Abstract
Purpose
Balanced chromosomal translocations are found in one out of 500 subjects in the general population. They usually do not carry any phenotypic consequences, except for possible infertility and for the production of unbalanced gametes leading to spontaneous abortions or chromosomal syndromes in the offspring. An association between chromosomal rearrangements and increased apoptosis markers has been demonstrated on a global scale in sperm samples of translocation and inversion carriers. In order to specify which kind of sperm cells is subject to an increased apoptosis process, this present study was aimed to analyse both chromosomal segregation and DNA fragmentation, sperm cell by sperm cell.
Methods
Six patients carrying a chromosomal rearrangement (three reciprocal translocations, two Robertsonian translocations, and one chromosomal pericentric inversion) were included in a retrospective manner. Both DNA fragmentation and chromosomal segregation in spermatozoa were evaluated simultaneously using a modified TUNEL assay associated with FISH. Two thousand spermatozoa were analysed for each patient.
Results
We showed a higher proportion of spermatozoa with fragmented DNA among the unbalanced sperm cells, compared to the balanced ones, in all six patients.
Conclusions
These results suggest an increased fragility of unbalanced spermatozoa to exogenous fragmentation factors. The exact mechanisms of those processes remain to be elucidated.
Keywords: Chromosome rearrangement, Translocation, Inversion, DNA fragmentation
Introduction
Although balanced chromosomal rearrangements are more commonly found in infertile men than in the general population [1], most carriers display normal or slightly decreased sperm counts [2], making these rearrangements frequently discovered after recurrent spontaneous abortions or after the birth of an affected child carrying the chromosomal rearrangement in an unbalanced state. Even in infertile carriers, spermatogenesis impairment is often partial, leading to oligozoospermia rather than to azoospermia and allowing pregnancies to occur using in vitro fertilization after intracytoplasmic sperm injection (ICSI). In this latter case, the chromosomal risk for the offspring is the same as that observed in spontaneous conceptions.
In reciprocal chromosomal translocation carriers, meiosis can complete only if translocated chromosomes can pair correctly by forming a quadrivalent whose segregation leads to several different chromosome combinations in gametes. The only balanced segregation pattern is the alternate mode which comprises the transmission of either the two normal chromosomes or the two translocated ones. The remaining segregation modes (adjacent-1, adjacent-2, 3:1 and exceptionally 4:0) lead to chromosomally unbalanced sperm cells and subsequent spontaneous abortions or chromosomal syndromes in the offspring. In Robertsonian translocations, chromosome pairing is achieved by the formation of a trivalent, the segregation of which produces either balanced or unbalanced gametes (adjacent or 3:0) leading to monosomies or trisomies in the offspring. The proportion of balanced spermatozoa is now easily estimated by fluorescent in situ hybridization (FISH) using appropriate DNA probes [5]. If usually predictable and around 80 % in Robertsonian translocations, it is rather unpredictable in reciprocal translocations, ranging from 18.6 % to 62.8 % [3]. In chromosomal inversions, correct pairing of homologous chromosomes can occur after the formation of a chromosomal loop whose size determines the proportion of recombinants, i.e. the rates of chromosomally balanced and unbalanced gametes. Proportions of each class are therefore even more variable [4].
DNA fragmentation is a parameter that has been thoroughly studied over the past years in reproductive medicine. Sperm DNA fragmentation can be both the cause and the consequence of apoptosis. Exogenous factors such as heat, radiations or a varicocele can affect sperm cells during their transit through the male genital tract, fragment their DNA and trigger a process of apoptosis leading to cell death [11]. Conversely, any defect in the meiosis process can lead to apoptosis which will itself lead to a number of cell modifications including DNA fragmentation.
DNA fragmentation has been related to male infertility as well as to lower pregnancy rates in natural conception, intra-uterine inseminations, in vitro fertilization (IVF) and ICSI [19–22]. One of the most commonly used test to assess DNA fragmentation in sperm is the TUNEL (Tdt-dUTP Nick End Labelling).
It has been shown that patients carrying a chromosomal rearrangement have significantly higher global sperm DNA fragmentation levels and expression of apoptosis markers than subjects with a normal karyotype [6,7,18]. However, in these studies, spermatozoa have been analyzed as a whole and no relationship between the level of DNA fragmentation and a particular nuclear content was evidenced. Based on the hypothesis that differences could exist in DNA fragmentation level according to the chromosomally balanced or unbalanced status of a gamete, Perrin et al. [18] studied four patients with a chromosomal translocation and showed a higher rate of DNA fragmentation in the unbalanced gametes than in the balanced ones. In order to confirm these findings, we studied six additional patients carrying various structural chromosomal rearrangements using a new technique which allows studying both chromosome segregation and DNA fragmentation simultaneously and cell by cell.
