Abstract
The tyrosine kinase Janus kinase 2 (JAK2) is activated by many cytokine receptors, including receptors for GH, leptin, and erythropoietin. However, very few proteins have been identified as binding partners for JAK2. Using a yeast 2-hybrid screen, we identified steroid-sensitive gene-1 (SSG1)/coiled-coil domain-containing protein 80 (Ccdc80) as a JAK2-binding partner. We demonstrate that Ccdc80 preferentially binds activated, tyrosyl-phosphorylated JAK2 but not kinase-inactive JAK2 (K882E) in both yeast and mammalian systems. Ccdc80 is tyrosyl phosphorylated in the presence of JAK2. The binding of Ccdc80 to JAK2 occurs via 1 or more of the 3 DUDES/SRPX (DRO1-URB-DRS-Equarin-SRPUL/sushi repeat containing protein, x-linked) domain 5 domains of Ccdc80. Mutagenesis of the second DUDES domain suggests that the N-terminal third of the DUDES domain is sufficient for JAK2 binding. Ccdc80 does not alter the kinase activity of JAK2. However, Ccdc80 increases GH-dependent phosphorylation of Stat (signal transducer and activator of transcription) 5b on Tyr699 and substantially enhances both basal and GH-dependent phosphorylation/activation of Stat3 on Tyr705. Furthermore, Ccdc80 belongs to the group of proteins that function both in the intracellular compartment and are secreted. Secreted Ccdc80 associates with the extracellular matrix and is also found in the medium. A substantial portion of the Ccdc80 detected in the medium is cleaved. Finally, consistent with the DUDES domain serving as a JAK2-binding domain, we also demonstrate that another protein that contains a DUDES domain, SRPX2, binds preferentially to the activated tyrosyl-phosphorylated form of JAK2.
The Janus family of tyrosine kinases, which includes Janus kinase 1 (JAK1) (1), JAK2, JAK3, and Tyk2, plays a critical role in signaling by members of the cytokine superfamily of receptors. JAK2 is activated by more than two-thirds of these receptors, including the receptors for GH, prolactin, erythropoietin, leptin, leukemia inhibitory factor, interferon-γ, and multiple ILs (1). JAK2 promotes the growth, proliferation, and/or differentiation of many cell types (2, 3), and dysregulation of JAK2 has been linked to various forms of cancer (4–10).
Despite the fact that JAK2 is necessary for signaling by multiple cytokines, hormones, and growth factors and is the most studied of the JAKs, only a handful of proteins have been identified that bind directly to JAK2. The negative regulators suppressor of cytokine signaling 1 (SOCS-1) (11), SOCS-3 (12), and protein tyrosine phosphatase-1B (13) bind to phosphorylated Tyr1007, the tyrosine critical for activation of JAK2. Suppressor of T cell receptor signaling-1 (Sts-1), a tyrosine phosphatase and negative regulator of Zap-70 that lacks a known phosphotyrosine (PY)-binding domain, binds preferentially to the activated form of JAK2 by an unknown mechanism (14–16). Casein kinase 2 (CK2), a serine/threonine that has been shown to enhance JAK2-signal transducer and activator of transcription (Stat) signaling also binds JAK2 by an unknown mechanism (17). Finally, the adapter proteins SH2B1, Src-homology 2B1 (SH2B1) (SH2-B, PSM [Pro-rich, PH, SH2 domain containing signal mediator]) (18), SH2B2 (adapter protein with pleckstrin homology and Src homology 2 [APS]) (19, 20), and SH2B3 (lymphocyte adapter protein [Lnk]) (21) bind to phosphorylated Tyr813 in JAK2. The fact that activated JAK2 is phosphorylated on more than 15 tyrosines (22) makes it seem likely that additional proteins will be found to be recruited to activated JAK2.
In this study, a yeast 2-hybrid screen of an adipocyte cDNA library identified steroid-sensitive gene-1 (SSG1) as a novel JAK2-binding protein. SSG1 was originally characterized as an estrogen-regulated gene based on its cloning using differential display of uterine tissues from ovariectomized and ovariectomized/estrogen-treated rats (23). Rat SSG1 was independently recloned as CL2 (clone 2), a protein up-regulated in rat thyroid PC Cl 3 cells infected with the adenovirus E1A gene (24) and DRO1 (down-regulated by oncogenes-1), a protein down-regulated in RK3E rat epithelial cells neoplastically transformed by β-catenin (25). Murine SSG1 was initially cloned as Urb (up-regulated in brown adipose tissue of bombesin receptor subtype-3-deficient mice) (26). A chicken ortholog of SSG1, Equarin-L, was cloned from a chick E6 lens cDNA library using a signal sequence trap screen and named for its expression in the equatorial region of the chick lens (27). This gene and its encoded protein have been renamed Ccdc80 (coiled-coil domain containing 80). Rat, murine, and human Ccdc80 have a wide tissue distribution (eg, fat, lung, ovary, uterus, mammary gland, testis, liver, spleen, pancreas, kidney, heart, stomach, bladder, skeletal muscle, skin, and brain) (23–26, 28–30). Roles for Ccdc80 in tumor suppression (24, 25, 31), energy metabolism (26, 30), and embryonic development and skeletal formation (28) have been suggested. However, the role of Ccdc80 at the cellular level, as well as how the cell regulates Ccdc80, is just beginning to be understood.
We show here that Ccdc80 binds to the active, tyrosyl-phosphorylated form of JAK2 but not the inactive form of JAK2. In the presence of active JAK2, Ccdc80 is phosphorylated. Binding to JAK2 requires the regions in Ccdc80 that are homologous to domain 5 of the sushi-repeat-containing protein, X-linked (SRPX). This domain (32) is common to a newly emerging group of proteins that includes Ccdc80 (23–26, 28), SRPX/drs (down-regulated by Src) (rat)/exon trapped x-chromosome clone 1 (ETX1) (human) (32), and SRPX2/SRPUL (sushi repeat protein up-regulated in leukemia) (33) and has been designated the DUDES (DRO1-URB-DRS-Equarin-SRPUL) domain (25). We show in mammalian systems that SRPX2, like Ccdc80, binds preferentially to activated JAK2. These results suggest that the DUDES protein interaction domain characterizes a novel class of proteins that bind activated JAK2. Regarding function, Ccdc80 does not enhance JAK2 kinase activity. However, Ccdc80 stimulates the phosphorylation of Stat5b on Tyr699, which is required for Stat5b activation, and enhances Stat3 phosphorylation on Tyr705, but not on Ser727. Finally, we show that Ccdc80 has a functional signal peptide and that a portion of Ccdc80 is secreted into the extracellular compartment where it both associates with the extracellular matrix and is released into the medium.
Materials and Methods
Cells and reagents
Saccharomyces cerevisiae EGY48 (MATα trp 1 ura3–52 his3 leu) and all yeast expression plasmids were from R. Brent (The Molecular Sciences Institute, Berkeley, California) (34). The 293T and COS-7 cells were from American Type Culture Collection (Manassas, Virginia). Chinese hamster ovary (CHO) cells expressing amino acids (aa) 1–454 of rat GH receptor (CHO-GHR [1–454]) were provided by G. Norstedt (Karolinska Institute, Stockholm, Sweden) (35). The stock of 3T3-F442A preadipocyte cells was provided by H. Green (Harvard University, Boston, Massachusetts). Recombinant 22 000-Da human GH was a gift from Eli Lilly & Co (Indianapolis, Indiana). DMEM was from Life Technologies (Carlsbad, California). Calf serum and fetal bovine serum were from Atlanta Biologicals (Lawrenceville, Georgia). BSA (CRG-7) was from Serologicals Corp (Norcross, Georgia). Aprotinin, leupeptin, and Triton X-100 were from Roche (Indianapolis, Indiana). Brefeldin A, polyvinylpyrrolidone (PVP-360), and sodium vanadate were from Sigma (St Louis, Missouri). Recombinant protein A-agarose was from Repligen (Waltham, Massachusetts). 4′6-diamidino-2-phenylindole (DAPI) was from Life Technologies. Paraformaldehyde was from Electron Microscopy Sciences (Hatfield, Pennsylvania). The enhanced chemiluminescence detection system and nitrocellulose paper were from GE Healthcare (Fairfield, Connecticut). X-ray film was from Kodak (Rochester, New York).
Antibodies
Anti-flag M2 antibody (α-flag, 1/1000 dilution) and anti-flag conjugated to agarose beads were from Sigma. Anti-green fluorescent protein (GFP) (αGFP; JL-8) (1/100 for immunoprecipitation and 1/5000 for immunoblotting) was from BD Biosciences (San Jose, California). Anti-GFP (1/100 for immunoprecipitation) was from Clontech (Palo Alto, California). Anti-giantin (1:500) and antibody to hemagglutinin (HA) tag (αHA; HA.11) (1/100 for immunoprecipitation; 1/10 000 for immunoblotting) were from Covance (Princeton, New Jersey). Anti-HA (3F10, 1/2000) was from Roche. Anti-calnexin (1/500) came from Assay Designs (Ann Arbor, Michigan). Anti-JAK2 (1/2000) was from Invitrogen or Millipore (Temecula, California). Anti-pY1007/pY1008 JAK2 (αpY1007/pY1008 JAK2, 1/2000), anti-PY (4G10, αPY; 1:7500), and anti–pY705-Stat3 (αpY705-Stat3, 1/1000) were from Millipore. Anti-Myc-tag (αmyc; 9E10) (1/100 for immunoprecipitation and 1/10,000 for immunoblotting), anti-myc (A14) (1/1000 dilution), anti-Stat3 (K-15, 1/1000), anti-Stat5b (αStat5b, G-2, 1/5000), horseradish peroxidase-conjugated antimouse IgG, and anti-rabbit IgG (1/7500 dilution) were from Santa Cruz Biotechnology (Santa Cruz, California). Anti–pS727-Stat3 (1/1000) and anti-pY694-Stat5 (anti-pY699 Stat5b) (1/7500) were from Cell Signaling Technology (Danvers, Massachusetts). AlexaFluor 488-conjugated anti-mouse IgG (1:500), AlexaFluor 594-conjugated anti-rabbit IgG (1/500), and Oregon green anti-rabbit IgG (1/1000) were from Molecular Probes. IRDye 680 and 800 conjugated to anti-mouse IgG or anti-rabbit IgG antibodies (1/20 000) were from Li-Cor Biosciences (Lincoln, Nebraska).
Two-hybrid library screening
Screening of the yeast 2-hybrid rat adipose cDNA library (from T. A. Gustafson, University of Maryland, Baltimore, Maryland) was conducted as described previously (36). Briefly, yeast (EGY48) cells were grown at 30°C in YPD medium (1% yeast extract, 2% polypeptone, 2% dextrose). Yeast cells were sequentially transformed with the lexAop-lacZ reporter plasmid encoding full-length rat JAK2, pEG202-JAK2, and rat adipose cDNA library prey plasmids by the lithium acetate method (37). Triple transformants were grown for 4 hours in liquid dropout medium lacking Trp, Ura, and His and plated on dropout medium containing 15% agar. The yeast cells were grown at 30°C for 4 days. Colonies were collected and replated on dropout medium lacking Trp, Ura, His, and Leu to select for Leu prototrophy. Dextrose was substituted with galactose to induce expression of the activation domain hybrids. After 5 days, colonies were subjected to the filter lift color assay to test for β-galactosidase activity. Positive clones were selected, and prey plasmids containing library cDNA inserts were isolated and transformed into DH5α bacterial strain for amplification. The inserts from the isolated cDNAs were sequenced.
Plasmids
The cDNA for SSG1/Ccdc80 (accession number AF223577) (23) was resequenced. The DNA sequence of rat SSG1/Ccdc80 was found to be identical to rat DRO1/Ccdc80 (accession number AY548105) (25). The open reading frame codes for a 949-aa protein.
