Abstract
Phytophthora root and stem rot caused by Phytophthora sojae Kaufmann and Gerdemann is one of the most severe soybean [Glycine max (L.) Merr] diseases in the USA. Partial resistance is as effective in managing this disease as single-gene (Rps gene)-mediated resistance and is more durable. The objective of this study was to identify quantitative trait loci (QTL) associated with partial resistance to P. sojae in PI 398841, which originated from South Korea. A population of 305 F7:8 recombinant inbred lines derived from a cross of OX20-8 × PI 398841 was used to evaluate partial resistance against P. sojae isolate C2S1 using a tray test. Composite interval mapping using a genome-wide logarithm of odd (LOD) threshold detected three QTL on chromosomes 1, 13, and 18, which individually explained 4–16 % of the phenotypic variance. Seven additional QTL, accounting for 2–3 % of phenotypic variance each, were identified using chromosome-wide LOD thresholds. Seven of the ten QTL for resistance to P. sojae were contributed by PI 398841. Seven QTL co-localized with known Rps genes and previously reported QTL for soil-borne root pathogens, isoflavone, and seed oil. Three QTL on chromosomes 3, 13, and 18 co-localized with known Rps genes, but PI 398841 did not exhibit an Rps gene-mediated resistance response following inoculation with 48 different isolates of P. sojae. PI 398841 is potentially a source of novel genes for improving soybean cultivars for partial resistance to P. sojae.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-013-2040-x) contains supplementary material, which is available to authorized users.
Introduction
Phytophthora root and stem rot is one of the most destructive diseases that suppresses soybean [Glycine max (L.) Merr] yield in the United States (Schmitthenner 1985). The causal agent, Phytophthora sojae (Kaufmann and Gerdemann), is a soilborne oomycete that germinates under wet conditions and causes seed rot or seedling damping-off of soybean at early growth stages. Under the same environmental conditions, a brown lesion begins on the lower taproot and extends up the stem in adult susceptible plants resulting in wilted and dead plants. Since it was first identified in Indiana in 1948, this disease has been found in all major soybean-growing regions around the world (Schmitthenner 1985; Anderson and Buzzell 1992; Yanchun and Chongyao 1993; Jee et al. 1998; Grau et al. 2004; Dorrance and Grünwald 2009). Annual soybean yield losses attributed to Phytophthora root and stem rot exceed $300 million in North America and $1–2 billion in worldwide production (Wrather and Koenning 2006).
Management of Phytophthora root and stem rot has primarily relied on single dominant resistance genes known as Rps genes. To date, 15 Rps genes have been identified at nine loci, and some of these have been deployed in modern soybean cultivars (Kilen et al. 1974; Mueller et al. 1978; Athow et al. 1980; Athow and Laviolette 1982; Anderson and Buzzell 1992; Buzzell and Anderson 1992; Diers et al. 1992b; Demirbas et al. 2001; Weng et al. 2001; Gordon et al. 2006; Sun et al. 2011). Rps gene resistance is race-specific, qualitatively inherited and confers an immune type of response following infection by P. sojae. However, this qualitative resistance tends to be short-lived, as R-genes are neutralized by adaptation of P. sojae populations (Schmitthenner 1985). Several mechanisms for the increasing diversity of P. sojae populations have been proposed, including mutation and outcrossing between different P. sojae strains (Förster et al. 1994). The emergence of new virulence pathotypes of P. sojae has limited the effectiveness of Rps genes deployed in commercial cultivars between 8 and 20 years (Grau et al. 2004).
Partial resistance is another type of genetic host resistance, also known as quantitative, rate-reducing, or field resistance (Tooley and Grau 1984; Walker and Schmitthenner 1984; Schmitthenner 1985). Partial resistance is quantitatively inherited, reduces the lesion development following root infection, and is effective against a wide range of P. sojae races (Tooley and Grau 1982; Schmitthenner 1985; Burnham et al. 2003; Mideros et al. 2007; Wang et al. 2010). Durable resistance in plants has been defined as resistance that remains effective, while it is widely used under environmental conditions favorable for disease development (Johnson 1984). For example, partial resistance in the barley (Hordeum vulgare L.) cultivars Minerva and Vada has been effective to barley leaf rust caused by Puccinia hordei Otth. for over 50 years (Parlevliet 2002), and the soybean cultivar Conrad has maintained its partial resistance in fields for over 30 years (Dorrance et al. unpublished). Soybean cultivars with high levels of partial resistance and Rps gene-mediated resistance were reported to be more stable for yield compared to those with either moderate or low levels of partial resistance and Rps gene-mediated resistance combined across a variety of environments (Dorrance et al. 2003). The combination of qualitative and quantitative resistance also prolonged effectiveness of resistance to Leptosphaeria maculans (Desm.) Ces. and de Not. in rapeseed (Brassica napus L.) (Brun et al. 2010). Moreover, partial resistance to P. sojae in soybean did not reduce soybean yield in the absence of disease pressure (Tooley and Grau 1984; St. Martin et al. 1994; Dorrance et al. 2003).
Several studies have mapped quantitative trait loci (QTL) for partial resistance to P. sojae in recombinant inbred line (RIL) populations. Two QTL for partial resistance were first mapped to chromosomes 2 and 13 in three populations, Conrad (high level of partial resistance R) × Sloan (susceptible, S), Conrad × Williams (S), and Conrad × Harosoy (S), with the alleles for partial resistance originating from Conrad (Burnham et al. 2003). Partial resistance was evaluated based on lesion length via root inoculation (tray test), and these two QTL explained a total of 42–50 % of the genotypic variance in each population (Burnham et al. 2003). Han et al. (2008) also identified QTL for partial resistance at similar locations on chromosomes 2 and 13 in a Conrad × OX760-6-1 (S) population using a different root inoculation procedure and isolates from Northeast China. Another QTL were mapped to chromosome 16 in Conrad × OX760-6-1 population based on the field-based disease incidence (Weng et al. 2007). Several QTL were detected on chromosomes 12, 13, 14, 17, 18, and 19 in a Conrad × Sloan population using a tray test or layer test. These QTL encompassed putative physiological trait genes, defense-related signaling genes, and an R-gene cluster (Wang et al. 2010, 2012). Four QTL were reported on chromosomes 13, 16, 18, and 20 in recombinant inbred lines (RILs) derived from an inter-specific cross between Glycine max V71-370 (R) and G. soja PI 407162 (S) (Tucker et al. 2010). Additional QTL were identified on chromosomes 2, 6, 8, 11, and 13 in a population from a cross between Conrad and Hefeng 25 (R) through field experiments conducted in multiple environments (Li et al. 2010) and on chromosomes 6, 10, and 15 in Su88-M21 (R) × Xinyixiaoheidou (S) population via a tray test (Wu et al. 2011). Recently, two more QTL on chromosomes 13 and 17 in a S99-2281(S) × PI 408105A (R) population were detected (Nguyen et al. 2012).
Very few sources of partial resistance have been used in QTL analyses and investigations into the molecular basis of partial resistance to P. sojae, with most studies conducted using Conrad as the source of partial resistance. Therefore, it is important to mine additional germplasm with partial resistance to P. sojae. More than 400 soybean accessions that have high levels of partial resistance have been identified and dozens of these accessions had higher levels of partial resistance than Conrad, including PI 398841 (Dorrance and Schmitthenner 2000). The objective of this study was to identify QTL for partial resistance to P. sojae using an advanced recombinant inbred (RI) population derived from the cross of OX20-8 × PI 398841, and to compare genetic locations of these QTL with previously mapped QTL.
Materials and methods
Plant materials and DNA extraction
A population of 305 F7:8 RI lines (RILs) derived from a cross of OX20-8 and PI 398841 was used for this study. Twenty-three F1 plants from this cross were self-fertilized to produce F2 seeds. The F2 plants were self-pollinated and each line was advanced up to the F7 generation by single-seed descent. OX20-8 is a breeding line developed in Ontario, Canada, which is highly susceptible (Buzzell and Anderson 1982) and PI 398841 has a high level of partial resistance to P. sojae (Dorrance and Schmitthenner 2000). PI 398841 was originally collected from Kwangju, South Korea.
