. Cell. 1995;83:101–110. doi: 10.1016/0092-8674(95)90238-4. [DOI] [PubMed] [Google Scholar]
- 71.Page AP, Johnstone IL. The cuticle. WormBook. 2007:1–15. doi: 10.1895/wormbook.1.138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujimoto D, Kanaya S. Cuticlin: a noncollagen structural protein from Ascaris cuticle. Arch Biochem Biophys. 1973;157:1–6. doi: 10.1016/0003-9861(73)90382-2. [DOI] [PubMed] [Google Scholar]
- 73.Sapio MR, Hilliard MA, Cermola M, Favre R, Bazzicalupo P. The Zona Pellucida domain containing proteins, CUT-1, CUT-3 and CUT-5, play essential roles in the development of the larval alae in Caenorhabditis elegans. Dev Biol. 2005;282:231–245. doi: 10.1016/j.ydbio.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 74.Sebastiano M, Lassandro F, Bazzicalupo P. cut-1, a Caenorhabditis elegans gene coding for a dauer-specific noncollagenous component of the cuticle. Dev Biol. 1991;146:519–530. doi: 10.1016/0012-1606(91)90253-y. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Y, Foster JM, Nelson LS, Ma D, Carlow CK. The chitin synthase genes chs-1 and chs-2 are essential for C. elegans development and responsible for chitin deposition in the eggshell and pharynx, respectively. Developmental biology. 2005;285:330–339. doi: 10.1016/j.ydbio.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 76.Edens WA, Sharling L, Cheng G, Shapira R, Kinkade JM, Lee T, Edens HA, Tang X, Sullards C, Flaherty DB, et al. Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91phox. J Cell Biol. 2001;154:879–891. doi: 10.1083/jcb.200103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thein MC, Winter AD, Stepek G, McCormack G, Stapleton G, Johnstone IL, Page AP. Combined extracellular matrix cross-linking activity of the peroxidase MLT-7 and the dual oxidase BLI-3 is critical for post-embryonic viability in Caenorhabditis elegans. J Biol Chem. 2009;284:17549–17563. doi: 10.1074/jbc.M900831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johnstone IL. Cuticle collagen genes. Expression in Caenorhabditis elegans. Trends Genet. 2000;16:21–27. doi: 10.1016/s0168-9525(99)01857-0. [DOI] [PubMed] [Google Scholar]
- 79.McMahon L, Muriel JM, Roberts B, Quinn M, Johnstone IL. Two sets of interacting collagens form functionally distinct substructures within a Caenorhabditis elegans extracellular matrix. Mol Biol Cell. 2003;14:1366–1378. doi: 10.1091/mbc.E02-08-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gilleard JS, McGhee JD. Activation of hypodermal differentiation in the Caenorhabditis elegans embryo by GATA transcription factors ELT-1 and ELT-3. Mol Cell Biol. 2001;21:2533–2544. doi: 10.1128/MCB.21.7.2533-2544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kouns NA, Nakielna J, Behensky F, Krause MW, Kostrouch Z, Kostrouchova M. NHR-23 dependent collagen and hedgehog-related genes required for molting. Biochem Biophys Res Commun. 2011;413:515–520. doi: 10.1016/j.bbrc.2011.08.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gissendanner CR, Crossgrove K, Kraus KA, Maina CV, Sluder AE. Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev Biol. 2004;266:399–416. doi: 10.1016/j.ydbio.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 83.Cai L, Phong BL, Fisher AL, Wang Z. Regulation of fertility, survival, and cuticle collagen function by the Caenorhabditis elegans eaf-1 and ell-1 genes. J Biol Chem. 2011;286:35915–35921. doi: 10.1074/jbc.M111.270454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu Z, Kirch S, Ambros V. The Caenorhabditis elegans heterochronic gene pathway controls stage-specific transcription of collagen genes. Development. 1995;121:2471–2478. doi: 10.1242/dev.121.8.2471. [DOI] [PubMed] [Google Scholar]
- 85.Rougvie AE, Ambros V. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development. 1995;121:2491–2500. doi: 10.1242/dev.121.8.2491. [DOI] [PubMed] [Google Scholar]
- 86.Blaxter M, Bird D. Parasitic Nematodes. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 87.Peixoto CA, De Souza W. Freeze-fracture and deep-etched view of the cuticle of Caenorhabditis elegans. Tissue Cell. 1995;27:561–568. doi: 10.1016/s0040-8166(05)80065-5. [DOI] [PubMed] [Google Scholar]