The cDNA encoding the C-terminal portion of rat Ccdc80 (aa 477–949) was cut from the yeast 2-hybrid prey plasmid using EcoRI and XhoI and subcloned in-frame into pcDNA3-HA (Invitrogen) (38) to produce N-terminal HA-tagged Ccdc80 (477–949) and into prk5-myc (BD Biosciences) to produce N-terminal myc-tagged Ccdc80 (477–949). PCR-based mutagenesis was performed using QuikChange Site-Directed Mutagenesis Kits from Stratagene (La Jolla, California). PCR products and junctions were verified by sequencing. The amino acids in Ccdc80 are numbered according to the rat DRO1 sequence (AAS66670). In the primers listed below, mutated bases are in lowercase; introduced stop codons and restriction sites are underlined; and sequences for myc, HA, and flag tags are in bold italic. Ccdc80 truncation mutants were created by introducing stop codons into HA-Ccdc80 (477–949) using the following primers: stop at 609, 5′-CCGAGTCCCAGGtAGTCAGTGGCC-3′; stop at 663: 5′-GGTTACTATCTTTtGaCCTGTCAACAACAGC-3′; stop at 689, 5′-CGTGTGGTGGATGACtAGGACTTGGTAGAC-3′; stop at 724, 5′-GGTCTCAGAGTCAAGtAATACTACGAAGTGC-3′; and stop at 763, 5′-ACCTGCAAGGAGGACtAGAGGCAGTCC-3′.
N-terminal myc-tagged Ccdc80 (472–788) was created by inserting EcoRI and XhoI sites into full-length Ccdc80 using 5′-tctatcccgagactcgaattcACCATGGAACATGGCCATCAGGACCCAAATGTGGTG-3′ and 5′-gatccgctcgagcggtctagagcaTCATTGGGAGCAGAGATCACCAGCAACCGCC-3′ and subcloned in-frame into prk5-myc.
Full-length Ccdc80 with a myc tag at the C terminus was created using 5′-tctatcccgatcgatagatctaggaattccaccATGACGTGGAAAATGGGACCCCACTTCACC-3′ and 5′-gatccggaattccccaagcttgggttacaggtcctcctcgctgatcagcttctgctccattccGTAAGGGTATCCATGGTGGTAACTTTCATGATG-3′ and subcloned into prk5.
Full-length Ccdc80-myc with an internal HA tag after the signal peptide at residue 30 (Ccdc80-HA-myc) was created using 5′-CAGTCTTCTGCCTTGGATgtCGAcGGCCGCCCAGG-3′ to insert a SalI site at residue 30, and 5′-tcgaggatgtcgacgcctacccatacgatgttcctgactatgcggtcgacggc-3′ and 5′-tcgaagcccgtcgaccgcatagtcaggaacatcgtatgggtaggcgtcgaccta-3′ were annealed and used to insert an HA tag.
The DUDES domains of Ccdc80 were inserted into XhoI and EcoRI restriction sites in pEGFP-C1 (Clontech) or prk5-myc using GFP-Ccdc80 (615–756), 5′-atcgatcgatctcgagccgatTTGGGGTCTTTCGAAGGCAAACGAAGACTC-3′ and 5′-atcgatcgatcgatcggatcccggaattcgctaGCCCTCCTTCTTCTGCTTTTCCATATCTTTGATTC-3′; myc-Ccdc80 (137–281), 5′-atcgatcgatctcgaggccgatAGTTCTCCCAATATCCTGGCCAGCTTTGC–3′ and 5′-atctgtcgacgaattcgctaACACTTTTGGACAAAACCCTTCTGCCTGATTTTCTC-3′; myc-Ccdc80 (615–756), 5′-atcgatcgatctcgaggccgatTTGGGGTCTTTCGAAGGCAAACGAAGACTC-3′ and 5′-atcgatcgatcgatcggatcccggaattcgctaGCCCTCCTTCTTCTGCTTTTCCATATCTTTGATT-3′; and myc-Ccdc80 (766–912), 5′-atcgatcgatctcgaggccgatTCCCTGGAGAATTTCCTATCCAGGTTCC-3′ and 5′-actgtcgacgaattcgctaACAGCGCATCCCCAGTGACTGCTG-3′.
Murine SRPX2 (accession number NM_026838) was obtained from OriGene. A NotI site and myc-tag was inserted into the BamHI and HindIII sites in prk5 using 5′-gctacgGGATCCgcggccgcagagcagaagctgatcagcgaggaggacctgtagAAGCTTacgcgt-3′. Full-length SRPX2 was then subcloned into BamHI and NotI sites in the modified myc-prk5 using (Kozak sequence and ATG in bold italic) 5′-gctacgggatccgccaccATGATGACCAGTCCACTGACTCAGAGAGGTGC-3′ and 5′-gctacgcggccgcCTCGCATAGGTCCCTCTGCTCCACTCTCC-3′.
Prk5 encoding murine JAK2 (39) or JAK2 (K882E) (40) in which the critical lysine (40) is mutated to glutamate were provided by J. Ihle (St Jude Children's Research Hospital, Memphis, Tennessee). N-terminal flag-tagged JAK2 (797–1132) and JAK2 (830–1132) were described previously (40). C-terminal HA-tagged JAK2 (JAK-HA), JAK2 lacking aa 548–804 (Δ JAK homology domain 2 [JH2]-HA), and the JH1-JH2 fragment JAK2 (535–1132) (JH1–2-ΗΑ) were gifts from O. Silvennoinen (University of Tampere, Tampere, Finland) (41). N-terminal flag-JAK2 was described previously (19). Flag-JAK2 (K882E) was prepared from N-terminal flag-JAK2 using 5′-GGTGGTCGCTGTGgAgAAACTCCAGCACAGCAC-3′. To create an N-terminal GFP-JAK2, a NotI site was inserted into pEGFP-C1 BamHI and XbaI sites using 5′-TCGAG GTCGAGGGATCCgatgcggccgcacgT-3′. JAK2 was then subcloned from prk5-JAK2 into the modified pEGFP-C1 using SalI and NotI restriction sites. GFP-JAK2 (K882E) was then prepared. Rat GH receptor in pLM108 was provided by G. Norstedt (Karolinska Institute, Stockholm, Sweden). Flag-Stat3 was provided by R. Jove (City of Hope National Medical Center, Duarte, California). The N-terminal tagged GFP-Stat5b was described previously (42).
Cell culture and transfection
The 3T3-F442A, 293T, and COS-7 cells were grown in DMEM supplemented with 1mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B, and either 8% calf serum (3T3-F442A and 293T) or 8% fetal bovine serum (COS-7). Both 293T and COS-7 cells were transiently transfected by calcium phosphate precipitation (43) and assayed 24 or 48 hours after transfection, respectively. The 3T3-F442A cells were electroporated using the Nucleofactor system (Amaxa). For experiments with GH, cells were incubated in DMEM containing 1% BSA for 18 hours before GH addition. For treatment with brefeldin A (BFA), cells were incubated in DMEM containing 1% calf serum and BFA for 16 hours. The amount (micrograms) of cDNA in all experiments was kept constant using empty vector.
Cell solubilization, immunoprecipitation, and immunoblotting
Cells were washed 3 times with PBSV (10mM sodium phosphate, 137 mM NaCl, 1 mM Na3VO4 [pH 7.4]) and solubilized in lysis buffer (50 mM Tris, 0.1% Triton X-100, 150 mM NaCl, 2 mM EGTA, 1 mM Na3VO4 [pH 7.5]) containing 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin. Lysates were centrifuged at 16 000 g at 4°C for 10 minutes. The supernatant (cell lysate) was incubated with the indicated antibodies on ice for 2 hours. The immune complexes were collected on protein A-agarose for 1 hour, washed 3 times with lysis buffer, and boiled for 5 minutes in a mixture (80:20) of lysis buffer and SDS-PAGE sample buffer (250 mM Tris-HCl, 10% SDS, 10% β-mercaptoethanol, 40% glycerol, 0.01% bromophenol blue [pH 6.8]). The solubilized proteins were separated on 5% to 12% SDS-PAGE gradient gels, transferred to nitrocellulose, immunoblotted, and visualized using enhanced chemiluminescence or the Odyssey Infrared Imaging System (Li-Cor Biosciences). To reduce nonspecific binding (see Figures 3–5), the lysis buffer was supplemented with 1% Triton X-100 plus 0.1% SDS; the protein A agarose beads were preincubated with 0.5% polyvinylpyrrolidone in 100 mM acetic acid (pH 6.2) at 4°C for 1 hour before incubation with the lysates; and the immobilized proteins washed in 500 mM NaCl before elution. To prepare whole-cell lysates, cells were washed with PBSV and boiling SDS-PAGE sample buffer was added to the cells. Samples were then boiled and diluted 1:4 with lysis buffer. Immunoblots were quantified using Odyssey version 2.1 software (LI-COR). Phosphorylated protein levels were normalized to the amount of total protein present. Experiments were performed at least 2 and usually 3 or more times with similar results. Means ± SEM are presented.
Figure 3.
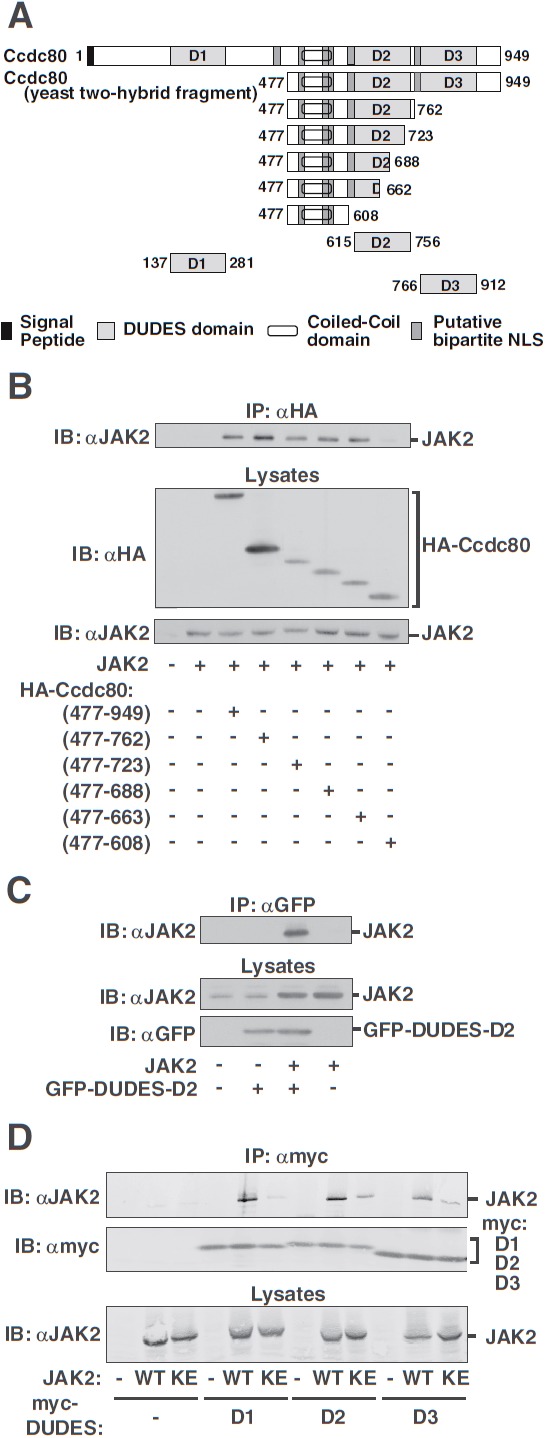
Identification of the 3 DUDES domains within Ccdc80 (D2) as regions of interaction with JAK2. A, Schematic of the N-terminally HA-tagged Ccdc80 truncation mutants. B, 293T cells expressing JAK2 and HA-Ccdc80 truncation mutants as indicated were immunoprecipitated (IP) with αHA, and immunoprecipitated proteins were immunoblotted (IB) with αJAK2. Cell lysates were immunoblotted with αHA or αJAK2. C, GFP-Ccdc80 (615–756) encoding the D2 domain of Ccdc80 (GFP-DUDES-D2) and JAK2 were expressed in 293T cells as indicated. GFP-Ccdc80 (615–756) was immunoprecipitated (IP) with αGFP, and immunoprecipitated proteins were immunoblotted (IB) with αJAK2. Cell lysates were immunoblotted with αJAK2 or αGFP as indicated. The migration of JAK2 and GFP-Ccdc80 (615–756) is indicated (n = 2). D, 293T cells expressing the designated myc-DUDES domains of Ccdc80 (D1, D2, or D3) and either JAK2 (WT) or JAK2 (K882E) (KE) were immunoprecipitated (IP) with αmyc, and immunoprecipitated proteins were immunoblotted (IB) with αJAK2. The blot was reprobed with αmyc. Lysates were immunoblotted with αJAK2. The migration of JAK2 and the myc-DUDES domains (D1, D2, D3) is indicated (n = 3).