Young leaf tissue was collected in a 2-ml tube at the V1 or V2 stage from each F7 plant grown in a field near the Ohio Agricultural Research and Development Center (OARDC), Wooster, OH, and flash frozen in liquid nitrogen. The frozen leaf tissue was lyophilized in a freeze drier (SP Industries Inc., Stone Ridge, NY, USA), then ground and homogenized using a Mixer Mill (Model MM301, Retsch, Hannover, Germany). DNA was extracted using a slightly modified CTAB method (Mian et al. 2008), and was dissolved in 200 μl of TE buffer (10 mM Tris–Cl, pH 7.5, 1 mM EDTA).
Molecular marker genotyping and linkage analysis
Parental genotypes for multiple mapping populations, including OX20-8 and PI 398841, were first genotyped using the Universal Soybean Linkage Panel (USLP) 1.0 containing 1,536 SNPs at Dr. Perry Cregan’s laboratory at the United States Department of Agriculture, Agricultural Research Service, in Beltsville, MD. From this parental data, 384 SNPs were selected and organized into the Oligo Pool All (OPA) assay to genotype multiple populations using Illumina GoldenGate® BeadXpress® SNP genotyping (Illumina Inc., San Diego, CA, USA). In this OPA assay, 239 SNPs were polymorphic between OX20-8 and PI 398841. The SNP marker genotypes of 305 RILs were determined using the OPA according to the protocol from Illumina. The genotyping was done at the Molecular and Cellular Imaging Center (MCIC) at the OARDC.
Selected SSR markers and SSR motifs from Song et al. (2010) were used to increase genome coverage by filling large gaps in the initial SNP linkage map of the population. These 20 μl PCRs were done with 50 ng of template DNA, 1 × PCR buffer, 1.0 mM of MgCl2, 50 μM of each of the dNTPs, 0.1 μM of each of forward and reverse primer (IDT Inc., Coralville, IA, USA), and 1.0 U of Taq polymerase (GeneScript Corp., Piscataway, NJ, USA) for the final concentration. The thermal cycles began at 95 °C for 5 min, followed by 32 cycles of denaturing at 95 °C for 30 s, annealing at 48–61 °C (according to the optimum temperature for the primer pair used) for 30 s, and extension at 72 °C for 45 s. Additional 10 min of extension at 72 °C followed at the end of the last cycle. The PCR product was resolved on a 4 % high-resolution agarose gel (Research Products International Corp., Mt. Prospect, IL, USA) by gel electrophoresis.
The genetic map was constructed with JoinMap® 4 (Van Ooijen 2006) using the Kosambi mapping function. Linkage was determined at the logarithm of odd (LOD) threshold of 3.0 with a maximum map distance of 50 centiMorgan (cM). The order of markers in linkage groups was compared with the Consensus Map 4.0 (Hyten et al. 2010).
Pathogenicity test of P. sojae isolates
The hypocotyl inoculation technique (Dorrance et al. 2008) was used to test pathogenicity of P. sojae isolates to identify isolates that were virulent or avirulent to OX20-8 and PI 398841. In brief, 10–15 seeds were placed on a germination paper, and the paper was then rolled and placed in a plastic bucket under dark conditions at 25 °C. Seven-day-old etiolated seedlings were inoculated with P. sojae inoculum slurry from 7-day-old cultures grown on lima bean agar (15 g agar/l). The papers were rolled and kept in a plastic bucket in darkness. Plants that have an R gene developed a hypersensitive reaction around the inoculation site, while susceptible plants developed an expanding brown lesion in 3–5 days. Plants were scored based on the percentage of susceptible interactions 7 days after inoculation as follows: <25 % plants killed as resistant; 25–75 % plants killed as intermediate; and >75 % plants killed as susceptible.
Those which induce the susceptible response on both parents are appropriate isolates for testing partial resistance, because the expression of partial resistance may be masked by the hypersensitive response caused by Rps genes if either parent has a Rps gene interacting with Avr genes of the same isolates. Alternatively, isolates that induced an Rps gene-mediated resistance response following inoculation of PI 398841, but virulent to OX20-8, could be used to detect and map Rps genes that may exist in PI 398841.
Partial resistance evaluation
Lesion lengths following P. sojae inoculation on roots were measured to evaluate levels of partial resistance among the RILs using a tray test assay which was previously published (Burnham et al. 2003; Tucker et al. 2010; Wang et al. 2010). Briefly, ten 7-day-old seedlings from each RIL were placed on a tray and a 1-cm wound was made on the tap root 20 mm below the crown with a scalpel. An agar-mycelial slurry from a 7-day-old culture of P. sojae isolate C2S1 (vir 1a, 1b, 1c, 1 k, 2, 3a, 3b, 3c, 4, 5, 6, and 7) was placed over the wound. After 7 days, lesion lengths were measured from the inoculation site to the edge of the lesion margin. The isolate C2S1 was one of the isolates which has a susceptible response to both parents, and the lesion length was significantly different between parents in a preliminary tray test compared to the other isolates that induced susceptible responses (data not shown).
A total of 305 RILs were separated into two sets consisting of 158 and 147 RILs. The first set was evaluated in 2010 (January to March) and the second in 2011 (January to March), respectively. Each set was divided into two incomplete blocks. An incomplete block consisted of six buckets, each of which included 16 RILs, two parents, and two checks, ‘Conrad’ and ‘Sloan’. Conrad and Sloan represent high level of partial resistance and moderate susceptibility, respectively (Burnham et al. 2003). There were three replications.
The data from both sets of RILs from the each experiment were combined and the mean lesion length of ten seedlings from each RIL was analyzed to obtain the best linear unbiased predictor (BLUP) using the PROC MIXED procedure in SAS (SAS 9.1, SAS Institute Inc., Cary, NC, USA) (Stroup 1989). The model was Y ijkl = μ + S i + R(S)ij + I(RS)ijk + B(IRS)ijkl + C m + G(C)mn + ε ijklmn, where μ is the overall mean, S i is the effect of ith set, R j is the effect of jth replication in ith set, I(SR)ijk is the effect of kth incomplete block in jth replication in ith set, B(SRI)ijkl is the effect of lth bucket in kth incomplete block and jth replication in ith set, C m is the effect of mth class of entry (l = 1, 2, 3, 4, and 5 for, OX20-8, PI 398841, Conrad, Sloan, and RIL, respectively), G(C)mn is the effect of nth genotype within class for recombinant inbred lines only (genotypic variance, σ 2G), ε ijklm is the experimental error (σ 2). Class of entry was assumed to be a fixed effect and all other terms random effects. Variance components were estimated using the restricted maximum likelihood (REML) method (Patterson and Thompson 1971). The heritability, on a line-mean basis, was calculated as σ 2G/(σ 2G + σ 2/r), where r is the number of replications per RIL.
QTL analysis, detection of epistasis and confirmation of QTL effects
Kruskal–Wallis analysis and interval mapping (IM) were initially performed to identify potential QTL, followed by composite interval mapping (CIM) using multiple-QTL method (MQM) with cofactors in MapQTL® 5 (Van Ooijen 2004). Walking speed was set to 1 cM for both IM and CIM. The genome-wide and chromosome-wide LOD threshold was determined by conducting a 1,000-permutation test at α = 0.05 (Churchill and Doerge 1994). Multiple regression analysis was conducted using PROC REG in SAS with the nearest markers to the QTL identified by CIM to calculate the total phenotypic variance (R 2). The LOD plots for the chromosomes on which a significant QTL were identified were graphically presented using the MapChart 2.2 software (Voorrips 2002). To test the interaction between identified QTL, genotypic data of the markers nearest to each QTL were analyzed by PROC GLM in SAS (SAS 9.1, SAS Institute Inc., Cary, NC, USA). The statistical model was Y ij = μ + M1i + M2j + (M1i × M2j) + ε ij, where Y ij is the phenotypic values (BLUP) for entries in the ith and jth marker classes for M1 and M2, μ is the overall mean, M1i is the effect of the ith marker class for M1, M2j is the effect of the jth marker class for M2, ε ij is the residual error. The appropriate F test for epistasis between molecular markers was F = (MSM1i × M2j)/(MSε ij).