- 88.Grenache DG, Caldicott I, Albert PS, Riddle DL, Politz SM. Environmental induction and genetic control of surface antigen switching in the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1996;93:12388–12393. doi: 10.1073/pnas.93.22.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Politz SM, Philipp M, Estevez M, O’Brien PJ, Chin KJ. Genes that can be mutated to unmask hidden antigenic determinants in the cuticle of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1990;87:2901–2905. doi: 10.1073/pnas.87.8.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Link CD, Silverman MA, Breen M, Watt KE, Dames SA. Characterization of Caenorhabditis elegans lectin-binding mutants. Genetics. 1992;131:867–881. doi: 10.1093/genetics/131.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gravato-Nobre MJ, Nicholas HR, Nijland R, O’Rourke D, Whittington DE, Yook KJ, Hodgkin J. Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 2005;171:1033–1045. doi: 10.1534/genetics.105.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Darby C, Chakraborti A, Politz SM, Daniels CC, Tan L, Drace K. Caenorhabditis elegans mutants resistant to attachment of Yersinia biofilms. Genetics. 2007;176:221–230. doi: 10.1534/genetics.106.067496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Drace K, McLaughlin S, Darby C. Caenorhabditis elegans BAH-1 is a DUF23 protein expressed in seam cells and required for microbial biofilm binding to the cuticle. PLoS One. 2009;4:e6741. doi: 10.1371/journal.pone.0006741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gravato-Nobre MJ, Stroud D, O’Rourke D, Darby C, Hodgkin J. Glycosylation genes expressed in seam cells determine complex surface properties and bacterial adhesion to the cuticle of Caenorhabditis elegans. Genetics. 2011;187:141–155. doi: 10.1534/genetics.110.122002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Plenefisch J, Xiao H, Mei B, Geng J, Komuniecki PR, Komuniecki R. Secretion of a novel class of iFABPs in nematodes: coordinate use of the Ascaris/Caenorhabditis model systems. Mol Biochem Parasitol. 2000;105:223–236. doi: 10.1016/s0166-6851(99)00179-6. [DOI] [PubMed] [Google Scholar]
- 96.Mancuso VP, Parry JM, Storer L, Poggioli C, Nguyen KC, Hall DH, Sundaram MV. Extracellular leucine-rich repeat proteins are required to organize the apical extracellular matrix and maintain epithelial junction integrity in C. elegans. Development. 2012 doi: 10.1242/dev.075135. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roberts B, Clucas C, Johnstone IL. Loss of SEC-23 in Caenorhabditis elegans causes defects in oogenesis, morphogenesis, and extracellular matrix secretion. Mol Biol Cell. 2003;14:4414–4426. doi: 10.1091/mbc.E03-03-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.White JG. The Anatomy. In: Wood WB, editor. The nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 81–122. [Google Scholar]
- 99.Liegeois S, Benedetto A, Garnier JM, Schwab Y, Labouesse M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J Cell Biol. 2006;173:949–961. doi: 10.1083/jcb.200511072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liegeois S, Benedetto A, Michaux G, Belliard G, Labouesse M. Genes required for osmoregulation and apical secretion in Caenorhabditis elegans. Genetics. 2007;175:709–724. doi: 10.1534/genetics.106.066035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kolotuev I, Apaydin A, Labouesse M. Secretion of Hedgehog-related peptides and WNT during Caenorhabditis elegans development. Traffic. 2009;10:803–810. doi: 10.1111/j.1600-0854.2008.00871.x. [DOI] [PubMed] [Google Scholar]
- 102.Frand AR, Russel S, Ruvkun G. Functional genomic analysis of C. elegans molting. PLoS Biol. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Magner DB, Antebi A. Caenorhabditis elegans nuclear receptors: insights into life traits. Trends Endocrinol Metab. 2008;19:153–160. doi: 10.1016/j.tem.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kurzchalia TV, Ward S. Why do worms need cholesterol? Nat Cell Biol. 2003;5:684–688. doi: 10.1038/ncb0803-684. [DOI] [PubMed] [Google Scholar]