Figure 4.
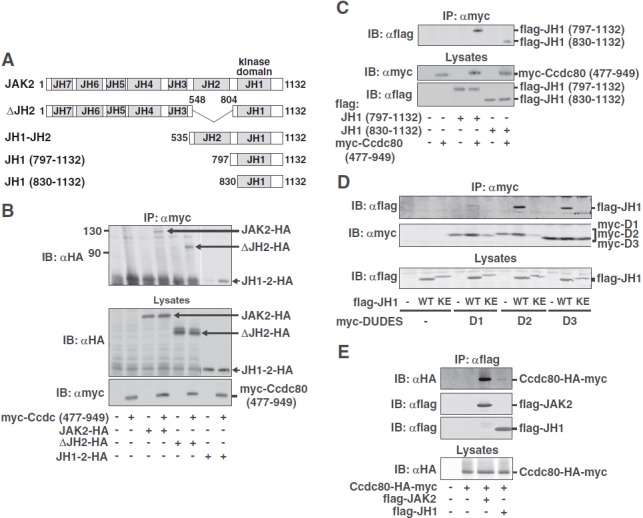
The D2 and the D3 DUDES domains of Ccdc80 interact with the kinase domain of JAK2. A, Domain structure of C-terminally HA-tagged JAK2 and JAK2 truncation mutants. B, 293T cells expressing JAK2-HA, ΔJH2-HA or JH1–2-HA, with or without myc-Ccdc80 (477–949) as indicated were immunoprecipitated (IP) with αmyc, and immunoprecipitated proteins were immunoblotted (IB) with αHA. Cell lysates were immunoblotted with αHA and αmyc. Because the JH1–2-HA construct expressed at very high levels, lanes 7 and 8 were exposed for a shorter period of time. Migration of JAK2-HA, ΔJH2-HA, JH1–2-HA, and myc-Ccdc80 (477–949) is indicated. C, 293T cells expressing myc-Ccdc80 (477–949) and flag-JH1 (797–1132) or flag-JH1 (830–1132) as indicated were immunoprecipitated (IP) with αmyc, and immunoprecipitated proteins were immunoblotted (IB) with αflag. Lysates were immunoblotted with αmyc and αflag. Migration of flag-JH1 (797–1132), flag-JH1 (830–1132), and myc-Ccdc80 (477–949) is indicated (n = 3–5). D, 293T cells expressing one of the myc-DUDES domains of Ccdc80 (D1, D2, or D3) with or without flag-JAK2 (797–1132) (flag-JH1 WT) or flag-JAK2 (797–1132, K882E) (flag-JH1 KE) were immunoprecipitated (IP) with αmyc, and immunoprecipitated proteins were immunoblotted (IB) with αflag. The blot was reprobed with αmyc. Lysates were immunoblotted with αflag. The migration of flag-JH1 and the myc-DUDES domains is indicated (n = 4). E, 293T cells expressing Ccdc80-HA-myc, flag-JAK2, and/or flag-JH1 as indicated were immunoprecipitated (IP) using αflag, and the immunoprecipitated proteins were immunoblotted (IB) with αHA. The blot was reprobed with αflag, and the regions of the blot containing flag-JAK2 and flag-JH1 are shown. Lysates were immunoblotted with αHA. The migration of Ccdc80-HA-myc, flag-JAK2, and flag-JH1 is indicated (n = 2).
Figure 5.
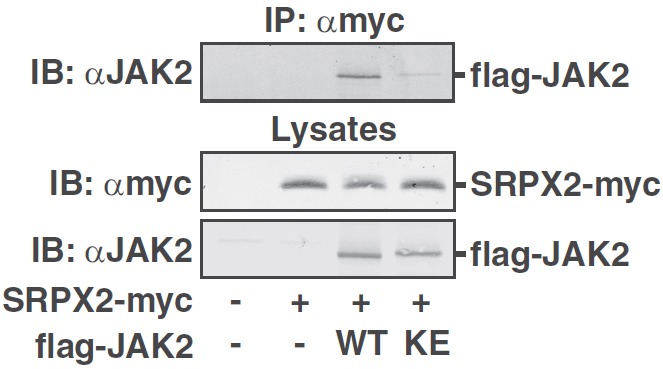
SRPX2, another DUDES-containing protein, binds kinase-active JAK2. 293T cells expressing SRPX2-myc with or without flag-JAK2 (WT) or JAK2 (K882E) (KE) as indicated were immunoprecipitated (IP) with αmyc, and immunoprecipitated proteins were immunoblotted (IB) with αJAK2. Lysates were immunoblotted with αmyc and reprobed with αJAK2. The migration of flag-JAK2 and SRPX2-myc is indicated (n = 3).
Trichloroacetic acid precipitation
Ten hours after transfection, medium was changed to 1% calf serum ± 1 μg/ml BFA. Medium was collected 16 hours later and centrifuged at 800 g for 2 minutes. The supernatant was transferred to a new tube, trichloroacetic acid (TCA) added (15% final concentration), and the tube incubated on ice for 5 minutes. Precipitated proteins were pelleted at 16 000g at room temperature for 5 minutes. The pellet was resuspended in 1% SDS, incubated at 37°C for 5 minutes, and neutralized with 10n NaOH.
Immunocytochemistry
Transfected COS-7 cells were grown on coverslips and fixed in medium containing 4% paraformaldehyde (PFA) at 37°C for 15 minutes. The cells were permeabilized in 0.1% Triton X-100 in PBS containing 4% PFA at room temperature for 7 minutes and then washed twice with PBS. Cells were incubated with 5% fat-free milk in PBS for 30 minutes and primary antibody in 5% milk in PBS for 30 minutes, rinsed twice with PBS, incubated with fluorescently labeled secondary antibody in 5% milk in PBS for 30 minutes, and washed with PBS. The cells were incubated with the nuclear stain DAPI (2 ng/ml) for 10 minutes and rinsed in distilled water. The coverslip was treated with prolong Gold antifade reagent (Life Technologies) and mounted on a microscope slide.
Microscopy
Fluorescence microscopy used a Nikon Eclipse TE200 microscope with a ×60 oil objective (Nikon, Melville, New York). Images were captured with a CoolSnap HQ digital camera (Roper Scientific, Martinsried, Germany) and visualized using MetaVue imaging software (Molecular Devices, Sunnyvale, California). When intracellular proteins were visualized, the microscope was focused within the cell. To visualize extracellular Ccdc80, the microscope was focused on the surface of the coverslip. The region of extracellular staining that surrounded the cell was manually traced, and MetaVue's region statistics function was used to quantify the signal intensity of the region selected. The trace was then dragged outside of the area of Ccdc80-myc secretion, and the signal intensity at this location was used to correct for background signal. Extracellular staining was assessed by a blinded observer. The observer randomly selected 8 to 10 isolated Ccdc80-myc–positive cells per experiment per condition. The experiment was performed 3 times. Means ± SEM are shown. The subcellular distribution of Ccdc80-myc and GFP-JAK2 was visualized with an Olympus FluoView 500 laser scanning confocal microscope using a ×60 oil-immersion objective and FluoView software, version 5.0 (Olympus, Center Valley, Pennsylvania). AlexaFluor 594 fluorescence was excited with a green helium neon laser at 543 nm. Emission was measured through a 560-nm long-pass filter (560 nm and above). GFP and/or Oregon green fluorescence was excited with an argon laser at 488 nm. Emission was measured through a 505- to 525-nm filter. Images to assess colocalization of Ccdc80-myc and GFP-JAK2 were captured using sequential scanning to avoid fluorescence cross talk.
Results
Identification of SSG1/Ccdc80 as a binding partner of JAK2 using the yeast 2-hybrid system
In an effort to identify novel cytokine receptor effector proteins that are recruited to activated JAK2, full-length JAK2 fused to the DNA-binding portion of LexA was used to screen for JAK2 interacting proteins in a yeast 2-hybrid system. Previous work showed that LexA-JAK2 is expressed in yeast, is active as a kinase, is tyrosyl phosphorylated, and does not auto-activate the system (36). An adipose tissue library was used because GH and a number of other cytokines (eg, IL-6, IL-11, leukemia inhibitory factor, leptin, interferon-γ, and oncostatin M) that activate JAK2 elicit multiple effects in adipocytes (44–49). Over 110 positive clones were isolated from 11 × 106 yeast transformants. Three clones, of the 9 first sequenced, encoded a protein that closely corresponded to the carboxyl-terminal portion of rat SSG1/Ccdc80. SSG1/Ccdc80 was initially cloned from rat uterus (GenBank accession number AF223677) and predicted to be a 385-aa, 41.8-kDa protein (23). Multiple mismatches were present between the sequence of our yeast 2-hybrid clone and the published sequence for SSG1. When we resequenced the full-length SSG1 cDNA of Marcantonio et al (23), we found that rat SSG1 cDNA codes for a 110-kDa protein of 949 aa and is identical to rat CL2 (24) and rat DRO1 (GenBank accession number AY548105) (25). The sequence of our yeast 2-hybrid clone corresponds to the C-terminal portion, aa 477–949, of the full-length protein (Figure 1A). The amino acid sequence of rat SSG1/Ccdc80 shares high identity with murine Urb (94%) (GenBank accession number AB075019), also cloned from adipose tissue (26), and human Urb (84%) (GenBank accession number AF506819). Rat SSG1/Ccdc80 also shares considerable amino acid sequence identity (69%) with chicken Equarin-L (GenBank accession number AB086824).
Figure 1.
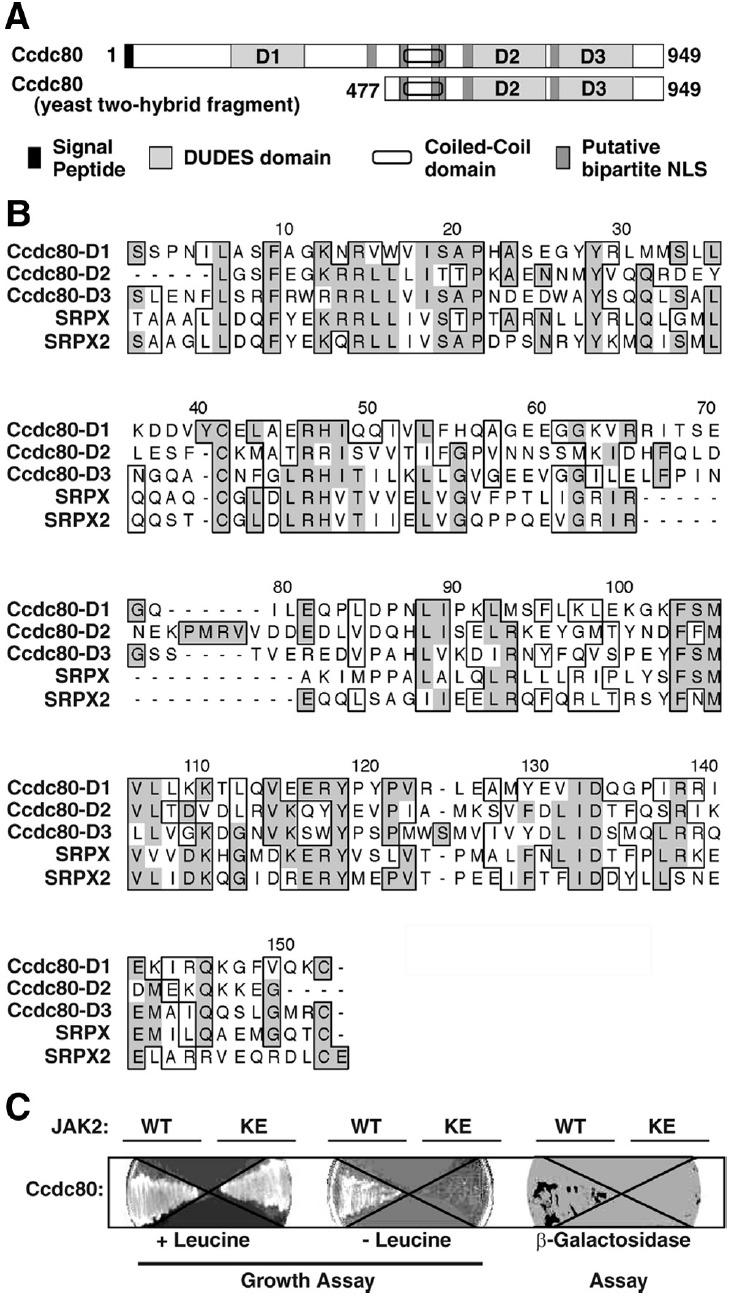
Identification of SSG1/Ccdc80 as a binding partner for JAK2 using the yeast 2-hybrid system. A, The domain structure for full-length Ccdc80 and the yeast 2-hybrid fragment Ccdc80 (477–949) are shown. The 3 DUDES domains (D1, D2, and D3) are noted. B, Clustal W plot of the 3 DUDES domains in Ccdc80 (D1, D2, and D3) and the DUDES domains in SRPX/drs/exon trapped x-chromosome clone 1 and SRPX2/SRPUL. Identical amino acids are shaded in gray; similar amino acids are boxed. C, Yeast cells transformed with cDNA encoding Ccdc80 (477–949) in the prey vector with cDNA encoding either JAK2 (WT) or JAK2 (K882E) (KE) fused to a LexA DNA-binding domain were streaked onto 15% agar-containing dropout medium in the absence (− leucine) or presence (+ leucine) of leucine or with X-gal (β-galactosidase assay).