Using the QTL identified with a genome-wide LOD threshold, the additivity of accumulated QTL effects on levels of partial resistance was verified. The cumulative additive effects (CAEs) of RILs provided by the alleles for the QTL were obtained based on the genotypes of the nearest markers to the QTL. The estimated additive effect of individual QTL was given to RIL that were homozygous for the resistance allele for the nearest marker to the corresponding QTL, while an effect of zero was given to those who were homozygous for the recessive alleles for the same marker. Then, a CAE of individual RIL was obtained by adding all given additive effects. Statistical significance of the regression of BLUP values on the CAEs was tested to confirm the trend of levels of partial resistance in relations with the cumulative additive effects using PROC REG in SAS.
Results
Partial resistance evaluation and statistical analysis
The mean lesion length was significantly different among the two parental lines and the two checks in all buckets (data not shown), indicating that PI 398841 had high levels of partial resistance to P. sojae. The mean lesion lengths averaged over all buckets from both sets were 50.1, 41.3, 29.6, and 25.5 mm for OX20-8, Sloan, Conrad, and PI 398841, respectively; the four means were separated from one another by Fisher’s least square difference (PROC GLM, P < 0.0001). PI 398841, the parent with partial resistance, had significantly shorter lesions than Conrad, which has been known to have high levels of partial resistance to P. sojae. However OX20-8, the susceptible parent, had significantly longer lesions than Sloan, which is highly susceptible. The mean lesion length of the individual RILs ranged from 12.6 to 67.6 mm, and the overall mean of all of the RILs was 39.1 mm.
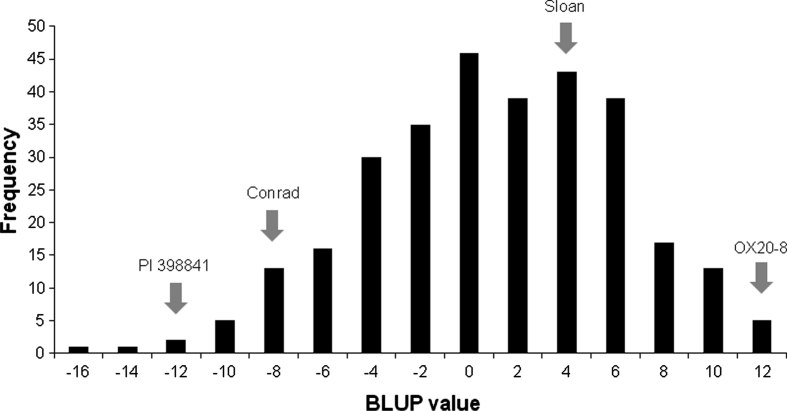
The BLUP values were calculated from the mean lesion length, and the frequency of BLUP values was normally distributed in this population (Fig. 1). Lower BLUP values indicate higher levels of partial resistance. The BLUP values estimated from the mixed model analysis were −13.6 and 11.1 for PI 398841 and OX20-8, respectively. The BLUP values of the checks were also separated with −9.4 and 2.3 for Conrad and Sloan, respectively. Two RILs had significantly lower BLUP values than PI 398841, the resistance parent, which indicates potential transgressive segregation. The mean lesion lengths of these two RILs were shorter than those of PI 398841 in each of the experiments across all replications, with one missing value for one of the RILs (data not shown). One-tailed paired t test with each RIL and PI 398841 in the same experimental units indicated one transgressive segregant (P < 0.05). Broad-sense heritability of mean lesion length was moderately high (0.77).
Fig. 1.
Frequency distribution of BLUP values for mean lesion length of RILs derived from OX20-8 × PI 398841 population. Estimates of two parents and two checks are indicated by arrows. A lower BLUP value means a higher level of partial resistance to P. sojae. OX20-8, the susceptible parent; PI 398841, the resistant parent; Conrad, the check for high level of partial resistance; Sloan, the check for low level of partial resistance
Linkage analysis
Two hundred and twenty SNP genotypes of 305 RILs were determined with the OPA containing 239 polymorphic SNPs for this population. Nineteen SNPs were not polymorphic nor scorable. Of these 220 SNP markers, 214 were mapped to construct the initial genetic map of the population. To increase the genome coverage by closing gaps found in the initial map, 59 SSR markers were added to the linkage map. A total of 214 SNPs and 59 SSRs were finally integrated into the genetic map of the OX20-8 × PI 398841 population for QTL analysis (Supplementary Fig 1). A total of 37 linkage fragments were generated, spanning 2,160 cM. This OX20-8 × PI 398841 genetic map corresponds to approximately 1,537 cM of the soybean Consensus Map 4.0, with 69 % coverage of the genome according to the genetic position (cM) of molecular markers shared between the two maps. Most of the shared markers were ordered in agreement with the Consensus Map 4.0 (Hyten et al. 2010) and only closely linked markers had an order which differed from that in the Consensus Map 4.0.
QTL identification by the genome-wide LOD threshold
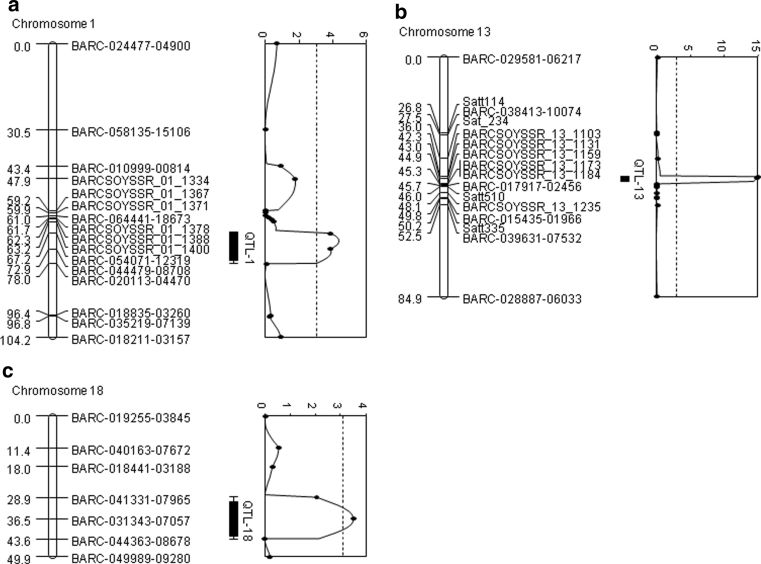
Three QTL associated with partial resistance to P. sojae were identified on chromosomes 1, 13, and 18 by composite interval mapping (Table 1; Fig. 2). The major QTL closely linked to BARCSOYSSR_13_1103 on the chromosome 13 (QTL-13) accounted for 16.1 % of the phenotypic variance (Table 1; Fig. 2). Two minor QTL adjacent to BARC-044479-08708 on chromosome 1 (QTL-1) and BARC-031343-07057 on chromosome 18 (QTL-18) explained 4.6 and 3.6 % of the phenotypic variance, respectively (Table 1; Fig. 2). The resistance alleles for all the QTL were contributed by PI 398841 as indicated by the additive effects of negative values (Table 1). A total of 28.8 % of the phenotypic variance were explained by the three QTL. Two-way ANOVA did not detect any epistatic allelic interactions between markers that were significant in Kruskal–Wallis analysis (data not shown).
Table 1.
Identification of three quantitative trait loci associated with partial resistance to P. sojae in the OX20-8 × PI 398841 population using the genome-wide LOD threshold
| Chr.a | Position (cM)b | Nearest marker | LODc | R 2 (%)d | Additive effect (contributor)e |
|---|---|---|---|---|---|
| 1 | 71 | BARC-044479-08708 | 4.4 | 4.6 | −1.26 (PI 398841) |
| 13 | 51 | BARCSOYSSR_13_1103 | 14.8 | 16.1 | −2.12 (PI 398841) |
| 18 | 91 | BARC-031343-07057 | 3.4 | 3.6 | −1.02 (PI 398841) |
aChromosome numbers
bPosition (cM) on Consensus Map 4.0 (Hyten et al. 2010). Since BARCSOYSSR_13_1103 was not integrated on the Consensus Map 4.0, an approximate position was denoted based on the adjacent marker, Satt334, on the Williams82 genome assembly Glyma1.01
cA genome-wide LOD threshold was 3.1 at 95 % of confidence level by a 1,000-permutation test
dPhenotypic variance explained by the QTL
eThe negative additive effects indicate that PI 398841 contributes favorable alleles for the QTL
Fig. 2.