- 105.Yochem J, Tuck S, Greenwald I, Han M. A gp330/megalin-related protein is required in the major epidermis of Caenorhabditis elegans for completion of molting. Development. 1999;126:597–606. doi: 10.1242/dev.126.3.597. [DOI] [PubMed] [Google Scholar]
- 106.Grigorenko AP, Moliaka YK, Soto MC, Mello CC, Rogaev EI. The Caenorhabditis elegans IMPAS gene, imp-2, is essential for development and is functionally distinct from related presenilins. Proc Natl Acad Sci U S A. 2004;101:14955–14960. doi: 10.1073/pnas.0406462101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roudier N, Lefebvre C, Legouis R. CeVPS-27 is an endosomal protein required for the molting and the endocytic trafficking of the low-density lipoprotein receptor-related protein 1 in Caenorhabditis elegans. Traffic. 2005;6:695–705. doi: 10.1111/j.1600-0854.2005.00309.x. [DOI] [PubMed] [Google Scholar]
- 108.Ruaud AF, Bessereau JL. Activation of nicotinic receptors uncouples a developmental timer from the molting timer in C. elegans. Development. 2006;133:2211–2222. doi: 10.1242/dev.02392. [DOI] [PubMed] [Google Scholar]
- 109.Ruaud AF, Bessereau JL. The P-type ATPase CATP-1 is a novel regulator of C. elegans developmental timing that acts independently of its predicted pump function. Development. 2007;134:867–879. doi: 10.1242/dev.02790. [DOI] [PubMed] [Google Scholar]
- 110.Stenvall J, Fierro-Gonzalez JC, Swoboda P, Saamarthy K, Cheng Q, Cacho-Valadez B, Arner ES, Persson OP, Miranda-Vizuete A, Tuck S. Selenoprotein TRXR-1 and GSR-1 are essential for removal of old cuticle during molting in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2011;108:1064–1069. doi: 10.1073/pnas.1006328108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Craig H, Isaac RE, Brooks DR. Unravelling the moulting degradome: new opportunities for chemotherapy? Trends Parasitol. 2007;23:248–253. doi: 10.1016/j.pt.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 112.Hashmi S, Zhang J, Oksov Y, Lustigman S. The Caenorhabditis elegans cathepsin Z-like cysteine protease, Ce-CPZ-1, has a multifunctional role during the worms’ development. J Biol Chem. 2004;279:6035–6045. doi: 10.1074/jbc.M312346200. [DOI] [PubMed] [Google Scholar]
- 113.Page AP, McCormack G, Birnie AJ. Biosynthesis and enzymology of the Caenorhabditis elegans cuticle: identification and characterization of a novel serine protease inhibitor. Int J Parasitol. 2006;36:681–689. doi: 10.1016/j.ijpara.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 114.Davis MW, Birnie AJ, Chan AC, Page AP, Jorgensen EM. A conserved metalloprotease mediates ecdysis in Caenorhabditis elegans. Development. 2004;131:6001–6008. doi: 10.1242/dev.01454. [DOI] [PubMed] [Google Scholar]
- 115.Stepek G, McCormack G, Birnie AJ, Page AP. The astacin metalloprotease moulting enzyme NAS-36 is required for normal cuticle ecdysis in free-living and parasitic nematodes. Parasitology. 2011;138:237–248. doi: 10.1017/S0031182010001113. [DOI] [PubMed] [Google Scholar]
- 116.Meli VS, Osuna B, Ruvkun G, Frand AR. MLT-10 defines a family of DUF644 and proline-rich repeat proteins involved in the molting cycle of Caenorhabditis elegans. Mol Biol Cell. 2010;21:1648–1661. doi: 10.1091/mbc.E08-07-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aspock G, Kagoshima H, Niklaus G, Burglin TR. Caenorhabditis elegans has scores of hedgehog-related genes: sequence and expression analysis. Genome Res. 1999;9:909–923. doi: 10.1101/gr.9.10.909. [DOI] [PubMed] [Google Scholar]
- 118.Hao L, Johnsen R, Lauter G, Baillie D, Burglin TR. Comprehensive analysis of gene expression patterns of hedgehog-related genes. BMC Genomics. 2006;7:280. doi: 10.1186/1471-2164-7-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hao L, Mukherjee K, Liegeois S, Baillie D, Labouesse M, Burglin TR. The hedgehog-related gene qua-1 is required for molting in Caenorhabditis elegans. Dev Dyn. 2006;235:1469–1481. doi: 10.1002/dvdy.20721. [DOI] [PubMed] [Google Scholar]