Materials and methods
Population
We conducted a retrospective study including six patients carrying a chromosomal rearrangement. Three of them (P1, P2, P3) carried a reciprocal translocation, two (P4, P5) a Robertsonian translocation, and one (P6) a pericentric inversion. P1 presented with a t(10;16)(q25;p12) reciprocal translocation. He was a 28 year-old single engineer, for whom the translocation was diagnosed as part of a familial study. His semen analysis showed normal parameters. Patient P2 presented with a t(6;17)(p21.3;q21) reciprocal translocation. He was a 39 year-old engineer, with a heavy tobacco and cannabis use. The translocation was diagnosed as part of an infertility work up, and the patient and his wife had had several unsuccessful IVF attempts, with failure of embryo development. Semen analysis showed oligoasthenoteratozoospermia. Patient P3 presented with a t(7;9)(p13q21) reciprocal translocation. He was a 40 year-old teacher, with a moderate five pack-year tobacco consumption. The translocation was discovered on the occasion of an amniocentesis during a past pregnancy. Semen analysis revealed teratozoospermia. Patient P4 presented with a rob(14;21)(p10;p10) Robertsonian translocation. He was a 40 year-old refuse collector. The couple had a history of infertlity, with several failed IVF attempts, and one interrupted pregnancy for a heart malformation. The semen analysis showed oligoasthenoteratozoospermia. Patient P5 presented with a rob(13;14)(p10;q10) Robertsonian translocation. He was a 30 year-old works foreman. The translocation was discovered on the occasion of an infertility workup, and a pregnancy was finally obtained through intrauterine insemination. A chorionic villus sampling showed that the fetus carried the paternal translocation. The semen analysis revealed oligoasthenoteratozoospermia. Patient P6, a hotel manager, carried a pericentric inversion of chromosome 4, inv(4)(p15.1q13.3). He and his partner had one miscarriage. Semen parameters were normal.
The global DNA fragmentation rate in rearrangement carriers was compared to that in six matched controls. Those latter were taking part in an insemination process, for female indications, and had normal semen parameters. The very same procedure was conducted on those samples and DNA fragmentation was evaluated with the normal unmodified TUNEL assay.
Sperm preparation
Each patient had a genetic consultation, with a thorough patient interview, and informed consent was obtained. Semen sample was obtained by masturbation after a 3 to 5 day abstinence period. We performed a regular semen analysis (volume, pH, concentration, motility, vitality, morphology), and spermatozoa were then rapidly fixed in ethanol-acetic acid (3:1) for 30 min. Sperm cells were then spread on microscope slides.
TUNEL assay
The TUNEL assay was performed first. We performed standard TUNEL procedure by using the In Situ Cell Death Detection Kit, Fluorescein (Roche, Indiana, USA). Since this procedure, as routinely used in IVF laboratories, resulted in an intense green fluorescence that prevented further FISH analysis, we made changes on two parameters: the TdT enzyme concentration was decreased from 1/10 to 1/50, and the incubation time was shortened from 1 h to 15 min. After being washed for 5 min twice in PBS, slides were put for 5 min at -20 °C in a permeabilization solution (0.1 % sodium citrate, 0.1 % Triton). The TdT enzyme was diluted at 1/50 in the buffer solution, and was put on the slides which were then left to incubate in a moist chamber at 37 °C for 15 min. Slides were finally washed twice for 5 min in PBS.
This modified TUNEL procedure was validated in five normospermic men: 500 spermatozoa were analyzed in each subject to compare the rate of spermatozoa with a fragmented DNA obtained either by the modified technique or by the non-modified one. Results were identical with both procedures (Data not shown).
FISH assay
A FISH assay was then performed on the same slides, immediately after the TUNEL technique. Indeed, when realized prior to the TUNEL, the FISH technique led to a global positivity of the TUNEL technique (Data not shown). According to the type of chromosomal rearrangement, a set of fluorescent probes was used to determine the segregation mode at meiosis and, therefore, the balanced or unbalanced status of the chromosomal nuclear content of each sperm cell analyzed (Fig. 1). For each patient with a reciprocal translocation (P1-P3), two non-commercial subtelomeric probes respectively labelled by Spectrum Green and Spectrum Orange and one commercial centromeric probe labelled by Spectrum Aqua (Vysis-Abott Molecular, Illinois, USA) were used. For Robertsonian translocations (P4, P5), only two subtelomeric probes were used but a centromeric probe for chromosome 18 was added to the probe mix in order to distinguish spermatozoa with a 3:0 segregation mode from diploid nuclei. For patient P6 carrying an inversion, only two probes located respectively in the subtelomeric regions of the short and long arms, and labelled respectively by Spectrum Green and Spectrum Orange, were used to discriminate normal sperm cells from recombined ones.