Sequence analysis indicates that Ccdc80 contains a putative signal peptide (aa 1–24), 6 putative bipartite nuclear localization signals, a coiled-coil domain (aa 546–586), and 3 consensus repeat domains (aa 137–281 [designated domain 1 or D1], aa 615–756 [D2], and aa 766–912 [D3]) (Figure 1A). The consensus repeat domains display approximately 30% amino acid identity and 40% to 50% amino acid similarity to each other (Figure 1B) and to the fifth domain of murine SRPX (GenBank accession number CAM18748) (32). These domains have been designated DUDES domains (25). The DUDES domains of Ccdc80 are highly conserved between species, with rat and chicken Ccdc80 showing 88%, 85%, and 89% amino acid identity for D1, D2, and D3 respectively.
Intracellular Ccdc80 associates preferentially with and is phosphorylated by kinase-active JAK2
To begin to characterize how Ccdc80 and JAK2 interact, we first determined whether JAK2 kinase activity and/or tyrosyl phosphorylation of JAK2 is required for the interaction of JAK2 with the fragment of Ccdc80 that bound JAK2 in the original yeast 2-hybrid screen. The Ccdc80 (477–949) prey plasmid was transformed into yeast with either LexA-JAK2 or the catalytically inactive LexA-JAK2 (K882E) in which the critical lysine in the ATP-binding site is mutated to glutamate. Ccdc80 (477–949) bound to LexA-JAK2 in the yeast 2-hybrid assay but not to LexA-JAK2 (K882E) (Figure 1C). This result suggested that Ccdc80 binds preferentially to tyrosyl-phosphorylated, kinase-active JAK2.
To verify that Ccdc80 and JAK2 interact in mammalian cells, HA-Ccdc80 (477–949) and JAK2 wild-type (WT), JAK2 (K882E), or JAK2 (Y1007F) were coexpressed in 293T cells. JAK2 (K882E) is completely inactive (40). In JAK2 (Y1007F), mutation of the activating tyrosine in the kinase domain of JAK2 markedly reduces, but does not eliminate, both the kinase activity and tyrosyl phosphorylation of JAK2 (50). When HA-Ccdc80 (477–949) was immunoprecipitated with αHA, immunoblotting with αJAK2 revealed that JAK2 coprecipitates with HA-Ccdc80 (477–949) to a much greater extent than either JAK2 (K882E) or JAK2 (Y1007F) (Figure 2A). Reprobing with αPY (Figure 2A) demonstrated that both HA-Ccdc80 (477–949) and the JAK2 that coprecipitated with HA-Ccdc80 (477–949) are tyrosyl phosphorylated when HA-Ccdc80 (477–949) is coexpressed with JAK2 but not with kinase-inactive JAK2 (K882E) or JAK2 (Y1007F). Immunoblotting the lysates with αJAK2 and αHA (Figure 2A) revealed that HA-Ccdc80 (477–949) migrates as a 60-kDa protein as predicted and that differences in coprecipitation of the various JAK2 proteins with HA-Ccdc80 (477–949) are not an artifact due to differences in expression of the various JAK2 proteins or of HA-Ccdc80 (477–949).
Figure 2.
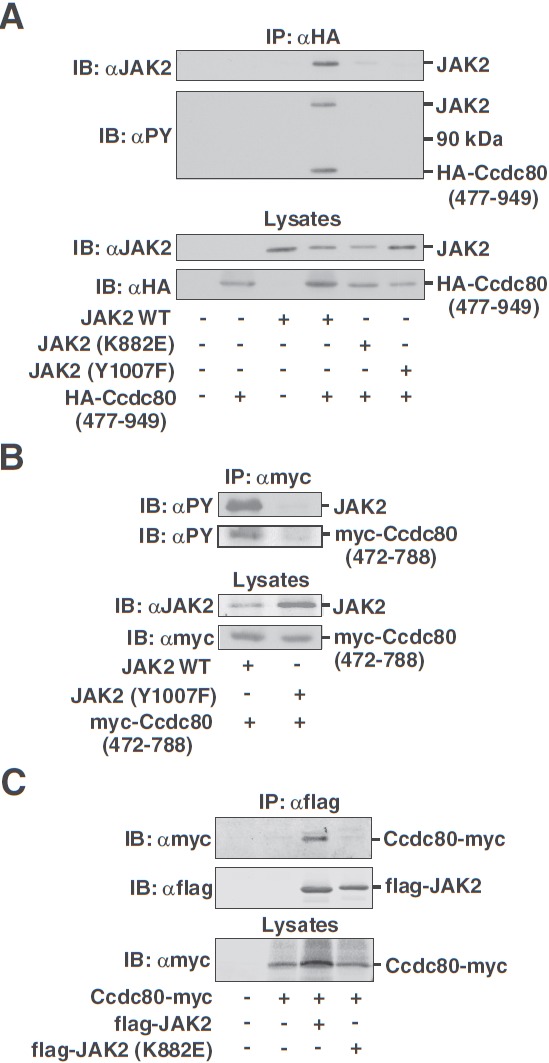
Ccdc80 (477–949) and full-length Ccdc80 bind to kinase-active, but not kinase-inactive, JAK2 in mammalian cells. JAK2 appears to phosphorylate tyrosines in the region between aa 472 and 788 of Ccdc80. A, 293T cells expressing HA-Ccdc80 (477–949) and JAK2, JAK2 (K882E), or JAK2 (Y1007F) as indicated were immunoprecipitated (IP) using αHA, and immunoprecipitated proteins were immunoblotted (IB) with αJAK2 or αPY. Cell lysates were immunoblotted with αJAK2 or αHA as indicated. The migration of JAK2, HA-Ccdc80 (477–949), and a 90-kDa molecular mass marker is indicated (n = 2–5). B, 293T cells expressing myc-Ccdc80 (472–788) and either JAK2 or JAK2 (Y1007F) were immunoprecipitated (IP) with αmyc, and immunoprecipitated proteins were immunoblotted (IB) with αPY. Cell lysates were immunoblotted with αJAK2 or αmyc as indicated. The migration of JAK2 and myc-Ccdc80 (472–788) is indicated (n = 2–5). C, 293T cells expressing Ccdc80-myc and either flag-JAK2 (WT) or flag-JAK2 (K882E) (KE) as indicated were immunoprecipitated (IP) using αflag, and immunoprecipitated proteins were immunoblotted (IB) with αmyc and reprobed with αflag. Cell lysates were immunoblotted with αmyc. The migration of the flag-tagged JAK2 constructs and Ccdc80-myc (at ∼110 kDa) is indicated (n = 5).
Because the HA tag contains tyrosines, to verify that Ccdc80 itself is tyrosyl phosphorylated, we repeated the assay using myc-tagged Ccdc80 (472–788). The myc tag contains no tyrosines. Myc-Ccdc80 (472–788) was tyrosyl phosphorylated when coexpressed with JAK2 WT, but not JAK2 (Y1007F) (Figure 2B), consistent with Ccdc80 being a substrate of JAK2. Furthermore, the JAK2 that coprecipitated with myc-Ccdc80 (472–788) was tyrosyl phosphorylated.
To determine whether full-length Ccdc80, like Ccdc80 (472–788) and Ccdc80 (477–949), preferentially binds kinase-active JAK2 in mammalian cells, Ccdc80-myc was coexpressed in 293T cells with flag-tagged JAK2 WT or JAK2 (K822E). Ccdc80-HA-myc coimmunoprecipitated with JAK2 WT but not with JAK2 (K882E) (Figure 2C). Together, the findings of Figure 2 suggest that Ccdc80 interacts preferentially with kinase-active, tyrosyl-phosphorylated JAK2 and that Ccdc80 is tyrosyl phosphorylated by JAK2.
All 3 DUDES domains of Ccdc80 interact with kinase-active JAK2
We next sought to define the regions in Ccdc80 (477–949) that bind JAK2. Carboxyl-terminal truncation mutants of HA-Ccdc80 (477–949) (Figure 3A) were coexpressed with JAK2 in 293T cells. JAK2 coimmunoprecipitated with all but HA-Ccdc80 (477–608) (Figure 3B), which suggests that aa 609–662 of Ccdc80 bind to JAK2. None of the Ccdc80 truncation mutants bound to the kinase-deficient JAK2 mutants, JAK2 (K882E) or JAK2 (Y1007F) (data not shown). Amino acids 609–662 of Ccdc80 contain a portion of the second DUDES domain (D2). When tested, the D2 domain alone was sufficient for binding to JAK2 (Figure 3C). These findings suggest that the second DUDES domain in Ccdc80 binds JAK2 and, furthermore, that the N-terminal third of that domain is sufficient for binding JAK2.
The truncation mutants used to identify the role of the D2 DUDES domain in JAK2 binding cannot exclude the possibility that the D1 and D3 DUDES domains also bind to JAK2. When coexpressed with either JAK2 or JAK2 (K882E) in 293T cells, all 3 myc-tagged DUDES domains coimmunoprecipitated JAK2 to a greater extent than JAK2 (K882E) (Figure 3D). Similar results were obtained with GFP-tagged D1, D2, and D3 domains (data not shown). These results indicate that all 3 DUDES domains interact preferentially with kinase-active JAK2.
The D2 and D3 domains in Ccdc80 bind the activated JH1 kinase domain of JAK2
We next sought to define the regions within JAK2 that bind Ccdc80. Myc-Ccdc80 (477–949) was coexpressed with HA-tagged JAK2 (JAK2-HA), JAK2 lacking its JH2 domain (ΔJH2-HA), or just the JH1 and JH2 domains of JAK2 (JH1–2-HA) (Figure 4A) in 293T cells and immunoprecipitated with αmyc. Precipitated proteins were immunoblotted with αHA (Figure 4B). All 3 forms of JAK2 coprecipitated with Myc-Ccdc80 (477–949), suggesting that the JH1 domain of JAK2 binds Myc-Ccdc80 (477–949). Further truncation of JAK2 to either JAK2 (797–1132) or JAK2 (830–1132), both of which contain just the JH1 domain and the small C-terminal tail of JAK2, confirmed that residues within the kinase domain (JH1) of JAK2 are likely to interact with Ccdc80 (477–949) (Figure 4C).