- 120.Dejima K, Murata D, Mizuguchi S, Nomura KH, Izumikawa T, Kitagawa H, Gengyo-Ando K, Yoshina S, Ichimiya T, Nishihara S, et al. Two Golgi-resident 3′-Phosphoadenosine 5′-phosphosulfate transporters play distinct roles in heparan sulfate modifications and embryonic and larval development in Caenorhabditis elegans. J Biol Chem. 2010;285:24717–24728. doi: 10.1074/jbc.M109.088229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zugasti O, Rajan J, Kuwabara PE. The function and expansion of the Patched- and Hedgehog-related homologs in C. elegans. Genome Res. 2005;15:1402–1410. doi: 10.1101/gr.3935405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hornsten A, Lieberthal J, Fadia S, Malins R, Ha L, Xu X, Daigle I, Markowitz M, O’Connor G, Plasterk R, et al. APL-1, a Caenorhabditis elegans protein related to the human beta-amyloid precursor protein, is essential for viability. Proc Natl Acad Sci U S A. 2007;104:1971–1976. doi: 10.1073/pnas.0603997104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Niwa R, Zhou F, Li C, Slack FJ. The expression of the Alzheimer’s amyloid precursor protein-like gene is regulated by developmental timing microRNAs and their targets in Caenorhabditis elegans. Dev Biol. 2008;315:418–425. doi: 10.1016/j.ydbio.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, Pasquinelli AE. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol. 2011;18:302–308. doi: 10.1038/nsmb.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hayes GD, Frand AR, Ruvkun G. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development. 2006;133:4631–4641. doi: 10.1242/dev.02655. [DOI] [PubMed] [Google Scholar]
- 126.Monsalve GC, Van Buskirk C, Frand AR. LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Curr Biol. 2011;21:2033–2045. doi: 10.1016/j.cub.2011.10.054. [DOI] [PubMed] [Google Scholar]
- 127.Jeon M, Gardner HF, Miller EA, Deshler J, Rougvie AE. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science. 1999;286:1141–1146. doi: 10.1126/science.286.5442.1141. [DOI] [PubMed] [Google Scholar]
- 128.Hammell CM, Karp X, Ambros V. A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18668–18673. doi: 10.1073/pnas.0908131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kostrouchova M, Krause M, Kostrouch Z, Rall JE. CHR3: a Caenorhabditis elegans orphan nuclear hormone receptor required for proper epidermal development and molting. Development. 1998;125:1617–1626. doi: 10.1242/dev.125.9.1617. [DOI] [PubMed] [Google Scholar]
- 131.Russel S, Frand AR, Ruvkun G. Regulation of the C. elegans molt by pqn-47. Dev Biol. 2011;360:297–309. doi: 10.1016/j.ydbio.2011.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Altun ZF, Chen B, Wang ZW, Hall DH. High resolution map of Caenorhabditis elegans gap junction proteins. Dev Dyn. 2009;238:1936–1950. doi: 10.1002/dvdy.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Van Buskirk C, Sternberg PW. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat Neurosci. 2007;10:1300–1307. doi: 10.1038/nn1981. [DOI] [PubMed] [Google Scholar]
- 134.Singh K, Chao MY, Somers GA, Komatsu H, Corkins ME, Larkins-Ford J, Tucey T, Dionne HM, Walsh MB, Beaumont EK, et al. C. elegans Notch Signaling Regulates Adult Chemosensory Response and Larval Molting Quiescence. Curr Biol. 2011;21:825–834. doi: 10.1016/j.cub.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Broday L, Hauser CA, Kolotuev I, Ronai Z. Muscle-epidermis interactions affect exoskeleton patterning in Caenorhabditis elegans. Dev Dyn. 2007;236:3129–3136. doi: 10.1002/dvdy.21341. [DOI] [PubMed] [Google Scholar]
- 136.Zaidel-Bar R, Miller S, Kaminsky R, Broday L. Molting-specific downregulation of C. elegans body-wall muscle attachment sites: the role of RNF-5 E3 ligase. Biochem Biophys Res Commun. 2010;395:509–514. doi: 10.1016/j.bbrc.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 137.Albert PS, Riddle DL. Developmental alterations in sensory neuroanatomy of the Caenorhabditis elegans dauer larva. J Comp Neurol. 1983;219:461–481. doi: 10.1002/cne.902190407. [DOI] [PubMed] [Google Scholar]
- 138.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 139.Cox GN, Staprans S, Edgar RS. The cuticle of Caenorhabditis elegans. II. Stage-specific changes in ultrastructure and protein composition during postembryonic development. Dev Biol. 1981;86:456–470. doi: 10.1016/0012-1606(81)90204-9. [DOI] [PubMed] [Google Scholar]