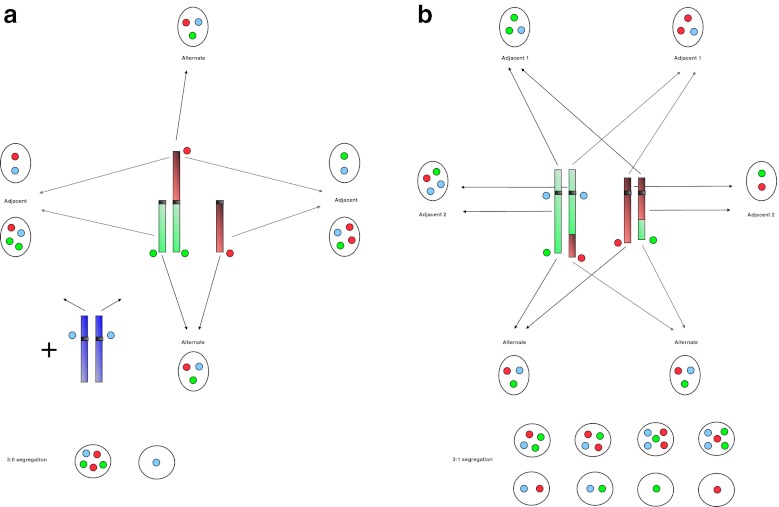
Fig. 1.
Sperm FISH patterns according to the type of translocation and the segregation modes. a Robertsonian translocations: one green and one red probes recognize the respective telomeric regions of the two acrocentric chromosomes involved. A third blue probe marks the centromere of another non-acrocentric chromosome, not involved in the translocation, in order to exclude diploid cells from the count. The alternate mode corresponds to the segregation of the two non translocated chromosomes in one sperm and of the translocated one in another sperm. The adjacent mode corresponds to the segregation of the translocated chromosomes with either one of the two normal ones. The 3:0 segregation mode leads to the segregation of the three involved chromosomes in one sperm, and none of them in another sperm. b Reciprocal translocations: one green and one red probes recognize the respective telomeric regions of the two chromosome arms involved, and a third blue probe marks the centromere of one of those two chromosomes. The alternate mode corresponds to the segregation of the two normal chromosomes in one sperm and of the two translocated ones in another sperm. The adjacent-1 mode corresponds to the segregation of one normal and one translocated chromosomes of each chromosomal pair in a sperm. The adjacent-2 mode corresponds to the segregation of the normal and the translocated chromosomes of the same chromosomal pair in a sperm. The 3:1 segregation mode leads to the segregation of three chromosomes in one sperm and the remaining chromosome in another sperm. In each case, the chromosomal content gives a specific combination of FISH signals
Non-commercial subtelomeric probes were prepared from BACs (bacterial artificial chromosomes) using an RCA (rolling circle amplification) technique. In order to attenuate the TUNEL fluorescence, slides were washed for 10 min at 73 °C in 0,4 SSC-NP40, and then rinsed for 1 min at 37 °C in 2-SSC-NP40. They were then dehydrated in alcohol (70°, 85°, 95°, 1 min each), denatured in sodium hydroxide (1 M) for 3 min, and dehydrated in alcohol again (70°, 85°, 95°, 1 min each). The three probes were then put on the slides and left to denaturate at 73 °C for 2 min. Hybridization was then performed at 37 °C for 24 h using a programmable hot plate (Slidebooster®, Beckman Coulter, FL, USA). The next day, slides were washed at 73 °C for 30 s in 0.4 SSC-NP40 and at 37 °C for 2 min in 2-SCC-NP40 and then counterstained with DAPI.
For each patient, probes were first tested on metaphase chromosomes using classical cytogenetic and FISH techniques.
Simultaneous microscopic observation
Slides were finally read on an Olympus BX-UCB microscope equipped with specific filters and 2000 sperms were analysed for each patient. Both TUNEL and FISH signals were observed in one run. First, sperm cells with a fragmented DNA were picked out within normal DAPI stained spermatozoa and then, by using specific filters, the FISH signals were analyzed on the same cells.