Graphical presentations of three quantitative trait loci for partial resistance to P. sojae identified using the genome-wide LOD threshold in the F7:8 RIL population derived from a cross of OX20-8 × PI 398841. Genetic distance (cM) and marker names are shown to the left and right of chromosomes, respectively. The LOD plots to the right of chromosomes indicate the most likely position of QTL conferring partial resistance to P. sojae. The hatched lines on the LOD plots indicate the genome-wide LOD threshold for CIM of 3.1. The 1- and 2-LOD intervals are displayed as black bars and solid lines, respectively
Effects of combining the resistant alleles for the QTL
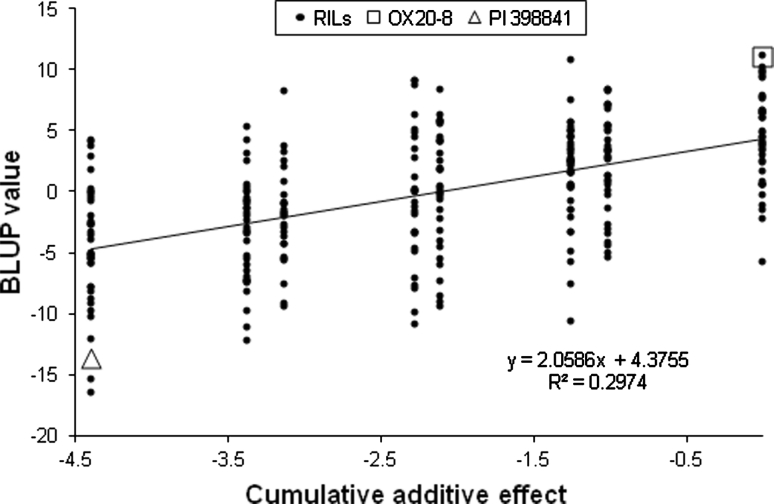
Eight different CAEs were obtained from the eight allelic combinations for the three QTL, which ranged −4.4 to 0. RILs that have homozygous PI 398841 alleles for all three QTL were given −4.4, and those with homozygous OX20-8 alleles for the three were given 0, since the resistance allele was contributed by PI 398841 for all three QTL. The BLUP values of RILs were visualized on their own CAEs (Fig. 3). RILs with the same CAE composed each vertical line of dots. The regression coefficient of 2.1 was significantly different from zero (P < 0.0001), indicating that levels of partial resistance to P. sojae decreased as the susceptible alleles were accumulated in a RIL. The R 2 value of the regression line was 0.29, which was somewhat higher than the sum of R 2 (%) values of three QTL. BLUP values of RILs were dispersed evenly on the same CAE since RILs were segregating for much more than three QTL used for this regression analysis and the three QTL accounted for the limited proportion of phenotypic variance, which implied that there was still a large proportion of phenotypic variance that was not explained.
Fig. 3.
Regression of the BLUP values on cumulative additive effects of pyramiding resistance alleles for QTL conferring partial resistance to P. sojae identified in this study. The accumulative additive effects were calculated based on three QTL, QTL-1, -13, and -18. X-axis indicates the sum of additive effects of combined resistance alleles for QTL in each RIL and two parental lines and Y-axis indicates BLUP values of parents and RILs. Lower BLUP values mean higher levels of resistance. The additive effects of resistance alleles for the three QTL identified in this study have negative values. The equation and R 2 of the regression line are shown on the scatter plots. The BLUP values of OX20-8, PI 398841, and RILs are plotted as square, triangle, and black circles
Indication of additional QTL by chromosome-wide LOD thresholds
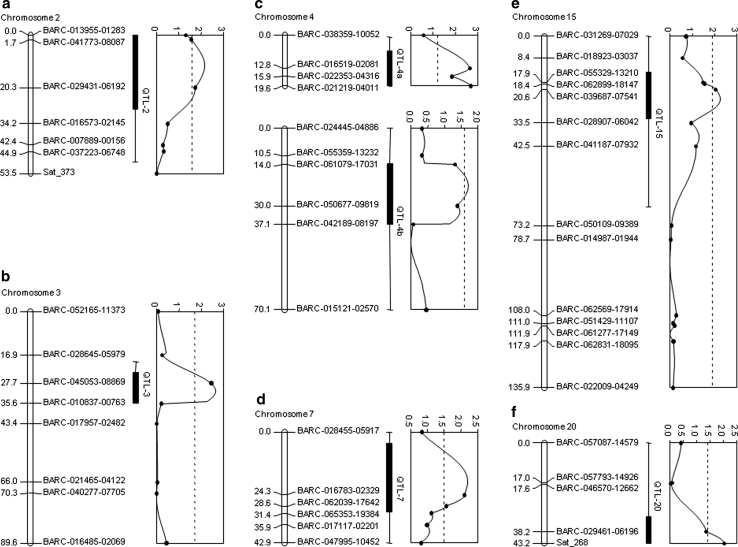
Chromosome-wide LOD thresholds (α = 0.05) allowed the detection of seven additional putative QTL (Table 2; Fig. 4). Four putative QTL were mapped near BARC-029431-01692, BARC-045053-08869, BARC-016783-02329, and Sat_268 on chromosomes 2, 3, 7, and 20 (QTL −2, −3, −7, and −20), respectively. These QTL individually explained 1.9–2.8 % of phenotypic variance and the resistance alleles were contributed by PI 398841. In concordance with the transgressive segregation observed (Fig. 1), susceptible genotype OX20-8 also contributed three putative QTL. They were identified near BARC-021219-04011 and BARC050677-09819 on chromosomes 4 and BARC-039687-07541 on chromosomes 15 (QTL-4a, QTL-4b, and QTL-15), which individually accounted for 1.9-2.6 % of phenotypic variance. The QTL-4a and -4b were located on two separate linkage groups from chromosome 4.
Table 2.
Seven putative quantitative trait loci for partial resistance to P. sojae in the OX20-8 × PI 398841 population using chromosome-wide LOD thresholds
| Chr.a | Position (cM)b | Nearest marker | LOD | LOD thresholdc | R 2 (%)d | Additive effect (contributor)e |
|---|---|---|---|---|---|---|
| 2 | 20 | BARC-029431-01692 | 2.2 | 1.6 | 2.3 | −0.88 (PI 398841) |
| 3 | 28 | BARC-045053-08869 | 2.6 | 1.7 | 2.8 | −0.92 (PI398841) |
| 4 | 20 | BARC-021219-04011 | 2.6 | 1.2 | 2.5 | 0.85 (OX20-8) |
| 4 | 64 | BARC-050677-09819 | 1.7 | 1.6 | 1.9 | 0.78 (OX20-8) |
| 7 | 50 | BARC-016783-02329 | 2.2 | 1.5 | 2.4 | −0.87 (PI 398841) |
| 15 | 19 | BARC-039687-07541 | 2.3 | 1.9 | 2.6 | 0.92 (OX20-8) |
| 20 | 48 | Sat_268 | 2.0 | 1.4 | 1.9 | −0.74 (PI 398841) |
aChromosome numbers
bPosition (cM) on Consensus Map 4.0 (Hyten et al. 2010)
cChromosome-wide LOD thresholds at α = 0.05 were calculated by a 1,000-permutation test
dPhenotypic variance explained by the QTL
eThe negative additive effects indicate that PI 398841 contributes favorable alleles for the QTL
Fig. 4.