- 140.Muriel JM, Brannan M, Taylor K, Johnstone IL, Lithgow GJ, Tuckwell D. M142. 2 (cut-6), a novel Caenorhabditis elegans matrix gene important for dauer body shape. Dev Biol. 2003;260:339–351. doi: 10.1016/s0012-1606(03)00237-9. [DOI] [PubMed] [Google Scholar]
- 141.Mak HY, Ruvkun G. Intercellular signaling of reproductive development by the C. elegans DAF-9 cytochrome P450. Development. 2004;131:1777–1786. doi: 10.1242/dev.01069. [DOI] [PubMed] [Google Scholar]
- 142.Gerisch B, Antebi A. Hormonal signals produced by DAF-9/cytochrome P450 regulate C. elegans dauer diapause in response to environmental cues. Development. 2004;131:1765–1776. doi: 10.1242/dev.01068. [DOI] [PubMed] [Google Scholar]
- 143.Antebi A. WormBook. 2006. Nuclear hormone receptors in C. elegans; pp. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hristova M, Birse D, Hong Y, Ambros V. The Caenorhabditis elegans heterochronic regulator LIN-14 is a novel transcription factor that controls the developmental timing of transcription from the insulin/insulin-like growth factor gene ins-33 by direct DNA binding. Mol Cell Biol. 2005;25:11059–11072. doi: 10.1128/MCB.25.24.11059-11072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Moribe H, Yochem J, Yamada H, Tabuse Y, Fujimoto T, Mekada E. Tetraspanin protein (TSP-15) is required for epidermal integrity in Caenorhabditis elegans. J Cell Sci. 2004;117:5209–5220. doi: 10.1242/jcs.01403. [DOI] [PubMed] [Google Scholar]
- 146.Kage-Nakadai E, Kobuna H, Kimura M, Gengyo-Ando K, Inoue T, Arai H, Mitani S. Two very long chain fatty acid acyl-CoA synthetase genes, acs-20 and acs-22, have roles in the cuticle surface barrier in Caenorhabditis elegans. PLoS One. 2010;5:e8857. doi: 10.1371/journal.pone.0008857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Partridge FA, Tearle AW, Gravato-Nobre MJ, Schafer WR, Hodgkin J. The C. elegans glycosyltransferase BUS-8 has two distinct and essential roles in epidermal morphogenesis. Dev Biol. 2008;317:549–559. doi: 10.1016/j.ydbio.2008.02.060. [DOI] [PubMed] [Google Scholar]
- 148.Yook K, Hodgkin J. Mos1 mutagenesis reveals a diversity of mechanisms affecting response of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 2007;175:681–697. doi: 10.1534/genetics.106.060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Loer CM, Davidson B, McKerrow J. A phenylalanine hydroxylase gene from the nematode C. elegans is expressed in the hypodermis. J Neurogenet. 1999;13:157–180. doi: 10.3109/01677069909083472. [DOI] [PubMed] [Google Scholar]
- 150.Calvo AC, Pey AL, Ying M, Loer CM, Martinez A. Anabolic function of phenylalanine hydroxylase in Caenorhabditis elegans. FASEB J. 2008;22:3046–3058. doi: 10.1096/fj.08-108522. [DOI] [PubMed] [Google Scholar]
- 151.Coomans A, Verschuren D, Vanderhaeghen R. The demanian system, traumatic insemination and reproductive strategy in Oncholaimus oxyuris Ditlevsen (Nematoda, Oncholaimina) Zool Scripta. 2005;17:15–23. [Google Scholar]
- 152.Hodgkin J, Kuwabara PE, Corneliussen B. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr Biol. 2000;10:1615–1618. doi: 10.1016/s0960-9822(00)00867-8. [DOI] [PubMed] [Google Scholar]
- 153.Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- 154.Pujol N, Zugasti O, Wong D, Couillault C, Kurz CL, Schulenburg H, Ewbank JJ. Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog. 2008;4:e1000105. doi: 10.1371/journal.ppat.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ziegler K, Kurz CL, Cypowyj S, Couillault C, Pophillat M, Pujol N, Ewbank JJ. Antifungal innate immunity in C. elegans: PKCδ links G protein signaling and a conserved p38 MAPK cascade. Cell Host Microbe. 2009;5:341–352. doi: 10.1016/j.chom.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 156.Dierking K, Polanowska J, Omi S, Engelmann I, Gut M, Lembo F, Ewbank JJ, Pujol N. Unusual Regulation of a STAT Protein by an SLC6 Family Transporter in C. elegans Epidermal Innate Immunity. Cell Host Microbe. 2011;9:425–435. doi: 10.1016/j.chom.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 157.Zugasti O, Ewbank JJ. Neuroimmune regulation of antimicrobial peptide expression by a noncanonical TGF-beta signaling pathway in Caenorhabditis elegans epidermis. Nat Immunol. 2009;10:249–256. doi: 10.1038/ni.1700. [DOI] [PubMed] [Google Scholar]