Spermatozoa were identified as carrying two signals of the same probe if the two fluorescent spots were: (i) of the same color and of similar size and intensity, (ii) separated by a distance of at least one spot, (iii) not connected by a fluorescent bridge and (iv) of similar size and intensity compared to neighboring germ cells.
FISH pattern of a sperm cell carrying a normal or a balanced translocated chromosomal content consists of one blue spot, one red spot and one green spot. On the other hand, FISH patterns of spermatozoa carrying unbalanced chromosomal contents vary from one translocation to the other according to the segregation mode and to the location of probes on the chromosomes, as previously described [3–5].
Statistical analysis
The global fragmentation rate in each of the six translocated patients was compared to that of one of the six normospermic controls using Chi2. When measuring DNA fragmentation in spermatozoa from translocation carriers and relating it to the balanced or unbalanced nuclear chromosomal content, each patient became his own control. Fragmentation rates in balanced and unbalanced gametes were compared using Chi2. Differences were considered as significant at p < 0.05.
Results
Modifications brought to the normal TUNEL protocol induced a faint but clearly visible green fluorescence of sperm heads which allowed asserting that a nucleus contained a fragmented DNA as well as counting easily the number of FISH signals (Fig. 2). Owing to the fact that the modified procedure did not modify the global fragmentation rate, as verified in five normal controls and 12 rearrangement carriers (data not shown), the relationship between DNA fragmentation level and chromosomal segregation was therefore possible to establish with our modified assay.
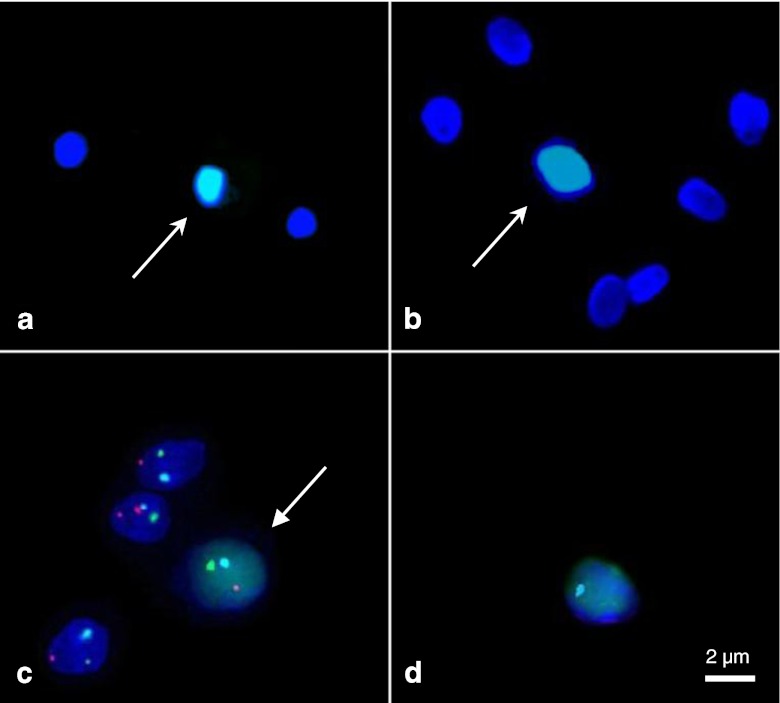
Fig. 2.
Simultaneous study of DNA fragmentation (faint green fluorescence of the whole sperm nucleus) and chromosomal segregation, in a reciprocal translocation carrier (P1). Green and red probes were directed to the telomeric regions of the chromosomal arms involved in the translocation, and a blue probe was directed to the centromere of another chromosome. aArrow: TUNEL-positive sperm, unmodified procedure. Note the bright green TUNEL fluorescence. bArrow: TUNEL-positive sperm, modified procedure. The TUNEL fluorescence is now faint, but clearly distinguishable from TUNEL-negative cells. cArrow: chromosomally balanced, TUNEL-positive sperm. d Chromosomally unbalanced, TUNEL-positive sperm. (Fluorescence microscopy, x1000)
Using this modified technique, the rate of apoptotic sperm cells was first evaluated for each patient and compared to that of the matched control subject (Table 1). Four of the six patients (P2, P4, P5, P6) had a significantly higher DNA fragmentation rate than the matched control (p = 0.01 and p < 0.001). One patient (P1) had a lower but non significantly different rate than that of the matched control (p = 0.12). The last patient (P3) had a slightly increased DNA fragmentation rate which was however not significantly different from that of his matched control (p = 0.09).