Graphical presentations of seven additional quantitative trait loci for partial resistance to P. sojae identified using chromosome-wide LOD thresholds in the F7:8 RIL population derived from a cross of OX20-8 × PI 398841. Genetic distance (cM) and marker names are shown to the left and right of chromosomes, respectively. The LOD plots to the right of chromosomes indicate the most likely position of QTL conferring partial resistance to P. sojae. The hatched lines on the LOD plots indicate the chromosome-wide LOD threshold for CIM. The 1- and 2-LOD intervals are displayed as black bars and solid lines, respectively
Presence of Rps genes
Forty-eight isolates of P. sojae were screened on parental genotypes and 15 differentials via hypocotyl inoculation technique to identify isolates that had a qualitative resistance response in PI 398841 which indicates the presence of Rps genes. Although a diverse collection of P. sojae pathotypes was used, PI 398841 did not exhibit Rps gene-mediated resistance to any of the isolates used in this study. PI 398841 had susceptible responses to 42 isolates of P. sojae and intermediate responses to six of the 48 isolates used to identify Rps genes in this study (Supplementary Table 1). OX20-8, which has Rps1a, had a susceptible, intermediate and resistant response following inoculation to 42, 3 and 3 isolates of P. sojae, respectively (Supplementary Table 1).
Discussion
In this study, a large number of highly homozygous RILs derived from a cross of OX20-8 and PI 398841 were evaluated via tray tests with three replications for resistance to P. sojae. A high broad-sense heritability was estimated for partial resistance to P. sojae in this population (H = 0.77), which is in agreement with several previous studies which had heritability estimates ranging from 0.59 to 0.92 (Burnham et al. 2003; Wang et al. 2010; Tucker et al. 2010; Wu et al. 2011). The moderate to high heritability indicates that partial resistance to P. sojae would be inherited stably, and could, therefore, be selected effectively in a breeding program. Moreover, PI 398841 has higher levels of partial resistance than Conrad in highly replicated experiments with the data collected from 12 independent tests (4 subsets × 3 replications). Therefore, PI 398841 is a potential source for high levels of partial resistance to P. sojae.
The QTL-13 contributed a major effect for partial resistance to P. sojae (Fig. 2). Based on the position of the closest marker BARCSOYSSR_13_1103, the QTL-13 was located near the markers associated with partial resistance to P. sojae previously reported in populations derived from a cross of V71-370 (R) × PI 407162 (S) (Tucker et al. 2010) and Conrad (R) × Sloan (S) (Wang et al. 2010). The QTL identified by Tucker et al. (2010) were flanked by Satt114 and Satt510, and the QTL-13 from the present study is within this interval. The QTL from Conrad × Sloan were located between Satt510 and Sct_033 (Wang et al. 2010), an interval outside of the one containing QTL-13, but partially overlapping with the QTL identified by Tucker et al. (2010) (Fig. 2). QTL-13 had a much higher LOD score (14.8) and explained a greater portion of the phenotypic variance (16.1 %) than QTL reported from at this location in two previous studies (Tucker et al. 2010; Wang et al. 2010). This genomic region is well known as an R gene and defense gene rich region. Two closely linked P. sojae resistance genes, Rps3 and Rps8, were mapped in this region (Gordon et al. 2006). In addition, Rpg1 for resistance to bacterial blight caused by Pseudomonas savastanoi pv. glycinea, Rsv1 for resistance to soybean mosaic virus, Rpv1 for resistance to peanut mottle virus, Rag2 for resistance to soybean aphid (Aphis glycines Matsumura), and a QTL for resistance to Sclerotinia sclerotiorum (Lib.) de Bary were mapped in this R gene cluster (Arahana et al. 2001; Ashfield et al. 1998; Gore et al. 2002; Jeong et al. 2001; Mian et al. 2008). Weak forms of R genes have been considered as a potential mechanism for expressing partial resistance, and quantitative disease resistance loci that co-localized with R genes have been reported in many crop species (Li et al. 1999, 2006; Poland et al. 2009; St. Clair 2010).
Two novel but minor QTL were also identified on chromosomes 1 and 18 (Table 1; Fig. 2a, c). QTL-1 is a novel locus associated with partial resistance to P. sojae and no Rps gene has been mapped to date on chromosome 1. The QTL for seed weight (Sd wt 15-2 and Sd wt 18-1.1) and oil (oil 24-21) have also been reported near this region (Hyten et al. 2004; Panthee et al. 2005; Qi et al. 2011). Coincidence of QTL for P. sojae partial resistance with QTL for seed yield and oil was described in an earlier study (Wu et al. 2011). Further studies are needed to explain any relationship between P. sojae partial resistance and seed quality-related traits. The QTL-18 is another novel QTL for partial resistance to P. sojae; though another QTL for partial resistance to P. sojae were mapped to a position 10 cM “below” and were consistently identified against multiple P. sojae isolates via tray tests and layer tests, explaining 11–23 % of phenotypic variance (Wang et al. 2012). The QTL-18 region was also located near Rps4, 5, and 6 (Demirbas et al. 2001). Rps4 and 6 have been mapped adjacent to Satt472 or Satt191 (Demirbas et al. 2001). The nearest marker BARC-031343-07057 to QTL-18 was located only 1 or 5 cM “above” Satt191 or Satt472, based on marker positions on the Consensus Map 4.0 (Hyten et al. 2010). In addition, a QTL for resistance to soybean cyst nematode (SCN), Heterodera glycines Ichinohe, was identified between Satt612 and Satt191 near the QTL-18 (Vuong et al. 2010).
Since the majority of partial resistance QTL identified in previous studies had small-effects and their significance was low, some minor QTL may be overlooked due to statistical insignificance for the given threshold. A genome-wide LOD threshold (i.e., experiment-wise error rate) at α = 0.05 is generally accepted for a standard to determine significant linkage in QTL mapping. Lander and Kruglyak (1995) proposed “suggestive linkage”, which denoted weak associations between genotype and phenotype with no statistical significance at the experiment-wise error rate. A chromosome-wide LOD threshold was proposed for determining “suggested linkage” as the analysis of the markers on a single chromosome can be considered as a separated experiment, and LOD thresholds are dependent on chromosome map lengths (Van Ooijen 1999). Chromosome-wide LOD thresholds (α = 0.05) allow for the identification of QTL which may contribute a minor proportion of the overall effect; consequently, type I errors may occur at a higher rate. The main advantage is enabling the identification of potential QTL with low levels of significance, but which may be real, over multiple experiments. Recurrent emergence of particular loci could allow for the assembly of complex genetic networks for complex traits such as partial resistance to P. sojae. Thus, this study also reported putative QTL based on significance at chromosome-wide LOD thresholds.
Seven putative QTL with relatively low LOD values (1.7–2.6) were identified based on chromosome-wide LOD thresholds (Table 2; Fig. 4). Near the QTL-2, one QTL for oil content was reported (Zhang et al. 2004). The QTL-3 overlapped to loci closely linked to Rps1 and 7 (Weng et al. 2001). QTL associated with isoflavone have been reported near the QTL-7 in previous studies (Primomo et al. 2005; Zeng et al. 2009). This phenomenon has also been described in Wu et al. (2011). In addition, QTL for sudden death syndrome (SDS) caused by Fusarium virguliforme as well as isoflavone had been mapped near the QTL-20 (Iqbal et al. 2001; Zeng et al. 2009). Isoflavone accumulation in soybean roots is thought to play an important role in resistance for soybean-P. sojae and -F. virguliforme interactions (Lozovaya et al. 2004; Subramanian et al. 2005). Interestingly, QTL where resistance alleles were contributed by the susceptible parent also co-localized with or neighbored QTL for soil-borne diseases. Two loci for resistance to SCN (SCN21-1 and 18-3) were previously reported within QTL-4a interval and QTL-4b was mapped within 5 cM of a QTL for resistance to SDS (SDS9-3) (Yue et al. 2001; Njiti and Lightfoot 2006). The QTL-15 region also overlapped QTL conferring resistance to P. sojae, S. sclerotiorum, and H. glycines (Qiu et al. 1999; Arahana et al. 2001; Wu et al. 2011). In addition, a few studies had identified the region within the QTL-15 interval as highly associated with seed oil (Diers et al. 1992a; Lee et al. 1996). The QTL for partial resistance to P. sojae identified in this study are coincident with QTL for isoflavone, oil, or resistance to other soil-borne pathogens. This co-localization indicates that a portion of the ten QTL may include genes involved in conserved roles in mediating resistance to various soil-borne pathogens and potentially the genes at these loci may have pleiotropic effects. Additional mapping of more sources of resistances for these traits as well as functional studies of the genes associated with these traits will be necessary.