- 158.Xu S, Chisholm AD. A Gαq-Ca(2+) signaling pathway promotes actin-mediated epidermal wound closure in C. elegans. Curr Biol. 2011;21:1960–1967. doi: 10.1016/j.cub.2011.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, Jin Y, Chisholm AD, Ewbank JJ. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr Biol. 2008;18:481–489. doi: 10.1016/j.cub.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tong A, Lynn G, Ngo V, Wong D, Moseley SL, Ewbank JJ, Goncharov A, Wu YC, Pujol N, Chisholm AD. Negative regulation of Caenorhabditis elegans epidermal damage responses by death-associated protein kinase. Proc Natl Acad Sci U S A. 2009;106:1457–1461. doi: 10.1073/pnas.0809339106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Van Epps H, Dai Y, Qi Y, Goncharov A, Jin Y. Nuclear pre-mRNA 3′-end processing regulates synapse and axon development in C. elegans. Development. 2010;137:2237–2250. doi: 10.1242/dev.049692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lamitina ST, Morrison R, Moeckel GW, Strange K. Adaptation of the nematode Caenorhabditis elegans to extreme osmotic stress. Am J Physiol Cell Physiol. 2004;286:C785–791. doi: 10.1152/ajpcell.00381.2003. [DOI] [PubMed] [Google Scholar]
- 163.Choe KP, Strange K. Evolutionarily conserved WNK and Ste20 kinases are essential for acute volume recovery and survival after hypertonic shrinkage in Caenorhabditis elegans. Am J Physiol Cell Physiol. 2007;293:C915–927. doi: 10.1152/ajpcell.00126.2007. [DOI] [PubMed] [Google Scholar]
- 164.Petalcorin MI, Oka T, Koga M, Ogura K, Wada Y, Ohshima Y, Futai M. Disruption of clh-1, a chloride channel gene, results in a wider body of Caenorhabditis elegans. J Mol Biol. 1999;294:347–355. doi: 10.1006/jmbi.1999.3241. [DOI] [PubMed] [Google Scholar]
- 165.Rohlfing AK, Miteva Y, Hannenhalli S, Lamitina T. Genetic and physiological activation of osmosensitive gene expression mimics transcriptional signatures of pathogen infection in C. elegans. PLoS One. 2010;5:e9010. doi: 10.1371/journal.pone.0009010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Solomon A, Bandhakavi S, Jabbar S, Shah R, Beitel GJ, Morimoto RI. Caenorhabditis elegans OSR-1 regulates behavioral and physiological responses to hyperosmotic environments. Genetics. 2004;167:161–170. doi: 10.1534/genetics.167.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wheeler JM, Thomas JH. Identification of a novel gene family involved in osmotic stress response in Caenorhabditis elegans. Genetics. 2006;174:1327–1336. doi: 10.1534/genetics.106.059089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rohlfing AK, Miteva Y, Moronetti L, He L, Lamitina T. The Caenorhabditis elegans mucin-like protein OSM-8 negatively regulates osmosensitive physiology via the transmembrane protein PTR-23. PLoS Genet. 2011;7:e1001267. doi: 10.1371/journal.pgen.1001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Soloviev A, Gallagher J, Marnef A, Kuwabara PE. C. elegans patched-3 is an essential gene implicated in osmoregulation and requiring an intact permease transporter domain. Dev Biol. 2011;351:242–253. doi: 10.1016/j.ydbio.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mizuno T, Fujiki K, Sasakawa A, Hisamoto N, Matsumoto K. Role of the Caenorhabditis elegans Shc adaptor protein in the c-Jun N-terminal kinase signaling pathway. Mol Cell Biol. 2008;28:7041–7049. doi: 10.1128/MCB.00938-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Budde MW, Roth MB. The response of Caenorhabditis elegans to hydrogen sulfide and hydrogen cyanide. Genetics. 2011;189:521–532. doi: 10.1534/genetics.111.129841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chen C, Samuel TK, Krause M, Dailey HA, Hamza I. Heme utilization in the Caenorhabditis elegans hypodermal cells is facilitated by Heme Responsive Gene-2. J Biol Chem. 2012 doi: 10.1074/jbc.M111.307694. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.O’Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hellerer T, Axang C, Brackmann C, Hillertz P, Pilon M, Enejder A. Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-Stokes Raman scattering (CARS) microscopy. Proc Natl Acad Sci U S A. 2007;104:14658–14663. doi: 10.1073/pnas.0703594104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Narbonne P, Roy R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature. 2009;457:210–214. doi: 10.1038/nature07536. [DOI] [PubMed] [Google Scholar]