To determine which DUDES domain binds the JH1 domain of JAK2, myc-tagged D1, D2, and D3 of Ccdc80 were coexpressed in 293T cells with either kinase-active flag-JAK2 (797–1132) (flag-JH1 WT) or kinase-inactive flag-JAK2 (797–1132, K882E) (flag-JH1 KE). Flag-JH1 WT coprecipitated with D3 to an extent that approached the interaction detected with D2 (Figure 4D). None of the DUDES domains coprecipitated with kinase-inactive flag-JH1 KE (Figure 4D). JH1 only weakly interacted with D1 (Figure 4D), suggesting that the primary binding site in JAK2 for DUDES-D1 may be located outside of the JH1 domain.
Because multiple DUDES domains in Ccdc80 can bind to JAK2, and D1 appears to bind to a site outside the JH1 domain, we reasoned that full-length Ccdc80 would bind to full-length JAK2 better than to the JH1 domain. To test this, flag-JAK2 or flag-JH1 WT was coexpressed in 293T cells with Ccdc80-HA-myc that contained an HA tag after the signal peptide and an N-terminal myc tag. Ccdc80-HA-myc bound full-length JAK2 to a much greater extent than JH1 (Figure 4E). The dramatic increase in Ccdc80 binding to full-length JAK2 suggests that the 3 DUDES domains in Ccdc80 interact with multiple sites in JAK2.
SRPX2, another DUDES domain-containing protein, also binds JAK2
We hypothesized that the DUDES domain may be a novel JAK2 or PY-binding domain. Two additional proteins, SRPX and SRPX2, are known to contain DUDES domains (25). SRPX2-myc and either flag-tagged JAK2 or JAK2 (K882E) were coexpressed in 293T cells. Like Ccdc80, SRPX2 preferentially bound to kinase-active JAK2 (Figure 5), providing further evidence that the DUDES domain is a JAK2-binding domain.
Ccdc80 has minimal effect on JAK2 kinase activity but enhances Stat3 and Stat5 phosphorylation
To determine whether Ccdc80 affects JAK2 signaling, we examined whether Ccdc80 alters the kinase activity of JAK2. Flag-JAK2 was coexpressed in 293T cells in the presence or absence of Ccdc80-HA-myc, immunoprecipitated with αflag, and immunoblotted with antibody to the activating PYs in JAK2, αpY1007/1008 JAK2, to assess JAK2 activity. Ccdc80-HA-myc had minimal effect on phosphorylation of tyrosines 1007/1008 in JAK2 (Figure 6A). Similar results were obtained using Ccdc80-myc (data not shown). We next examined whether Ccdc80 affects the ability of GH to stimulate more distal signaling. Stat proteins are well-known GH signaling molecules. Phosphorylation of Stat3 at Tyr705 and Stat5b at Tyr699 is a necessary first step in the activation of Stats 3 and Stat5b, respectively (51). Stat3, rat GH receptor, and Ccdc80-myc were coexpressed in 293T cells. Cells were stimulated with or without 500 ng/ml GH for 15 minutes. Overexpression of Ccdc80 stimulated both basal and GH-dependent Stat3 phosphorylation at Tyr705 with GH-dependent phosphorylation increasing to 4.3 ± 0.6 times control levels (mean ± SEM, P < .05, n = 4) (Figure 6B). In contrast, when Stat5b signaling was monitored, overexpression of Ccdc80 only modestly increased GH-dependent phosphorylation of Stat5b at Tyr694 (28% ± 11%, P < .05, n = 5) (data not shown). In case activation of Stat5b was already maximal at 500 ng/ml GH, thus preventing further activation by Ccdc80, we tested whether Ccdc80 affects GH-dependent Stat5b phosphorylation when the cells were treated with a lower concentration of GH. When cells were stimulated with or without 50 ng/ml GH, overexpression of Ccdc80 increased GH-dependent phosphorylation of Stat5b at Tyr699 to 2.5 ± 0.75 (P < .05, n = 3) times control levels after 30 minutes of treatment with GH (Figure 6C).
Figure 6.
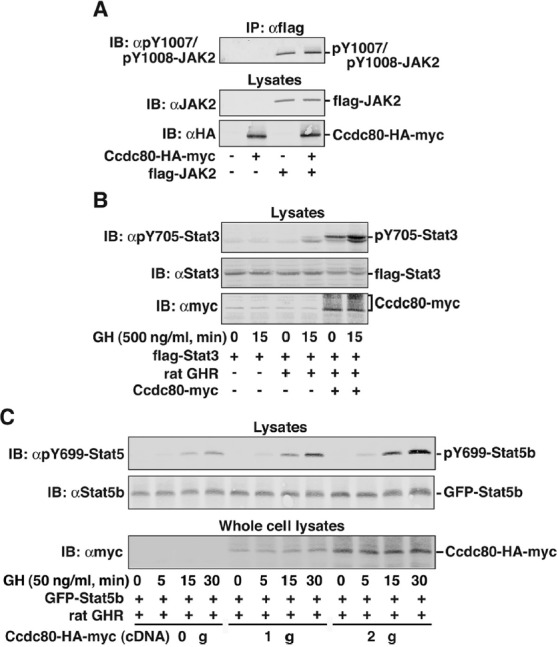
Ccdc80 has minimal effect upon JAK2 kinase activity but stimulates the tyrosyl phosphorylation of Stat3 and Stat5b. A, 293T cells expressing Ccdc80-HA-myc and flag-JAK2 as indicated were immunoprecipitated (IP) with αflag agarose beads, and immunoprecipitated proteins were immunoblotted (IB) with αpY1007/pY1008 JAK2. Cell lysates were immunoblotted with αHA. The blot was reprobed with αJAK2. The migration of flag-JAK2 and Ccdc80-HA-myc is indicated (n = 4). B, 293T cells expressing flag-Stat3, rat GH receptor (GHR), and Ccdc80-myc were serum-starved overnight and stimulated with 500 ng/ml GH for 15 minutes. Cell lysates were immunoblotted (IB) with αpY705-Stat3 and reprobed with αStat3. A parallel set of cell lysates was probed with αmyc. The migration of pY705-Stat3, Stat3 and Ccdc80-myc is indicated (n = 4–6). C, 293T cells expressing GFP-Stat5b, rat GH receptor (GHR), and Ccdc80-HA-myc as indicated were serum-starved overnight and stimulated with 50 ng/ml GH for the indicated times. Cell lysates were immunoblotted (IB) with αpY699-Stat5b and reprobed with αStat5b. Whole-cell lysates were immunoblotted with αmyc. The migration of pY699-Stat5b, GFP-Stat5b and Ccdc80-HA-myc is indicated (n = 3).
In the presence of Ccdc80, the Stat3 bands in the αpY705-Stat3 blot migrate slightly higher than the corresponding bands observed in the absence of Ccdc80 (Figure 6B and data not shown). An upward shift in migration of proteins is consistent with the presence of an additional posttranslational modification. Stat3 is known to be phosphorylated on Ser727 (52); however, immunoblotting with a phosphospecific antibody to Ser727 indicated that overexpression of Ccdc80 does not affect the phosphorylation of Stat3 Ser727 (data not shown). Therefore, phosphorylation if present occurs on another residue, or perhaps the shift in migration is due to another type of posttranslational modification.
We next wished to investigate whether Ccdc80 affected GH signaling in a naturally GH-responsive cell line. 3T3-F442A preadipocytes support robust GH signaling and are frequently used as a model system for GH signaling (22, 36). We were able to express Ccdc80-HA-myc in 3T3-F442A preadipocytes. However, additional Ccdc80 did not enhance the ability of GH (25–50 ng/ml) to stimulate the phosphorylation of endogenous Stat3, Stat5, Erk1, or Erk2 (data not shown). The inability of Ccdc80-HA-myc expression to enhance phosphorylation of Stat3 or Stat5 in these experiments could arise if the level of Ccdc80 was not altered substantially by expression of additional Ccdc80 in the cells. In support of this possibility, the GEO profile (NCBI) for Ccdc80 in 3T3-F442A cells suggests that Ccdc80 mRNA is relatively abundant.
Cellular localization of Ccdc80
Ccdc80 contains a signal peptide as well as several putative nuclear localization sequences (Figure 1), which suggests that Ccdc80 may localize to and possibly move between multiple cellular compartments. To determine the localization of Ccdc80, Ccdc80-HA-myc or Ccdc80-myc was overexpressed in COS-7 cells, immunostained using αHA or αmyc, and visualized by fluorescence microscopy (Figure 7, A–I). The perinuclear region was intensely stained in a pattern suggestive of the endoplasmic reticulum and/or Golgi apparatus. Only rarely was Ccdc80 seen in the nucleus (data not shown), suggesting that the nuclear localization sequences do not serve to localize Ccdc80 to the nucleus or that the cells or culture conditions used did not favor nuclear accumulation. Colocalization of the HA and myc stains to the perinuclear region precludes the possibility that an N- or C-terminal portion of a cleaved Ccdc80 localizes to the nucleus. Counterstaining with DAPI confirmed that Ccdc80-myc strongly localized to the perinuclear region (Figure 7F). Furthermore, Ccdc80 colocalized with the Golgi marker giantin (Figure 7, D–F) and the endoplasmic membrane marker calnexin (Figure 7, G–I). Darker exposures and exposures focusing on different planes revealed Ccdc80 throughout the cytoplasm (see Figure 9, data not shown).
Figure 7.
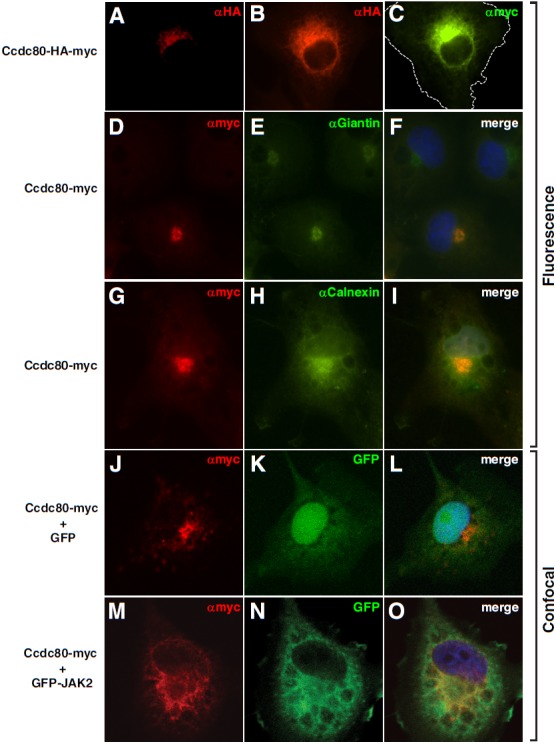
Localization of Ccdc80. COS-7 cells expressing Ccdc80-HA-myc, Ccdc80-myc, GFP, and/or GFP-JAK2 as indicated were fixed with 4% PFA and immunostained using αmyc or αHA followed by staining with AlexaFluor594 (A, B, D, F, G, and I) or AlexaFluor488 (C). Cells were stained for the Golgi marker giantin (E and F) or the endoplasmic reticulum marker calnexin (H and I) followed by staining with Oregon green. Nuclei were visualized with DAPI (F, I, L, and O). Images A–I are fluorescent images. Images J–O are confocal images. Exposure times were selected to avoid saturation where possible. The dotted line in image C represents the cell boundary. Image B is a longer exposure of the cell in image A.
Figure 9.