In previous studies, most of the QTL associated with partial resistance to P. sojae have been mapped in regions distinct from those known to contain Rps genes. In addition to the Rps3 and 8 cluster, the Rps1 and 7 cluster as well as the Rps4, 5, and 6 cluster were first reported to be associated with partial resistance to P. sojae in PI 398841. One of the hypotheses of quantitative resistance summarized by Poland et al. (2009) is that defeated or alternative forms of R-genes may contribute to the expression of quantitative disease resistance. The mapping of Rps genes in the OX20-8 × PI 398841 population would provide further evidence for or against this hypothesis. To find isolates causing a resistant reaction in PI 398841, but a susceptible reaction in OX20-8, 48 isolates of P. sojae were used to inoculate parents and 15 differentials via the hypocotyl inoculation technique. PI 398841, however, did not exhibit a resistant response to any of the isolates with diverse pathotypes, including OH 1, which has one virulence gene to Rps7 (Supplementary Table 1). Some possible interpretations of this result are that (1) Rps genes may exist in PI 398841 that may have incomplete expression similar to Rps2; (2) PI 398841 may have alleles of Rps genes that were not effective towards the pathotypes tested in this study; or (3) there are no Rps genes present in PI 398841.
There was a considerable gap between the phenotypic variance (0.41) when all of the QTL were considered and the estimated heritability (0.77). Possible explanations for the remainder of genetic components include the following: (1) some QTL are too small to detect via designated experiments, (2) unknown QTL may be located on the regions not covered in the genetic map used in the study, (3) multi-gene interactions were unable to be significantly detected in this study, or (4) the heritability may be somewhat overestimated. Recent studies noted the importance of epistatic QTL, which are interacting loci without significant individual effects on the given trait, in highly complex traits such as fatty acid composition and drought tolerance (Ravi et al. 2011; Yang et al. 2010). Though no significant QTL × QTL interaction was detected by two-way ANOVA, genome-wide scans for epistatic effects could provide more information about genetic network of partial resistance to P. sojae.
This study reported a total of ten QTL associated with partial resistance to P. sojae from a OX20-8 × PI 398841 population. Seven QTL of the ten detected in this study are reported for the first time here. In comparison with previously reported QTL, the majority of the ten QTL were located near R gene rich regions, including Rps genes, or regions for resistance to soil-borne fungal pathogens. Three were mapped near QTL regions associated with oil or isoflavone. Further studies will confirm the QTL in the next generation and with near-isogenic lines. Detection of any isolate-specificity of QTL will be especially important due to the ability of pathogen populations to rapidly evolve. Thus, the comparison of QTL identified against multiple isolates in multiple populations sharing a common parent will be critical to identify durable QTL for partial resistance.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 A genetic map of OX20-8 × PI 398841 population (PDF 74 kb)
Supplementary material 2 The response of OX20-8, PI 398841, and fifteen differentials following inoculations with forty-eight isolates of Phytophthora sojae (DOCX 29 kb)
Acknowledgments
This study was funded in part by United Soybean Board and Ohio Soybean Council through soybean producer’s check-off dollars. We would like to acknowledge Dr. Perry Cregan’s laboratory (USDA-ARS, Beltsville, MA) for the preliminary SNP genotyping of parental genotypes with 1,536 SNPs. We also thank T. Mendiola, S. A. Berry, M. Ellis, M. Ortega, Z. Zhang, L. Zelaya, A. Gunadi, D. Wickramasinghe, and C. Phelan for technical assistance and J. Whittier for conducting Illumina’s GoldenGate® SNP genotyping at the MCIC/OARDC and the Ohio BioProducts Innovation Center (OBIC) for funding of genotyping equipment. Salaries and research support for this project were provided by State and Federal Funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University.
References
- Anderson TR, Buzzell RI. Inheritance and linkage of the Rps7 gene for resistance to Phytophthora rot of soybean. Plant Dis. 1992;76:958–959. doi: 10.1094/PD-76-0958. [DOI] [Google Scholar]
- Arahana VS, Graef GL, Specht JE, Steadman JR, Eskridge KM. Identification of QTLs for resistance to Sclerotinia sclerotiorum in soybean. Crop Sci. 2001;41:180–188. doi: 10.2135/cropsci2001.411180x. [DOI] [Google Scholar]
- Ashfield T, Danzer JR, Held D, Clayton K, Keim P, Saghai Maroof MA, Webb DM, Innes RW. Rpg1, a soybean gene effective against races of bacterial blight, maps to a cluster of previously identified disease resistance genes. Theor Appl Genet. 1998;96:1013–1021. doi: 10.1007/s001220050833. [DOI] [Google Scholar]
- Athow KL, Laviolette FA. Rps6, a major gene for resistance to Phytophthora megasperma f-sp glycinea in soybean. Phytopathology. 1982;72:1564–1567. doi: 10.1094/Phyto-72-1564. [DOI] [Google Scholar]
- Athow KL, Laviolette FA, Mueller EH, Wilcox JR. A new major gene for resistance to Phytophthora megasperma var. sojae in soybean. Phytopathology. 1980;70:977–980. doi: 10.1094/Phyto-70-977. [DOI] [Google Scholar]
- Brun H, Chevre AM, Fitt BDL, Powers S, Besnard AL, Ermel M, Huteau V, Marquer B, Eber F, Renard M, Andrivon D. Quantitative resistance increases the durability of qualitative resistance to Leptosphaeria maculans in Brassica napus. New Phytol. 2010;185:285–299. doi: 10.1111/j.1469-8137.2009.03049.x. [DOI] [PubMed] [Google Scholar]
- Burnham KD, Dorrance AE, VanToai TT, St. Martin SK. Quantitative trait loci for partial resistance to Phytophthora sojae in soybean. Crop Sci. 2003;43:1610–1617. doi: 10.2135/cropsci2003.1610. [DOI] [Google Scholar]
- Buzzell RI, Anderson TR. Plant loss response of soybean cultivars to Phytophthora megasperma f. sp. glycinea under field conditions. Plant Dis. 1982;66:1146–1148. doi: 10.1094/PD-66-1146. [DOI] [Google Scholar]
- Buzzell RI, Anderson TR. Inheritance and race reaction of a new soybean Rps1 allele. Plant Dis. 1992;76:600–601. doi: 10.1094/PD-76-0600. [DOI] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirbas A, Rector BG, Lohnes DG, Fioritto RJ, Graef GL, Cregan PB, Shoemaker RC, Specht JE. Simple sequence repeat markers linked to the soybean genes for Phytophthora resistance. Crop Sci. 2001;41:1220–1227. doi: 10.2135/cropsci2001.4141220x. [DOI] [Google Scholar]
- Diers BW, Keim P, Fehr WR, Shoemaker RC. RFLP analysis of soybean seed protein and oil content. Theor Appl Genet. 1992;83:608–612. doi: 10.1007/BF00226905. [DOI] [PubMed] [Google Scholar]
- Diers BW, Mansur L, Imsande J, Shoemaker RC. Mapping Phytophthora resistance loci in soybean with restriction-fragment-length-polymorphism markers. Crop Sci. 1992;32:377–383. doi: 10.2135/cropsci1992.0011183X003200020020x. [DOI] [Google Scholar]