Table 1.
Percentages of TUNEL-positive spermatozoa, for each patient (P1-P6) and control (C1-C6). Chi2 results with p < 0.05 were considered as statistically significant
| Patient | % of TUNEL-positive sperm | Control | % of TUNEL-positive sperm | p |
|---|---|---|---|---|
| P1 | 1.2 % | C1 | 1.8 % | NS |
| P2 | 12.3 % | C2 | 0.6 % | p < 0.001 |
| P3 | 2 % | C3 | 0.8 % | NS |
| P4 | 4 % | C4 | 1.4 % | p < 0.001 |
| P5 | 4.8 % | C5 | 1.8 % | p = 0.01 |
| P6 | 4.4 % | C6 | 1.2 % | p < 0.001 |
We then analyzed chromosomal segregation and DNA fragmentation status for approximatively 2000 spermatozoa in each patient. Sperm cells were divided into “balanced” and “unbalanced” categories, and DNA fragmentation was compared between the two groups for each patient (Table 2). Our results showed a significantly higher proportion (p = 0,015 to p < 0,004) of TUNEL-positive spermatozoa in unbalanced spermatozoa than in balanced spermatozoa. In other words, unbalanced spermatozoa were more likely to have a fragmented DNA than their balanced counterparts. This was the case for both reciprocal and Robertsonian translocations, as well as for the inversion.
Table 2.
Chromosomally balanced or unbalanced status and DNA fragmentation, for each patient. Chi2 results with p < 0.05 were considered as statistically significant
| Balanced | Unbalanced | ||
|---|---|---|---|
| P1 | Segregation | 47.7 % | 52.3 % |
| TUNEL-positive | 0.5 % | 2 % * (p = 0.003) | |
| P2 | Segregation | 30 % | 70 % |
| TUNEL-positive | 5.4 % | 8.9 % * (p = 0.015) | |
| P3 | Segregation | 44.8 % | 55.2 % |
| TUNEL-positive | 2.3 % | 4.9 % * (p = 0.003) | |
| P4 | Segregation | 70 % | 30 % |
| TUNEL-positive | 2.5 % | 6.5 % * (p < 0.01) | |
| P5 | Segregation | 80.7 % | 19.3 % |
| TUNEL-positive | 1.1 % | 3.6 % * (p = 0.001) | |
| P6 | Segregation | 97.9 % | 2.1 % |
| TUNEL-positive | 4.2 % | 14.3 % * (p = 0.004) | |
For translocation carriers, we then subdivided the unbalanced category into the different unbalanced segregation modes: adjacent-1, adjacent-2, and 3:1 (ignoring the exceptional 4:0) for reciprocal translocations, and adjacent and 3:0 for Robertsonian translocations. We compared DNA fragmentation levels between the different modes (Table 3). Although this result was not statistically significant in all cases, it seemed that the adjacent II mode carried the highest DNA fragmentation rate.
Table 3.
Segregation pattern and DNA fragmentation, for each patient. Chi2 results with p < 0.05 were considered as statistically significant
| Alternate | Adjacent-I | Adjacent-II | 3:1 | ||
| P1 | Segregation | 47.7 % | 25.9 % | 9 % | 17.4 % |
| TUNEL-positive | 0.5 % | 1.5 % | 3.75 % | 2.2 % | |
| P2 | Segregation | 30 % | 16.8 % | 8.4 % | 44.7 % |
| TUNEL-positive | 5.4 % | 8.2 % | 11.5 % | 8.7 % | |
| P3 | Segregation | 44.8 % | 9.3 % | 8.2 % | 37.7 % |
| TUNEL-positive | 2.3 % | 9.9 % | 9.2 % | 2.7 % | |
| Alternate | Adjacent | 3:0 | |||
| P4 | Segregation | 70 % | 27.6 % | 2.3 % | |
| TUNEL-positive | 2.5 % | 6.2 % | 10.6 % | ||
| P5 | Segregation | 80.7 % | 10.8 % | 8.5 % | |
| TUNEL-positive | 1.1 % | 3.3 % | 4.1 % | ||
Discussion
Our results show that the proportion of spermatozoa with fragmented DNA is higher in unbalanced than in balanced sperm cells. This is true for every patient of the study and in agreement with the results published by Perrin et al. [18]. However, our procedure is far more simple and less time consuming than theirs which needed the relocalization of each sperm cell analyzed successively for both parameters. Indeed, our technique allows the cell by cell and simultaneous detection of both DNA fragmentation and chromosomal segregation in spermatozoa from translocation or inversion male carriers. Muriel et al. [14] also performed a simultaneous analysis of both DNA fragmentation and sperm chromosomal content. However, their study used a different DNA fragmentation assessment test (the sperm chromatin dispersion assay) which is less widely used, technically more complex than TUNEL, and does not detect DNA fragmentation directly but after a denaturation step [13]. Furthermore their study only concerned the aneuploidy rate, which is quite different from the segregation process in translocation carriers.