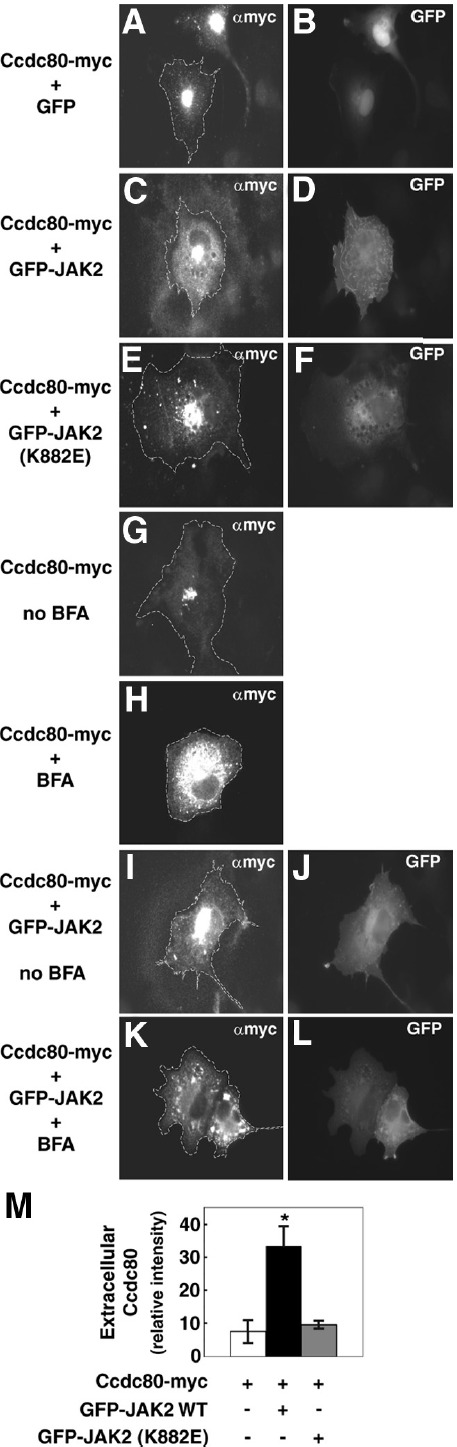
JAK2 increases the amount of Ccdc80 that binds to the extracellular matrix. A–L, COS-7 cells expressing Ccdc80-myc and GFP, GFP-JAK2, or GFP-JAK2 (K882E) as indicated were treated with or without 10 μg/ml BFA for 16 hours and fixed with 4% PFA in the presence of medium. Ccdc80-myc was immunostained with αmyc followed by AlexaFluor594-conjugated anti-rabbit IgG. The cells were visualized using a Nikon Eclipse TE200 fluorescent microscope with the microscope focused on the region of the cells adjacent to the coverslip. The dotted lines represent cell boundaries. M, The intensity of the region of extracellular staining that surrounded the cell was calculated as described in Materials and Methods. The data are expressed as means ± SEM of 3 independent experiments in which 8–10 cells were assessed for each condition. *, P < .05.
To monitor whether JAK2 colocalizes with Ccdc80 or alters the localization of Ccdc80, confocal microscopy was used to visualize the subcellular localization of Ccdc80-myc in the absence and presence of GFP-JAK2. The localization of Ccdc80 in the absence of coexpressed JAK2 as visualized by confocal microscopy (Figure 7J) was similar to that visualized by fluorescence microscopy (Figure 7, A–I). No major shifts in subcellular localization were detected in the presence of JAK2. Low level staining of Ccdc80 was easily detectable throughout the cytoplasm with prominent perinuclear Ccdc80 (Figure 7, M–O). The localization of Ccdc80 to the cytoplasmic region is consistent with the ability of Ccdc80 to affect intracellular signaling (eg, bind JAK2 or stimulate Stat3 and Stat5 phosphorylation). However, the localization of Ccdc80 to the ER/Golgi suggests that at least a portion of Ccdc80 is likely to be secreted.
Secretion of Ccdc80
Ccdc80 and the various Ccdc80 orthologs have been reported to be secreted in some studies (28, 29) and not secreted in others (25, 53). To determine whether Ccdc80 is secreted in the system used in the current study, Ccdc80-myc was expressed in 293T cells, the proteins in the medium precipitated with TCA, and the proteins in the lysate and the proteins precipitated from the medium were blotted with αmyc. A tight, ∼120-kDa band and a lighter diffuse 125- to 175-kDa band are evident in the cell lysates (Figure 8A). Myc-Ccdc80 is also detected in the medium (Figure 8A). The secreted Ccdc80-myc migrates as a diffuse 125- to 175-kDa band, with cleavage products detected between 50 and 70 kDa. Notably, Ccdc80 cleavage seems to be restricted to the Ccdc80 that is exported; cleavage products are not detected in the cell lysate. Supporting the conclusion that myc-Ccdc80 is exported via an active secretion process, BFA, an inhibitor of protein secretion (54), blocks Ccdc80 secretion (Figure 8A). These data suggest that in 293T cells, a portion of Ccdc80 undergoes posttranslational modification with the modified, presumably glycosylated, Ccdc80 secreted into the medium via a BFA-sensitive secretion pathway. A portion of the secreted Ccdc80 is cleaved. Secretion of the larger, diffuse form of Ccdc80 is also detected in cultured 3T3-F442A cells ectopically expressing Ccdc80-myc (Figure 8B).
Figure 8.
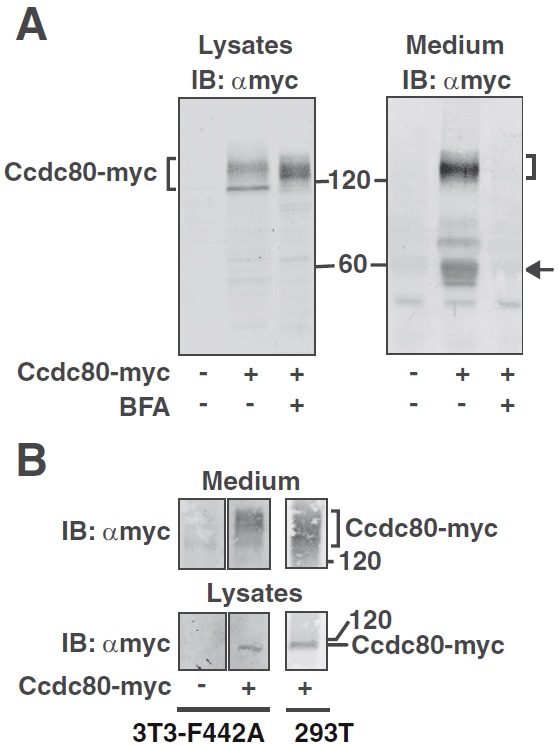
Ccdc80 is secreted. A, 293T cells expressing Ccdc80-myc were incubated in medium containing 1% calf serum with or without 1 μg/ml BFA for 16 hours as indicated. Only minimal cell death was detected. Cell lysates and proteins precipitated from the medium using TCA were immunoblotted (IB) with αmyc (n = 3). B, cDNA encoding empty vector or Ccdc80-myc was introduced into 3T3-F442A preadipocytes by electroporation. As a positive control, Ccdc80-myc overexpressed in 293T cells was similarly processed and included as a reference. Cell lysates and the proteins precipitated from the medium were immunoblotted with αmyc (n = 2). The migration of the 60- and 120-kDa molecular mass standards, Ccdc80-myc, and cleavage products of Ccdc80-myc (arrow) is indicated.
JAK2 increases the amount of Ccdc80 that binds to the extracellular matrix
The extracellular matrix is known to sequester growth factors and other proteins secreted from cells (55, 56). Thus, we investigated whether the secreted Ccdc80 associates with the extracellular matrix in the region surrounding the cell. Ccdc80-myc was coexpressed with or without GFP-JAK2 in COS-7 cells and immunostained with αmyc, and the region of the cell that contacts the glass slide was visualized by fluorescence microscopy. In cells expressing Ccdc80-myc alone, we observed faint αmyc staining in insoluble particulate in the area surrounding the outside of the cells (Figure 9, A and G) (best seen by comparing the gray background of Figure 9, A or G, with the black background in Figure 9H). In the presence of GFP-JAK2, a halo of Ccdc80-myc surrounding the cell is readily visible (Figure 9, C, I, and M). Minimal if any extracellular Ccdc80-myc was detected in the presence of GFP-JAK2 (K882E) (Figure 9, E and M). In contrast to Ccdc80-myc, GFP-JAK remained intracellular (Figure 9, D and J). As expected, inhibition of protein secretion with BFA suppressed the Ccdc80 in the extracellular region (Figure 9, H and K), which confirms that Ccdc80 secretion is required for Ccdc80 to associate with the extracellular matrix.
Discussion
In this study using a yeast 2-hybrid screen of a rat adipose cDNA library, we identified SSG1/Ccdc80 as a JAK2-binding protein. Although SSG1/Ccdc80 had been previously cloned (23), its size and sequence were incorrect. Here, we provide evidence that rat SSG1/Ccdc80 is a protein of 949 aa (∼110 kDa) that is identical to rat DRO1 (25) and rat CL2 (24), binds to activated JAK2, and is phosphorylated by JAK2.
Little is known about Ccdc80 function. It is expressed in a number of tissues in both the embryo and adult. It is down-regulated in thyroid, ovarian, pancreatic, and colon cancer cell lines and tumors (29). Overexpression of Ccdc80 in colorectal and pancreatic cancer cell lines inhibits malignant growth and suppresses anchorage-independent growth (25), suggesting that Ccdc80 may be a tumor suppressor. Murine Ccdc80 (Urb) is highly expressed in adipose tissue (57), the tissue used to prepare the library used in our yeast 2-hybrid screen. Ccdc80 mRNA expression was found to be down-regulated in obese mouse models, including ob/ob, KKAy, and diet-induced obese mice. In 3T3-L1 adipocytes, insulin, TNF-α, H2O2, and hypoxia decrease Ccdc80 mRNA levels, suggesting that these factors may contribute to the down-regulation of Ccdc80 seen with obesity (29). When mice lacking Ccdc80 are fed a high-fat diet, the absence of Ccdc80 in these mice is associated with a worsening of diet-induced glucose intolerance and hyperglycemia and reduced glucose-stimulated insulin secretion (58). In 3T3-L1 cells, the expression of Ccdc80 fluctuates during differentiation into adipocytes. Before differentiation, the expression of Ccdc80 increases as the cells become confluent. When differentiation is induced with dexamethasone or 3-isobutyl-1-methylxanthine, Ccdc80 levels are rapidly suppressed. Ccdc80 is expressed again during the late stages of differentiation. Knockdown of Ccdc80 confirmed that Ccdc80 is required for differentiation to adipocytes, apparently by a mechanism involving the inhibition of T-cell factor-mediated transcriptional activity and wnt/β-catenin signaling (30). Because Ccdc80 has a wide tissue distribution (23–25), and JAK2 is activated by nearly two-thirds of the cytokine receptors, it seems likely that Ccdc80 plays a role in not only GH signaling but also signaling by other ligands that bind to members of the cytokine receptor family. Consistent with this hypothesis, in the bombesin-receptor-subtype (BRS)-3–deficient model of obesity, the brown adipose tissue is characterized by elevated Ccdc80 levels, suppressed GH levels, and dramatically elevated leptin levels (26, 59). An inhibitory role for Ccdc80 on leptin signaling would be consistent with the apparent leptin resistance detected in these mice.
Because there is considerable homology between the 4 members of the JAK family, we were also interested in whether Ccdc80 binds to any of the other members of the JAK family. Ccdc80 appears to bind to JAK3 but only minimally to JAK1 (data not shown). Therefore, there appears to be some specificity in the ability of the various JAK kinases to bind and thus signal through Ccdc80.
When we investigated the mechanism by which Ccdc80 and JAK2 interact, we found that the 3 DUDES domains of Ccdc80 bind JAK2. Maximal binding required activated, tyrosyl-phosphorylated JAK2. Further analysis of the second (D2) DUDES domain suggested that for at least the D2 domain, the N-terminal third of the domain is sufficient for JAK2 binding. The second (D2) and third (D3) DUDES domains preferentially bind to the kinase (JH1) domain of JAK2. This finding adds Ccdc80 to the growing list of proteins (eg, SOCS-1 [11], SOCS-3 [12], protein tyrosine phosphatase-1B [13], and suppressor of T cell receptor signaling-1 [14]) that bind to the kinase domain of JAK2. In contrast, DUDES domain D1 in Ccdc80 appears to bind to JAK2 outside the kinase domain. Because the 3 DUDES domains in Ccdc80 bind to 2 distinct regions in activated, tyrosyl-phosphorylated JAK2, the DUDES domain may be a novel PY-binding domain. However, we have not been able to identify specific tyrosines in JAK2 that bind to the DUDES domains in Ccdc80 (data not shown), raising the possibility that each DUDES domain can bind to multiple PYs in JAK2.