- 176.Robertson AMG, Thomson JN. Morphology of programmed cell death in the ventral nerve cord of Caenorhabditis elegans larvae. J Embryol Exp Morph. 1982;67:89–100. [Google Scholar]
- 177.Hall DH, Gu G, Garcia-Anoveros J, Gong L, Chalfie M, Driscoll M. Neuropathology of degenerative cell death in Caenorhabditis elegans. J Neurosci. 1997;17:1033–1045. doi: 10.1523/JNEUROSCI.17-03-01033.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chung S, Gumienny TL, Hengartner MO, Driscoll M. A common set of engulfment genes mediates removal of both apoptotic and necrotic cell corpses in C. elegans. Nat Cell Biol. 2000;2:931–937. doi: 10.1038/35046585. [DOI] [PubMed] [Google Scholar]
- 179.Wu Z, Ghosh-Roy A, Yanik MF, Zhang JZ, Jin Y, Chisholm AD. Caenorhabditis elegans neuronal regeneration is influenced by life stage, ephrin signaling, and synaptic branching. Proc Natl Acad Sci U S A. 2007;104:15132–15137. doi: 10.1073/pnas.0707001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hammarlund M, Jorgensen EM, Bastiani MJ. Axons break in animals lacking β-spectrin. J Cell Biol. 2007;176:269–275. doi: 10.1083/jcb.200611117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.MacDonald JM, Beach MG, Porpiglia E, Sheehan AE, Watts RJ, Freeman MR. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50:869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 182.Maurer CW, Chiorazzi M, Shaham S. Timing of the onset of a developmental cell death is controlled by transcriptional induction of the C. elegans ced-3 caspase-encoding gene. Development. 2007;134:1357–1368. doi: 10.1242/dev.02818. [DOI] [PubMed] [Google Scholar]
- 183.Sommer RJ, Eizinger A, Lee KZ, Jungblut B, Bubeck A, Schlak I. The Pristionchus HOX gene Ppa-lin-39 inhibits programmed cell death to specify the vulva equivalence group and is not required during vulval induction. Development. 1998;125:3865–3873. doi: 10.1242/dev.125.19.3865. [DOI] [PubMed] [Google Scholar]
- 184.Galvin BD, Kim S, Horvitz HR. Caenorhabditis elegans genes required for the engulfment of apoptotic corpses function in the cytotoxic cell deaths induced by mutations in lin-24 and lin-33. Genetics. 2008;179:403–417. doi: 10.1534/genetics.108.087221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Joshi P, Eisenmann DM. The Caenorhabditis elegans pvl-5 gene protects hypodermal cells from ced-3-dependent, ced-4-independent cell death. Genetics. 2004;167:673–685. doi: 10.1534/genetics.103.020503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Aladzsity I, Toth ML, Sigmond T, Szabo E, Bicsak B, Barna J, Regos A, Orosz L, Kovacs AL, Vellai T. Autophagy genes unc-51 and bec-1 are required for normal cell size in Caenorhabditis elegans. Genetics. 2007;177:655–660. doi: 10.1534/genetics.107.075762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Borsos E, Erdelyi P, Vellai T. Autophagy and apoptosis are redundantly required for C. elegans embryogenesis. Autophagy. 2011;7:557–559. doi: 10.4161/auto.7.5.14685. [DOI] [PubMed] [Google Scholar]
- 188.Cinar HN, Chisholm AD. Genetic analysis of the Caenorhabditis elegans pax-6 locus: roles of paired domain-containing and nonpaired domain-containing isoforms. Genetics. 2004;168:1307–1322. doi: 10.1534/genetics.104.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Emtage L, Gu G, Hartwieg E, Chalfie M. Extracellular proteins organize the mechanosensory channel complex in C. elegans touch receptor neurons. Neuron. 2004;44:795–807. doi: 10.1016/j.neuron.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 190.Vogel BE, Hedgecock EM. Hemicentin, a conserved extracellular member of the immunoglobulin superfamily, organizes epithelial and other cell attachments into oriented line-shaped junctions. Development. 2001;128:883–894. doi: 10.1242/dev.128.6.883. [DOI] [PubMed] [Google Scholar]