It can be argued that this association between chromosomal imbalance and DNA fragmentation has only been shown in ten patients so far (four by Perrin et al. in [18], and six in the present study). However, since the association between DNA fragmentation, apoptosis markers, and chromosomal rearrangements has been evidenced in multiple studies [6,7,17], it is possible than in most if not all of the patients analyzed in those studies, DNA fragmentation is more often found among unbalanced spermatozoa. Therefore, it is likely that our population of six patients is representative of the chromosomal events in sperm from translocations carriers. Furthermore, we analyzed an important number of spermatozoa, which allowed us to show the association between DNA fragmentation and chromosomal imbalance with a strong statistical significance.
Frequencies of the different segregation modes in our patients are in agreement with those published in the literature [5,12,15], with a majority of balanced spermatozoa in Robertsonian translocations carriers and much more variable results in reciprocal translocations carriers. Indeed, in our patients the proportion of balanced sperm was around 70–80 % in the former (P4, P5) and varied from 30 % to 47.7 % in the latter (P1, P2, P3). In P6, the presence of a pericentric inversion on the chromosome 4 led to 2.1 % of recombinant forms. This low proportion is related to the short distance between the two breakpoints, thus leading to a small inversion loop at meiosis with a low probability for a crossing over to occur within this loop.
In our study the inclusion of patients with chromosomal rearrangements was made regardless of the initial complaint that led to the diagnosis of the rearrangement. Previous studies on the subject stated that chromosomal rearrangements were associated with elevated global levels of DNA fragmentation and apoptosis [6,7,17]. However those studies were carried out in reproductive medicine centers, and most subjects initially consulted for infertility. Among our patients, two (P1, P3) had normal DNA fragmentation levels and none of them consulted for infertility. Their semen analysis only showed moderate alterations. This observation suggests that chromosomal rearrangements could induce DNA fragmentation only when associated with spermatogenesis failure in infertile patients. Further studies should aim at finding a correlation between chromosomal rearrangements, DNA fragmentation and sperm count in fertile asymptomatic carriers as well as in infertile oligozoospermic patients.
With the exception of the study by Perrin et al. [18], these studies report a global analysis of the semen in these men, thus preventing the distinction between balanced and unbalanced sperm cells [6,7,17]. Eaker et al. [9] showed increased apoptosis markers and asynapsis in unbalanced sperm in the mouse. Increased apoptosis markers were shown in humans as well [6,7]. The nature of the rearrangement, i.e. the position of the chromosomal breakpoints and consequently the length of the translocated chromosomal segments, could be a physical explanation to the increased rate of DNA fragmentation in spermatozoa carrying an unbalanced karyotype. Because complete synapsis of all chromosomes is prerequisite for meiosis ending, meiotic cells that do not achieve a particular configuration of the translocated chromosomes, named quadrivalent, are eliminated through an apoptotic process at the pachytene stage of meiosis. Perrin et al. [18] suggest that this phenomenon of abortive apoptosis is responsible for the increased DNA fragmentation in unbalanced sperm. They hypothesise that the abnormal quantity of DNA in unbalanced cells could trigger an apoptosis process between the spermatocyte and spermatid stages. However, incomplete synapsis cannot explain gamete DNA fragmentation because it occurs several weeks before the mature spermatozoa are ejaculated. Thus, we hypothesize that, while meiotic checkpoint mechanisms probably do play a role in eliminating a fraction of the unbalanced sperm cells within the seminiferous tubules, the DNA fragmentation observed in the ejaculate has another origin.