The DUDES domain was first identified in the protein SRPX (32) and subsequently identified in SRPX2 (33) and Ccdc80 (26). DUDES domains were also identified in numerous additional proteins of prokaryotic origin via a PSI-Blast search (60). Bioinformatics analysis assigned most the identified domains to the Pfam thioredoxin-like clan of protein domains, although the canonical cysteine residue in the active site is not conserved (60). The DUDES domains in Ccdc80, SRPX, and SRPX2 share roughly 30% amino acid identity. Our finding that the DUDES domains of Ccdc80 bind JAK2 is the first function attributed to this domain. We hypothesized that SRPX2 and SRPX might also bind JAK2. Significantly, we showed that SRPX2, like Ccdc80, binds activated, but not kinase-dead, JAK2. Although not tested, we predict that SRPX will also bind JAK2.
SRPX2 and its cellular function are just beginning to be studied. SRPX2 mRNA is expressed in heart, ovary, and placenta (33), and SRPX2 protein has been visualized in brain (61). SRPX2 gene expression appears to be induced by the E2A-HLF oncoprotein (33). Mutations in SRPX2 have been associated with a rolandic/sylvian person affected by epilepsy seizures and perisylvian polymicrogyria (61, 62). The N372S mutation results in partial gain of N-glycosylation resulting in increased SRPX2 protein secretion and protein misfolding (61). The R75K mutation in SRPX2 plays a role in 2 disorders of the rolandic/sylvian speech area in the brain and is hypothesized to be important for protein-protein interaction (62). Recently, SRPX2 was determined to be the ligand for urokinase-type plasminogen activator receptor, important for regulating cell adhesion, degradation of the extracellular matrix, migration, angiogenesis, and apoptosis (63). The determination that JAK2 binds SRPX2 suggests that SRPX2, like Ccdc80, may function in both the intracellular and extracellular compartments. Furthermore, the binding of JAK2 to SRPX2 suggests that investigations of ligands that activate JAK2 in affected tissues might provide insight into the mechanisms underlying these medical conditions.
Our investigations of how Ccdc80 affects signaling downstream of JAK2 revealed that Ccdc80 does not seem to regulate JAK2 kinase activity. However, Ccdc80 activates both Stat3 and Stat5 as monitored by phosphorylation of critical tyrosines required for activation and appears to promote additional modifications on Stat3 at presently unknown sites. Together, these results suggest that although Ccdc80 has only a minimal effect upon JAK2 activation and is therefore unlikely to globally stimulate the JAK2 signaling network, Ccdc80 may stimulate a specific subset of the signaling molecules in the JAK2 network.
Our findings that Ccdc80 binds JAK2 and that coexpression of Ccdc80 with JAK2 elevates phosphorylation of Stat3 and Stat5 suggest that Ccdc80 functions in the intracellular compartment. Furthermore, immunocytochemistry presented in The Human Protein Atlas (http://www.proteinatlas.org/) indicates that in some cells, Ccdc80 is primarily nuclear. Visconti and coworkers (24) report that in Cos cells expressing GFP-Ccdc80, in some cells, cytosolic and nuclear-nucleolar Ccdc80 staining is detected. These findings further support an intracellular role for Ccdc80. However, Ccdc80 contains a putative signal peptide, the hallmark of a protein destined for processing by the endoplasmic reticulum/Golgi and secretion. Indeed, we and others observed that Ccdc80 localizes to the perinuclear region of the cell and comigrates with markers for the endoplasmic reticulum and Golgi (25, 31), is secreted from the cell (28, 29), binds to the extracellular matrix (27, 28, 64), and, once secreted, is cleaved into several fragments (25, 27, 29, 30). However, not all cells that express Ccdc80 secrete Ccdc80. Intracellular, but not secreted, Ccdc80 was detected in dermal papilla cells (53). Tremblay et al (30) detected flag-Ccdc80 in conditioned medium of 293T cells expressing flag-Ccdc80 and determined that the secreted, soluble form of Ccdc80 stimulates adipogenesis. In our studies, because at least a portion of Ccdc80 is secreted, one could argue that the effects we observed on Stat3 and Stat5 were indirect effects of secreted Ccdc80 acting in an autocrine fashion. However, conditioned medium from cells overexpressing Ccdc80 did not mimic the effects we observed in cells overexpressing Ccdc80 (data not shown).
The trafficking of proteins containing a signal peptide is only partially understood; some glycoproteins are present in the cytosol, and numerous proteins are located in the cytosol as well as the endoplasmic reticulum and/or the extracellular compartment (65–67). For example, stromal interaction molecule 2 is present in both the cytoplasmic and endoplasmic reticulum, and both forms are essential for stromal interaction molecule 2 (STIM2) to regulate the influx of calcium into the cell (67). In U937 monocytes, plasminogen activator inhibitor-2 exists as both a cytoplasmic form and a secreted form. Synthesis of both forms is initiated from the same AUG codon. The fact that the relative amount of each form depends upon the differentiation state of the cell suggests that the process is regulated (68–70).
In yeast, the presence of a small or acidic amino acid at the second residue of a protein increases the cleavage of the N-terminal methionine, which increases the likelihood of N-terminal acetylation. N-terminal acetylation interferes with the function of the signal peptide and reduces transport to the endoplasmic reticulum (71). In mammals, the effect of N-terminal processing is less pronounced but is still evident (71). Significantly, the second residue in human Ccdc80 and SRPX2 is threonine, and in SRPX, the second residue is glycine. If human methionine aminopeptidase and N-α-acetyl transferase have similar specificities to their counterparts in yeast, Ccdc80, SRPX, and SRPX2 would be fair to good substrates for cleavage and acetylation, and the presence of both cytoplasmic and secreted Ccdc80, SRPX, and SRPX2 would be probable.
In summary, we identified Ccdc80/SSG1/DRO1/CL2/Urb as a novel JAK2-binding protein with a molecular mass of ∼110 000 Da. JAK2 interacts with Ccdc80 via the DUDES domains of Ccdc80. Consistent with DUDES domains serving as JAK2-binding domains, we demonstrated that SRPX2, another protein that contains a DUDES domain, binds activated tyrosyl-phosphorylated JAK2. We also observed that Ccdc80 is tyrosyl phosphorylated in the presence of activated JAK2, that in the presence of Ccdc80 both basal and GH-stimulated Stat3 and Stat5 phosphorylation is increased, and that when JAK2 levels increase, the amount of Ccdc80 associated with the extracellular matrix is elevated. The findings that Ccdc80 associates with and is likely phosphorylated by JAK2 and is expressed in a large number of tissues both in the embryo and adult, that expression of Ccdc80 appears to be highly regulated, and that Ccdc80 has been implicated in a wide range of functions, including tumor suppression and obesity, suggests that Ccdc80 may play an important and fundamental role in cell function.
Acknowledgments
We thank Joel Cline, Baljeet Deo, Xiaqing Wang, and Alison Su for technical assistance; Barbara Hawkins for assistance with the manuscript and figures; Dr Olli Silvennoinen for cDNA encoding truncated versions of JAK2; Dr Thomas Gustafson for the yeast 2-hybrid rat adipose cDNA library; and Drs Anne Vojtek, Stephen Ethier, Jessica Schwartz, Ram Menon, Liangyou Rui, and Benjamin Margolis for helpful discussions and suggestions.
This work was supported by National Institutes of Health (NIH) Grant DK34171 (to C.C.-S.). A.M.M.-M. and E.E.O'L. were supported by a Predoctoral Fellowship from the Cellular and Molecular Biology Training Grant (NIH T32-GM 073115). E.E.O. was also supported by a Predoctoral Fellowship from the Systems and Integrative Biology Training Grant (NIH T32-GM008322). cDNA sequencing was supported by the Cellular and Molecular Biology Core of the Michigan Diabetes Research and Training Center (NIH P60-DK20572), University of Michigan Rheumatologic Diseases Comprehensive Center (NIH P60-AR20557), and the University of Michigan Comprehensive Cancer Center (NIH P30-CA46592). Confocal microscopy was supported by the Morphology and Image Analysis Core of the Michigan Diabetes Research and Training Center (NIH P60-DK20572).
Disclosure Summary: The authors have nothing to declare.
Footnotes
- aa
- amino acids
- BFA
- brefeldin A
- Ccdc80
- coiled-coil domain-containing protein 80
- CL2
- clone 2
- DAPI
- 4′6-diamidino-2-phenylindole
- DRO1
- down-regulated by oncogenes-1
- drs
- down-regulated by Src
- DUDES
- DRO1-URB-DRS-Equarin-SRPUL
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- JAK1
- Janus kinase 1
- JH2
- JAK homology domain 2
- PFA
- paraformaldehyde
- PY
- phosphotyrosine
- SH2B1
- Src-homology 2B1
- SOCS-1
- suppressor of cytokine signaling 1
- SRPUL
- sushi repeat protein up-regulated in leukemia
- SRPX
- sushi repeat containing protein, X-linked
- SSG1
- steroid-sensitive gene-1
- Stat
- signal transducer and activator of transcription
- TCA
- trichloroacetic acid
- Urb
- up-regulated in brown adipose tissue of bombesin-receptor-subtype-3-deficient mice
- WT
- wild-type.