- Dorrance AE, Grünwald NJ. Phytophthora sojae: Diveristy among and within populations. In: Lamour K, Kamoun S, editors. Oomycete genetics and genomics. New Jersey: Wiley; 2009. pp. 197–212. [Google Scholar]
- Dorrance AE, Schmitthenner AF. New sources of resistance to Phytophthora sojae in the soybean plant introductions. Plant Dis. 2000;84:1303–1308. doi: 10.1094/PDIS.2000.84.12.1303. [DOI] [PubMed] [Google Scholar]
- Dorrance AE, McClure SA, St. Martin SK. Effect of partial resistance on Phytophthora stem rot incidence and yield of soybean in Ohio. Plant Dis. 2003;87:308–312. doi: 10.1094/PDIS.2003.87.3.308. [DOI] [PubMed] [Google Scholar]
- Dorrance AE, Berry SA, Anderson TR, Meharg C. Isolation, storage, pathotype characterization, and evaluation of resistance for Phytophthora sojae in soybean. Plant Health Progress. 2008 [Google Scholar]
- Förster H, Tyler BM, Coffey MD. Phytophthora sojae races have arisen by clonal evolution and by rare outcrosses. Mol Plant-Microbe Interact. 1994;7:780–791. doi: 10.1094/MPMI-7-0780. [DOI] [Google Scholar]
- Gordon SG, St. Martin SK, Dorrance AE. Rps8 maps to a resistance gene rich region on soybean molecular linkage group F. Crop Sci. 2006;46:168–173. doi: 10.2135/cropsci2004.04-0024. [DOI] [Google Scholar]
- Gore MA, Hayes AJ, Jeong SC, Yue YG, Buss GR, Maroof MAS. Mapping tightly linked genes controlling potyvirus infection at the Rsv1 and Rpv1 region in soybean. Genome. 2002;45:592–599. doi: 10.1139/g02-009. [DOI] [PubMed] [Google Scholar]
- Grau CR, Dorrance AE, Bond J, Russin JS. Fungal diseases. In: Boerma HR, Specht JE, editors. Soybeans: improvement, production, and uses. 3. Wisconsin: American Society of Agronomy—Crop Science Society of America—Soil Science Society of America; 2004. pp. 679–763. [Google Scholar]
- Han YP, Teng WL, Yu KF, Poysa V, Anderson T, Qiu LJ, Lightfoot DA, Li WB. Mapping QTL tolerance to Phytophthora root rot in soybean using microsatellite and RAPD/SCAR derived markers. Euphytica. 2008;162:231–239. doi: 10.1007/s10681-007-9558-4. [DOI] [Google Scholar]
- Hyten DL, Pantalone VR, Sams CE, Saxton AM, Landau-Ellis D, Stefaniak TR, Schmidt ME. Seed quality QTL in a prominent soybean population. Theor Appl Genet. 2004;109:552–561. doi: 10.1007/s00122-004-1661-5. [DOI] [PubMed] [Google Scholar]
- Hyten DL, Choi IY, Song QJ, Specht JE, Carter TE, Shoemaker RC, Hwang EY, Matukumalli LK, Cregan PB. A high density integrated genetic linkage map of soybean and the development of a 1536 universal soy linkage panel for quantitative trait locus mapping. Crop Sci. 2010;50:960–968. doi: 10.2135/cropsci2009.06.0360. [DOI] [Google Scholar]
- Iqbal MJ, Meksem K, Njiti VN, Kassem MA, Lightfoot DA. Microsatellite markers identify three additional quantitative trait loci for resistance to soybean sudden-death syndrome (SDS) in Essex × Forrest RILs. Theor Appl Genet. 2001;102:187–192. doi: 10.1007/s001220051634. [DOI] [Google Scholar]
- Jee H, Kim W, Cho W. Occurance of Phytophthora root rot on soybean (Glycine max) and identification of the causal fungus. Crop Prot. 1998;40:16–22. [Google Scholar]
- Jeong SC, Hayes AJ, Biyashev RM, Maroof MAS. Diversity and evolution of a non-TIR-NBS sequence family that clusters to a chromosomal “hotspot” for disease resistance genes in soybean. Theor Appl Genet. 2001;103:406–414. doi: 10.1007/s001220100567. [DOI] [Google Scholar]
- Johnson R. A critical analysis of durable resistance. Annu Rev Phytopathol. 1984;22:309–330. doi: 10.1146/annurev.py.22.090184.001521. [DOI] [Google Scholar]
- Kilen TC, Hartwig EE, Keeling BL. Inheritance of a second major gene for resistance to Phytophthora rot in soybeans. Crop Sci. 1974;14:260–262. doi: 10.2135/cropsci1974.0011183X001400020027x. [DOI] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lee SH, Bailey MA, Mian MAR, Carter TE, Shipe ER, Ashley DA, Parrott WA, Hussey RS, Boerma HR. RFLP loci associated with soybean seed protein and oil content across populations and locations. Theor Appl Genet. 1996;93:649–657. doi: 10.1007/BF00224058. [DOI] [PubMed] [Google Scholar]
- Li ZK, Luo LJ, Mei HW, Paterson AH, Zhao XZ, Zhong DB, Wang YP, Yu XQ, Zhu L, Tabien R, Stansel JW, Ying CS. A “defeated” rice resistance gene acts as a QTL against a virulent strain of Xanthomonas oryzae pv. oryzae. Mol Gen Genet. 1999;261:58–63. doi: 10.1007/s004380050941. [DOI] [PubMed] [Google Scholar]
- Li ZK, Arif M, Zhong DB, Fu BY, Xu JL, Domingo-Rey J, Ali J, Vijayakumar CHM, Yu SB, Khush GS. Complex genetic networks underlying the defensive system of rice (Oryza sativa L.) to Xanthomonas oryzae pv. oryzae. Proc Natl Acad Sci USA. 2006;103:7994–7999. doi: 10.1073/pnas.0507492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Han Y, Teng W, Zhang S, Yu K, Poysa V, Anderson T, Ding J, Li W. Pyramided QTL underlying tolerance to Phytophthora root rot in mega-environments from soybean cultivars ‘Conrad’ and ‘Hefeng 25’. Theor Appl Genet. 2010;121:651–658. doi: 10.1007/s00122-010-1337-2. [DOI] [PubMed] [Google Scholar]
- Lozovaya VV, Lygin AV, Zernova OV, Li S, Hartman GL, Widholm JM. Isoflavonoid accumulation in soybean hairy roots upon treatment with Fusarium solani. Plant Physiol Biochem. 2004;42:671–679. doi: 10.1016/j.plaphy.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Mian MAR, Kang S-T, Beil S, Hammond R. Genetic linkage mapping of the soybean aphid resistance gene in PI 243540. Theor Appl Genet. 2008;117:955–962. doi: 10.1007/s00122-008-0835-y. [DOI] [PubMed] [Google Scholar]
- Mideros S, Nita M, Dorrance AE. Characterization of components of partial resistance, Rps2, and root resistance to Phytophthora sojae in soybean. Phytopathology. 2007;97:655–662. doi: 10.1094/PHYTO-97-5-0655. [DOI] [PubMed] [Google Scholar]
- Mueller EH, Athow KL, Laviolette FA. Inheritance of resistance to four physiologic races of Phytophthora megasperma var sojae. Phytopathology. 1978;68:1318–1322. doi: 10.1094/Phyto-68-1318. [DOI] [Google Scholar]
- Nguyen VT, Vuong TD, VanToai T, Lee JD, Wu X, Rouf Mian MA, Dorrance AE, Shannon JG, Nguyen HT. Mapping of quantitative trait loci associated with resistance to Phytophthora sojae and flooding tolerance in soybean. Crop Sci. 2012;52:2481–2493. doi: 10.2135/cropsci2011.09.0466. [DOI] [Google Scholar]
- Njiti V, Lightfoot D. Genetic analysis infers Dt loci resistance to Fusarium solani f. sp. Glycines in indeterminate soybeans. Can J Plant Sci. 2006;86:83–90. doi: 10.4141/P05-046. [DOI] [Google Scholar]
- Panthee DR, Pantalone VR, West DR, Saxton AM, Sams CE. Quantitative trait loci for seed protein and oil concentration, and seed size in soybean. Crop Sci. 2005;45:2015–2022. doi: 10.2135/cropsci2004.0720. [DOI] [Google Scholar]
- Parlevliet JE. Durability of resistance against fungal, bacterial and viral pathogens; present situation. Euphytica. 2002;124:147–156. doi: 10.1023/A:1015601731446. [DOI] [Google Scholar]
- Patterson HD, Thompson R. Recovery of inter-block information when block sizes are unequal. Biometrika. 1971;58:545–554. doi: 10.1093/biomet/58.3.545. [DOI] [Google Scholar]
- Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ. Shades of gray: the world of quantitative disease resistance. Trends in Plant Sci. 2009;14:21–29. doi: 10.1016/j.tplants.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Primomo V, Poysa V, Ablett G, Jackson C, Gijzen M, Rajcan I. Mapping QTL for individual and total isoflavone content in soybean seeds. Crop Sci. 2005;45:2545–2564. [Google Scholar]