It is known that part of the DNA fragmentation observed in the ejaculate of a normal patient is acquired during the transit of spermatozoa along the male genital tract where it is subject to a number of exogenous damages, such as oxidative stress, heat, or radiations [16]. Furthermore, at this step, DNA maturation and compaction by protamines are not fully completed yet [19]. Spermatozoa from translocation carriers cannot be classified as balanced or unbalanced on the basis of their morphology, even at high magnification [8]. We suggest that an abnormal chromosomal content could disturb the fine three-dimensional architecture of the sperm nucleus, leading to its higher sensitivity to exogenous factors of apoptosis and to an increase in the DNA fragmentation process. Indeed, it has been shown that chromosomes present a well defined and probably highly regulated organization within the sperm nucleus [10]. Any disturbance to the chromosomal assembly, due to chromosome segments changes of localization or of quantity, could lead to a late apoptotic elimination of abnormal gametes during their transit through the genital tract. According to this hypothesis, translocated but balanced spermatozoa could also be affected by a spatial disorganization of their chromosomes within their nucleus and thus be subjected to a similar elimination process. Unfortunately, FISH with centromeric and telomeric probes cannot distinguish between balanced spermatozoa carrying the translocation or not. Further studies, using appropriate probes spanning chromosomal breakpoints would be necessary to answer this question. Although our study was limited to the relationship between DNA fragmentation and chromosomal content, it is likely that DNA fragmented spermatozoa are apoptotic cells. Other markers of DNA alteration, such as the sperm chromatin structure assay or the COMET assay as well as markers of apoptosis such as the expression of activated caspases and the externalization of phosphatidyl serine could have been used for this study. However, most of these procedures do not allow subsequent FISH analysis.
In conclusion, we confirm the relationship between unbalanced chromosomal content and DNA fragmentation in spermatozoa from translocation carriers. These findings could be the very first step in developping a new, compatible with assisted reproductive techniques, method which would eliminate unbalanced sperm cells, thus increasing the chances for affected couples to successfully procreate.
Acknowledgments
Disclosure statement
The authors have nothing to disclose, and declare that they have no conflict of interest.
Footnotes
Capsule DNA fragmentation is elevated in chromosomal translocation carriers, and predominantly affects unbalanced spermatozoa.
References
- 1.Meschede D, Lemcke B, Exeler JR, et al. Chromosome abnormalities in 447 couples undergoing intracytoplasmic sperm injection–prevalence, types, sex distribution and reproductive relevance. Hum Reprod. 1998;13(3):576–582. doi: 10.1093/humrep/13.3.576. [DOI] [PubMed] [Google Scholar]
- 2.Ravel C, Berthaut I, Bresson JL, Siffroi JP. Prevalence of chromosomal abnormalities in phenotypically normal and fertile adult males: large-scale survey of over 10,000 sperm donor karyotypes. Hum Reprod. 2006;21(6):1484–1489. doi: 10.1093/humrep/del024. [DOI] [PubMed] [Google Scholar]
- 3.Anton E, Blanco J, Egozcue J, Vidal F. Sperm FISH studies in seven male carriers of Robertsonian translocation t(13;14)(q10;q10) Hum Reprod. 2004;19(6):1345–1351. doi: 10.1093/humrep/deh232. [DOI] [PubMed] [Google Scholar]
- 4.Anton E, Vidal F, Blanco J. Role of sperm FISH studies in the genetic reproductive advice of structural reorganization carriers. Hum Reprod. 2007;22(8):2088–2092. doi: 10.1093/humrep/dem152. [DOI] [PubMed] [Google Scholar]
- 5.Benet J, Oliver-Bonet M, Cifuentes P, Templado C, Navarro J. Segregation of chromosomes in sperm of reciprocal translocation carriers: a review. Cytogenet Genome Res. 2005;111(3–4):281–290. doi: 10.1159/000086901. [DOI] [PubMed] [Google Scholar]
- 6.Brugnon F, Van Assche E, Verheyen G, et al. Study of two markers of apoptosis and meiotic segregation in ejaculated sperm of chromosomal translocation carrier patients. Hum Reprod. 2006;21(3):685–693. doi: 10.1093/humrep/dei401. [DOI] [PubMed] [Google Scholar]