- 191.Smith CJ, Watson JD, Spencer WC, O’Brien T, Cha B, Albeg A, Treinin M, Miller DM., 3rd Time-lapse imaging and cell-specific expression profiling reveal dynamic branching and molecular determinants of a multi-dendritic nociceptor in C. elegans. Dev Biol. 2010;345:18–33. doi: 10.1016/j.ydbio.2010.05.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Albeg A, Smith CJ, Chatzigeorgiou M, Feitelson DG, Hall DH, Schafer WR, Miller DM, 3rd, Treinin M. C. elegans multi-dendritic sensory neurons: morphology and function. Mol Cell Neurosci. 2011;46:308–317. doi: 10.1016/j.mcn.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rosenbluth J. Ultrastructure of somatic muscle cells in Ascaris lumbricoides. II. Intermuscular junctions, neuromuscular junctions, and glycogen stores. J Cell Biol. 1965;26:579–591. doi: 10.1083/jcb.26.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wright KA. Some Neuron-Hypodermis Relationships in the Parasitic Nematode Trichuris myocastoris Enigk. J Nematol. 1970;2:152–160. [PMC free article] [PubMed] [Google Scholar]
- 195.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 196.Stawicki TM, Zhou K, Yochem J, Chen L, Jin Y. TRPM Channels Modulate Epileptic-like Convulsions via Systemic Ion Homeostasis. Curr Biol. 2011;21:883–888. doi: 10.1016/j.cub.2011.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Searcy DG, Kisiel MJ, Zuckerman BM. Age-related increase of cuticle permeability in the nematode Caenorhabditis briggsae. Exp Aging Res. 1976;2:293–301. doi: 10.1080/03610737608257987. [DOI] [PubMed] [Google Scholar]
- 198.Himmelhoch S, Zuckerman BM. Caenorhabditis briggsae: aging and the structural turnover of the outer cuticle surface and the intestine. Exp Parasitol. 1978;45:208–214. doi: 10.1016/0014-4894(78)90061-9. [DOI] [PubMed] [Google Scholar]
- 199.Kisiel MJ, Castillo JM, Zuckerman LS, Zuckerman BM. Studies on ageing in Turbatrix aceti. Mech Ageing Dev. 1975;4:81–88. doi: 10.1016/0047-6374(75)90009-3. [DOI] [PubMed] [Google Scholar]
- 200.Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 201.Luo S, Kleemann GA, Ashraf JM, Shaw WM, Murphy CT. TGF-β and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell. 2010;143:299–312. doi: 10.1016/j.cell.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. doi: 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Qadota H, Inoue M, Hikita T, Koppen M, Hardin JD, Amano M, Moerman DG, Kaibuchi K. Establishment of a tissue-specific RNAi system in C. elegans. Gene. 2007;400:166–173. doi: 10.1016/j.gene.2007.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Spencer WC, Zeller G, Watson JD, Henz SR, Watkins KL, McWhirter RD, Petersen S, Sreedharan VT, Widmer C, Jo J, et al. A spatial and temporal map of C. elegans gene expression. Genome Res. 2011;21:325–341. doi: 10.1101/gr.114595.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Harris JE, Crofton HD. Structure and function the nematodes: internal pressure and cuticular structure in. Ascaris J Exp Biol. 1957;34:116–130. [Google Scholar]
- 206.Park SJ, Goodman MB, Pruitt BL. Analysis of nematode mechanics by piezoresistive displacement clamp. Proc Natl Acad Sci U S A. 2007;104:17376–17381. doi: 10.1073/pnas.0702138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Petzold BC, Park SJ, Ponce P, Roozeboom C, Powell C, Goodman MB, Pruitt BL. Caenorhabditis elegans Body Mechanics Are Regulated by Body Wall Muscle Tone. Biophys J. 2011;100:1977–1985. doi: 10.1016/j.bpj.2011.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Van De Velde MC, Coomans A. A putative new hydrostatic skeletal function for the epidermis in Monhysterids (Nematoda) Tissue Cell. 1989;21:525–533. doi: 10.1016/0040-8166(89)90005-0. [DOI] [PubMed] [Google Scholar]
- 209.Jansson HB. Adhesion of Conidia of Drechmeria coniospora to Caenorhabditis elegans Wild Type and Mutants. J Nematol. 1994;26:430–435. [PMC free article] [PubMed] [Google Scholar]
- 210.O’Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 2006;16:1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Nicholas HR, Hodgkin J. The ERK MAP kinase cascade mediates tail swelling and a protective response to rectal infection in C. elegans. Curr Biol. 2004;14:1256–1261. doi: 10.1016/j.cub.2004.07.022. [DOI] [PubMed] [Google Scholar]