References
- 1. Smit LS, Meyer DJ, Argetsinger LS, Schwartz J, Carter-Su C. Molecular events in growth hormone-receptor interaction and signaling. In: Kostyo JL, ed. Handbook of Physiology. New York, NY: Oxford University Press; 1999:445–480 [Google Scholar]
- 2. Aaronson DS, Horvath CM. A road map for those who know JAK-STAT. Science. 2002;296:1653–1655 [DOI] [PubMed] [Google Scholar]
- 3. Hou SX, Zheng Z, Chen X, Perrimon N. The Jak/STAT pathway in model organisms: emerging roles in cell movement. Dev Cell. 2002;3:765–778 [DOI] [PubMed] [Google Scholar]
- 4. Lacronique V, Boureux A, Valle VD, et al. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–1312 [DOI] [PubMed] [Google Scholar]
- 5. Peeters P, Raynaud SD, Cools J, et al. Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15:12) in a myeloid leukemia. Blood. 1997;90:2535–2540 [PubMed] [Google Scholar]
- 6. Xie S, Lin H, Sun T, Arlinghaus RB. Jak2 is involved in c-Myc induction by Bcr-Abl. Oncogene. 2002;21:7137–7146 [DOI] [PubMed] [Google Scholar]
- 7. Levine RL, Wadleigh M, Cools J, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397 [DOI] [PubMed] [Google Scholar]
- 8. Kralovics R, Passamonti F, Buser AS, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790 [DOI] [PubMed] [Google Scholar]
- 9. Baxter EJ, Scott LM, Campbell PJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061 [DOI] [PubMed] [Google Scholar]
- 10. James C, Ugo V, Le Couedic JP, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148 [DOI] [PubMed] [Google Scholar]
- 11. Yasukawa H, Misawa H, Sakamoto H, et al. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. EMBO J. 1999;18:1309–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sasaki A, Yasukawa H, Suzuki A, et al. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes Cells. 1999;4:339–351 [DOI] [PubMed] [Google Scholar]
- 13. Myers MP, Andersen JN, Cheng A, et al. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J Biol Chem. 2001;276:47771–47774 [DOI] [PubMed] [Google Scholar]
- 14. Carpino N, Kobayashi R, Zang H, et al. Identification, cDNA cloning, and targeted deletion of p70, a novel, ubiquitously expressed SH3 domain-containing protein. Mol Cell Biol. 2002;22:7491–7500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carpino N, Turner S, Mekala D, et al. Regulation of ZAP-70 activation and TCR signaling by two related proteins, Sts-1 and Sts-2. Immunity. 2004;20:37–46 [DOI] [PubMed] [Google Scholar]
- 16. Mikhailik A, Ford B, Keller J, Chen Y, Nassar N, Carpino N. A phosphatase activity of Sts-1 contributes to the suppression of TCR signaling. Mol Cell. 2007;27:486–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng Y, Qin H, Frank SJ, et al. A CK2-dependent mechanism for activation of the JAK-STAT signaling pathwayy. Blood. 2011;118:156–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kurzer JH, Argetsinger LS, Zhou YJ, Kouadio JL, O'Shea JJ, Carter-Su C. Tyrosine 813 is a site of JAK2 autophosphorylation critical for activation of JAK2 by SH2-Bβ. Mol Cell Biol. 2004;24:4557–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kurzer JH, Saharinen P, Silvennoinen O, Carter-Su C. Binding of SH2-B family members within a potential negative regulatory region maintains JAK2 in an active state. Mol Cell Biol. 2006;26:6381–6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nishi M, Werner ED, Oh BC, et al. Kinase activation through dimerization by human SH2-B. Mol Cell Biol. 2005;25:2607–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bersenev A, Wu C, Balcerek J, Tong W. Lnk controls mouse hematopoietic stem cell self-renewal and quiescence through direct interactions with JAK2. J Clin Invest. 2008;118:2832–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Argetsinger LS, Stuckey JA, Robertson SA, et al. Tyrosines 868, 966, and 972 in the kinase domain of JAK2 are autophosphorylated and required for maximal JAK2 kinase activity. Mol Endocrinol. 2010;24:1062–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marcantonio D, Chalifour LE, Alaoui-Jamali MA, Alpert L, Huynh HT. Cloning and characterization of a novel gene that is regulated by estrogen and is associated with mammary gland carcinogenesis. Endocrinology. 2001;142:2409–2418 [DOI] [PubMed] [Google Scholar]
- 24. Visconti R, Schepis F, Iuliano R, et al. Cloning and molecular characterization of a novel gene strongly induced by the adenovirus E1A gene in rat thyroid cells. Oncogene. 2003;22:1087–1097 [DOI] [PubMed] [Google Scholar]
- 25. Bommer GT, Jager C, Durr EM, et al. DRO1, a gene down-regulated by oncogenes, mediates growth inhibition in colon and pancreatic cancer cells. J Biol Chem. 2005;280:7962–7975 [DOI] [PubMed] [Google Scholar]
- 26. Aoki K, Sun YJ, Aoki S, Wada K, Wada E. Cloning, expression, and mapping of a gene that is upregulated in adipose tissue of mice deficient in bombesin receptor subtype-3. Biochem Biophys Res Commun. 2002;290:1282–1288 [DOI] [PubMed] [Google Scholar]
- 27. Mu H, Ohta K, Kuriyama S, et al. Equarin, a novel soluble molecule expressed with polarity at chick embryonic lens equator, is involved in eye formation. Mech Dev. 2003;120:143–155 [DOI] [PubMed] [Google Scholar]
- 28. Liu Y, Monticone M, Tonachini L, et al. URB expression in human bone marrow stromal cells and during mouse development. Biochem Biophys Res Commun. 2004;322:497–507 [DOI] [PubMed] [Google Scholar]
- 29. Okada T, Nishizawa H, Kurata A, et al. URB is abundantly expressed in adipose tissue and dysregulated in obesity. Biochem Biophys Res Commun. 2008;367:370–376 [DOI] [PubMed] [Google Scholar]
- 30. Tremblay F, Revett T, Huard C, et al. Bidirectional modulation of adipogenesis by the secreted protein Ccdc80/DRO1/URB. J Biol Chem. 2009;284:8136–8147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ferragud J, Avivar-Valderas A, Pla A, De Las Rivas J, de Mora JF. Transcriptional repression of the tumor suppressor DRO1 by AIB1. FEBS lett. 2011;585:3041–3046 [DOI] [PubMed] [Google Scholar]
- 32. Meindl A, Carvalho MR, Herrmann K, et al. A gene (SRPX) encoding a sushi-repeat-containing protein is deleted in patients with X-linked retinitis pigmentosa. Hum Mol Genet. 1995;4:2339–2346 [DOI] [PubMed] [Google Scholar]
- 33. Kurosawa H, Goi K, Inukai T, et al. Two candidate downstream target genes for E2A-HLF. Blood. 1999;93:321–332 [PubMed] [Google Scholar]
- 34. Golemis EA, Serebriiskii I, Finley RL, Jr, Kolonin MG, Gyuris J, Brent R. Interaction trap/two-hybrid system to identify interacting proteins. Curr Protoc Mol Biol. 2008;82:20.1.1–20.1.35 [DOI] [PubMed] [Google Scholar]
- 35. Moller C, Hansson A, Enberg B, Lobie PE, Norstedt G. Growth hormone (GH) induction of tyrosine phosphorylation and activation of mitogen activated protein kinases in cells transfected with rat GH receptor cDNA. J Biol Chem. 1992;267:23403–23408 [PubMed] [Google Scholar]
- 36. Rui L, Mathews LS, Hotta K, Gustafson TA, Carter-Su C. Identification of SH2-Bβ as a substrate of the tyrosine kinase JAK2 involved in growth hormone signaling. Mol Cell Biol. 1997;17:6633–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346 [DOI] [PubMed] [Google Scholar]
- 38. Sugimoto T, Stewart S, Han M, Guan KL. The kinase suppressor of Ras (KSR) modulates growth factor and Ras signaling by uncoupling Elk-1 phosphorylation from MAP kinase activation. EMBO J. 1998;17:1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Silvennoinen O, Witthuhn B, Quelle FW, Cleveland JL, Yi T, Ihle JN. Structure of the murine JAK2 protein-tyrosine kinase and its role in interleukin 3 signal transduction. Proc Natl Acad Sci U S A. 1993;90:8429–8433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Watling D, Guschin D, Muller M, et al. Complementation by the protein tyrosine kinase JAK2 of a mutant cell line defective in the interferon-γ signal transduction pathway. Nature. 1993;366:166–170 [DOI] [PubMed] [Google Scholar]
- 41. Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol Cell Biol. 2000;20:3387–3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Herrington J, Rui L, Luo G, Yu-Lee L, Carter-Su C. A functional DNA-binding domain is required for growth hormone-induced nuclear localization of Stat5B. J Biol Chem. 1999;274:5138–5145 [DOI] [PubMed] [Google Scholar]
- 43. Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yin T, Yang YC. Mitogen-activated protein kinases and ribosomal S6 protein kinases are involved in signaling pathways shared by interleukin-11, interleukin-6, leukemia inhibitory factor, and oncostatin M in mouse 3T3-L1 cells. J Biol Chem. 1994;269:3731–3738 [PubMed] [Google Scholar]
- 45. Doerrler W, Feingold KR, Grunfeld C. Cytokines induce catabolic effects in cultured adipocytes by multiple mechanisms. Cytokine. 1994;6:478–484 [DOI] [PubMed] [Google Scholar]
- 46. Fruhbeck G, Aguado M, Martinez JA. In vitro lipolytic effect of leptin on mouse adipocytes: evidence for a possible autocrine/paracrine role of leptin. Biochem Biophys Res Commun. 1997;240:590–594 [DOI] [PubMed] [Google Scholar]
- 47. Aubert J, Dessolin S, Belmonte N, et al. Leukemia inhibitory factor and its receptor promote adipocyte differentiation via the mitogen-activated protein kinase cascade. J Biol Chem. 1999;274:24965–24972 [DOI] [PubMed] [Google Scholar]
- 48. Kaplan SL. Hormonal regulation of growth and metabolic effects of growth hormone. In: Kostyo JL, ed. Handbook of Physiology. New York, NY: Oxford University Press; 1999:129–143 [Google Scholar]
- 49. Scanes CG. Hormones and growth in domestic animals. In: Kostyo JL, ed. Handbook of Physiology. New York, NY: Oxford University Press; 1999:99–127 [Google Scholar]
- 50. Feng J, Witthuhn BA, Matsuda T, Kohlhuber F, Kerr IM, Ihle JN. Activation of Jak2 catalytic activity requires phosphorylation of Y1007 in the kinase activation loop. Mol Cell Biol. 1997;17:2497–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kaptein A, Paillard V, Saunders M. Dominant negative stat3 mutant inhibits interleukin-6-induced Jak-STAT signal transduction. J Biol Chem. 1996;271:5961–5964 [DOI] [PubMed] [Google Scholar]
- 52. Wen Z, Zhong Z, Darnell JE., Jr 1995. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 82:241–250 [DOI] [PubMed] [Google Scholar]
- 53. Cha SY, Sung YK, Im S, Kwack MH, Kim MK, Kim JC. URB expression in human dermal papilla cells. J Dermatol Sci. 2005;39:128–130 [DOI] [PubMed] [Google Scholar]
- 54. Misumi Y, Miki K, Takatsuki A, Tamura G, Ikehara Y. Novel blockade by brefeldin A of intracellular transport of secretory proteins in cultured rat hepatocytes. J Biol Chem. 1986;261:11398–11403 [PubMed] [Google Scholar]
- 55. Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGFβ and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14:163–176 [PMC free article] [PubMed] [Google Scholar]
- 56. Taipale J, Keski-Oja J. Growth factors in the extracellular matrix. FASEB J. 1997;11:51–59 [DOI] [PubMed] [Google Scholar]
- 57. Aoki N, Matsuda T. A nuclear protein tyrosine phosphatase TC-PTP is a potential negative regulator of the PRL-mediated signaling pathway: dephosphorylation and deactivation of signal transducer and activator of transcription 5a and 5b by TC-PTP in nucleus. Mol Endocrinol. 2002;16:58–69 [DOI] [PubMed] [Google Scholar]
- 58. Tremblay F, Huard C, Dow J, et al. Loss of coiled-coil domain containing 80 negatively modulates glucose homeostasis in diet-induced obese mice. Endocrinology. 2012;153:4290–4303 [DOI] [PubMed] [Google Scholar]
- 59. Ohki-Hamazaki H, Watase K, Yamamoto K, et al. Mice lacking bombesin receptor subtype-3 develop metabolic defects and obesity. Nature. 1997;390:165–169 [DOI] [PubMed] [Google Scholar]
- 60. Pawlowski K, Muszewska A, Lenart A, Szczepinska T, Godzik A, Grynberg M. A widespread peroxiredoxin-like domain present in tumor suppression- and progression-implicated proteins. BMC Genomics. 2010;11:590–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Roll P, Rudolf G, Pereira S, et al. SRPX2 mutations in disorders of language cortex and cognition. Hum Mol Genet. 2006;15:1195–1207 [DOI] [PubMed] [Google Scholar]
- 62. Royer B, Soares DC, Barlow PN, et al. Molecular evolution of the human SRPX2 gene that causes brain disorders of the Rolandic and Sylvian speech areas. BMC Genet. 2007;8:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Royer-Zemmour B, Ponsole-Lenfant M, Gara H, et al. Epileptic and developmental disorders of the speech cortex: ligand/receptor interaction of wild-type and mutant SRPX2 with the plasminogen activator receptor uPAR. Hum Mol Genet. 2008;17:3617–3530 [DOI] [PubMed] [Google Scholar]
- 64. Manabe R, Tsutsui K, Yamada T, et al. Transcriptome-based systematic identification of extracellular matrix proteins. Proc Natl Acad Sci U S A. 2008;105:12849–12854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Danpure CJ. How can the products of a single gene be localized to more than one intracellular compartment? Trends Cell Biol. 1995;5:230–238 [DOI] [PubMed] [Google Scholar]
- 66. Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol. 2009;10:148–155 [DOI] [PubMed] [Google Scholar]
- 67. Graham SJ, Dziadek MA, Johnstone LS. A cytosolic STIM2 preprotein created by signal peptide inefficiency activates ORAI1 in a store-independent manner. J Biol Chem. 2011;286:16174–16185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Belin D, Bost S, Vassalli JD, Strub K. A two-step recognition of signal sequences determines the translocation efficiency of proteins. EMBO J. 1996;15:468–478 [PMC free article] [PubMed] [Google Scholar]
- 69. Lin P, Le-Niculescu H, Hofmeister R, et al. The mammalian calcium-binding protein, nucleobindin (CALNUC), is a Golgi resident protein. J Cell Biol. 1998;141:1515–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lin P, Fischer T, Weiss T, Farquhar MG. Calnuc, an EF-hand Ca2+ binding protein, specifically interacts with the C-terminal α5-helix of Gαi3. Proc Nat Acad Sci U S A. 2000;97:674–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Forte GM, Pool MR, Stirling CJ. N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol. 2011;9:e1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]