- 7.Brugnon F, Janny L, Communal Y, et al. Apoptosis and meiotic segregation in ejaculated sperm from Robertsonian translocation carrier patients. Hum Reprod. 2010;25(7):1631–1642. doi: 10.1093/humrep/deq113. [DOI] [PubMed] [Google Scholar]
- 8.Cassuto NG, Le Foll N, Chantot-Bastaraud S, et al. Sperm fluorescence in situ hybridization study in nine men carrying a Robertsonian or a reciprocal translocation: relationship between segregation modes and high-magnification sperm morphology examination. Fertil Steril. 2011;96(4):826–832. doi: 10.1016/j.fertnstert.2011.07.1143. [DOI] [PubMed] [Google Scholar]
- 9.Eaker S, Pyle A, Cobb J, Handel MA. Evidence for meiotic spindle checkpoint from analysis of spermatocytes from Robertsonian-chromosome heterozygous mice. J Cell Sci. 2001;114(Pt 16):2953–2965. doi: 10.1242/jcs.114.16.2953. [DOI] [PubMed] [Google Scholar]
- 10.Kasinsky HE, Eirín-López JM, Ausió J. Protamines: structural complexity, evolution and chromatin patterning. Protein Pept Lett. 2011;18(8):755–771. doi: 10.2174/092986611795713989. [DOI] [PubMed] [Google Scholar]
- 11.Kaufmann SH, Earnshaw WC. Induction of apoptosis by cancer chemotherapy. Exp Cell Res. 2000;256(1):42–49. doi: 10.1006/excr.2000.4838. [DOI] [PubMed] [Google Scholar]
- 12.Morel F, Douet-Guilbert N, Le Bris M-J, et al. Meiotic segregation of translocations during male gametogenesis. Int J Androl. 2004;27(4):200–212. doi: 10.1111/j.1365-2605.2004.00490.x. [DOI] [PubMed] [Google Scholar]
- 13.Larson KL, DeJonge CJ, Barnes AM, Jost LK, Evenson DP. Sperm chromatin structure assay parameters as predictors of failed pregnancy following assisted reproductive techniques. Hum Reprod. 2000;15(8):1717–1722. doi: 10.1093/humrep/15.8.1717. [DOI] [PubMed] [Google Scholar]
- 14.Muriel L, Goyanes V, Segrelles E, et al. Increased aneuploidy rate in sperm with fragmented DNA as determined by the sperm chromatin dispersion (SCD) test and FISH analysis. J Androl. 2007;28(1):38–49. doi: 10.2164/jandrol.106.000067. [DOI] [PubMed] [Google Scholar]
- 15.Ogur G, Van Assche E, Vegetti W, et al. Chromosomal segregation in spermatozoa of 14 Robertsonian translocation carriers. Mol Hum Reprod. 2006;12(3):209–215. doi: 10.1093/molehr/gah253. [DOI] [PubMed] [Google Scholar]
- 16.Ollero M, Gil-Guzman E, Lopez MC, et al. Characterization of subsets of human spermatozoa at different stages of maturation: implications in the diagnosis and treatment of male infertility. Hum Reprod. 2001;16(9):1912–1921. doi: 10.1093/humrep/16.9.1912. [DOI] [PubMed] [Google Scholar]
- 17.Perrin A, Caer E, Oliver-Bonet M, et al. DNA fragmentation and meiotic segregation in sperm of carriers of a chromosomal structural abnormality. Fertil Steril. 2009;92(2):583–589. doi: 10.1016/j.fertnstert.2008.06.052. [DOI] [PubMed] [Google Scholar]
- 18.Perrin A, Basinko A, Douet-Guilbert N, et al. Aneuploidy and DNA fragmentation in sperm of carriers of a constitutional chromosomal abnormality. Cytogenet Genome Res. 2011;133(2–4):100–106. doi: 10.1159/000323980. [DOI] [PubMed] [Google Scholar]
- 19.Sakkas D, Alvarez JG. Sperm DNA fragmentation: mechanisms of origin, impact on reproductive outcome, and analysis. Fertil Steril. 2010;93(4):1027–1036. doi: 10.1016/j.fertnstert.2009.10.046. [DOI] [PubMed] [Google Scholar]
- 20.Spanò M, Bonde JP, Hjøllund HI, et al. Sperm chromatin damage impairs human fertility. The danish first pregnancy planner study team. Fertil Steril. 2000;73(1):43–50. doi: 10.1016/S0015-0282(99)00462-8. [DOI] [PubMed] [Google Scholar]
- 21.Thomson LK, Zieschang J-A, Clark AM. Oxidative deoxyribonucleic acid damage in sperm has a negative impact on clinical pregnancy rate in intrauterine insemination but not intracytoplasmic sperm injection cycles. Fertil Steril. 2011;96(4):843–847. doi: 10.1016/j.fertnstert.2011.07.356. [DOI] [PubMed] [Google Scholar]
- 22.Zini A, Jamal W, Cowan L, Al-Hathal N. Is sperm DNA damage associated with IVF embryo quality? A systematic review. J Assist Reprod Genet. 2011;28(5):391–397. doi: 10.1007/s10815-011-9544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]