- Qi ZM, Wu Q, Han X, Sun Y, Du X, Liu C, Jiang H, Hu G, Chen Q. Soybean oil content QTL mapping and integrating with meta-analysis method for mining genes. Euphytica. 2011;179:499–514. doi: 10.1007/s10681-011-0386-1. [DOI] [Google Scholar]
- Qiu BX, Arelli PR, Sleper DA. RFLP markers associated with soybean cyst nematode resistance and seed composition in a ‘Peking’ × ’Essex’ population. Theor Appl Genet. 1999;98:356–364. doi: 10.1007/s001220051080. [DOI] [Google Scholar]
- Ravi K, Vadez V, Isobe S, Mir R, Guo Y, Nigam S, Gowda M, Radhakrishnan T, Bertioli D, Knapp S, Varshney R. Identification of several small main-effect QTLs and a large number of epistatic QTLs for drought tolerance related traits in groundnut (Arachis hypogaea L.) Theor Appl Genet. 2011;122:1119–1132. doi: 10.1007/s00122-010-1517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitthenner AF. Problems and progress in control of Phytophthora root rot of soybean. Plant Dis. 1985;69:362–368. doi: 10.1094/PD-69-362. [DOI] [Google Scholar]
- Song Q, Jia G, Zhu Y, Grant D, Nelson RT, Hwang E-Y, Hyten DL, Cregan PB. Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR_1.0) in soybean. Crop Sci. 2010;50:1950–1960. doi: 10.2135/cropsci2009.10.0607. [DOI] [Google Scholar]
- St. Clair DA. Quantitative disease resistance and quantitative resistance loci in breeding. Annu Rev Phytopathol. 2010;48:247–268. doi: 10.1146/annurev-phyto-080508-081904. [DOI] [PubMed] [Google Scholar]
- St. Martin SK, Scott DR, Schmitthenner AF, Mcblain BA. Relationship between tolerance to Phytophthora rot and soybean yield. Plant Breed. 1994;113:331–334. doi: 10.1111/j.1439-0523.1994.tb00745.x. [DOI] [Google Scholar]
- Stroup WW (1989) Why mixed models in applications of mixed models in agriculture and related disciplines. South Coop Ser Bull 343. Louisiana Agricultural Experiment Station, Baton Rouge, pp 1–8
- Subramanian S, Graham MY, Yu O, Graham TL. RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae. Plant Physiol. 2005;137:1345–1353. doi: 10.1104/pp.104.057257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Wu XL, Zhao JM, Wang YC, Tang QH, Yu DY, Gai JY, Xing H. Characterization and mapping of RpsYu25, a novel resistance gene to Phytophthora sojae. Plant Breed. 2011;30:139–143. doi: 10.1111/j.1439-0523.2010.01794.x. [DOI] [Google Scholar]
- Tooley PW, Grau CR. Field characterization of rate-reducing resistance to Phytophthora megasperma f sp glycinea in soybean. Phytopathology. 1982;72:1140. doi: 10.1094/Phyto-72-727. [DOI] [Google Scholar]
- Tooley PW, Grau CR. The relationship between rate-reducing resistance to Phytophthora megasperma f sp glycinea and yield of soybean. Phytopathology. 1984;74:1209–1216. doi: 10.1094/Phyto-74-1209. [DOI] [Google Scholar]
- Tucker DM, Saghai Maroof MA, Mideros S, Skoneczka JA, Nabati DA, Buss GR, Hoeschele I, Tyler BM, St. Martin SK, Dorrance AE. Mapping quantitative trait loci for partial resistance to Phytophthora sojae in a soybean interspecific cross. Crop Sci. 2010;50:628–635. doi: 10.2135/cropsci2009.03.0161. [DOI] [Google Scholar]
- Van Ooijen JW. LOD significance thresholds for QTL analysis in experimental populations of diploid species. Hered. 1999;83:613–624. doi: 10.1038/sj.hdy.6886230. [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW (2004) MapQTL® 5, Software for the mapping of quantitative trait loci in experimental populations. Kyazma B.V., Wageningen
- Van Ooijen JW (2006) JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V., Wageningen
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Vuong T, Sleper D, Shannon J, Nguyen H. Novel quantitative trait loci for broad-based resistance to soybean cyst nematode (Heterodera glycines Ichinohe) in soybean PI 567516C. Theor Appl Genet. 2010;121:1253–1266. doi: 10.1007/s00122-010-1385-7. [DOI] [PubMed] [Google Scholar]
- Walker AK, Schmitthenner AF. Heritability of tolerance to Phytophthora rot in soybean. Crop Sci. 1984;24:490–491. doi: 10.2135/cropsci1984.0011183X002400030014x. [DOI] [Google Scholar]
- Wang H, Waller L, Tripathy S, St. Martin SK, Zhou L, Krampis K, Tucker DM, Mao Y, Hoeschele I, Saghai Maroof MA, Tyler BM, Dorrance AE. Analysis of genes underlying soybean quantitative trait loci conferring partial resistance to Phytophthora sojae. Plant Genome. 2010;3:23–40. doi: 10.3835/plantgenome2009.12.0029. [DOI] [Google Scholar]
- Wang H, St. Martin SK, Dorrance AE. Comparison of phenotypic methods and yield contributions of quantitative trait loci for partial resistance to Phytophthora sojae in soybean. Crop Sci. 2012;52:609–622. doi: 10.2135/cropsci2011.06.0323. [DOI] [Google Scholar]
- Weng C, Yu K, Anderson TR, Poysa V. Mapping genes conferring resistance to Phytophthora root rot of soybean, Rps1a and Rps7. J Hered. 2001;92:442–446. doi: 10.1093/jhered/92.5.442. [DOI] [PubMed] [Google Scholar]
- Weng C, Yu K, Anderson T, Poysa V. A quantitative trait locus influencing tolerance to Phytophthora root rot in the soybean cultivar ‘Conrad’. Euphytica. 2007;158:81–86. doi: 10.1007/s10681-007-9428-0. [DOI] [Google Scholar]
- Wrather JA, Koenning SR. Estimates of disease effects on soybean yields in the United States 2003 to 2005. J Nematol. 2006;38:173–180. [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou B, Zhao J, Guo N, Zhang B, Yang F, Chen S, Gai J, Xing H. Identification of quantitative trait loci for partial resistance to Phytophthora sojae in soybean. Plant Breed. 2011;130:144–149. doi: 10.1111/j.1439-0523.2010.01799.x. [DOI] [Google Scholar]
- Yanchun S, Chongyao S. The discovery and biological characteristics studies of Phytophthora megasperma f.sp. glycinea on soybean in China. Acta Phytopathol Sin. 1993;23:341–347. [Google Scholar]
- Yang X, Guo Y, Yan J, Zhang J, Song T, Rocheford T, Li JS. Major and minor QTL and epistasis contribute to fatty acid compositions and oil concentration in high-oil maize. Theor Appl Genet. 2010;120:665–678. doi: 10.1007/s00122-009-1184-1. [DOI] [PubMed] [Google Scholar]
- Yue P, Arelli P, Sleper DA. Molecular characterization of resistance to Heterodera glycines in soybean PI 438489B. Theor Appl Genet. 2001;102:921–928. doi: 10.1007/s001220000453. [DOI] [Google Scholar]
- Zeng G, Li D, Han Y, Teng W, Wang J, Qiu L, Li W. Identification of QTL underlying isoflavone contents in soybean seeds among multiple environments. Theor Appl Genet. 2009;118:1455–1463. doi: 10.1007/s00122-009-0994-5. [DOI] [PubMed] [Google Scholar]
- Zhang W, Wang Y, Luo G, Zhang J, He C, Wu X, Gai J, Chen S. QTL mapping of ten agronomic traits on the soybean (Glycine max L. Merr) genetic map and their association with EST markers. Theor Appl Genet. 2004;108:1131–1139. doi: 10.1007/s00122-003-1527-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 A genetic map of OX20-8 × PI 398841 population (PDF 74 kb)
Supplementary material 2 The response of OX20-8, PI 398841, and fifteen differentials following inoculations with forty-eight isolates of Phytophthora sojae (DOCX 29 kb)